Abstract
Nitrogen (N) cycles have been directly linked to the functional stability of ecosystems because N is an essential element for life. Furthermore, the supply of N to organisms regulates primary productivity in many natural ecosystems. Microbial communities have been shown to significantly contribute to N cycles because many N-cycling processes are microbially mediated. Only particular groups of microbes were implicated in N-cycling processes, such as nitrogen fixation, nitrification, and denitrification, until a few decades ago. However, recent advances in high-throughput sequencing technologies and sophisticated isolation techniques have enabled microbiologists to discover that N-cycling microbes are unexpectedly diverse in their functions and phylogenies. Therefore, elucidating the link between biogeochemical N-cycling processes and microbial community dynamics can provide a more mechanistic understanding of N cycles than the direct observation of N dynamics. In this review, we summarized recent findings that characterized the microbes governing novel N-cycling processes. We also discussed the ecological role of N-cycling microbial community dynamics, which is essential for advancing our understanding of the functional stability of ecosystems.
Keywords: nitrogen cycle, ecosystem functions, microbial community dynamics
The fundamental issues of ecosystem ecology are related to understanding how ecosystems maintain functional stability and predicting how ecosystems respond to environmental changes. An ecosystem can be defined as an interacting system composed of an environment and all the organisms involved in it. Many ecosystem ecologists have focused on the N cycle or the dynamics of N transformation in various ecosystems because N (along with H, C, O, S and P), as a major component of proteins and nucleic acids, is an essential element for life and its supply can limit primary productivity in many natural terrestrial and marine ecosystems (12, 122). The N cycle has also been the focus of debate in nitrogen-rich ecosystems, such as fertilized agricultural fields or eutrophic rivers and coasts that are affected by anthropogenic N input (48, 53, 85, 90, 101, 110, 114, 115, 134, 135). Wastewater treatment systems are also examples of artificial ecosystems in which N removal is frequently studied (29, 78, 116, 121). Studies on the pathways and rates of input, output, and internal cycle of N and its interactions with other elements can provide insights into the fundamental issues related to ecosystem ecology.
Nitrogen is a versatile element that forms compounds in various oxidation states, ranging from −3 (ammonium and amino-nitrogen) to +5 (nitrate) (Fig. 1). N transitions between compounds with different oxidation states are largely driven by thermodynamically constrained redox reactions and are typically catalyzed by microbes (24). Given the ubiquity and biogeochemical contributions of microbes, microbial community dynamics may be directly associated with temporal and spatial variations in internal N-cycling pathways and rates in ecosystems. Microbiologists have already demonstrated the critical ecological roles that microbes play in N-cycling pathways and rates by integrating microbial community dynamics into N biogeochemical phenomena. Microbial community dynamics may also ultimately affect the functional stability of ecosystems. Ecosystem ecologists have frequently reported non-linear alterations in N dynamics and sometimes identified the thresholds at which these alterations occurred as ecosystems responded to perturbations or disturbances. For example, Aber et al. (1) proposed a conceptual model of N saturation in temperate forests in which the response of the forest to chronic atmospheric N deposition could be quantitatively classified into three progressive stages. The first stage was characterized by an increase in net soil N mineralization and tree growth. Although net soil N mineralization decreased in the second stage, nitrification was induced, resulting in more NO3− leaching. Finally, the uptake of N by plants and tree growth declined, whereas nitrification and NO3− loss continued to increase. Non-linear alterations in the N dynamics of ecosystems may be largely due to the non-linear responses of microbial community dynamics or the physiological constraints of the community. Therefore, ecosystem ecologists are beginning to explicitly consider the ecological roles of microbial community dynamics (2, 95). Few studies have investigated the ecological roles of an entire microbial community’s dynamics in the functional stability of the ecosystem, and this has been attributed to microbiologists being more inclined to focus on functionally equivalent microbial groups that are involved in specific (single or a few) N-cycling processes. However, ecological studies by microbiologists may provide valuable insights into the mechanisms of not only the N cycle of the ecosystem, but also its functional stability, and can ultimately permit predictions of the functional stability in a changing environment with an unprecedented level of detail.
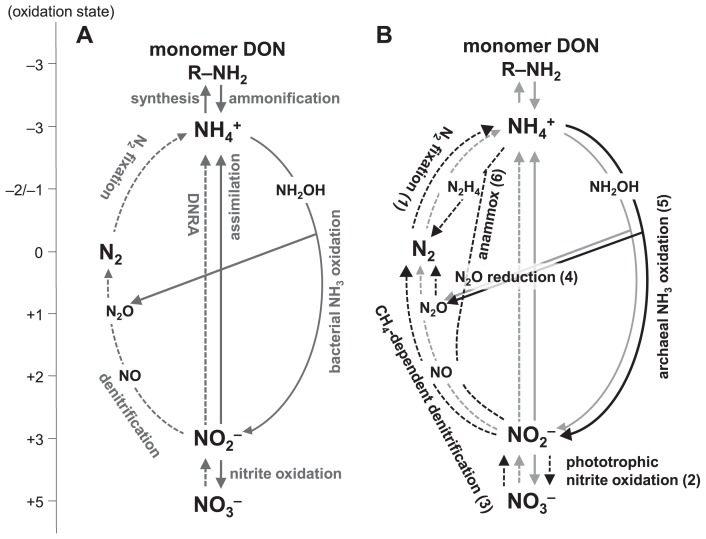
Fig. 1.
Schematic representation highlighting the main processes in the microbial N cycle, with a focus on (A, gray) the classical processes and (B, black) recently discovered processes discussed in the text. Processes mainly occurring under oxic and anoxic conditions are shown as solid and dashed arrows, respectively. Detailed reactions of recently discovered processes are described in Table 1. Note: denitrification (A), denitrification coupled to the oxidation of organic matter, hydrogen, reduced iron, or reduced sulfur species; nitrite oxidation (A), aerobic chemolithotrophic nitrite oxidation; N2 fixation(B), N2 fixation by unicellular cyanobacteria; N2O reduction (B), N2O reduction by non-denitrifying bacteria. Abbreviations: DON, dissolved organic N; DNRA, dissimilatory nitrate reduction to ammonia.
We described recent findings on microbes governing novel N transformations in this brief review. We also discussed the relationship between microbial community dynamics and N biogeochemistry as well as the ecological roles of microbial community dynamics in the N-cycling rates and processes in ecosystems. We lastly highlighted pressing topics in microbiology that may advance our understanding of the role of microbial community dynamics in the functional stability of the ecosystem with a focus on N dynamics.
New pathways and players
The primary ecological function of the N cycle is to provide N to organisms such as microbes, plants, and animals (118). Almost all prokaryotes (except for nitrogen-fixing bacteria and archaea) and eukaryotes require fixed forms of N (such as ammonium, nitrate, and monomer-dissolved N, which include amino acids and amino sugars) for their growth (Fig. 1). The physiology of the dissimilatory oxidative and reductive reactions involved in N-cycling has been studied extensively with isolated strains; however, these isolates have mainly been limited to N-fixing bacteria (e.g., the genera Azotobacter and Bradyrhizobium), ammonia- oxidizing bacteria (AOB; e.g., the genera Nitrosomonas and Nitrosospira), nitrite-oxidizing bacteria (NOB; e.g., the genera Nitrobacter and Nitrospira), and heterotrophic-denitrifying bacteria (e.g., the genera Pseudomonas and Azospirillum) (29, 41, 42, 59, 64, 91, 121). In addition to these culture-dependent studies (20, 88), molecular analyses (such as clone libraries and qPCR) have been performed with 16S rRNA genes (28, 48) as well as the “functional” genes involved in N-cycling: nifH (nitrogenase gene subunit H), amoA (ammonia monooxygenase gene subunit A), nirS and nirK (heme cd1-containing and copper-containing nitrite reductase genes), and nosZ (nitrous oxide reductase gene subunit Z) (39, 42, 43, 46, 70, 79, 91, 130). These studies have increased our understanding of the physiology and population dynamics of N2-fixers, AOB, NOB, and denitrifiers in both natural and artificial environments. Many microbes have also been identified as key players in novel N-cycling processes in the past few decades. In this section, we discussed recent studies on N-cycling microbes and their ecological functions from a biogeochemical perspective.
1) Unicellular N2 fixing cyanobacteria (Fig. 1B [1], Table 1)
Table 1.
Recently characterized N cycling pathways and representatives of relevant organisms, as discussed in this review
| Process | Reaction | Environments | Representative organisms | Refs* |
|---|---|---|---|---|
| Unicellular cyanobacterial N2 fixation | N2+ 8H+ + 8e− + 16ATP → 2NH3 + H2 + 16ADP + 16Pi | ocean | Uncultured cyanobacteria (UCYN-A group) Crocosphaera watsonii (UCYN-B group) |
83, 119, 120, 138, 140 |
| Anaerobic phototrophic nitrite oxidation | NO2− + H2O → NO3− + 2H+ + 2e− | sediment/sewage |
Rhodopseudomonas sp. (class Alphaproteobacteria) Thiocapsa sp. (class Gammaproteobacteria) |
33, 108 |
| CH4-dependent denitrification | 4NO3− + CH4 → 4NO2− + CO2 + 2H2O 8NO2− + 3CH4 + 8H+ → 4N2 + 3CO2 + 10H2O |
soil/sediment/ wastewater sludge | “Methanoperedens nitroreducens” (phylum Euryarchaeota) “Methylomirabilis oxyfera” (phylum “NC10”) |
23, 35, 96 |
| Non-denitrifying N2O reduction | N2O + 2H+ + 2e− → N2+ H2O | soil | Anaeromyxobacter sp. (class Deltaproteobacteria) | 51, 102 |
| Aerobic archaeal NH3 oxidation | NH3+ O2 + 2H+ + 2e− → NH2OH + H2O NH2OH + H2O → HNO2 + 4H+ + 4e− |
soil/ocean/lake/ sediment/ hot spring/ wastewater sludge | “Nitrosopumilus maritimus”/ “Nitrososphaera viennensis”/ “N. gargensis”/ “Nitrosocaldus yellowstonii”/ “Nitrosotalea devanaterra” (phylum Thaumarchaeota) |
34, 65, 68, 75, 141 |
| Anaerobic NH3-oxidation (nitrite-dependent NH3 oxidation) | NO2− + NH4+ → N2 + 2H2O NO2− + 2H+ + e− → NO + H2O NO + NH4+ + 2H+ + 3e− → N2H4 + H2O N2H4 → N2 + 4H+ + 4e− |
soil/ocean/lake/ sediment/ wastewater sludge | “Brocadia anammoxidans”/ “B. fulgida”/“B. sinica”/ “Kuenenia stuttgartiensis”/ “Jettenia asiatica”/ “Anammoxoglobus propionicus”/ “Scalindua brodae”/“S. sorokinii”/ “S. wagneri”/“S. profunda” (phylum Planctomycetes) |
56, 112, 113 |
Other references are described in the text.
Biological N2 fixation is the main process that controls the supply of N to organisms in the ocean (137). The filamentous nonheterocystous cyanobacteria of the genus Trichodesmium were believed to be the principal N2 fixers and suppliers of nitrogen compounds in oceanic N-cycling (13). They were also shown to be capable of CO2 fixation via the oxygenic photosynthetic pathway. However, the oxygen (O2) generated by these bacteria may inhibit the activity of nitrogenase, which is the key enzyme in N2 fixation, and the reason for this has yet to be determined (8). In an attempt to resolve this issue, Finzi-Hart et al. (27) analyzed the quantitative metabolic uptake patterns of NaH13CO3 and 15N2 in individual Trichodesmium cells using nanometer-scale secondary ion mass spectrometry (NanoSIMS). They found that the segregation of CO2 and N2 fixation in Trichodesmium was regulated in part by temporal factors.
In addition to Trichodesmium populations, two unicellular diazotrophic cyanobacteria (UCYN groups A and B) have been identified as major N-fixers in oceans (14, 15, 83). Although UCYN-A-type organisms have yet to be cultivated, Zehr’s group screened UCYN-A-type organisms using flow cytometry combined with a UCYN-A-specific qPCR assay and revealed that they did not have an oxygen-evolving photosystem II, RubisCo, or tricarboxylic acid (TCA) cycle in UCYN-A-type organism through a metagenomic analysis of the UCYN-A genomes (120, 138). Thompson et al. subsequently analyzed the quantitative metabolic uptake of NaH13CO3 and 15N2 in cells that were sorted by flow cytometry using halogenated in situ hybridization NanoSIMS (HISH-SIMS), and demonstrated that the UCYN-A-type organism formed a symbiotic partnership with a prymnesiophyte to maintain its growth. In this partnership, the UCYN-A-type organism received fixed carbon in exchange for fixed N, which was transferred to the prymnesiophyte (119). A previous study reported that Crocosphaera watsonii of the UCYN-B group possessed typical cyanobacterial photosynthetic machinery and its N2 fixation rates were the highest at night (137). These filamentous and unicellular diazotrophic cyanobacteria phylotypes are found in oligotrophic tropical oceans, and N2 fixation by UCYN-A/B-type cyanobacteria has been shown to account for a large fraction (>50%) of total N2 fixation in some locations (83, 139, 140).
Farnelid et al. (25) analyzed nifH genes in seawater samples collected from 10 different geographic locations using 454 pyrotag sequencing, and demonstrated that nifH gene clusters related to Alpha-, Beta-, and Gamma-proteobacteria were the most common and had distinct geographic distributions. These non-cyanobacteria groups may also play significant roles in global N-cycling (50). Thus, both cyanobacteria and non-cyanobacteria N-fixers are broadly distributed in marine environments, have unique ecophysiological traits, and may strongly influence the marine N budget (82, 84).
2) Phototrophic and unrealized chemolithotrophic nitrite-oxidizing bacteria (Fig. 1B [2], Table 1)
Nitrite functions as a substrate or intermediate in many N transformation processes, but does not generally accumulate in natural ecosystems (47). Nitrite produced via ammonia oxidation is readily oxidized to nitrate under oxic conditions. Therefore, the phylogeny, physiology, and ecological niches of nitrite-oxidizing bacteria are thought to be diverse. Only five genera of aerobic chemolithotrophic nitrite had been described until recently: Nitrobacter, Nitrotoga, and Nitrococcus within the Alpha-, Beta-, and Gamma-proteobacteria, respectively, and Nitrospira within the Nitrospirae and Nitrospina (66). The genus Nitrospina was provisionally assigned to the Deltaproteobacteria based on its 16S rRNA-based phylogenetic inference (107, 117); however, based on detailed phylogenetic analyses using concatenated marker genes in the Nitrospina gracilis genome, Lücker et al. (73) suggested that Nitrospina may form a novel bacterial phylum distinct from the Proteobacteria, and proposed the name Nitrospinae.
Recent findings have expanded the known physiology and phylogeny of nitrite oxidizers. Griffin et al. (33) enriched phototrophic nitrite oxidizers from freshwater sediments and sewage that could use nitrite as an electron donor for anoxygenic photosynthesis and stoichiometrically oxidized nitrite to nitrate. Two phototrophic nitrite-oxidizing strains, namely Rhodopseudomonas sp. strain LQ17 and Thiocapsa sp. strain KS1 within the Alpha- and Gamma-proteobacteria, respectively, were isolated from sewage (108). Although phototrophs generally have direct impacts on the N cycle through reductive processes such as nitrogen fixation, assimilation, and respiration, this discovery demonstrated that oxidation in N-cycling can be driven by photosynthesis. The numbers of these nitrite-oxidizing phototrophs were low in the most-probable-number (MPN) dilution assay, and the two isolates could use many reductants other than nitrite (organic compounds, H2, HS−, and Fe2+) as electron donors (108). These findings suggest that their functional importance in nitrite oxidation in natural environments may be limited. Sorokin et al. (111) isolated a chemolithotrophic nitrite oxidizer (Nitrolancetus hollandicus) belonging to the widespread phylum Chloroflexi from a bioreactor. N. hollandicus tolerates a broad temperature range (25–63°C) and high nitrite concentration (75 mM, half saturation constant Ks=1 mM) and can grow mixotrophically on nitrite and formate, which distinguishes it from all other known nitrite oxidizers. However, because most conventional nitrifying wastewater treatment plants are operated at lower temperatures and lower nitrite concentrations than optimal conditions for the growth of N. hollandicus, it is unlikely to contribute to nitrite oxidation during the treatment of wastewater. Although the functional importance of both nitrite-oxidizing phototrophs and chemolithotrophic nitrite oxidizers within Chloroflexi remains unclear, even in the environments from which they were isolated, these discoveries have provided an insight into the evolution of nitrite-oxidizing bacteria. A phototrophic origin had previously been suggested for Nitrobacter and Nitrococcus based on their cell morphology and 16S rRNA-based phylogenetic inference (117), and this hypothesis was strongly supported by the discovery of phototrophic nitrite-oxidizing bacteria in the genera Rhodopseudomonas and Thiocapsa, which are closely affiliated with Nitrobacter and Nitrococcus, respectively. Moreover, comparative genomic analysis of nitrite oxidoreductase (Nxr) loci indicated lateral gene transfer events between Nitrolancetus and other nitrite-oxidizing bacteria carrying cytoplasmic Nxr including Nitrobacter and Nitrococcus, which suggested that the horizontal transfer of the Nxr module allowed the spread of nitrite oxidation ability during bacterial evolution.
3) CH4-dependent denitrifying bacteria (Fig. 1B [3], Table 1)
Microbes that couple the formation of N2 with the oxidation of organic carbon (C) (organotrophic denitrification) have been examined in detail; however, some microbes can couple the formation of N2 with the oxidation of various reductants other than organic C (Fig. 2). The coupling reaction with CH4 is currently receiving attention because of its biogeochemical and evolutionary importance (23). The anaerobic oxidation of CH4 coupled with denitrification is thermodynamically feasible; therefore, it has been speculated that this reaction could occur in nature. Raghoebarsing et al. (96) provided empirical evidence from an enrichment culture from sediment in a Dutch canal, which consisted of a co- culture of a dominant bacterial phylotype of the candidate phylum NC10 and archaea that were phylogenetically positioned between Methanosaeta (methanogenesis) and ANME-2 (anaerobic methanotrophs), later named ANME-2d. Ettwig et al. (22, 23) subsequently showed that the complete anaerobic oxidation of CH4 coupled with the reduction of nitrite to N2 could be achieved using bacteria identified as “Candidatus Methylomirabilis oxyfera” in the absence of archaea. Genome analysis of “Ca. M. oxyfera” revealed that this bacterium possessed a well-established aerobic pathway for CH4 oxidation, whereas it lacked known genes for N2 production (the gene cluster encoding enzymes for the reduction of N2O to N2 [nosZDFY]). Isotopic labeling experiments also revealed that “Ca. M. oxyfera” bypassed the denitrification intermediate, N2O, by converting two NO molecules to N2 and O2, which was then used to oxidize CH4. The proposal of this metabolic pathway had a significant impact on the “evolution of organisms” debate because the suggested intra-aerobic metabolism allowed for the possibility that oxygen was available for microbial metabolism before the evolution of oxygenic photosynthesis. The metabolic mechanisms of “Ca. M. oxyfera” may be controversial because the presence and nature of the oxygen-producing enzyme are unknown. However, the functional importance of CH4-dependent denitrifiers in both natural and engineered environments has been suggested by the detection of 16S rRNA genes or pmoA of NC10 bacteria with high similarity to “Ca. M. oxyfera” in wastewater sludge (74), paddy soil (125), and oligotrophic lake sediments (63). Haroon et al. (35) more recently revealed that archaea belonging to ANME-2d, named “Ca. Methanoperedens nitroreducens”, which were co-enriched with bacteria of the candidate phylum NC10 in their consortia, exhibited CH4-dependent nitrate reduction to N2 (40) through metagenomic, single-cell genomic and metatranscriptomic analyses combined with isotopic labeling experiments. “Candidatus M. nitroreducens” possesses genes that reduce nitrate to nitrite (narGH), but lacks genes for the subsequent steps in denitrification, and can supply nitrite to “Ca. M. oxyfera” in the consortia by coupling nitrate reduction to nitrite with anaerobic CH4 oxidation in a reverse methanogenesis pathway.
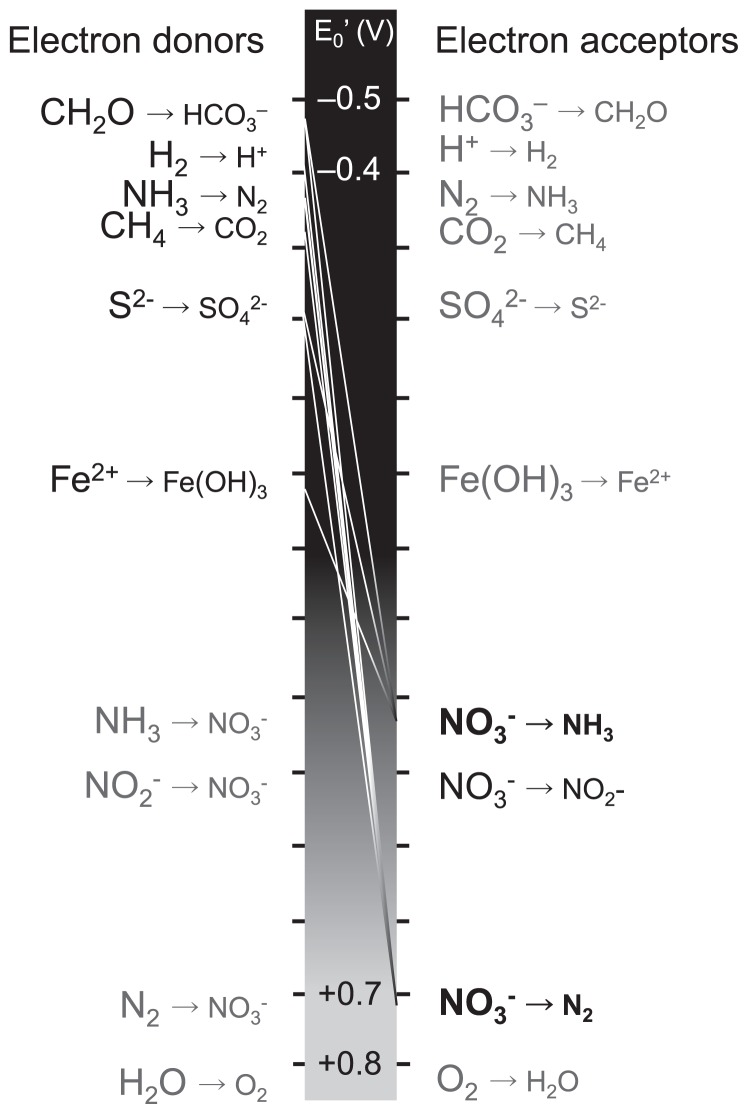
Fig. 2.
Representative reactions that have been confirmed with bacterial isolates of nitrate reduction to N2 or NH3 coupled with the oxidation of reductants ranging from organic carbon to Fe (II) along the redox tower.
4) Non-denitrifying N2O-reducing bacteria (Fig. 1B [4], Table 1)
Nitrous oxide is a greenhouse gas that is controlled under the Kyoto Protocol. Among non-CO2 greenhouse gasses, the contribution of N2O to climate forcing is second only to methane, and has a global warming potential that is ca. 300 times greater than an equivalent amount of CO2 (97). It has also been shown to be the single most dominant ozone-depleting substance (97). The production and consumption (reduction) of N2O are largely governed by microbial activities. N2O is produced mainly through denitrification and nitrification. Genome analysis of Agrobacterium tumefacience first revealed that denitrifying bacteria can lack the Nos gene, which codes for the N2O reductase that catalyzes the reduction of N2O to N2 (129). Approximately one-third of the genomes that possess Nir genes, which encode the nitrite reductases necessary for denitrification, are known to lack nosZ. Genome analysis revealed that aerobic ammonia-oxidizing bacteria and archaea also lacked nosZ. The reduction of N2O to N2 has been attributed to nos-possessing denitrifiers (94). However, recent studies demonstrated that N2O reducers were not always the denitrifiers. Sanford et al. (102) and Jones et al. (51) performed comprehensive phylogenetic analyses of the full-length nosZ in genomes retrieved from a public database and discovered that nosZ phylogeny formed two distinct clades (clades I and II). Clade II has not yet been detected with widely used primer sets or accounted for in studies on N2O-reducing communities. Jones et al. (51) designed primers to detect nosZ in clade II and showed that nosZ from clade II was at least as abundant as that from clade I in various environments using quantitative PCR. In addition, Sanford et al. (102) showed that approximately half of the genomes from phylogenetically diverse microbes containing nosZ from clade II (Delta- and Epsilon-proteobacteria, Verrucomicrobia, Bacteroidetes, Chlorobi, Firmicutes, Deferribacteres, and Euryarchaota) lacked nirK or nirS. They subsequently verified the physiological function of nosZ from clade II as an N2O reductase in growth experiments using the non-denitrifying species, Anaeromixobacter dehalogenans, which is widely and abundantly distributed on land, and N2O as an electron acceptor. Accordingly, these sequential studies showed that nosZ was more diverse than previously thought and that non-denitrifiers possessing nosZ were widely distributed, at least on land (51, 102). Denitrification is not always the result of successive reactions carried out in a single cell, but can result from successive reactions in microbial communities. Non-denitrifying populations with a broad range of metabolisms and habitats may be significant contributors to the mitigation of N 2O emissions.
5) Aerobic ammonia-oxidizing archaea (Fig. 1 [5], Table 1)
Ammonia oxidation, the first and rate-limiting step of nitrification, was considered to be performed mostly by certain groups of chemolithoautotrophic Proteobacteria (genera Nitrosospira, Nitrosomonas, and Nitrosococcus) for more than one hundred years (44). The recent discovery of homologs of ammonia monooxygenase (Amo) genes in archaea of the phylum Thaumarchaeota and the cultivation of thaumarchaeal ammonia oxidizers (“Candidatus Nitrosopumilus maritimus”, a marine group I.1a representative; “Candidatus Nitrososphaera viennensis” and “Candidatus Nitrososphaera gargensis”, soil group I.1b representatives isolated from soil and enriched from a hot spring, respectively; “Candidatus Nitrosocaldus yellowstonii”, thermophilic ThAOA or HWCGIII representatives enriched from a hot spring; “Candidatus Nitrosotalea devanaterra”, a soil group I.1a-associated representative enriched from soil) has radically changed this view, indicating that an additional, predominant group of microbes is also able to perform this process.
Accurate estimates of nitrification activity may revise our understanding of oceanic productivity (76, 133). Nitrate is the most abundant form of fixed N in open oceans (124), and nitrification was, until recently, believed to occur almost entirely in deep waters (133) because ammonia oxidation is inhibited by light and ammonia concentrations in surface waters are generally markedly lower than those estimated to represent the growth threshold of AOB. Therefore, nitrate was thought to be a non-regenerated nutrient form in the euphotic zone, and nitrate uptake in surface waters was generally ascribed to new primary production. However, physiological studies on “Ca. N. maritimus” (75) have suggested that marine ammonia-oxidizing archaea (AOA) may be adapted to these low ammonia levels. Both its extremely low substrate threshold and half-saturation constant are unprecedented, but consistent with the conditions found in oligotrophic open oceans and effectively compete with bacterio- and phytoplankton. Advances in 15N measurement techniques have also revealed the occurrence of nitrification in the euphotic zone or at the bottom of this zone, most likely by the thaumarchaeal ammonia oxidizers (16). Accurately quantifying the role of nitrification in the production of nitrate in oligotrophic surface waters will contribute to more realistic model predictions of ocean productivity.
The successful enrichment of acidophilic “Ca. N. devanaterra” within thaumarchaeota group I.1a-associated has provided a new solution to the longstanding paradox of nitrification in terrestrials. Approximately 30% of the world’s soils are acidic (pH <5.5) and autotrophic ammonia oxidation can occur in acidic soils. However, all cultivated aerobic AOB readily enriched from acid soils are neutrophilic, and none grow in liquid batch-cultures with pH below 6.5 (18). Therefore, some ammonia oxidation mechanisms have been proposed in acidic soils (e.g., the presence of a neutrophilic space or the ureolytic growth of AOB in soils) (18). However, 13CO2-DNA-stable-isotope probing (SIP) experiments convincingly linked the autotrophic nitrification activity in acidic soils to thaumarchaeal ammonia oxidizers (68, 141). Additionally, the growth of “Ca. N. devanaterra” was shown to be chemolithotrophic and optimal in a pH range between 4 and 5, unlike all previously cultivated ammonia oxidizers. Moreover, the pH selection of soil thaumarchaeal ammonia oxidizers was demonstrated using 454 barcoded pyrosequencing, which identified group I.1a-associated thaumarchaeal amoA lineages with specific adaptations to acidic soils (34). These studies have provided a plausible explanation for the high rates of nitrification in acidic soils and also confirmed the vital role played by thaumarchaea in meditating ammonia oxidation in acidic soils.
6) Anaerobic ammonia-oxidizing bacteria (Fig. 1 [6], Table 1)
The discovery of anammox (anaerobic ammonium oxidation) filled in certain knowledge gaps in the N loss pathway (19). Oceanographers previously reported a pervasive loss in ammonium in highly stratifiedanoxic basins since the mid-1960s from analyses of the N balance, which indicated that ammonium was removed by anaerobic microbial activity (99). According to a thermodynamic perspective, the physicist Broda (10) also proposed the existence of lithotrophic microbes that could derive their energy for growth from the oxidation of ammonia coupled with the reduction of nitrate or nitrite to produce N2. This empirical discovery was made in the bioreactors of wastewater treatment plants in the 1990s. Strous et al. (113) obtained a highly enriched culture of anammox bacteria, named “Candidatus Brocadia anammoxidans” within the order Planctomycetales by density gradient centrifugation. The culture produced N2 from ammonium and nitrite and was capable of CO2 fixation (86, 112, 113). Since then, five genera of anammox bacteria have been (provisionally) described: “Brocadia”, “Kuenenia”, “Anammoxoglobus”, “Jettenia” (all fresh water species), and “Scalindua” (marine species). The anammox reaction has been detected not only in anoxic wastewater, but also in natural environments such as marine, coastal, and estuarine sediments, anoxic basins, mangrove sediments, oceanic oxygen-depleted zones, freshwater sediments, and even in agricultural soils (3, 4, 60, 67, 69, 103, 132, 136, 142). The control of denitrification and anammox, which are the main N loss processes and fundamentally rely on different organisms and metabolic pathways, is receiving particular attention in the ocean and is discussed below.
Relationship between microbial community dynamics and N biogeochemistry
As described above, the discovery of novel processes and players has greatly broadened our knowledge of how N is transformed and utilized in ecosystems. In this section, we have discussed attempts to elucidate the relationship between microbial community dynamics and N biogeochemistry. We also highlighted how such approaches have advanced our understanding of the biogeochemical roles of microbial communities in N cycles.
Microbiologists have attempted to identify the specific microbes responsible for N transformation processes and describe how their population dynamics impact N transformations. However, it is generally difficult to identify the biogeochemical roles of specific microbes involved in the assemblages of diverse microbial communities. Therefore, microbiologists often make rough estimates of their biogeochemical roles by correlating microbial community dynamics with environmental gradients and/or changes in N biogeochemical properties. The basic concept of linking microbial community dynamics with N biogeochemistry is based on individual processes in the N cycle being mediated by a certain group of the microbial community, with the population dynamics of this group being likely to affect the rate of the corresponding process (106). This response is most likely to be observed when the process is physiologically defined and when the responsible microbes are metabolically and phylogenetically limited. Ammonia oxidation, for example, has physiologically been defined as ammonia oxidation to nitrite via hydroxylamine. The microbes responsible are metabolically and phylogenetically limited groups (genera Nitrosospira, Nitrosomonas, and Nitrosococcus within the phylum Proteobacteria and Candidatus genera “Nitrosopumilus”, “Nitrososphaera”, “Nitrosocaldus”, and “Nitrosotalea” within the phylum “Thaumarchaeota”). The positive correlation between the quantity of amoA and rate of gross ammonia oxidation (nitrification) has frequently been observed in many environments such as forests, agricultural soils, and the ocean (7, 36, 46, 127), and has permitted us to identify the groups involved in ammonia oxidation (e.g., proteobacterial or thaumarchaeal ammonia oxidizers). For example, Di et al. (21) suggested that ammonia oxidation in N-rich grassland soils was mainly driven by proteobacterial ammonia oxidizers by showing that the amoA quantity of Proteobacteria in soil was related to the net nitrification rate, whereas the amoA quantity of “Thaumarchaeota” was not. The observed correlation also allowed us to explain changes in the nitrification rates along environmental gradients by changes in the population size or community compositions of ammonia oxidizers (127). Hawkes et al. (36) showed that changes in the gross nitrification rate along with changes in the plant community after exotic plant invasion in grasslands could be explained by the population size of AOB. The basic concept linking microbial community dynamics with N biogeochemistry apply to the majority of other dissimilating processes in N-cycling, including N-fixation, nitrite oxidation, denitrification, N 2O reduction, and anammox (5, 38, 43, 58, 67, 100, 135). Spatial variations in N2O/N2 emission potential in a grassland pasture can be described using spatial variations in the relative abundance of nirS and nirK in denitrifiers to nosZ in denitrifiers and non-denitrifies (93). Differences in the N loss pathway in oceans could be explained by the population size or community composition of denitrifiers and anammox bacteria (123). However, we need to be aware that only a rough estimate of their biogeochemical roles can be made because of limitations and biases in the basic concept. Other environmental factors (e.g., resource supply) may have a greater impact on the rates of N transformations than microbial community dynamics. Microbes are generally metabolically versatile; therefore, population dynamics do not always cause changes in the specific process rate. We have generally focused on sensitive microbial groups, the population size of which may change, with less attention been given to the functional importance of insensitive groups. Additionally, there may be a discrepancy between the presence of a gene (or even mRNA) and in situ activity. Therefore, RNA- or protein-based analyses such as metatranscriptomic or proteomic analyses combined with substrate uptake assays such as SIP, microautoradiography combined with fluorescence in situ hybridization (MAR-FISH), and NanoSIMS are powerful tools that can be used to refine rough estimates. The series of attempts to link microbiological community dynamics with N biogeochemistry discussed here have provided a more mechanistic understanding of N dynamics than a direct observation. These findings should also contribute to more realistic model predictions of the N cycle.
Highlighted Topics
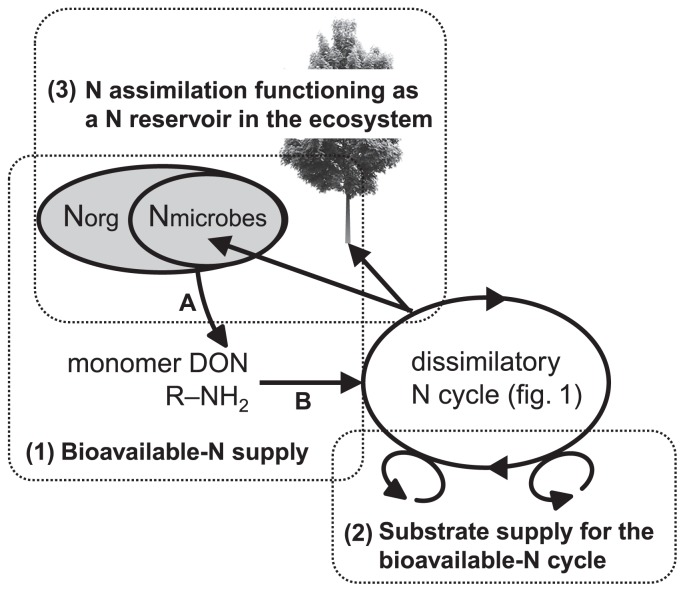
We here highlighted three topics that have not been examined in detail, but are essential for identifying the roles of microbial community dynamics in the functional stability of the ecosystem. These topics are fundamental for understanding the control of ecosystem functioning, including (1) how bioavailable N is microbially supplied, (2) how the N supplied is dissimilatorily transformed to yield energy, and (3) how the storage of N in an ecosystem through microbial N assimilation using energy contributes to the stability of the ecosystem.
1) N mineralization and the supply of available N (Fig. 3 [1])
Fig. 3.
Schematic outline of N flows and microbial involvements at the ecosystem level, as discussed in this review. (1) bioavailable N is microbially supplied through the mineralization of organic N including microbe-derived N, (2) supplied bioavailable N is dissimilatorily transformed in the redox reaction with other oxidants/reductants to yield energy, and (3) the storage of N in an ecosystem through microbial N assimilation using this energy contributes to the stability of the ecosystem. A: depolymerization; B: ammonification.
The first topic discussed is the mineralization of N-containing organic compounds to NH4+ and the microbes responsible for this transformation. The mineralization of dissolved organic N (DON) to NH4+, which represents a bottleneck in the subsequent N-cycling processes and the supply of available N to organisms in various environments, occurs through microbial enzymatic activity. However, little progress has been made in understanding the relationship between N mineralization and microbial community dynamics. The main reason for this may be that the basic concept linking microbial community dynamics with N biogeochemistry discussed above cannot be sufficiently applied to N mineralization (106). We generally measure N mineralization in the form of ammonium production as a single process; however, it is actually the sum of multiple distinct physiological processes. Therefore, N mineralization can involve diverse microbial communities that contribute to the process; N mineralization rate may be insensitive to the dynamics of the microbial community involved in the mineralization (106).
Biogeochemists are currently attempting to break down the entire process of mineralization into individual processes, which may then be physiologically defined and more sensitive to microbial community dynamics. Schimel and Bennet (105) suggested that a single mineralization step in soil should be separated into at least two processes that are under different microbial control, namely, depolymerization (proteolysis and aminization; organic-N polymers to R-NH2) (Fig. 3 [A]) and ammonification (deamination and deamidation; R-NH2 to NH3 + H2O) (Fig. 3 [B]). Ocean biogeochemists have agreed that this fractionation should be performed to enable N regenerated production to be accurately described (54, 80). Ammonium can potentially be produced by the direct enzymatic cleavage of a free amino group, either amine- or amide-N (R-NH2). Because deaminase and deamidase enzymes are intercellular in nature, are active inside living organisms, and can be produced by most microbes, the microbial production of NH4+ can depend on the N status of cells taking up small organic N compounds (R-NH2), which will subsequently determine if N is sequestered or excreted as NH4+. Small organic N monomers such as amino acids, amino sugars, and nucleotides are produced by the activities of several extracellular enzymes, which break down high-molecular-weight polymeric organic compounds, including proteins, cell wall polymers (aminopolysaccharides), and nucleic acids. For example, proteins are broken down by a wide variety of proteinases and peptidases. Proteinases break down large proteins, while peptidases may cleave tri- or dipeptides or split off an individual amino acid, which is then taken up by a microbe. Amino-polysaccharides are also broken down by extracellular enzymes. Chitinase depolymerizes chitin (a polymer of N-acetylglucosamine), which forms the cell walls of many fungi and is also a part of insect exoskeletons. Chitinase breaks chitin into dimers of chitobiose. N-acetylglucosaminidase subsequently cleaves chitobiose into two molecules of N-acetylglucosamine. Several enzymes degrade the peptidoglycan portion of bacterial cell walls. For example, lysozyme breaks the β-1,4 linkage between N-acetylmuramic acid and N-acetylglucosamine. The end products of extracellular enzymes that degrade microbial cell walls are individual amino sugars, which are taken up by microbes. Because each of the extracellular enzymes used in proteolysis and aminization may be synthesized by a more limited group of microbes than the intercellular enzymes used in deamination and deamidation, this process may become more physiologically defined and, thus, more sensitive to the dynamics of the microbial community involved in the extra-cellular enzyme steps.
Microbiologists are currently attempting to identify the microbial groups that contribute to the extracellular enzyme steps in order to link this process with microbial community dynamics. Depolymerization occurs, to some degree, in most environments; therefore, if this process is carried out by specialized or phylogenetically limited microbial groups, such groups may be ubiquitous. Zimmerman et al. (143) recently analyzed 3,058 annotated prokaryotic genomes to identify taxa with the genetic potential to produce chitinase and N-acetylglucosaminidase, and found a non-random correlation between genetic potential and 16S rRNA-based phylogeny. Chitinase- and N-acetylglucosaminidase-positive genotypes were detected in 12 and 19 of the 30 phyla. All genomes of the phylum Acidobacteria, which are ubiquitous and abundant in soil, but have unknown ecological characteristics (49, 52), possessed both chitinase and N-acetylglucosaminidase-encoding genes, which allowed the complete hydrolysis of chitin substrates by individual organisms. Most of the genomes of the genus Vibrio, which can be found in a range of aquatic habitats, also possess these genes. These ubiquitous groups may be actively involved in the depolymerization process in terrestrial or aquatic environments. Identifying and studying the ecology of the microbial communities responsible for N mineralization combined with enzyme activity analyses and proteomics will provide a deeper insight into how the supply of bioavailable N can be microbially controlled to maintain the functional stability of ecosystems.
2) N-dissimilating reactions and substrate availability/limitation (Fig. 3 [2])
The second topic relates to the supply of substrates for dissimilating N reactions. Previous studies on dissimilating N transformations have primarily focused on the flow of N. However, other substrates in N transformation redox reactions that are reduced when N is oxidized (or oxidized when N is reduced) are also regulating factors that drive the flow of N. The functioning of an ecosystem can primitively be supported by the energy yielded by microbial N-dissimilating reactions because each microbial function can be maintained by this energy, and the sum of these functions may represent the principal functions of the ecosystem. Therefore, the nature of available, but limited substrates that drive energy-yielding N transformations can characterize ecosystems. For example, we may estimate the denitrifier’s control on the denitrification rate in an ecosystem by tracking the genes of denitrifiers (such as nirS, nirK, and nosZ) and N-gas emissions, as discussed in the previous section. However, the nature of available substrates other than nitrate may have a stronger influence on denitrification rates by altering denitrifying microbial communities. Moreover, the diversification of the substrate that microbial communities in ecosystems utilize for energy-yielding N transformations is strongly relevant to the robustness of the ecosystem.
Many pairs of electron donors and acceptors have been observed in each N dissimilation reaction using enrichment cultures or isolates. The recent findings described above have also revealed new electron donors/acceptors and energy sources (phototrophic nitrite-oxidizing bacteria, CH4-dependent denitrifying bacteria, non-denitrifying N2O reducing bacteria, and anaerobic AOB) for N transformations. The microbial reduction of nitrate, for example, is often considered to be coupled with the oxidation of organic-C (e.g. acetate, succinate, and glucose) on land; however, this reduction coupled with the oxidation of reductants ranging from organic C to Fe (II) along the redox tower has been reported previously in bacterial isolates (11) (Fig. 2). The constituents of organic C may be selectively used for microbial growth. Additionally, reactions observed in a single cell can also be performed in the surrounding cells if there is a flow of electrons between these cells (57, 98).
The spatial distribution and ecological importance of these nitrate-reducing reactions coupled with reductants other than organic carbon have not been sufficiently studied at the ecosystem level. However, several recent studies have suggested that the supply of available substrates could regulate the pathway of N loss through nitrate reduction in an ecosystem. Kuyper’s group reported that N was lost in the Black Sea suboxic zone and Peruvian oxygen-minimum zones (OMZs), which mainly occurred through an ammonia-dependent denitrification (anammox) reaction, and also that anammox was coupled with an ammonia production process known as dissimilatory nitrate reduction to ammonium (DNRA) (Fig. 1A) (67) or DON ammonification (55) using 15N-labelling experiments corroborated by functional gene expression analyses. On the other hand, Ward’s group showed that N was lost in the OMZs in the Arabian Sea mainly via DOC-dependent denitrification under high DOC production conditions (126). Dalsgaad et al. (17) more recently showed that both anammox and denitrification were detected in a transect along the coast of South America (the eastern tropical South Pacific OMZ), and that anammox occurred at low rates at almost every station, whereas denitrification was less commonly detected, but occurred at very high rates at a few stations. These findings suggest that the timing and magnitude of the DOC supply may regulate the relative contribution of anammox and denitrification in the anoxic region of oceans. Regarding the other mechanisms, Hayakawa et al. (37) revealed from the stoichiometric analysis of sulfur and nitrogen that N was lost through sulfur-dependent denitrification in FeS-rich sediments. Yang et al. (131) proposed a new N loss pathway from tropical upland soils, anaerobic ammonium oxidation to N2 coupled with iron (III) reduction, otherwise known as Feammox, based on the results of their 15N-labeling experiments. Although this reaction can be thermodynamically favorable (Fig. 2), the microbes that could govern this reaction have not yet been identified and the possibility of a corporative reaction with organotrophic denitrification should not be ruled out. As described here, studies on new electron donors or acceptors in N loss reactions as well as the importance of new N loss pathways are being conducted at the ecosystem level (Fig. 2). Further studies on the diversity and variability of the substrates utilized in N transformations are needed. Understanding the diversity and variability of substrates and related microbial community dynamics may allow us to understand the nature of the available, but limiting resources of the ecosystem and also characterize ecosystems.
3) N-assimilating reactions and their functions as a N reservoir (Fig. 3 [3])
Ecologists have historically considered that communities of organisms are primarily structured by available resources and, in particular, by the nature of the limiting resource (9). A variant in the substrate partitioning theory, which was built around the Monod model of microbial growth, has effectively been used to explain the microbial community dynamics (9, 128). The properties associated with substrate partitioning have been expressed by the terms copiotrophy and oligotrophy in microbiology (r-strategy and K-strategy in ecology, respectively) (26, 62). These terms have primarily been used to describe the relationship between growth rates and carbon concentrations (typically in the medium in cultivation) (62). Fierer et al. (26) showed that growth responses to sucrose amendments in soils slightly differed among taxa at the phylum/class level (phylum Acidobacteria and Bacteroidetes, class Betaproteobacteria). Martiny et al. (77) analyzed the distribution of 89 functional traits, most of which were associated with simple carbon utilization, across a broad range of prokaryotes using genome and phenotypic carbon substrate utilization data, and found that the capacity for simple carbon utilization correlated with the 16S rRNA-based phylogeny. However, carbon is not always the limiting substrate for microbial growth. Previous studies have shown that many natural environments, including terrestrial areas, lakes, and oceans, are frequently N-limited. Microbial community dynamics may be reflected in both the nature of the carbon sources and supply of N available for their growth in such environments. However, few studies have explored the N assimilation kinetics of each microbial taxon in such environments and the link between N assimilation properties and microbial community dynamics has not been elucidated in detail (30, 61). Taxon-specific mechanisms are difficult to isolate because all microbes are involved in the N assimilation process. Furthermore, measuring the N assimilation rate is methodologically difficult (particularly in the case of soils) because of the difficulty involved in separating microbial cells from soil particles (45). Therefore, we can describe community dynamics, but do not have an appropriate theory or model to explain these dynamics in N-limiting environments.
Another important aspect of N assimilation by microbial communities and microbial growth is their function as an N reservoir in an ecosystem. Microbial biomass N is the main component of organic N in an ecosystem and can, thus, be a source of bioavailable N. The lysis of microbial cells and subsequent release of N (depolymerization and ammonification) can supply bioavailable N. For example, we emphasized the importance of microbial N assimilation and community dynamics in forest ecosystems. Plants in many boreal and temperate forests strongly demand N for their growth and increased productivity in late spring and summer. Therefore, the supply of N available to plants must be greater during the growing season. Bacterial growth and an increase in the biomass N in summer and autumn and bacterial death and subsequent release of biomass N in winter may be important for meeting the N demands of plants in the coming spring and summer. The activity of the bacterial community in soil has generally been correlated with the soil temperature; thus, it is low in winter. Seasonal variations have been reported in bacterial communities, with the bacterial population size being smaller in the winter. Furthermore, bacterial death can be accelerated when soils are frozen and thawed repeatedly. Therefore, a large quantity of N from bacterial biomass can be released into the soil in the winter. The degree to which bacteria-derived organic N can be utilized by plants in the plant-growing season remains unclear. However, Shibata et al. (109) recently showed that increases in freeze-thaw cycles, which accelerate bacterial death and the physical degradation of organic N, significantly enhanced the net production of ammonium in soil during the winter, which indicated that bacteria-derived organic N may be an important source of NH4+. Litters that fall in winter and fine roots that are killed in winter provide additional N. The depolymerization and/or ammonification of bacteria-derived organic N can be accelerated by increases in temperature or snow melts in the spring, but may also occur in soil in the winter, particularly when it is not frozen. The Schmidt group (71, 72, 104) used light microscopy to show that fungal biomass reached its annual peak with high diversity under snow in tundra soils, although their function in the mineralization of bacteria-derived organic N remains unknown. To date, bacterial growth and metabolism have been confirmed at −15°C and −32°C, respectively (6, 87). Understanding 1) the microbial community dynamics involved in bacterial growth before winter, 2) bacterial death and mineralization of the released N in winter, and 3) the competition with plants for N assimilation in the plant-growing season will allow us to identify the role of microbial community dynamics in the functional stability of the forest ecosystem.
Regional variations in the seasonal succession of the roles of microbial community dynamics in the functional stability of an ecosystem should not be ignored. For example, forests located in the monsoon climate region in Japan generally have the highest water discharge rate during the plant-growing season (early summer) because precipitation inputs are typically high (89). The high discharge rates during this season are in marked contrast to forests in most regions of the United States and Europe, in which the summer period is characterized by high evapotranspiration, but also by low or moderate precipitation inputs that result in low water discharge rates (81, 89). High water discharge causes the marked loss of nitrate, which is the main source of available N for plants, through leaching from the soil. Thus, the magnitude and timing of the mineralization of microbe-derived organic N and subsequent nitrification may be more strictly regulated based on the season in forests in Japan than in the United States and Europe. Studies that can evaluate such regional differences in the ecological consequences of microbial community dynamics will allow us to characterize regional variations in ecosystems and should also provide a deeper insight into how ecosystems will respond to environmental changes.
Concluding remarks
We have presented three topics that could further our understanding on the role of microbial community dynamics in the functional stability of ecosystems. These topics are not limited to functionally equivalent microbial groups or to specific (a single or few) N transformation processes. Therefore, the basic concepts that are discussed in the previous section and used to link microbial community dynamics with N biogeochemistry are not pertinent. Moreover, advances cannot be expected simply by using high-throughput sequencing techniques to examine the assembly of individual genes, even though these techniques have led to great successes in the heuristic search for gene diversity. Substrates that drive N transformations, the energy efficiency of N transformation redox reactions, competition for available N, and mutual interactions between micro- and macro-organisms must also be considered. These kinds of studies by microbiologists can potentially provide a more mechanistic understanding of the fundamental issues of ecosystem ecology in unprecedented levels of detail.
Another topic that ecosystem ecologists have focused on is how a diverse range of organisms can contribute to the resilience of an ecosystem (2, 31). Phylogenetically limited ammonia oxidizers are known to be responsible for critical functions in an ecosystem (nitrification). The ammonia oxidation rate is more likely to be influenced by environmental changes or ecosystem perturbations than the mineralization rate (32, 92), which may be attributed to the markedly lower phylogenetic diversity of ammonia oxidizers than that of microbial communities involved in mineralization. Thus, the associations between microbial diversity and stability of the biogeochemical functions of the microbial community may directly impact the resilience of an ecosystem.
Recent technological advances have permitted microbiologists to assess these topics in ecosystem ecology. Advances in high-throughput sequencing have allowed microbiologists to better delineate microbial phylogenetic and functional diversities through meta-genomic, transcriptomic, and proteomic analyses. The use of 15N can also improve our understanding on microbial functional metabolism and diversity as well as ecosystem functions through Nano-SIMS, SIP, and N isotope tracer techniques. As described here, there are many fields for microbiologists to be involved in in ecosystem ecology, and they can now play indispensable roles in the fundamental issues of ecosystem ecology in collaborations with ecologists, geochemists, and geologists.
Acknowledgements
We are grateful to Drs. Takuhei Shiozaki and Mark Green for providing helpful comments. This work was supported by Grants-in-Aid for Scientific Research from the Japanese Society for the Promotion of Science (Nos. 24658133, 24780055 and 25252026) and the GRENE/Ecoinformatics project (PI: Motomi Itoh) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
References
- 1.Aber J, McDowell W, Nadelhoffer K, Magill A, Berntson G, Kamakea M, McNulty S, Currie W, Rustad L, Fernandez I. Nitrogen saturation in temperate forest ecosystems. BioScience. 1998;48:921–934. [Google Scholar]
- 2.Allison SD, Martiny JBH. Colloquium paper: resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci USA. 2008;105:11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amano T, Yoshinaga I, Okada K, Yamagishi T, Ueda S, Obuchi A, Sako Y, Suwa Y. Detection of anammox activity and diversity of anammox bacteria-related 16S rRNA genes in coastal marine sediment in Japan. Microbes Environ. 2007;22:232–242. [Google Scholar]
- 4.Amano T, Yoshinaga I, Yamagishi T, Van Thuoc C, Thu PT, Ueda S, Kato K, Sako Y, Suwa Y. Contribution of anammox bacteria to benthic nitrogen cycling in a mangrove forest and shrimp ponds, Haiphong, Vietnam. Microbes Environ. 2011;26:1–6. doi: 10.1264/jsme2.me10150. [DOI] [PubMed] [Google Scholar]
- 5.Attard E, Poly F, Commeaux C, Laurent F, Terada A, Smets BF, Recous S, Le Roux X. Shifts between Nitrospira- and Nitrobacter-like nitrite oxidizers underlie the response of soil potential nitrite oxidation to changes in tillage practices. Environ Microbiol. 2010;12:315–326. doi: 10.1111/j.1462-2920.2009.02070.x. [DOI] [PubMed] [Google Scholar]
- 6.Bakermans C, Skidmore M. Microbial respiration in ice at subzero temperatures (−4°C to −33°C) Environ Microbiol Rep. 2011;3:774–782. doi: 10.1111/j.1758-2229.2011.00298.x. [DOI] [PubMed] [Google Scholar]
- 7.Beman JM, Popp BN, Francis CA. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2008;2:429–441. doi: 10.1038/ismej.2007.118. [DOI] [PubMed] [Google Scholar]
- 8.Berman-Frank I, Lundgren P, Chen YB, Küpper H, Kolber Z, Bergman B, Falkowski P. Segregation of nitrogen fixation and oxygenic photosynthesis in the marine cyanobacterium Trichodesmium. Science. 2001;294:1534–1537. doi: 10.1126/science.1064082. [DOI] [PubMed] [Google Scholar]
- 9.Bever JD, Platt TG, Morton ER. Microbial population and community dynamics on plant roots and their feedbacks on plant communities. Annu Rev Microbiol. 2012;66:265–283. doi: 10.1146/annurev-micro-092611-150107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bronda E. Two kinds of lithotrophs missing in nature. Z Allg Mikrobiol. 1977;17:491–493. doi: 10.1002/jobm.3630170611. [DOI] [PubMed] [Google Scholar]
- 11.Burgin AJ, Hamilton SK. Have we overemphasized the role of denitrification in aquatic ecosystems? A review of nitrate removal pathways. Front Ecol Environ. 2007;5:89–96. [Google Scholar]
- 12.Canfield DE, Glazer AN, Falkowski PG. The evolution and future of earth’s nitrogen cycle. Science. 2010;330:192–196. doi: 10.1126/science.1186120. [DOI] [PubMed] [Google Scholar]
- 13.Capone DG. Trichodesmium, a globally significant marine cyanobacterium. Science. 1997;276:1221–1229. [Google Scholar]
- 14.Church M, Jenkins B, Karl D, Zehr J. Vertical distributions of nitrogen-fixing phylotypes at Stn Aloha in the oligotrophic North Pacific Ocean. Aqua Microbial Ecol. 2005;38:3–14. [Google Scholar]
- 15.Church MJ, Short CM, Jenkins BD, Karl DM, Zehr JP. Temporal patterns of nitrogenase gene (nifH) expression in the oligotrophic North Pacific Ocean. Appl Environ Microbiol. 2005;71:5362–5370. doi: 10.1128/AEM.71.9.5362-5370.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark DR, Rees AP, Joint I. Ammonium regeneration and nitrification rates in the oligotrophic Atlantic Ocean: implications for new production estimates. Limnol Oceanogr. 2008;53:52–62. [Google Scholar]
- 17.Dalsgaard T, Thamdrup B, Farías L, Peter Revsbech N. Anammox and denitrification in the oxygen minimum zone of the eastern South Pacific. Limnol Oceanogr. 2012;57:1331–1346. [Google Scholar]
- 18.De Boer W, Kowalchuk GA. Nitrification in acid soils: micro-organisms and mechanisms. Soil Biol Biochem. 2001;33:853–866. [Google Scholar]
- 19.Devol AH. Nitrogen cycle: solution to a marine mystery. Nature. 2003;422:575–576. doi: 10.1038/422575a. [DOI] [PubMed] [Google Scholar]
- 20.Dewi Puspita I, Kamagata Y, Tanaka M, Asano K, Nakatsu CH. Are uncultivated bacteria really uncultivable? Microbes Environ. 2012;27:356–366. doi: 10.1264/jsme2.ME12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di HJ, Cameron KC, Shen JP, Winefield CS, O’Callaghan M, Bowatte S, He JZ. Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat Geosci. 2009;2:621–624. [Google Scholar]
- 22.Ettwig KF, van Alen T, van de Pas-Schoonen KT, Jetten MSM, Strous M. Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl Environ Microbiol. 2009;75:3656–3662. doi: 10.1128/AEM.00067-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ettwig KF, Butler MK, Le Paslier D, et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 24.Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive earth’s biogeochemical cycles. Science. 2008;320:1034–1039. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- 25.Farnelid H, Andersson AF, Bertilsson S, Al-Soud WA, Hansen LH, Sørensen S, Steward GF, Hagström Å, Riemann L. Nitrogenase gene amplicons from global marine surface waters are dominated by genes of non-cyanobacteria. PLoS One. 2011;6:e19223. doi: 10.1371/journal.pone.0019223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology. 2007;88:1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- 27.Finzi-Hart JA, Pett-Ridge J, Weber PK, Popa R, Fallon SJ, Gunderson T, Hutcheon ID, Nealson KH, Capone DG. Fixation and fate of C and N in the cyanobacterium Trichodesmium using nanometer-scale secondary ion mass spectrometry. Proc Natl Acad Sci USA. 2009;106:6345–6350. doi: 10.1073/pnas.0810547106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujimura R, Sato Y, Nishizawa T, Nanba K, Oshima K, Hattori M, Kamijo T, Ohta H. Analysis of early bacterial communities on volcanic deposits on the Island of Miyake (Miyakejima), Japan: a 6-year study at a fixed site. Microbes Environ. 2012;27:19–29. doi: 10.1264/jsme2.ME11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujitani H, Aoi Y, Tsuneda S. Selective enrichment of two different types of Nitrospira-like nitrite-oxidizing bacteria from a wastewater treatment plant. Microbes Environ. 2013;28:236–243. doi: 10.1264/jsme2.ME12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geisseler D, Horwath WR, Joergensen RG, Ludwig B. Pathways of nitrogen utilization by soil microorganisms—a review. Soil Biol Biochem. 2010;42:2058–2067. [Google Scholar]
- 31.Girvan MS, Campbell CD, Killham K, Prosser JI, Glover LA. Bacterial diversity promotes community stability and functional resilience after perturbation. Environ Microbiol. 2005;7:301–313. doi: 10.1111/j.1462-2920.2005.00695.x. [DOI] [PubMed] [Google Scholar]
- 32.Graham DW, Knapp CW, Van Vleck ES, Bloor K, Lane TB, Graham CE. Experimental demonstration of chaotic instability in biological nitrification. ISME J. 2007;1:385–393. doi: 10.1038/ismej.2007.45. [DOI] [PubMed] [Google Scholar]
- 33.Griffin BM, Schott J, Schink B. Nitrite, an electron donor for anoxygenic photosynthesis. Science. 2007;316:1870. doi: 10.1126/science.1139478. [DOI] [PubMed] [Google Scholar]
- 34.Gubry-Rangin C, Hai B, Quince C, Engel M, Thomson BC, James P, Schloter M, Griffiths RI, Prosser JI, Nicol GW. Niche specialization of terrestrial archaeal ammonia oxidizers. Proc Natl Acad Sci USA. 2011;108:21206–21211. doi: 10.1073/pnas.1109000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, Yuan Z, Tyson GW. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature. 2013;500:567–570. doi: 10.1038/nature12375. [DOI] [PubMed] [Google Scholar]
- 36.Hawkes CV, Wren IF, Herman DJ, Firestone MK. Plant invasion alters nitrogen cycling by modifying the soil nitrifying community. Ecol Lett. 2005;8:976–985. doi: 10.1111/j.1461-0248.2005.00802.x. [DOI] [PubMed] [Google Scholar]
- 37.Hayakawa A, Hatakeyama M, Asano R, Ishikawa Y, Hidaka S. Nitrate reduction coupled with pyrite oxidation in the surface sediments of a sulfide-rich ecosystem. J. Geophys. Research: Biogeosci. 2013;118:639–649. [Google Scholar]
- 38.Henderson SL, Dandie CE, Patten CL, Zebarth BJ, Burton DL, Trevors JT, Goyer C. Changes in denitrifier abundance, denitrification gene mRNA levels, nitrous oxide emissions, and denitrification in anoxic soil microcosms amended with glucose and plant residues. Appl Environ Microbiol. 2010;76:2155–2164. doi: 10.1128/AEM.02993-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hong Y, Youshao W, Chen F. Archaea dominate ammonia oxidizers in the Permian water ecosystem of midland basin. Microbes Environ. 2013;28:396–399. doi: 10.1264/jsme2.ME13022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu S, Zeng RJ, Burow LC, Lant P, Keller J, Yuan Z. Enrichment of denitrifying anaerobic methane oxidizing micro-organisms. Environ Microbiol Rep. 2009;1:377–384. doi: 10.1111/j.1758-2229.2009.00083.x. [DOI] [PubMed] [Google Scholar]
- 41.Inaba S, Ikenishi F, Itakura M, Kikuchi M, Eda S, Chiba N, Katsuyama C, Suwa Y, Mitsui H, Minamisawa K. N2O emission from degraded soybean nodules depends on denitrification by Bradyrhizobium japonicum and other microbes in the rhizosphere. Microbes Environ. 2012;27:470–476. doi: 10.1264/jsme2.ME12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishii S, Ikeda S, Minamisawa K, Senoo K. Nitrogen cycling in rice paddy environments: past achievements and future challenges. Microbes Environ. 2011;26:282–292. doi: 10.1264/jsme2.me11293. [DOI] [PubMed] [Google Scholar]
- 43.Ishii S, Ohno H, Tsuboi M, Otsuka S, Senoo K. Identification and isolation of active N 2O reducers in rice paddy soil. ISME J. 2011;5:1936–1945. doi: 10.1038/ismej.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Isobe K, Koba K, Otsuka S, Senoo K. Nitrification and nitrifying microbial communities in forest soils. J Fore Res. 2011;16:351–362. [Google Scholar]
- 45.Isobe K, Suwa Y, Ikutani J, et al. Analytical techniques for quantifying 15N/14N of nitrate, nitrite, total dissolved nitrogen and ammonium in environmental samples using a gas chromatograph equipped with a quadrupole mass spectrometer. Microbes Environ. 2011;26:46–53. doi: 10.1264/jsme2.me10159. [DOI] [PubMed] [Google Scholar]
- 46.Isobe K, Koba K, Suwa Y, Ikutani J, Fang Y, Yoh M, Mo J, Otsuka S, Senoo K. High abundance of ammonia-oxidizing archaea in acidified subtropical forest soils in southern China after long-term N deposition. FEMS Microbiol Ecol. 2012;80:193–203. doi: 10.1111/j.1574-6941.2011.01294.x. [DOI] [PubMed] [Google Scholar]
- 47.Isobe K, Koba K, Suwa Y, et al. Nitrite transformations in an N-saturated forest soil. Soil Biol Biochem. 2012;52:61–63. [Google Scholar]
- 48.Itoh H, Ishii S, Shiratori Y, Oshima K, Otsuka S, Hattori M, Senoo K. Seasonal transition of active bacterial and archaeal communities in relation to water management in paddy soils. Microbes Environ. 2013;28:370–380. doi: 10.1264/jsme2.ME13030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Janssen PH. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol. 2006;72:1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jayakumar A, Al-Rshaidat MMD, Ward BB, Mulholland MR. Diversity, distribution, and expression of diazotroph nifH genes in oxygen-deficient waters of the Arabian Sea. FEMS Microbiol Ecol. 2012;82:597–606. doi: 10.1111/j.1574-6941.2012.01430.x. [DOI] [PubMed] [Google Scholar]
- 51.Jones CM, Graf DRH, Bru D, Philippot L, Hallin S. The unaccounted yet abundant nitrous oxide-reducing microbial community: a potential nitrous oxide sink. ISME J. 2013;7:417–726. doi: 10.1038/ismej.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 2009;3:442–453. doi: 10.1038/ismej.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jumadi O, Hala Y, Muis A, Ali A, Palennari M, Yagi K, Inubushi K. Influences of chemical fertilizers and a nitrification inhibitor on greenhouse gas fluxes in a corn (Zea mays L.) field in Indonesia. Microbes Environ. 2008;23:29–34. doi: 10.1264/jsme2.23.29. [DOI] [PubMed] [Google Scholar]
- 54.Kaiser K, Benner R. Major bacterial contribution to the ocean reservoir of detrital organic carbon and nitrogen. Limnol Oceanogr. 2008;53:99–112. [Google Scholar]
- 55.Kalvelage T, Jensen MM, Contreras S, Revsbeth NP, Lam P, Günter M, LaRoche J, Lavik G, Kuypers MMM. Oxygen sensitivity of anammox and coupled N-cycle processes in oxygen minimum zones. PLoS One. 2011;6:e29299. doi: 10.1371/journal.pone.0029299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kartal B, Maalcke WJ, de Almeida NM, et al. Molecular mechanism of anaerobic ammonium oxidation. Nature. 2011;479:127–130. doi: 10.1038/nature10453. [DOI] [PubMed] [Google Scholar]
- 57.Kato S, Hashimoto K, Watanabe K. Microbial inter-species electron transfer via electric currents through conductive minerals. Proc Natl Acad Sci USA. 2012;109:10042–10046. doi: 10.1073/pnas.1117592109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katsuyama C, Kondo N, Suwa Y, Yamagishi T, Itoh M, Ohte N, Kimura H, Nagaosa K, Kato K. Denitrification activity and relevant bacteria revealed by nitrite reductase gene fragments in soil of temperate mixed forest. Microbes Environ. 2008;23:337–345. doi: 10.1264/jsme2.me08541. [DOI] [PubMed] [Google Scholar]
- 59.Kaur K, Goyal S, Kapoor KK. Impact of organic fertilizers with and without chemical fertilizers on soil chemical properties and the establishment of nitrogen-fixing bacteria in the rhizosphere. Microbes Environ. 2008;23:313–316. doi: 10.1264/jsme2.me08524. [DOI] [PubMed] [Google Scholar]
- 60.Kindaichi T, Awata T, Suzuki Y, Tanabe K, Hatamoto M, Ozaki N, Ohashi A. Enrichment using an up-flow column reactor and community structure of marine anammox bacteria from coastal sediment. Microbes Environ. 2011;26:67–73. doi: 10.1264/jsme2.me10158. [DOI] [PubMed] [Google Scholar]
- 61.Kirchman DL. The uptake of inorganic nutrients by heterotrophic bacteria. Microbial Ecol. 1994;28:255–271. doi: 10.1007/BF00166816. [DOI] [PubMed] [Google Scholar]
- 62.Koch AL. Oligotrophs versus copiotrophs. BioEssays. 2001;23:657–661. doi: 10.1002/bies.1091. [DOI] [PubMed] [Google Scholar]
- 63.Kojima H, Tsutsumi M, Ishikawa K, Iwata T, Mußmann M, Fukui M. Distribution of putative denitrifying methane oxidizing bacteria in sediment of a freshwater lake, Lake Biwa. Syst Appl Microbiol. 2012;35:233–238. doi: 10.1016/j.syapm.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 64.Kondo K, Yoshimatsu K, Fujiwara T. Expression, and molecular and enzymatic characterization of Cu-containing nitrite reductase from a marine ammonia-oxidizing gammaproteo-bacterium, Nitrosococcus oceani. Microbes Environ. 2012;27:407–412. doi: 10.1264/jsme2.ME11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Könneke M, Bernhard AE, de La Torre JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 66.Kowalchuk GA, Stephen JR. Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu Rev Microbiol. 2001;55:485–529. doi: 10.1146/annurev.micro.55.1.485. [DOI] [PubMed] [Google Scholar]
- 67.Lam P, Lavik G, Jensen MM, van de Vossenberg J, Schmid M, Woebken D, Gutierrez D, Amann R, Jetten MSM, Kuypers MMM. Revising the nitrogen cycle in the Peruvian oxygen minimum zone. Proc Natl Acad Sci USA. 2009;106:4752–4757. doi: 10.1073/pnas.0812444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lehtovirta-Morley LE, Stoecker K, Vilcinskas A, Prosser JI, Nicol GW. Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc Natl Acad Sci USA. 2011;108:15892–15897. doi: 10.1073/pnas.1107196108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li M, Cao H, Hong YG, Gu JD. Seasonal dynamics of anammox bacteria in estuarial sediment of the Mai Po Nature Reserve revealed by analyzing the 16S rRNA and hydrazine oxidoreductase (hzo) genes. Microbes Environ. 2011;26:15–22. doi: 10.1264/jsme2.me10131. [DOI] [PubMed] [Google Scholar]
- 70.Lin L, Li Z, Hu C, Zhang X, Chang S, Yang L, Li Y, An Q. Plant growth-promoting nitrogen-fixing enterobacteria are in association with sugarcane plants growing in Guangxi, China. Microbes Environ. 2012;27:391–398. doi: 10.1264/jsme2.ME11275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lipson DA, Schmidt SK. Seasonal changes in an alpine soil bacterial community in the Colorado rocky mountains. Appl Environ Microbiol. 2004;70:2867–2879. doi: 10.1128/AEM.70.5.2867-2879.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lipson DA, Schadt CW, Schmidt SK. Changes in soil microbial community structure and function in an alpine dry meadow following spring snow melt. Microbial Ecol. 2002;43:307–314. doi: 10.1007/s00248-001-1057-x. [DOI] [PubMed] [Google Scholar]
- 73.Lücker S, Nowka B, Rattei T, Spieck E, Daims H. The genome of Nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer. Front Microbiol. 2013;4:27. doi: 10.3389/fmicb.2013.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luesken FA, Wu ML, Op den Camp HJM, Keltjens JT, Stunnenberg H, Francoijs KJ, Strous M, Jetten MS. Effect of oxygen on the anaerobic methanotroph ‘Candidatus Methylomirabilis oxyfera’: kinetic and transcriptional analysis. Environ Microbiol. 2012;14:1024–1034. doi: 10.1111/j.1462-2920.2011.02682.x. [DOI] [PubMed] [Google Scholar]
- 75.Martens-Habbena W, Berube PM, Urakawa H, de La Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- 76.Martin AP, Pondaven P. New primary production and nitrification in the western subtropical North Atlantic: a modeling study. Global Biogeochem Cycles. 2006;20:GB4014. [Google Scholar]
- 77.Martiny AC, Treseder K, Pusch G. Phylogenetic conservatism of functional traits in microorganisms. ISME J. 2013;7:830–838. doi: 10.1038/ismej.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matsumoto S, Ishikawa D, Saeki G, Aoi Y, Tsuneda S. Microbial population dynamics and community structure during the formation of nitrifying granules to treat ammonia-rich inorganic wastewater. Microbes Environ. 2010;25:164–170. doi: 10.1264/jsme2.me10107. [DOI] [PubMed] [Google Scholar]
- 79.Matsutani N, Nakagawa T, Nakamura K, Takahashi R, Yoshihara K, Tokuyama T. Enrichment of a novel marine ammonia-oxidizing archaeon obtained from sand of an Eelgrass Zone. Microbes Environ. 2011;26:23–29. doi: 10.1264/jsme2.me10156. [DOI] [PubMed] [Google Scholar]
- 80.McCarthy MD. Major bacterial contribution to marine dissolved organic nitrogen. Science. 1998;281:231–234. doi: 10.1126/science.281.5374.231. [DOI] [PubMed] [Google Scholar]
- 81.Mitchell MJ, Raynal DJ, Driscoll CT. Biogeochemistry of a forested watershed in the central Adirondack Mountains: temporal changes and mass balances. Water, Air, Soil Pollu. 1996;88:355–369. [Google Scholar]
- 82.Moisander PH, Beinart RA, Hewson I, White AE, Johnson KS, Carlson CA, Montoya JP, Zehr JP. Unicellular cyanobacterial distributions broaden the oceanic N2 fixation domain. Science. 2010;327:1512–1514. doi: 10.1126/science.1185468. [DOI] [PubMed] [Google Scholar]
- 83.Montoya JP, Holl CM, Zehr JP, Hansen A, Villareal TA, Capone DG. High rates of N2 fixation by unicellular diazotrophs in the oligotrophic Pacific Ocean. Nature. 2004;430:1027–1032. doi: 10.1038/nature02824. [DOI] [PubMed] [Google Scholar]
- 84.Moore RM, Punshon S, Mahaffey C, Karl D. The relationship between dissolved hydrogen and nitrogen fixation in ocean waters. Deep Sea Res. Part I: Oceanogr Res Pap. 2009;56:1449–1458. [Google Scholar]
- 85.Morimoto S, Hayatsu M, Takada Hoshino Y, Nagaoka K, Yamazaki M, Karasawa T, Takenaka M, Akiyama H. Quantitative analyses of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) in fields with different soil types. Microbes Environ. 2011;26:248–253. doi: 10.1264/jsme2.me11127. [DOI] [PubMed] [Google Scholar]
- 86.Mulder A, Graaf AA, Robertson LA, Kuenen JG. Anaerobic ammonium oxidation discovered in a denitrifying fluidized bed reactor. FEMS Microbiol Ecol. 1995;16:177–184. [Google Scholar]
- 87.Mykytczuk NCS, Foote SJ, Omelon CR, Southam G, Greer CW, Whyte LG. Bacterial growth at −15°C; molecular insights from the permafrost bacterium Planococcus halocryophilus Or1. ISME J. 2013;7:1211–1126. doi: 10.1038/ismej.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Narihiro T, Kamagata Y. Cultivating yet-to-be cultivated microbes: the challenge continues. Microbes Environ. 2013;28:163–165. doi: 10.1264/jsme2.ME2802rh. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ohte N, Mitchell M, Shibata H, Tokuchi N, Toda H, Iwatsubo G. Water, Air, Soil Pollu. Vol. 130. Springer; Netherlands: 2001. Comparative evaluation on nitrogen saturation of forest catchments in Japan and Northeastern United States; pp. 649–654. [Google Scholar]
- 90.Okabe S, Nakamura Y, Satoh H. Community structure and in situ activity of nitrifying bacteria in Phragmites root- associated biofilms. Microbes Environ. 2012;27:242–249. doi: 10.1264/jsme2.ME11314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Okubo T, Tsukui T, Maita H, et al. Complete genome sequence of Bradyrhizobium sp. S23321: insights into symbiosis evolution in soil oligotrophs. Microbes Environ. 2012;27:306–315. doi: 10.1264/jsme2.ME11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pett-Ridge J, Silver WL, Firestone MK. Redox fluctuations frame microbial community impacts on N-cycling rates in a humid tropical forest soil. Biogeochem. 2006;8:95–110. [Google Scholar]
- 93.Philippot L, Cuhel J, Saby NPA, Chèneby D, Chronáková A, Bru D, Arrouays D, Martin-Laurent F, Simek M. Mapping field-scale spatial patterns of size and activity of the denitrifier community. Environ Microbiol. 2009;11:1518–1526. doi: 10.1111/j.1462-2920.2009.01879.x. [DOI] [PubMed] [Google Scholar]
- 94.Philippot L, Andert J, Jones CM, Bru D, Hallin S. Importance of denitrifiers lacking the genes encoding the nitrous oxide reductase for N2O emissions from soil. Glob Change Biol. 2010;17:1497–1504. [Google Scholar]
- 95.Raes J, Bork P. Molecular eco-systems biology: towards an understanding of community function. Nature Rev Microbiol. 2008;6:693–699. doi: 10.1038/nrmicro1935. [DOI] [PubMed] [Google Scholar]
- 96.Raghoebarsing AA, Pol A, van de Pas-Schoonen KT, et al. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature. 2006;440:918–921. doi: 10.1038/nature04617. [DOI] [PubMed] [Google Scholar]
- 97.Ravishankara AR, Daniel JS, Portmann RW. Nitrous oxide (N2O): the dominant ozone-depleting substance emitted in the 21st century. Science. 2009;326:123–125. doi: 10.1126/science.1176985. [DOI] [PubMed] [Google Scholar]
- 98.Reguera G, McCarthy KD, Mehta T, Nicoll JS, Tuominen MT, Lovley DR. Extracellular electron transfer via microbial nanowires. Nature. 2005;435:1098–1101. doi: 10.1038/nature03661. [DOI] [PubMed] [Google Scholar]
- 99.Richards F. Anoxic basins and fjords. In: Riley J, Skirrow G, editors. Chemical Oceanography. Academic Press; London: 1965. pp. 611–645. [Google Scholar]
- 100.Saito T, Ishii S, Otsuka S, Nishiyama M, Senoo K. Identification of novel Betaproteobacteria in a succinate-assimilating population in denitrifying rice paddy soil by using stable isotope probing. Microbes Environ. 2008;23:192–200. doi: 10.1264/jsme2.23.192. [DOI] [PubMed] [Google Scholar]
- 101.Sakami T. Distribution of ammonia-oxidizing archaea and bacteria in the surface sediments of Matsushima Bay in relation to environmental variables. Microbes Environ. 2012;27:61–66. doi: 10.1264/jsme2.ME11218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sanford RA, Wagner DD, Wu Q, et al. Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils. Proc Natl Acad Sci USA. 2012;109:19709–19714. doi: 10.1073/pnas.1211238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sato Y, Ohta H, Yamagishi T, et al. Detection of anammox activity and 16S rRNA genes in ravine paddy field soil. Microbes Environ. 2012;27:316–319. doi: 10.1264/jsme2.ME11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schadt CW, Martin AP, Lipson DA, Schmidt SK. Seasonal dynamics of previously unknown fungal lineages in tundra soils. Science. 2003;301:1359–1361. doi: 10.1126/science.1086940. [DOI] [PubMed] [Google Scholar]
- 105.Schimel JP, Bennett J. Nitrogen mineralization: challenges of a changing paradigm. Ecology. 2004;85:591–602. [Google Scholar]
- 106.Schimel JP, Bennet J, Fierer N. Microbial community composition and soil nitrogen cycling: is there really a connection? In: Bardgett RD, Usher MB, Hopkins DW, editors. Biological Diversity and Function in Soil. Cambridge University Press; USA: 2005. pp. 171–188. [Google Scholar]
- 107.Schloss PD, Handelsman J. Status of the microbial census. Microbiol Mol Biol Rev. 2004;68:686–691. doi: 10.1128/MMBR.68.4.686-691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schott J, Griffin BM, Schink B. Anaerobic phototrophic nitrite oxidation by Thiocapsa sp. strain KS1 and Rhodopseudomonas sp. strain LQ17. Microbiol. 2010;156:2428–2437. doi: 10.1099/mic.0.036004-0. [DOI] [PubMed] [Google Scholar]
- 109.Shibata H, Hasegawa Y, Watanabe T, Fukuzawa K. Impact of snowpack decrease on net nitrogen mineralization and nitrification in forest soil of northern Japan. Biogeochemistry. 2013;116:69–82. [Google Scholar]
- 110.Shimomura Y, Morimoto S, Takada Hoshino Y, Uchida Y, Akiyama H, Hayatsu M. Comparison among amoA primers suited for quantification and diversity analyses of ammonia-oxidizing bacteria in soil. Microbes Environ. 2012;27:94–98. doi: 10.1264/jsme2.ME11230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sorokin DY, Lücker S, Vejmelkova D, et al. Nitrification expanded: discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. ISME J. 2012;6:2245–2256. doi: 10.1038/ismej.2012.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Strous M, Pelletier E, Mangenot S, et al. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature. 2006;440:790–794. doi: 10.1038/nature04647. [DOI] [PubMed] [Google Scholar]
- 113.Strous M, Fuerst JA, Kramer EH, Logemann S, Muyzer G, van de Pas-Schoonen KT, Webb R, Kuenen JG, Jetten MS. Missing lithotroph identified as new planctomycete. Nature. 1999;400:446–449. doi: 10.1038/22749. [DOI] [PubMed] [Google Scholar]
- 114.Tago K, Ishii S, Nishizawa T, Otsuka S, Senoo K. Phylogenetic and functional diversity of denitrifying bacteria isolated from various rice paddy and rice-soybean rotation fields. Microbes Environ. 2011;26:30–35. doi: 10.1264/jsme2.me10167. [DOI] [PubMed] [Google Scholar]
- 115.Takada Hoshino Y, Morimoto S, Hayatsu M, Nagaoka K, Suzuki C, Karasawa T, Takenaka M, Akiyama H. Effect of soil type and fertilizer management on archaeal community in upland field soils. Microbes Environ. 2011;26:307–316. doi: 10.1264/jsme2.me11131. [DOI] [PubMed] [Google Scholar]
- 116.Takahashi M, Yamada T, Tanno M, Tsuji H, Hiraishi A. Nitrate removal efficiency and bacterial community dynamics in denitrification processes using poly (L-lactic acid) as the solid substrate. Microbes Environ. 2011;26:212–219. doi: 10.1264/jsme2.me11107. [DOI] [PubMed] [Google Scholar]
- 117.Teske A, Alm E, Regan JM, Toze S, Rittmann BE, Stahl DA. Evolutionary relationships among ammonia- and nitrite-oxidizing bacteria. J Bacteriol. 1994;176:6623–6630. doi: 10.1128/jb.176.21.6623-6630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Thamdrup B. New pathways and processes in the global nitrogen cycle. Annu Rev Ecol Evol Syst. 2012;43:407–428. [Google Scholar]
- 119.Thompson AW, Foster RA, Krupke A, Carter BJ, Musat N, Vaulot D, Kuypers MMM, Zehr JP. Unicellular cyanobacterium symbiotic with a single-celled eukaryotic alga. Science. 2012;337:1546–1550. doi: 10.1126/science.1222700. [DOI] [PubMed] [Google Scholar]
- 120.Tripp HJ, Bench SR, Turk KA, Foster RA, Desany BA, Niazi F, Affourtit JP, Zehr JP. Metabolic streamlining in an open-ocean nitrogen-fixing cyanobacterium. Nature. 2010;464:90–94. doi: 10.1038/nature08786. [DOI] [PubMed] [Google Scholar]
- 121.Ushiki N, Fujitani H, Aoi Y, Tsuneda S. Isolation of Nitrospira belonging to sublineage II from a wastewater treatment plant. Microbes Environ. 2013;28:346–353. doi: 10.1264/jsme2.ME13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vitousek P, Howarth R. Nitrogen limitation on land and in the sea: how can it occur? Biogeochem. 1991;13:87–115. [Google Scholar]
- 123.Voss M, Montoya JP. Nitrogen cycle: oceans apart. Nature. 2009;461:49–50. doi: 10.1038/461049a. [DOI] [PubMed] [Google Scholar]
- 124.Wada E, Hattori A. Nitrogen in the Sea: Forms, Abundances, and Rate Processes. CRC Press; Llc, USA: 1991. [Google Scholar]
- 125.Wang Y, Zhu G, Harhangi HR, Zhu B, Jetten MSM, Yin C, Op den Camp HJM. Co-occurrence and distribution of nitrite-dependent anaerobic ammonium and methane-oxidizing bacteria in a paddy soil. FEMS microbial Lett. 2012;336:79–88. doi: 10.1111/j.1574-6968.2012.02654.x. [DOI] [PubMed] [Google Scholar]
- 126.Ward BB, Devol AH, Rich JJ, Chang BX, Bulow SE, Naik H, Pratihary A, Jayakumar A. Denitrification as the dominant nitrogen loss process in the Arabian Sea. Nature. 2009;461:78–81. doi: 10.1038/nature08276. [DOI] [PubMed] [Google Scholar]
- 127.Wieder WR, Cleveland CC, Taylor PG, Nemergut DR, Hinckley EL, Philippot L, Bru D, Weintraub SR, Martin M, Townsend AR. Experimental removal and addition of leaf litter inputs reduces nitrate production and loss in a lowland tropical forest. Biogeochem. 2012;113:629–642. [Google Scholar]
- 128.Wilson JB, Spijkerman E, Huisman J. Is there really insufficient support for Tilman’s R* concept? A comment on Miller et al. American Nat. 2007;169:700–706. doi: 10.1086/513113. [DOI] [PubMed] [Google Scholar]
- 129.Wood DW, Setubal JC, Kaul R, et al. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science. 2001;294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- 130.Yamamoto N, Oishi R, Suyama Y, Tada C, Nakai Y. Ammonia-oxidizing bacteria rather than ammonia-oxidizing archaea were widely distributed in animal manure composts from field-scale facilities. Microbes Environ. 2012;27:519–524. doi: 10.1264/jsme2.ME12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yang WH, Weber KA, Silver WL. Nitrogen loss from soil through anaerobic ammonium oxidation coupled to iron reduction. Nature Geosci. 2012;5:538–541. [Google Scholar]
- 132.Yasuda T, Waki M, Yoshinaga I, Amano T, Suzuki K, Tanaka Y, Yamagishi T, Suwa Y. Evidence of exponential growth of an anammox population in an anaerobic batch culture. Microbes Environ. 2011;26:266–269. doi: 10.1264/jsme2.me10181. [DOI] [PubMed] [Google Scholar]
- 133.Yool A, Martin AP, Fernández C, Clark DR. The significance of nitrification for oceanic new production. Nature. 2007;447:999–1002. doi: 10.1038/nature05885. [DOI] [PubMed] [Google Scholar]
- 134.Yoshida M, Ishii S, Otsuka S, Senoo K. nirK-harboring denitrifiers are more responsive to denitrification-inducing conditions in rice paddy soil than nirS-harboring bacteria. Microbes Environ. 2010;25:45–48. doi: 10.1264/jsme2.me09160. [DOI] [PubMed] [Google Scholar]
- 135.Yoshida M, Ishii S, Fujii D, Otsuka S, Senoo K. Identification of active denitrifiers in rice paddy soil by DNA- and RNA-based analyses. Microbes Environ. 2012;27:456–461. doi: 10.1264/jsme2.ME12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yoshinaga I, Amano T, Yamagishi T, Okada K, Ueda S, Sako Y, Suwa Y. Distribution and diversity of anaerobic ammonium oxidation (anammox) bacteria in the sediment of a eutrophic freshwater lake, Lake Kitaura, Japan. Microbes Environ. 2011;26:189–197. doi: 10.1264/jsme2.me10184. [DOI] [PubMed] [Google Scholar]
- 137.Zehr JP. Nitrogen fixation by marine cyanobacteria. Trends Microbiol. 2011;19:162–173. doi: 10.1016/j.tim.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 138.Zehr JP, Bench SR, Carter BJ, Hewson I, Niazi F, Shi T, Tripp HJ, Affourtit JP. Globally distributed uncultivated oceanic N2-fixing cyanobacteria lack oxygenic photosystem II. Science. 2008;322:1110–1112. doi: 10.1126/science.1165340. [DOI] [PubMed] [Google Scholar]
- 139.Zehr JP, Montoya JP, Hewson I, et al. Nitrogenase gene expression and N2fixation in the North Pacific Subtropical Gyre. Limnol Oceanogr. 2007;52:169–183. [Google Scholar]
- 140.Zehr JP, Waterbury JB, Turner PJ, Montoya JP, Omoregie E, Steward GF, Hansen A, Karl DM. Unicellular cyanobacteria fix N2 in the subtropical North Pacific Ocean. Nature. 2001;412:635–638. doi: 10.1038/35088063. [DOI] [PubMed] [Google Scholar]
- 141.Zhang LM, Offre PR, He JZ, Verhamme DT, Nicol GW, Prosser JI. Autotrophic ammonia oxidation by soil thaumarchaea. Proc Natl Acad Sci USA. 2010:17240–17245. doi: 10.1073/pnas.1004947107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhu G, Wang S, Wang Y, Wang C, Risgaard-Petersen N, Jetten MS, Yin C. Anaerobic ammonia oxidation in a fertilized paddy soil. ISME J. 2011;5:1905–1912. doi: 10.1038/ismej.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zimmerman AE, Martiny AC, Allison SD. Microdiversity of extracellular enzyme genes among sequenced prokaryotic genomes. ISME J. 2013;7:1187–1199. doi: 10.1038/ismej.2012.176. [DOI] [PMC free article] [PubMed] [Google Scholar]