Abstract
Viroids, subviral noncoding RNAs, replicate, move, and incite diseases in plants. Viroids replicate through a rolling-circle mechanism in which oligomeric RNAs of one or both polarities are cleaved and ligated into the circular monomers. Attempts to transmit viroids to Arabidopsis have failed for unknown reasons. To tackle this question, Arabidopsis was transformed with cDNAs expressing dimeric (+) transcripts of representative species of the families Pospiviroidae and Avsunviroidae, which replicate in the nucleus and the chloroplast, respectively. Correct processing to the circular (+) monomers was always observed, demonstrating that Arabidopsis has the appropriate RNase and RNA ligase. Northern blot hybridization also revealed the multimeric (-) RNAs of Citrus exocortis viroid and Hop stunt viroid (HSVd) of the family Pospiviroidae, but not of Avocado sunblotch viroid of the family Avsunviroidae, showing that the first RNA-RNA transcription of the rolling-circle mechanism occurs in Arabidopsis for the two nuclear viroids and that their multimeric (-) RNAs remain unprocessed as in typical hosts. Moreover, transgenic Arabidopsis expressing HSVd dimeric (-) transcripts accumulated the circular (+) monomers, although at low levels, together with the unprocessed primary transcript that served as the template for the second RNA-RNA transcription. Agroinoculation of Arabidopsis with the dimeric (+) Citrus exocortis viroid, HSVd, and Coleus blumei viroid 1 cDNAs showed that these viroids could not move to distal plant parts, in contrast with the situation observed in their experimental hosts. Therefore, deficiencies in movement or low replication appear to be the factors limiting infectivity of some viroids in Arabidopsis.
Since Arabidopsis thaliana was adopted as the model organism for higher plants, multiple tools, resources, and experimental approaches have been developed that facilitate research with this system (1). Principal among them is the availability of the complete sequence of the Arabidopsis genome (2). Research on plant viruses has also benefited from the use of such a versatile system. In particular, studies with viruses that naturally or experimentally infect Arabidopsis have led to the identification of host factors involved in their amplification (3, 4) and movement (5, 6) and in posttranscriptional gene silencing-mediated phenomena that include disease induction (7) and plant defense responses (8, 9). However, no viroid has been reported to infect Arabidopsis so far.
Viroids are small, noncoding circular RNAs of 246-401 nucleotides able to replicate autonomously in certain plants (see refs. 10-12 for reviews). Despite this minimal genome, viroids, in addition to their replication, are able to direct their intracellular, intercellular, and long-distance movement (13-15) and to activate host defense mechanisms (16-18) that in some cases are insufficient to block the induction of pathogenic effects. For these functions, viroids need to interact with multiple host factors, the identification of which would be greatly facilitated by an Arabidopsis-based system.
Thirty different viroid species infecting higher plants have been characterized to date and classified into two families (11). Twenty-six viroid species, which contain in their RNAs a central conserved region, belong to the family Pospiviroidae, whereas four viroid species that lack a central conserved region but are able to self-cleave through hammerhead ribozymes are grouped in the family Avsunviroidae (12, 19). Viroid replication occurs through an RNA-based rolling-circle mechanism (20) in which the infecting monomeric (+) circular RNA is transcribed by an RNA polymerase into head-to-tail (-) multimers that serve as templates for a second RNA-RNA transcription step. The resulting head-to-tail (+) multimers are cleaved into unit-length strands and subsequently ligated into the final progeny of monomeric (+) circular RNAs by an RNase and an RNA ligase, respectively. This asymmetric version of the rolling-circle mechanism is followed by Potato spindle tuber viroid (PSTVd) (21, 22) and other members of the family Pospiviroidae, which replicate in the nucleus (23-28). In contrast, Avocado sunblotch viroid (ASBVd) and other members of the family Avsunviroidae, which replicate in the chloroplast, follow a symmetric version in which the (-) multimers are processed to the monomeric (-) circular forms, the template for the second half of the replication cycle that is symmetric to the first (29-32). Remarkably, cleavage of (+) and (-) multimers is autocatalytic in the family Avsunviroidae and mediated by hammerhead ribozymes (refs. 33-35; see ref. 36 for a review). The RNA ligase-catalyzing circularization of linear monomeric forms is presumably a host enzyme (37-39), although for a member of the family Avsunviroidae it has been proposed that not only cleavage but also ligation is autocatalytic and produces atypical 2′-5′ phosphodiester bonds (40).
Here, we report the establishment and properties of an Arabidopsis-based system for the study of viroid-host interactions. Our data indicate that, when introduced transgenically in Arabidopsis, Hop stunt viroid (HSVd) and most likely other members of the family Pospiviroidae are able to generate the typical intermediates and the final replication products. However, when agroinoculated, they could not spread systemically. These results suggest that the infectivity of some viroids in Arabidopsis may be limited by deficiencies in movement or in reaching an accumulation level above a minimum threshold.
Materials and Methods
Viroid Sequence Variants. Citrus exocortis viroid (CEVd) variant was M34917 having a deletion of one G between positions 70 and 74; HSVd variant was Y09352; Coconut cadang-cadang viroid (CCCVd) variant was J02050, differing in a point mutation (C31→ U); Apple scar skin viroid (ASSVd) variant was AF421195; Coleus blumei viroid 1 (CbVd-1) variant was X69293 but with a G inserted between positions 146 and 147; and ASBVd variant was J02020, differing in a point mutation (C213→U).
Plasmid Constructs. A series of recombinant plasmids harboring different dimeric viroid cDNAs were constructed from pBluescript II KS (+) (Stratagene, GenBank accession no. X52327). pBdCEVd, pBdHSVd, and pBdCCCVd, contained a PstI dimeric CEVd cDNA, a ClaI dimeric HSVd cDNA, and a BamHI dimeric CCCVd cDNA cloned in the PstI, ClaI, and BamHI vector sites, respectively. pBdASSVd contained a dimeric ASSVd cDNA (starting at position 90 and ending at position 89) cloned in the SmaI site of the vector. pBdCbVd-1 contained a dimeric CbVd-1 cDNA (starting at position 68 and ending at position 67) cloned in the SmaI site of the vector. pBdASBVd contained a Sau3AI dimeric ASBVd cDNA cloned in the BamHI site of the vector. Binary plasmids pCdCEVd, pCdHSVd(+), pCdHSVd(-), pCdCCCVd, pCdASSVd, pCdCbVd-1, and pCdASBVd were constructed by replacing the GFP cDNA of the binary vector pCAMBIA-1302 (GenBank accession no. AF234298) by the corresponding dimeric viroid cDNAs from the pBluescript II KS (+)-derived plasmids.
Transgenic Plants. Agrobacterium tumefaciens (strain C58C1) was transformed with plasmids pCdCEVd, pCdHSVd(+), pCdHSVd(-), pCdCCCVd, pCdASSVd, pCdCbVd-1, and pCdASBVd. Transformation of A. thaliana (ecotype Col-0) was performed by the f loral dip method with midlog grown Agrobacterium cultures (41). Seeds from dipped Arabidopsis were germinated in plates containing 20 μg/ml hygromycin B, 300 μg/ml cefotaxime, and 10 μg/ml benomyl, and plants able to grow were selected for further analysis.
RNA Extraction and Analysis. Tissue was ground in buffer B (0.1 M Tris·HCl, pH 9.0, containing 5 M urea, 0.1 M NaCl, 0.1 M 2-mercaptoethanol, and 10 mM EDTA) at a ratio of 5 ml/g of fresh weight. The homogenate was clarified by centrifugation, and the supernatant was extracted with 0.5 vol of phenol/chloroform (1:1). Total RNA in the aqueous phase was recovered by ethanol precipitation and resuspended in 0.1 vol of 10 mM Tris·HCl (pH 8.0) containing 1 mM EDTA and 98% formamide. Alternatively, viroid-enriched RNAs were obtained by chromatography on nonionic cellulose (CF11, Whatman) (42). For dot-blot analysis, RNA preparations (5 μl), boiled for 1.5 min and chilled on ice, were spotted on positively charged Nylon membranes (Roche Diagnostics) and fixed by UV irradiation. For Northern blot analysis, aliquots (20 μl) of the RNA preparations were separated either by double PAGE under nondenaturing and denaturing conditions (43) or by single denaturing PAGE (44). Double PAGE was performed first in a 5% gel made with TAE buffer (40 mM Tris/20 mM sodium acetate/1 mM EDTA, pH 7.2) and then in a 5% gel containing 8 M urea in 0.25× TBE buffer. Single denaturing PAGE was performed in 5% gels containing 8 M urea in 1× TBE buffer (89 mM Tris/89 mM boric acid/2.5 mM EDTA, pH 8.3). After electrophoresis, the RNAs were electroblotted to membranes and UV-fixed. Strand-specific 32P-labeled riboprobes were obtained by transcription of plasmids pBdCEVd, pBdHSVd, pBdCCCVd, pBdASSVd, pBdCbVd-1, and pBdASBVd linearized with appropriate enzymes. Membranes were hybridized at 70°C in the presence of 50% formamide and autoradiographed (30).
Purification of Viroid Circular RNA, Cloning, and Sequencing. Monomeric circular viroid RNAs were separated by two consecutive PAGE steps under nondenaturing and denaturing conditions (see above). The second denaturing gel was stained by ethidium bromide, and the zones where the monomeric circular viroid RNAs migrate were located with appropriate markers and cut, with the RNAs being eluted by diffusion in 0.1 M Tris·HCl (pH 9.0) containing 0.1 M 2-mercaptoethanol, 10 mM EDTA, and 1% SDS. The RNAs were recovered by ethanol precipitation and were RT-PCR-amplified with avian myeloblastosis virus reverse transcriptase (Invitrogen) and Pfu DNA polymerase (Stratagene) by using different pairs of viroid-specific primers (Table 1). The amplified cDNAs were ligated into plasmid pUC18, opened in the SmaI site, and used to transform Escherichia coli DH5α cells. The cDNAs from the resulting recombinant plasmids were sequenced with an ABI PRISM 377 apparatus (Perkin-Elmer).
Table 1. Sequence of monomeric circular (+) RNAs accumulating in transgenic Arabidopsis expressing viroid dimeric (+) RNAs.
| Viroid species | Primers* | Clones sequenced | Observed mutations |
|---|---|---|---|
| CEVd | 75-95, 96-113 | 10 | Two clones: C38→U, G287→U |
| HSVd | 64-87, 88-109 | 6 | None |
| 276-1, 2-23 | 6 | Two clones: +A(39-45) | |
| CCCVd | 41-65, 66-90 | 9 | None |
| ASSVd | 79-89, 90-110 | 10 | One clone: G29→Δ, C31→U, C210→U |
| ASBVd | 42-57, 58-78 | 10 | One clone: U148→C, C151→Δ |
| One clone: G196→A |
Positions of the viroid sequence covered by primers of (−) and (+) polarities, respectively, used for RT-PCR amplification.
Agroinoculation. Agrobacterium tumefaciens strain C58C1 carrying the virulence helper plasmid pCH32 (45) was transformed with the binary plasmids pCdCEVd, pCdHSVd(+), pCdHSVd(-), pCdCCCVd, pCdASSVd, pCdCbVd-1, and pCdASBVd described above. Agrobacterium cultures, grown to midlog phase, were resuspended in 10 mM Mes-NaOH (pH 5.6) containing 10 mM MgCl2 and 150 μM acetosiringone, incubated for 2 h at 28°C, and infiltrated into two leaves of A. thaliana (ecotype Col-0), Gynura aurantiaca, cucumber (Cucumis sativus L.) cv. Suyo, and Coleus blumei plants as described (46). One month after infiltration, upper uninoculated leaves were harvested for viroid RNA analysis by Northern blot hybridization.
Results
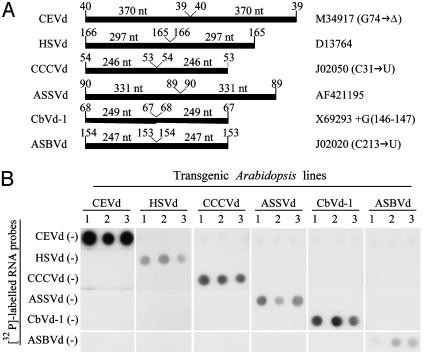
Transgenic Arabidopsis Expressing Viroid RNAs. Previous results have shown that in vitro-synthesized dimeric (+) transcripts of several viroids are infectious when inoculated mechanically into their hosts, most likely because these transcripts are processed in vivo to their monomeric (+) circular forms that subsequently initiate replication (47, 48). Although experiments of this type and others using as inocula nucleic acid preparations from viroid-infected plants have failed to promote viroid infection of Arabidopsis (49), they do not exclude the possibility that some viroids could replicate in the inoculated cells but were unable to move to neighboring cells and distal plant parts. To screen for Arabidopsis susceptibility to viroid infection under the most favorable conditions, a series of Arabidopsis transgenic lines expressing dimeric viroid transcripts were constructed. Selected viroids were CEVd (50, 51), HSVd (52), CCCVd (53), ASSVd (54), and CbVd-1 (55), the type (or a representative) species of the five genera of the family Pospiviroidae, and ASBVd, the type species of the family Avsunviroidae (34, 56). Monomeric cDNAs were ligated, and the resulting dimeric head-to-tail cDNAs (Fig. 1A) were subcloned in a binary vector so that transcription of the dimeric (+) RNAs was under the control of the cauliflower mosaic virus 35S promoter and the nopaline synthase terminator. Ti plasmid-mediated transformation of Arabidopsis and analysis by dot-blot hybridization of 20 putative transformants (growing in plates containing hygromycin B) of each construction revealed that most of them were indeed expressing viroid RNAs, although the intensity of the signals was variable. Three independent expressing lines of each construction (Fig. 1B) were selected to further confirm the presence of the viroid dimeric (+) RNAs and their resulting derivatives if processing had occurred.
Fig. 1.
(A) Bar diagram representing the viroid sequences embedded in the transcripts expressed in the transgenic Arabidopsis. Viroid species are indicated on the left, and the database accession numbers of the particular sequence variants used are on the right. Whenever sequence variants do not exactly match a database entry, the differences are indicated in parentheses after the accession number. Positions where the viroid sequences start and end in the transcript and the size of each monomeric unit are also indicated. (B) Dot-blot hybridization analysis of transgenic Arabidopsis expressing different dimeric (+) viroid RNAs. Total RNAs from three independent transgenic lines (columns 1, 2, and 3) for each of the six constructs (CEVd, HSVd, CCCVd, ASSVd, CbVd-1, and ASBVd) were hybridized with each of the six 32P-labeled complementary riboprobes indicated on the left.
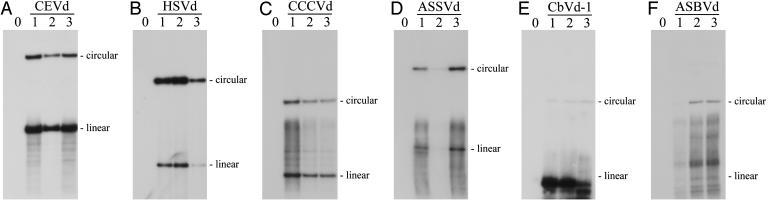
Dimeric (+) Viroid Transcripts Appear to Be Properly Processed to Their Monomeric Circular Forms in Arabidopsis. As a first step to screen for viroid replication, the monomeric (+) circular RNAs, the final product of the viroid replicative cycle, were looked for in the transgenic Arabidopsis. For this purpose, total RNA preparations were fractionated by two consecutive PAGE steps, the first under nondenaturing and the second under denaturing conditions (43). With this approach the viroid circular RNAs can be easily separated from the linear RNAs of similar size, which display a considerably faster mobility in the second gel. Analysis of the denaturing gels by Northern blot hybridization with viroid-specific riboprobes revealed bands with the electrophoretic mobilities expected for the monomeric (+) circular and linear forms of CEVd, HSVd, CCCVd, ASSVd, and ASBVd (Fig. 2 A-D and F, respectively). The signals were particularly intense in CEVd and HSVd (Fig. 2 A and B). Detection of the presumed monomeric (+) circular forms of CbVd-1 was not possible in total RNA extracts and required viroid-enriched preparations (obtained by chromatography on nonionic cellulose), indicating a low accumulation level of this species in the transgenic plants (Fig. 2E). No signals were observed in nontransgenic Arabidopsis controls (Fig. 2). These results strongly suggested that Arabidopsis contains the RNase and RNA ligase activities that catalyze the cleavage of dimeric (+) viroid RNAs and the subsequent circularization of the resulting unit-length strands. Considering the self-cleavage ability of the dimeric (+) ASBVd RNA, it was surprising to find that the presumed monomeric (+) circular and, in particular, linear forms were not accumulating at high levels (Fig. 2F).
Fig. 2.
Northern blot hybridization analysis of transgenic Arabidopsis expressing different dimeric (+) viroid RNAs. Nucleic acid preparations from a nontranformed Arabidopsis control (lane 0) and three independent transgenic lines (lanes 1, 2, and 3) expressing dimeric (+) RNAs of CEVd (A), HSVd (B), CCCVd (C), ASSVd (D), CbVd-1 (E), and ASBVd (F) were separated by two consecutive PAGE steps. A segment of the first nondenaturing gel, which included the RNAs with sizes between ≈200 and ≈400 nucleotides, was cut and applied on top of the second denaturing gel. After separation in this second gel, RNAs were blotted to nylon membranes and hybridized with 32P-labeled riboprobes complementary to each of the viroid RNAs. The analysis was performed with total RNA preparations except for CbVd-1, in which a viroid-enriched preparation obtained by chromatography on nonionic cellulose was used. Bands with the mobilities of the corresponding monomeric circular and linear viroid RNAs are indicated on the right (positions of linear CbVd-1 and ASBVd forms are only tentative). The exposure time was not the same in all autoradiographs.
Correct Monomeric Circular Viroid (+) RNAs Are Found in Transgenic Arabidopsis Expressing Dimeric (+) RNAs. To verify that the bands with the slow mobilities in the denaturing gel were indeed generated by viroid circular (+) RNAs that were accurately processed, these RNAs were eluted and subjected to RT-PCR amplification by using pairs of adjacent viroid-specific primers of complementary polarity. Cloning and sequencing of the resulting amplification products (between 9 and 12 recombinant plasmids per viroid, except in CbVd-1 in which no amplification product could be obtained), showed no changes in the nine clones from CCCVd (0/9), whereas 2/10 of CEVd, 2/12 of HSVd, 1/10 of ASSVd, and 2/10 of ASBVd held minor mutations (Table 1). These results confirmed that transgenic Arabidopsis expressing different dimeric viroid (+) RNAs accumulate bona fide viroid (+) circular RNAs, which could arise from cleavage and ligation of the primary transgene transcript, from viroid replication in transgenic Arabidopsis, or from a combination of both. Even if only processing of the primary transcripts is considered, it was unexpected to find that Arabidopsis has the appropriate enzymes required for cleavage and ligation of a series of viroid RNAs with very different sequences.
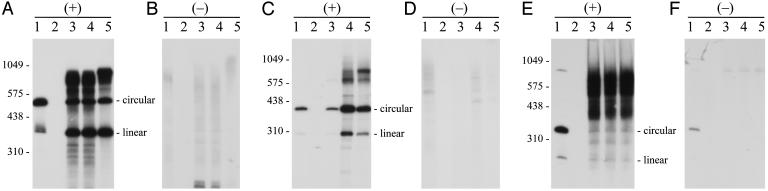
Further Analysis of Viroid (+) and (-) RNAs Accumulating in Transgenic Arabidopsis Expressing CEVd, HSVd, and ASBVd Dimeric (+) RNAs. As indicated previously, viroids replicate through a rolling-circle mechanism with multimeric RNA intermediates of both polarities. To search for these intermediates, total nucleic acid preparations enriched in viroid RNAs by chromatography on nonionic cellulose were obtained from transgenic Arabidopsis expressing dimeric CEVd, HSVd, and ASBVd (+) RNAs. The two first viroids were selected as representatives of the family Pospiviroidae, and the third viroid was selected as representative of the family Avsunviroidae. For comparative purposes, parallel RNA preparations were also obtained from known natural or experimental hosts infected by these viroids: CEVd from gynura, HSVd from cucumber, and ASBVd from avocado. Analysis of transgenic Arabidopsis expressing dimeric CEVd and HSVd (+) RNAs by single denaturing PAGE and Northern blot hybridization with viroid-specific riboprobes for both polarity strands showed prominent bands corresponding to the monomeric (+) circular and linear RNAs, with the linear forms predominating over their circular counterparts in CEVd and the opposite situation occurring in HSVd. Bands with the same mobility were also prominent in preparations of CEVd-infected gynura and HSVd-infected cucumber (Fig. 3). Additional bands were detected in the transgenic Arabidopsis, which, according to their position in the blot, should mostly correspond to the primary dimeric (+) transcripts and to some products resulting from their partial processing (Fig. 3). From their relative band intensity, it could be concluded that the CEVd and HSVd dimeric (+) RNAs were processed to a significant extent in Arabidopsis. However, the situation was different in ASBVd. Whereas two major bands corresponding to the monomeric (+) circular and linear viroid RNA were detected in ASBVd-infected avocado, the intensity of these bands was weak in transgenic Arabidopsis expressing the dimeric ASBVd (+) RNAs, in which the most prominent bands were presumably generated by the transcripts from the transgene and their degradation products (the possibility that they were the consequence of RNA-RNA transcription seems less tenable, because no minus strands were observed, see below) (Fig. 3). These results confirmed that processing of the dimeric (+) transcripts of ASBVd, despite their ability to self-cleave through hammerhead structures, is rather inefficient in an Arabidopsis context, thus establishing a distinction from the two members of the family Pospiviroidae.
Fig. 3.
Northern blot hybridization analysis of transgenic Arabidopsis expressing different dimeric (+) viroid RNAs. Viroid-enriched RNA preparations, obtained by chromatography on nonionic cellulose, were fractionated by single denaturing PAGE in duplicated gels and blotted to nylon membranes that were hybridized with 32P-labeled riboprobes for detecting the (+) and (-) strands of CEVd (A and B), HSVd (C and D), and ASBVd (E and F), respectively. Lanes 1, controls of CEVd-infected gynura (A and B), HSVd-infected cucumber (C and D), and ASBVd-infected avocado (E and F); lanes 2, nontransformed Arabidopsis control; lanes 3-5, transgenic Arabidopsis expressing dimeric (+) RNAs of CEVd (A and B), HSVd (C and D), and ASBVd (E and F). Positions of linear RNA markers, with their size in nucleotides, are indicated on the left in A, C, and E. Positions of circular and linear CEVd, HSVd, and ASBVd monomeric RNAs are indicated on the right in A, C, and E, respectively. Both riboprobes for each viroid were equalized in acid-precipitable counts, and the films were exposed for the same time. For facilitating detection of CEVd and HSVd (-) strands, the volume of applied extract was 10-fold higher.
Faint hybridization signals corresponding to longer-than-unit viroid (-) strands were also observed in transgenic Arabidopsis expressing dimeric CEVd and HSVd (+) RNAs. The profile of these bands, which in size and intensity paralleled those of CEVd-infected gynura and HSVd-infected cucumber (Fig. 3), were consistent with their emergence from RNA-RNA transcription of the primary dimeric (+) RNAs or the resulting monomeric (+) circular RNA. The situation was again different in ASBVd (Fig. 3). Whereas viroid RNAs of (-) polarity were visible in the ASBVd-infected avocado control, no significant hybridization signals were observed in transgenic Arabidopsis expressing the dimeric ASBVd (+) RNAs; the weak signals in the upper part of the gel were artifacts because a signal in the same position was also detected in the nontransformed control when it was overexposed (data not shown).
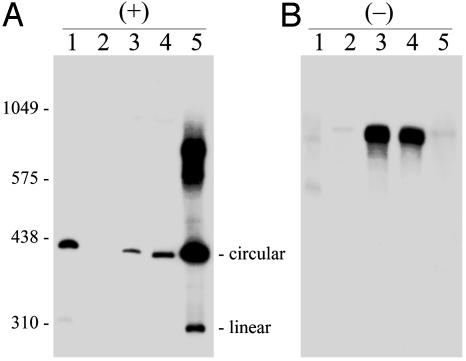
Transgenic Arabidopsis Expressing HSVd Dimeric (-) RNAs Accumulate the Monomeric Viroid (+) Circular Forms. To distinguish between processing and replication in Arabidopsis of members of the family Pospiviroidae, the Arabidopsis-HSVd system was further explored by constructing transgenic lines expressing HSVd dimeric (-) RNAs. Analysis of nucleic acid preparations from two of these plants by single denaturing PAGE and Northern blot hybridization, with a HSVd riboprobe specific for (+) strands, revealed a band with the electrophoretic mobility characteristic of the monomeric (+) circular form (Fig. 4A, compare lanes 3 and 4 with lanes 1 and 5). The most direct interpretation of these results is that the primary HSVd dimeric (-) transcript served as a template for synthesis of its complementary counterpart, which was then processed to the monomeric (+) circular form. Analysis of the same preparations with a HSVd riboprobe specific for (-) strands failed to reveal the monomeric forms but showed a major band in the position of the primary HSVd dimeric (-) transcript (Fig. 4B, lanes 3 and 4), indicating that this RNA, in contrast to the HSVd dimeric (+) transcript (Fig. 3), is a very poor substrate (if any) for the Arabidopsis enzymes catalyzing cleavage and ligation or is located in a different subnuclear compartment (see below). Therefore, processing of HSVd dimeric transcripts appears to be a polarity intrinsic property, which dictates the susceptibility to and the specificity of the reactions mediated by the host enzymes. The recent finding that in infected cultured cells and plants, PSTVd (-) strands accumulate in the nucleoplasm, whereas the (+) strands are localized in the nucleolus and in the nucleoplasm, provides an explanation for this different behavior and suggests that processing of the (+) strands occurs in the nucleolus (28), where processing of the precursors of rRNAs and tRNAs also takes place (see ref. 57 for a review).
Fig. 4.
Northern blot hybridization analysis of transgenic Arabidopsis expressing dimeric (+) and (-) HSVd RNAs. Viroid-enriched RNA preparations, obtained by chromatography on nonionic cellulose, were fractionated by single denaturing PAGE in duplicated gels and blotted to nylon membranes that were hybridized with 32P-labeled riboprobes for detecting the (+) and (-) HSVd strands (A and B, respectively). Lanes 1, control of HSVd-infected cucumber; lanes 2, control of nontransformed Arabidopsis; lanes 3 and 4, two independent transgenic Arabidopsis expressing dimeric (-) HSVd RNA; and lanes 5, transgenic Arabidopsis expressing dimeric (+) HSVd RNA. Positions of linear RNA markers, with their size in nucleotides, are indicated on the left. Positions of circular and linear HSVd RNAs are indicated on the right in A. Both riboprobes were equalized in acid-precipitable counts, and the films were exposed for the same time. For facilitating detection of HSVd strands accumulating at lower levels, the volume of applied extract was 10-fold higher in lanes 3 and 4 in A, and in lanes 1 and 5 in B.
Agroinoculation of Arabidopsis with Viroid RNAs. If transgenic Arabidopsis expressing certain dimeric viroid transcripts accumulate RNAs with the characteristic properties of the intermediates and final products of the replication cycle, why does Arabidopsis appear to be recalcitrant to viroid infection? In an attempt to provide an answer to this question, wild-type Arabidopsis were infiltrated with cultures of Agrobacterium tumefaciens bearing binary plasmids expressing the dimeric (+) CEVd, HSVd, CCCVd, ASSVd, CbVd-1, and ASBVd RNAs described above (Fig. 1 A). One month after infiltration, analysis by PAGE and Northern blot hybridization with viroid-specific riboprobes failed to detect any of the viroids in upper uninoculated leaves. By contrast, CEVd, HSVd, and CbVd-1 were readily detected in upper uninoculated leaves of CEVd-infiltrated gynura, HSVd-infiltrated cucumber, and CbVd-1-infiltrated coleus (data not shown). These results and others showing that mechanical inoculations with HSVd dimeric (+) RNAs led to systemic infection of cucumber but not of Arabidopsis suggest that CEVd, HSVd, and CbVd-1, and presumably other members of the family Pospiviroidae, may not systemically infect Arabidopsis because they are defective in movement, although lack of efficient replication may also explain this behavior.
Phenotype of Arabidopsis Expressing HSVd Dimeric (+) RNAs. To explore whether accumulation of viroid RNAs induced any symptoms in Arabidopsis, three independent homozygous lines were selected from the progeny of plants expressing the HSVd dimeric (+) RNA. Inspection of the growing pattern of five plants from each of these lines showed a slight stunting as compared with nontransformed controls. However, in attempts to reproduce this observation under the same experimental conditions, only one of the lines consistently displayed the slight stunting, making it difficult to conclude whether the expression of HSVd sequences has any phenotypic effect.
Discussion
The identification of host factors involved in the biological cycle of viroids would be greatly facilitated by the development of an Arabidopsis-based system. However, no viroid has been reported to occur naturally in Arabidopsis or in any other species of the family Brassicaceae (11), and previous efforts aimed at experimentally infecting this model plant with PSTVd, CEVd, and Chrysanthemum stunt viroid have been unsuccessful (49). Although the number of viroids (and sequence variants thereof) and of Arabidopsis ecotypes tested in this and in other assays, which because of their negative results were not reported, may have been low, the available evidence suggests that viroids cannot systemically infect Arabidopsis. However, this does not exclude the possibility that at least certain viroids could replicate in the inoculated cells but were defective in movement and consequently unable to invade distal plant parts. This possibility has been tested here by constructing a series of transgenic Arabidopsis lines expressing different dimeric viroid RNAs and determining whether the characteristic intermediate and final products accumulate in these plants. The potential of transgenic-based approaches in viroid research has been illustrated with the demonstration that HSVd can infect tobacco (58), a plant considered a nonhost for this viroid up to that time.
Six viroid species were chosen for the present study, five (CEVd, HSVd, CCCVd, ASSVd, and CbVd-1) representing the established genera of the family Pospiviroidae, and one (ASBVd) of the family Avsunviroidae (11). Transgenic Arabidopsis expressing dimeric (+) transcripts of these viroids accumulated the correct monomeric (+) circular and linear RNAs as revealed by double PAGE and Northern blot hybridization and by RTPCR amplification, cloning, and sequencing. In all instances the viroid variants corresponding to the transgenes were predominant; the less abundant variants could be artifacts of the RT-PCR amplification or result from RNA-RNA transcription (see below). Moreover, examination by single denaturing PAGE and Northern blot hybridization of the relative band intensities showed that the CEVd and HSVd dimeric (+) RNAs were processed to a significant extent in Arabidopsis and that ligation was particularly efficient in HSVd. These results indicate that at least some steps of the replication cycle of representative members of the family Pospiviroidae, specifically the correct cleavage and ligation of dimeric (+) RNAs to produce the monomeric circular (+) RNA, take place in Arabidopsis, which must therefore contain the enzymes and auxiliary factors required for these reactions. Although processing of the dimeric (+) RNAs of some of the five tested species of the family Pospiviroidae was inefficient, possibly reflecting different affinities of the Arabidopsis machinery, it was precise in all cases despite the very different RNA sequences involved. This finding suggests that the specificity for the cleavage reaction is provided by a particular conformation of the RNA that makes a certain phosphodiester bond particularly susceptible to one or more RNases. In line with this view, a heterologous RNase (the fungal RNase T1) has been shown to accurately process linear oligomeric PSTVd RNAs in vitro (59). Therefore, the specific cleavage of the oligomeric intermediates of both viroid families, whether enzyme- or ribozyme-mediated, appears to be an intrinsic property of the RNA. Regarding ligation, some plant RNA ligases such as the wheat germ ligase are not substrate-specific and only require proper termini (60). The low level of processing of the ASBVd (+) dimeric RNA observed in Arabidopsis most likely results from the replication site of this viroid, the chloroplast instead of the nucleus, and from its cleavage mechanism, catalyzed by ribozymes instead of by enzymes, although it was assisted by chloroplastic proteins (61) whose functional equivalents may not exist in the nucleus.
In addition to correct processing of the primary transcripts, transgenic Arabidopsis expressing dimeric CEVd and HSVd (+) RNAs also accumulated the corresponding (-) strands, which remained essentially unprocessed as in typical host plants infected by the two viroids. These results strongly suggest that the dimeric CEVd and HSVd (+) transcripts, or their resulting monomeric circular forms, can serve as templates for synthesis of the complementary (-) strands. However, (-) polarity strands were not detected in Arabidopsis expressing the dimeric ASBVd (+) RNA, underlining again the distinct behavior of representative members of both viroid families in an Arabidopsis nuclear habitat. The possibility that the CEVd and HSVd (-) RNAs could result from transcription of the antisense strand of the transgene driven by a plant promoter close to the integration site, or by duplication and reorganization of the transgene during the integration, appears unlikely, because three independent transgenic Arabidopsis lines expressing the dimeric CEVd and HSVd (+) RNAs showed the same accumulation pattern of their corresponding (-) polarity strands. Furthermore, the monomeric circular HSVd (+) RNA was also detected in transgenic Arabidopsis expressing the dimeric HSVd (-) RNA, indicating that this RNA may serve as the template for the synthesis of the second RNA-RNA transcription step.
Altogether these results support the view that Arabidopsis has the enzymatic machinery for replicating representative viroid species of the family Pospiviroidae and that the same may happen with additional plants not considered to be viroid hosts. Other results with certain PSTVd mutants obtained at high temperature also indicate that they are able to replicate in Arabidopsis (J. Matousek, personal communication). However, when compared with typical experimental hosts, the replication efficiency of these viroids in Arabidopsis appears to be considerably lower and, most importantly, the agroinoculation experiments show that they are unable to move to distal plant parts. Therefore, movement and low replication or accumulation may be the limiting steps of viroid infection in different plant species. In any case, the transgenic Arabidopsis lines generated in this work and others that may be developed in the future could be a valuable tool in combination with microarray and proteomic technologies for the identification of host factors involved in the biological cycle of some viroids.
Acknowledgments
We thank A. Ahuir and M. Bordás for excellent technical assistance, Dr. M. A. Pérez-Amador for advice during preparation of transgenic Arabidopsis lines, and Drs. V. Pallás and C. Hernández for critical reading and suggestions. This work was supported, in part, by Ministerio de Ciencia y Tecnología of Spain Grant BMC2002-03694. J.-A.D. was the recipient of a postdoctoral “Ramón y Cajal” contract of the Ministerio de Ciencia y Tecnología of Spain.
Abbreviations: ASSVd, apple scar skin viroid; ASBVd, avocado sunblotch viroid; CEVd, citrus exocortis viroid; CCCVd, coconut cadang-cadang viroid; CbVd-1, coleus blumei viroid 1; HSVd, hop stunt viroid; PSTVd, potato spindle tuber viroid.
References
- 1.Somerville, C. & Koornneef, M. (2002) Nat. Rev. Genet. 3, 883-889. [DOI] [PubMed] [Google Scholar]
- 2.Arabidopsis Genomic Initiative. (2000) Nature 408, 796-815.11130711 [Google Scholar]
- 3.Yamanaka, T., Ohta, T., Takahashi, M., Meshi, T., Schmidt, R., Dean, C., Naito, S. & Ishikawa, M. (2000) Proc. Natl. Acad. Sci. USA 97, 10107-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lellis, A. D., Kasschau, K. D., Whitham, S. A. & Carrington, J. C. (2002) Curr. Biol. 25, 1046-1051. [DOI] [PubMed] [Google Scholar]
- 5.Chisholm, S. T., Mahajan, S. K., Whitham, S. A., Yamamoto, M. L. & Carrington, J. C. (2000) Proc. Natl. Acad. Sci. USA 97, 489-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitham, S. A., Anderberg, R. J., Chisholm, S. T. & Carrington, J. C. (2000) Plant Cell 12, 569-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasschau, K. D., Xie, Z., Allen, E., Llave, C., Chapman, E. J., Krizan, K. A. & Carrington, J. C. (2003) Dev. Cell 4, 205-217. [DOI] [PubMed] [Google Scholar]
- 8.Mourrain, P., Beclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J. B., Jouette, D., Lacombe, A. M., Nikic, S., Picault, N., et al. (2000) Cell 26, 533-542. [DOI] [PubMed] [Google Scholar]
- 9.Dalmay, T., Horsefield, R., Braunstein, T. H. & Baulcombe, D. C. (2001) EMBO J. 20, 2069-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diener, T. O. (2001) Adv. Virus Res. 57, 137-184. [DOI] [PubMed] [Google Scholar]
- 11.Flores, R., Randles, J. W., Bar-Joseph, M. & Diener, T. O. (2000) in Virus Taxonomy, Seventh Report of the International Committee on Taxonomy of Viruses, eds. van Regenmortel, M. H. V., Fauquet, C. M., Bishop, D. H. L., Carstens, E. B., Estes, M. K., Lemon, S. M., Maniloff, J., Mayo, M. A., McGeoch, D. J., Pringle, C. R. & Wickner, R. B. (Academic, San Diego), pp. 1009-1024.
- 12.Flores, R., Daròs, J. A. & Hernández, C. (2000) Adv. Virus Res. 55, 271-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palukaitis, P. (1987) Virology 158, 239-241. [DOI] [PubMed] [Google Scholar]
- 14.Woo, Y.-M., Itaya, A., Owens, R. A., Tang, L., Hammons, R. W., Chou, H.-C., Lai, M. M. C. & Ding, B. (1999) Plant J. 17, 627-635. [Google Scholar]
- 15.Zhu, Y., Green, L., Woo, Y.-M., Owens, R. A. & Ding, B. (2001) Virology 279, 69-77. [DOI] [PubMed] [Google Scholar]
- 16.Itaya, A., Folimonov, A., Matsuda, Y., Nelson, R. & Ding, B. (2001) Mol. Plant-Microbe Interact. 14, 1332-1334. [DOI] [PubMed] [Google Scholar]
- 17.Papaefthimiou, I., Hamilton, A. J., Denti, M. A., Baulcombe, D. C., Tsagris, M. & Tabler, M. (2001) Nucleic Acids Res. 29, 2395-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martínez de Alba, A. E., Flores, R. & Hernández, C. (2002) J. Virol. 76, 13094-13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fadda, Z., Daròs, J. A., Fagoaga, C., Flores, R. & Durán-Vila, N. (2003) J. Virol. 77, 6528-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Branch, A. D. & Robertson, H. D. (1984) Science 223, 450-455. [DOI] [PubMed] [Google Scholar]
- 21.Diener, T. O. (1972) Virology 50, 606-609. [DOI] [PubMed] [Google Scholar]
- 22.Gross, H. J., Domdey, H., Lossow, C., Jank, P., Raba, M., Alberty, H. & Sänger, H. L. (1978) Nature 273, 203-208. [DOI] [PubMed] [Google Scholar]
- 23.Owens, R. A. & Diener, T. O. (1982) Proc. Natl. Acad. Sci. USA 79, 113-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spiesmacher, E., Mühlbach, H. P., Schnölzer, M., Haas, B. & Sänger, H. L. (1983) Biosci. Rep. 3, 767-774. [DOI] [PubMed] [Google Scholar]
- 25.Ishikawa, M., Meshi, T., Ohno, T., Okada, Y., Sano, T., Ueda, I. & Shikata, E. (1984) Mol. Gen. Genet. 196, 421-428. [DOI] [PubMed] [Google Scholar]
- 26.Branch, A. D., Benenfeld, B. J. & Robertson. H. D. (1988) Proc. Natl. Acad. Sci. USA 85, 9128-9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feldstein, P. A., Hu, Y. & Owens, R. A. (1998) Proc. Natl. Acad. Sci. USA 95, 6560-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi, Y. & Ding, B. (2003) Plant Cell 15, 2566-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutchins, C. J., Keese, P., Visvader, J. E., Rathjen, P. D., McInnes, J. L. & Symons, R. H. (1985) Plant Mol. Biol. 4, 293-304. [DOI] [PubMed] [Google Scholar]
- 30.Daròs, J. A., Marcos, J. F., Hernández, C. & Flores, R. (1994) Proc. Natl. Acad. Sci. USA 91, 12813-12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Navarro, J. A., Daròs, J. A. & Flores, R. (1999) Virology 253, 77-85. [DOI] [PubMed] [Google Scholar]
- 32.Bussière, F., Lehoux, J., Thompson, D. A., Skrzeczkowski, L. J. & Perreault, J. P. (1999) J. Virol. 73, 6353-6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prody, G. A., Bakos, J. T., Buzayan, J. M., Schneider, I. R. & Bruening, G. (1986) Science 231, 1577-1580. [DOI] [PubMed] [Google Scholar]
- 34.Hutchins, C., Rathjen, P. D., Forster, A. C. & Symons, R. H. (1986) Nucleic Acids Res. 14, 3627-3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forster, A. C. & Symons, R. H. (1987) Cell 49, 211-220. [DOI] [PubMed] [Google Scholar]
- 36.Flores, R., Hernández, C., De la Peña, M., Vera, A. & Daròs, J. A. (2001) Methods Enzymol. 341, 540-552. [DOI] [PubMed] [Google Scholar]
- 37.Branch, A. D., Robertson, H. D., Greer, C., Gegenheimer, P., Peebles, C. & Abelson, J. (1982) Science 217, 1147-1149. [DOI] [PubMed] [Google Scholar]
- 38.Tsagris M., Tabler M., Mühlbach H. P. & Sänger H. L. (1987) EMBO J. 6, 2173-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baumstark, T., Schröder, A. R. W. & Riesner, D. (1997) EMBO J. 16, 599-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Côté F., Lévesque D. & Perreault, J. P. (2001) J. Virol. 75, 19-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clough, S. J. & Bent, A. F. (1998) Plant J. 16, 735-743. [DOI] [PubMed] [Google Scholar]
- 42.Pallás, V., Navarro, A. & Flores, R. (1987) J. Gen. Virol. 68, 3201-3205. [Google Scholar]
- 43.Flores, R., Duran-Vila, N., Pallás, V. & Semancik, J. S. (1985) J. Gen. Virol. 66, 2095-2102. [Google Scholar]
- 44.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 45.Hamilton, C. M., Frary, A., Lewis, C. & Tanksley, S. D. (1996) Proc. Natl. Acad. Sci. USA 93, 9975-9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scofield, S. R., Tobias, C. M., Rathjen, J. P., Chang, J. H., Lavelle, D. T., Michelmore, R. W. & Staskawicz, B. J. (1996) Science 274, 2063-2065. [DOI] [PubMed] [Google Scholar]
- 47.Cress, D. E., Kiefer, M. C. & Owens, R. A. (1983) Nucleic Acids Res. 11, 6821-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tabler, M. & Sanger, H. L. (1984) EMBO J. 3, 3055-3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martínez-Herrera, D. (1994) Ph.D. thesis (Universidad Complutense de Madrid, Madrid).
- 50.Semancik, J. S. & Weathers, L. G. (1972) Nat. New Biol. 237, 242-244. [DOI] [PubMed] [Google Scholar]
- 51.Gross, H. J., Krupp, G., Domdey, H., Raba, M., Jank, P., Lossow, C., Alberty, H., Ramm, K. & Sanger, H. L. (1982) Eur. J. Biochem. 121, 249-257. [DOI] [PubMed] [Google Scholar]
- 52.Ohno, T., Takamatsu, N., Meshi, T & Okada, Y. (1983) Nucleic Acids Res. 11, 6185-6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haseloff, J., Mohamed, N. A. & Symons, R. H. (1982) Nature 229, 316-321. [Google Scholar]
- 54.Hashimoto, J. & Koganezawa, H. (1987) Nucleic Acids Res. 15, 7045-7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spieker, R. L., Haas, B., Charng, Y.-C., Freimüller, K. & Sänger, H. L. (1990) Nucleic Acids Res. 18, 3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Symons, R. H. (1981) Nucleic Acids Res. 9, 6527-6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lewis, J. D. & Tollervey, D. (2000) Science 288, 1385-1389. [DOI] [PubMed] [Google Scholar]
- 58.Yamaya, J., Yoshioka, M., Sano, T., Shikata, E. & Okada, Y. (1989) Mol. Plant-Microbe Interact. 2, 169-174. [Google Scholar]
- 59.Tabler, M., Tzortzakaki, S. & Tsagris M. (1992) Virology 190, 746-753. [DOI] [PubMed] [Google Scholar]
- 60.Arn, E. A. & Abelson, J. (1998) in RNA Structure and Function (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 695-726.
- 61.Daròs, J. A. & Flores, R. (2002) EMBO J. 21, 749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]