Abstract
The differentiation of pluripotent or totipotent cells into various differentiated cell types is accompanied by a restriction of gene expression patterns, alteration in histone and DNA methylation, and changes in the gross nuclear organization of eu- and heterochromatic domains. Several recent studies have coupled genome-wide mapping of histone modifications with changes in gene expression. Other studies have examined changes in the subnuclear positioning of tissue-specific genes upon transcriptional induction or repression. Here we summarize intriguing correlations of the three phenomena, which suggest that in some cases causal relationships may exist.
In the last decade, interest in embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) has increased sharply due to their potential for medical application [1•]. A number of studies link specific chromatin modifications and the spatial organization of the genome with cellular pluripotency, defined as the capacity of ESCs and iPSCs to generate differentiated tissues. Here we review recent data on chromatin and DNA modifications and their relationship to nuclear organization during cell fate acquisition and differentiation. We explore evidence that argues for a causal link between the 3D organization of the genome and cell type commitment in model organisms.
Genomic marks: pluripotent versus differentiated epigenomic landscapes
During the course of development and differentiation, cells acquire specific fates by altering their transcriptional profiles. Not surprisingly, differentiation also correlates with changes in the distribution of epigenetic marks. These changes alter the repartition and abundance of both repressive and active histone tail modifications, DNA methylation and the accessibility of transcription factor binding sites to their ligands [2–6].
Genome-wide methods such as ChIP-chip or ChIP-seq have begun to map systematically the human and mouse epigenomes of both undifferentiated (e.g. ESCs) and differentiated cells, and have followed changes in these marks during induced differentiation. A frequently used model system exploits the differentiation of mouse ESCs first into neural progenitors (neural precursor cells, NPCs) and then into various neural cell types [7]. Alternatively, ESCs can be compared with independently obtained differentiated cell lines. Although a complete epigenomic analysis through a differentiation pathway is still lacking, some general conclusions can be extracted from these studies.
One uncontested change is that DNA methylation generally increases on tissue-specific promoters that become silenced during tissue differentiation; intriguingly, in the committed precursor stage these de novo methylation targets often are bivalent for active and repressive marks (i.e. carry both active H3K4 and silent H3K27 methylation) [8]. Similar bivalent promoters have been found in zebrafish embryos at the onset of genome activation [9•]. A second conclusion from these studies is that the global amount of the repressive Polycomb-associated mark H3K27 trimethylation does not change significantly during the differentiation of ESCs to neuronal cells, even though H3K27me3 levels fluctuate a great deal at individual promoters. Hundreds of promoters gain this Polycomb-deposited mark and hundreds also lose it during the transitions from ESC to committed precursor (NPCs) and from NPC to differentiated neuron [10]. In many cases a loss of H3K27me3 coincides with loss of bivalency. Third, in zebrafish, mouse and human pluripotent cells many promoters appear to be marked with H3K4 trimethylation, even in the absence of detectable polymerase [9•,11–14], Upon differentiation, on the other hand, most promoters with H3K4 trimethylation become transcriptionally active. Importantly, the methylation of both H3K27 and H3K4 is enriched within CpG islands, providing one explanation for the overlap. It has been shown that the CpG-binding protein Cfp1 induces H3K4 trimethylation at CpG islands that lack DNA methylation, even in the absence of a promoter [15]. On the repressive side, many Polycomb-binding sites reside within hyperconserved CpG islands, and introduction of exogenous CpG sequences is sufficient to recruit PRC2 [8,14,16,17].
There is less consensus on the behavior of the histone modification associated with constitutive heterochromatin; that is, it is unclear whether the amount of histone H3K9 dimethylation and trimethylation increases during differentiation of ESCs [18,19]. In many organisms this mark is associated with the repetitive DNA at centromeres, and would not be expected to change with differentiation. However, H3K9me is also associated with the binding of heterochromatin protein 1 variants (HP-1 α, (β, or γ), which are linked to both gene repression and transcriptional elongation during tissue development [20–22]. Immunofluorescence studies showed increase in the number of HP1-α containing foci and the intensity of H3K9me staining during conversion of mouse ESCs to NPCs [23]. However, genome-wide changes in histone marks were not quantitatively as large one might have expected given the major reorganization of the genome that occurs during differentiation (see below).
Multiple classes of chromatin during differentiation
Many of the chromatin marks analyzed in the mapping studies are spatially segregated in domains of similarly marked chromatin within the nucleus. Although there is no comprehensive study of the nuclear morphology of histone marks as yet, anecdotal evidence shows that the shape, number and size of such chromatin domains often change with differentiation [24–27]. Indeed, electron-dense and DAPI-staining heterochromatin foci appear to increase during the course of differentiation [24,28,29•].
For decades, electron microscopic images of osmium-stained nuclei allowed one to distinguish euchromatin (lightly stained) from heterochromatin (darkly stained) with the latter often tightly sequestered by the inner nuclear envelope [30]. Two recent studies, using principal component analysis and/or Hidden Markov Models (HMMs) to define chromatin classes on the basis of nonhistone protein enrichment [31••] or enrichment of specific histone marks [32••], now suggest that this binary classification scheme is too simple to describe the relevant classes of chromatin. At least five distinct types of chromatin, classified by the abundance of specific nonhistone binding factors, were identified in Drosophila Kc167 cells [31••]. This remarkable study by the van Steensel laboratory used fusions of the bacterial DNA methylase (Dam) to 53 different chromatin associated factors, to score their distribution over the genome through the quantitative mapping of adenine methylation (DamID [33]). They identified three distinct classes of silent chromatin, namely, the simple-repeat associated HP1-binding chromatin found at centromeres, the H1-associated and lamin-associated chromatin that is enriched for silent tissue-specific genes, and finally silent domains enriched for Polycomb [31••]. They further identified two classes of transcriptionally active chromatin: one distinguished by an enrichment for H3K36 methylation and its ligand, Mrg15, and a second that is replicated very early and is enriched for large regulatory protein complexes.
A distinct HMM approach was applied to histone modifications mapped in human CD4+ T cells [32••,34].This study also identified five classes of euchromatin and heterochromatin, and in contrast to the van Steensel study, could distinguish upstream regulatory sequences from coding regions based on their histone mark profiles [32••]. These classifications will surely be exploited in future studies that characterize chromatin changes during cellular differentiation. Even if a few more chromatin types are found, the fact that a limited number of categories of chromatin can be robustly identified by HMM analysis, argues that large domains of the genome share structural characteristics.
Consistent with the notion that at least one class of heterochromatin accumulates at the nuclear rim during differentiation, the progressive association of repressed pluripotency genes and silent tissue-specific genes with the nuclear lamin has been scored during fly differentiation or as neurons are generated from ESCs [35••,36]. Analysis of the genomic sequences lying close to the nuclear envelope has shown that lamin-interacting domains tend to be transcriptionally silent [36,37]. In both flies and mouse cells the overall percentage of the genome attached to lamin is large (40–48% of the probed genome) [36,37], yet only about 1000 of over 17 000 genes scored (12%) show a significant increase in lamin association during the mouse ESC to neuronal differentiation [35••]. Importantly, these 1000 are enriched for pluripotency genes, which become repressed as cells differentiate, and silent non-neuronal tissue-specific genes. Nonetheless, 30% of the genes that became lamin-bound did not change in expression, indicating that the nuclear periphery does not necessarily impose transcriptional repression [38]. In the other direction the correlation was somewhat more robust: many of the genes that were released from the lamina upon differentiation were shown to be ‘unlocked’ or ‘open’ for lineage-specific transcription, even though active transcription might only occur much later. In conclusion, DamID profiles on in vitro neuronal differentiation showed surprisingly few changes in lamin-associated domains (LADs), despite the seemingly major increase in chromatin at the nuclear rim in differentiated cells [24,28]. Moreover, although genes shifted to the nuclear envelope are enriched for repressed loci, it is clear that gene repression and peripheral sequestration are not fully congruent.
Increase of genome compaction correlates with decrease of genome plasticity and stabilization of cell fate
While tissue culture cells and in vitro differentiation systems have opened the door to genome-wide chromatin studies, recent work in the nematode Caenorhabditis gans has allowed one to explore how changes in nuclear organization influence differentiation events on an organismal level. Consistent with the lack of centromeric repeats and pericentric heterochromatin, C. elegans embryos show little electron-dense chromatin at the earliest stages of development [39]. Yet, compact, transcriptionally silent heterochromatin appears progressively as cells differentiate in late embryonic stages and is found enriched at the nuclear periphery or close to the nucleolus (Figure 1 and [39]). Similarly, mouse ESCs were also reported to have low levels of heterochromatin, which increase during differentiation [24,28,29•]. In worms and mice, this accumulation of heterochromatin correlates with a loss of both pluripotency and the capacity to be reprogrammed to another cell fate. Inactivation of Polycomb in worms both prolongs both developmental plasticity and an open chromatin configuration [40••]. While the role of Polycomb for chromatin compaction in ES cells has not been investigated, inactivation of Polycomb interferes with differentiation, similar to worms [41].
Figure 1.
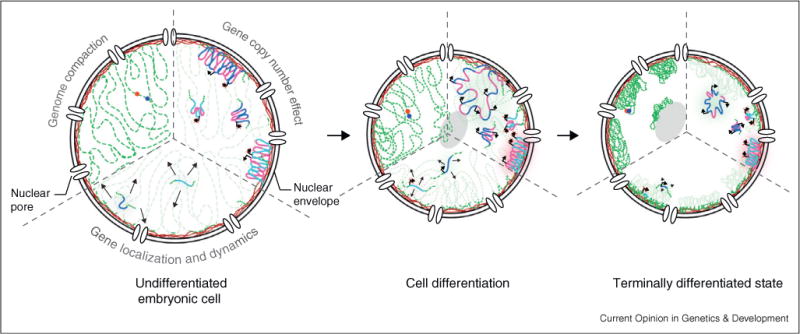
Major nuclear reorganization events over the course of cell fate acquisition. During differentiation, the genome gets globally compacted (upper left third), with clusters of compact chromatin being located close to the lamina (red meshwork) at the nuclear periphery or the nucleolus (light grey), leaving gaps or channels between large chromatin masses. Based on C. elegans studies, gene copy number is seen to influence nuclear localization (upper right third). In embryonic cells, heterochromatic high copy number repeats are located at the nuclear rim while low-copy repeats do not show preferential localization. During differentiation, activation of promoters is able to overcome the anchoring at the nuclear periphery. Preferential localization at the nuclear lamina is observed for silent tissue-specific promoters, while active tissue-specific promoters are more likely to be internal. Gene and chromatin associated dynamics decrease progressively as cells differentiate (lower third). This is correlated to the disappearance of some remodelers and the increase of anchored compact chromatin domains.
Nuclear reprogramming to an alternative cell fate can be readily induced during worm development by the ectopic expression of ‘selector genes’ or ‘master regulatory’ transcription factors. If artificially induced at early stages of worm development, these master regulators are able to transform an entire embryo into a single tissue [42–44]. The efficiency of this transformation, or the degree of ‘cell plasticity’, decreases as worms develop and its cells acquire specific cell fates [40••]. A similar drop in the efficiency of reprogramming has also been characterized in mouse by the use of nuclear transplantation. As early as the blastocyst stage, mouse nuclei injected into enucleated oocytes or into early embryos showed a reduced ability to support organismal growth [45]. This same inefficiency for dedifferentiation or reprogramming is manifest when nuclei from differentiated tissues are fused with ESCs [46].
This loss of developmental plasticity correlates with a compaction of genomic chromatin — not only into domains associated with H3K27 methylation [40••], but also with the nuclear lamina [35••]. Consistent with these observations, chromosomes 6 and 8 appeared to undergo compaction during human ESC differentiation [25]. Electron microscopic studies suggest that the mouse embryonic genome similarly undergoes progressive compaction between the one cell stage and E5.5 [29•].
Given this apparent reduction of genome volume, it is relevant to ask what fraction of nuclear space is occupied by DNA, and how this changes through development. Using serial block face electron microscopy, a technique that allows reconstruction of entire nuclei at electron microscopy resolution, differentiated hepatocytes and endothelial nuclei were estimated to have between 40 and 60% of their volume free of DNA [47]. The compaction of the genome through differentiation may leave space for transcription, splicing, replication and repair events in ‘empty zones’ or ‘perichromatin’ channels. Indeed, transcription occurs primarily external to dense chromatin territories and can be visualized by the looping out of activated genes from the bulk of a chromosomal domain into less dense interchromosomal space [48,49].
What could be the function of this genomic subcompartmentalization into domains of compacted heterochromatic and more open euchromatin? One hypothesis is that compartmentalization would reduce the complexity of the genome by hiding a number of possible binding sites for transcription factors in silenced heterochromatin. This is particularly important for higher eukaryotes in which most of the genome is non-coding. Alternatively, the sequestration of inactive chromatin may by default help target the transcription machinery to sites of transcription [5]. Finally, the formation of heterochromatin clusters could sequester general repressors from promiscuous binding and repression as shown in yeast [50]. All three hypotheses share the virtue of reinforcing cell fate restrictions. In contrast, gene induction events leading to cell fate changes appear to be more stochastic.
In support of the model that transcription maintains an open chromatin state in undifferentiated cells, a microarray analysis suggested that ESCs have widespread transcription throughout their genomes, including intergenic sequences, whereas differentiated cells did not [28]. This finding, however, has been challenged by recent RNA-seq experiments which suggest that transcription is largely confined to annotated genes and is not widely different between ESCs and differentiated cells ([51] and D Schuebeler, personal communication). Clearly this topic merits further analysis. Intriguingly, chemical inhibition of heterochromatin formation by the histone deacetylase inhibitor TSA was found to impair ES cell differentiation [52], and both TSA and other chemicals that interfere with heterochromatin structure were shown to improve the efficiency of iPSCs generation (from 0.001% to 0.15%) when dedifferentiation is induced by a subset of the Yamanaka factors in differentiated cells [53,54].
Reorganizing chromosomes and genes in three dimensions
A small number of studies have directly examined the subnuclear organization of genes and chromosomes during mouse ESC differentiation. From the evidence available it seems that most chromosomes do not change their radial positioning during differentiation [27,55]. This suggests either that radial positioning is established as early as the stage represented by ESCs, or that position depends primarily on gene density and not on activity [27,56]. Nonetheless, some mouse chromosomes do show some degree of reorganization during differentiation. For example, centromeres are found more frequently associated with the nuclear rim or the nucleolus after differentiation [27,55]. Similarly, the silent X chromosome in female cells relocates toward the nuclear periphery when female hESCs are induced to differentiate [27]. Still, global chromosome positioning is unlikely to be a major regulatory feature in development, since developmentally regulated genes can be translocated or expressed ectopically with only minor effects on development.
In attempts to correlate chromosome position with the regulation of single genes during differentiation, genes encoding the pluripotency factors NANOG and OCT3/4 have been a major focus. These genes, together with SOX2, c-MYC and KLF4, are highly expressed in ESCs, and their overexpression in differentiated cells can induce dedifferentiation and a pluripotent state (iPS cells) [57,58]. Consistent with results from other genes, the NANOG gene on the short arm of chromosome 12, gets repositioned upon repression, moving from the nuclear center in hESCs to the nuclear periphery in lymphoblastoid cells (hLCLs). Other studies documented the inverse switch for nonpluripotency genes: the β-globin gene gets relocated internally upon activation during erythroid differentiation, as does the proneural gene Mash1 upon activation [59,60]. The monoallelically expressed astrocyte-specific GFAP gene also shows a slight inward shift, which again correlates with activity (internally active and peripherally inactive) [61]. Finally, the complete Hox locus was seen to reposition upon activation, by looping away from its chromosome territory [48,49]. These observations are anecdotal, however, and exceptions also exist: for example, the OCT4 pluripotency gene, in contrast to NANOG, does not show changes in radial positioning upon differentiation [25,55]. This may reflect the fact that OCT4 is situated much further from the edge of its chromosomal territory in LCLs than NANOG [55].
Unfortunately, these correlative data do not address what drives relocation nor do they prove that a shift in position is necessary to ensure gene activity. For this the C. elegans system proved more adept: by monitoring transgene insertions of different size, it could be shown that two major parameters dictate gene positioning in vivo: the size of a repetitive array and the transcriptional status of developmentally regulated promoters (Figure 1 and [62••]). Silent promoters in low-copy-number are randomly localized in undifferentiated embryonic cell nuclei, while the same sequence integrated as a high copy number array is recruited to the nuclear periphery [62••,63]. This copy-number dependence is correlated with the presence of inactive chromatin marks, H3K9me3 and H3K27me3, although neither modification is sufficient for perinuclear anchoring, as they are found in foci throughout the nucleus. On a genome-wide level, recent modENCODE data have shown a striking correlation in worms between peripheral localization, high repeat density and H3K9 methylation [64,65•,66•]. Similarly, LADs are associated with H3K9me2 and H3K27me3 histone marks in mammalian cells [37].
As differentiation progresses in C. elegans, tissue-specific genes become positioned, driven by the activity of their promoters, independently of copy number (Figure 1 and [62••]). This developmental control of promoter position is strongly reminiscent of the hypothesis drawn from differences in nuclear organization between ESCs and differentiated cells; that is, heterochromatin was seen to accumulate at the nuclear periphery [28] and chromosomes became increasingly compacted as cells acquired a differentiated cell fate [25]. Similar to mammalian cells, relocalization and decompaction could be correlated with the binding of master regulatory transcription factors [62••,67]. As proposed for mammalian cells [60,61,68,69], the acquisition of an altered nuclear organization, following from an altered chromatin structure, could be triggered by the opening of promoter domains by a master regulator [62••,67]. However, the nuclear lamina-associated protein emerin could impede the DNA binding and decompaction activities of the endoderm determining factor PHA-4 in a subpopulation of embryonic cells [67], suggesting a tug of war between repressive activities associated with the lamina and activation by tissue-determining transcription factors.
Pluripotent chromatin: moving genes and proteins to keep all options open
Further evidence that the chromatin structure in ESCs is different from that in differentiated cells arose from fluorescence recovery after photobleaching (FRAP) experiments carried out with GFP-tagged nuclear proteins. GFP fusions to core histones H2A and H3, linker histone H1 and HP1 all showed higher turnover rates in ESCs than in committed neuronal precursor cells [70]. This suggests that nucleosome stability changes during differentiation, yet the removal of a histone chaperone, HirA had the surprising effect that ESCs differentiated faster, and not more slowly. Thus a simple correlation of higher turnover rates and increased pluripotency cannot be drawn, although decreased exchange of the linker histone H1 appeared to inhibit NPC cell fate acquisition, consistent with a reduction in cell plasticity [70].
The enhanced turnover of chromatin-bound proteins may also in part reflect the action of ATP-dependent chromatin remodeling. Chromatin remodelers are a family of multiprotein complexes containing an AAA+ ATPases domain, which is able to reposition or remove nucleosomes or subcomplexes of histones. Their activities facilitate both the induction and the repression of genes [71]. A number of remodelers have been shown to be essential for the maintenance of pluripotency, while others have been implicated in cellular transformation, with genes for specific subunits being identified as tumor suppressor genes [28,72–75]. For example, the chromatin remodeling factor Chd1 promotes open chromatin and a pluripotent state in mouse ES and iPS cells [75]. Moreover, it has been shown that some remodeling complexes are expressed uniquely in undifferentiated cells and are essential for pluripotent state maintenance, although their mechanism of action and target genes is still unclear [72,73]. Thus, much remains to be resolved as to how chromatin remodelers contribute to pluripotent states [28,72–75].
The effect of removing chromatin remodelers has been studied in yeast, since none is absolutely essential for vegetative growth on rich media. This redundancy suggests that no single nucleosome remodeler will be sufficient to induce pluripotency, nor is one likely to be essential for a specific differentiation pathway. Rather they can be considered to be instruments that ‘grease the wheels’ of change from one state to another. This may be achieved by facilitating the binding of transcription factors to their cognate sites, or by promoting the removal of certain histone variants or modified forms. With this perspective it is likely that the loss of a remodeler might limit access of tissue-specific transcription factors to their cognate sites, thus indirectly promoting the heterochromatinization of adjacent genes, to restrict genome-wide transcription of uncommitted cells [28].
In yeast, an unexpected effect of remodelers was monitored when they were targeted to a fluorescently tagged genomic locus: they were seen to increase gross chromatin mobility as monitored with time lapse microscopy, without changing its transcription level (F Neumann, SM Gasser, personal communication). In this context it is plausible that chromatin remodelers play a key role in the repositioning of genes and chromosomes during the transition from a pluripotent to a restricted potency state coincident with cell type differentiation [76]. This gene mobility, like histone eviction or repositioning, is an active process that requires ATP and responds to nutrient levels [77]. The space that a given locus can explore inside the nucleus is restrained by its physical link to a chromosome [78]. This constraint is further reinforced if the locus is located close to sequences which are anchored to nuclear structures, like the nuclear envelope or the nucleolus [78,79]. Thus it is probable, although not yet proven, that the relocalization of activated promoters is driven by chromatin remodeling.
In conclusion, systematic mapping techniques over the course of development have revealed relatively few large-scale changes in comparison with the extensive reorganization of specific chromatin loci inside the nucleus, which can be observed microscopically. A careful comparative study using both approaches will be important to understand both sets of data. Only by characterizing the molecular players involved in genome three-dimensional reorganization, will we be able to understand how the spatial arrangement of genes impacts differentiation and the maintenance of cell fate.
Acknowledgments
We thank J Lis and D Schubeler for communicating unpublished data, G Chew for comments on the manuscript. PM is a fellow of the Fondation Suisse de Recherche sur les Maladies Musculaires, the Gasser laboratory is supported by the Novartis Research Foundation, the NCCR Frontiers in Genetics, the FP7 Network of Excellence, ‘Epigenome’. SEM is funded by NIH R01GM056264, the John D and Catherine T MacArthur Foundation and Harvard University.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1•.Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24:2239–2263. doi: 10.1101/gad.1963910. This very detailed and up to date review encompasses all aspects of induced pluripotent cells generation, as well as the characteristics of iPSCs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cedar H, Bergman Y. Linking DNA, methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 3.Meshorer E. Chromatin in embryonic stem cell neuronal differentiation. Histol Histopathol. 2007;22:311–319. doi: 10.14670/HH-22.311. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 5.Mohn F, Schubeler D. Genetics and epigenetics: stability and plasticity during cellular differentiation. Trends Genet. 2009;25:129–136. doi: 10.1016/j.tig.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 6.Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- 7.Bibel M, Richter J, Schrenk K, Tucker KL, Staiger V, Korte M, Goetz M, Barde YA. Differentiation of mouse embryonic stem cells into a defined neuronal lineage. Nat Neurosci. 2004;7:1003–1009. doi: 10.1038/nn1301. [DOI] [PubMed] [Google Scholar]
- 8.Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 9•.Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. 2010;464:922–926. doi: 10.1038/nature08866. Using zebrafish as a model of early embryogenesis, the authors ask when and where H3K27 and H3K4 methylation marks appear in the genome. They show that these modifications appear at the maternal-zygotic transition, when the genome becomes transcriptionally active. Similar to ES cells, one third of the genes show bivalent chromatin marks. Another third is unmarked, while the last third shows only H3K4 methylation. This last category is present but has been overlooked in ES cells and most surprisingly, most genes are silent and not bound by RNA polymerase II. It is proposed that many promoters get the H3K4 methylation mark during zygotic genome activation, which poises them for activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 11.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, Bernstein BE. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 2010;6:e1001244. doi: 10.1371/journal.pgen.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanay A, O’Donnell AH, Damelin M, Bestor TH. Hyperconserved CpG domains underlie Polycomb-binding sites. Proc Natl Acad Sci U S A. 2007;104:5521–5526. doi: 10.1073/pnas.0609746104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filion GJ, van Steensel B. Reassessing the abundance of H3K9me2 chromatin domains in embryonic stem cells. Nat Genet. 2010;42:4. doi: 10.1038/ng0110-4. (author reply 5–6) [DOI] [PubMed] [Google Scholar]
- 20.de Wit E, Braunschweig U, Greil F, Bussemaker HJ, van Steensel B. Global chromatin domain organization of the Drosophila genome. PLoS Genet. 2008;4:e1000045. doi: 10.1371/journal.pgen.1000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaudin V, Libault M, Pouteau S, Juul T, Zhao G, Lefebvre D, Grandjean O. Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis. Development. 2001;128:4847–4858. doi: 10.1242/dev.128.23.4847. [DOI] [PubMed] [Google Scholar]
- 22.Hediger F, Gasser SM. Heterochromatin protein 1: don’t judge the book by its cover! Curr Opin Genet Dev. 2006;16:143–150. doi: 10.1016/j.gde.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayakawa S, Miike K, Nakao M, Abe K. Dynamic changes in the epigenomic state and nuclear organization of differentiating mouse embryonic stem cells. Genes Cells. 2007;12:447–460. doi: 10.1111/j.1365-2443.2007.01063.x. [DOI] [PubMed] [Google Scholar]
- 25.Bartova E, Krejci J, Harnicarova A, Kozubek S. Differentiation of human embryonic stem cells induces condensation of chromosome territories and formation of heterochromatin protein 1 foci. Differentiation. 2008;76:24–32. doi: 10.1111/j.1432-0436.2007.00192.x. [DOI] [PubMed] [Google Scholar]
- 26.Aoto T, Saitoh N, Ichimura T, Niwa H, Nakao M. Nuclear and chromatin reorganization in the MHC-Oct3/4 locus at developmental phases of embryonic stem cell differentiation. Dev Biol. 2006;298:354–367. doi: 10.1016/j.ydbio.2006.04.450. [DOI] [PubMed] [Google Scholar]
- 27.Bartova E, Galiova G, Krejci J, Harnicarova A, Strasak L, Kozubek S. Epigenome and chromatin structure in human embryonic stem cells undergoing differentiation. Dev Dyn. 2008;237:3690–3702. doi: 10.1002/dvdy.21773. [DOI] [PubMed] [Google Scholar]
- 28.Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH, et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Ahmed K, Dehghani H, Rugg-Gunn P, Fussner E, Rossant J, Bazett-Jones DP. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS One. 2010;5:e10531. doi: 10.1371/journal.pone.0010531. This paper describes chromatin in embryonic, differentiated and induced pluripotent cells using electron spectroscopic imaging. This electron microscopy technique allows to identify phosphate or nitrogen-enriched regions under the electron beam, giving a high-resolution image of where chromatin and proteins are located inside the nucleus. The authors show extensive alterations in nuclear organization. ES and iPSC cells nuclei show a highly dispersed chromatin mesh, while differentiated cells nuclei contain compact chromatin domains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heitz E. Das Heterochromatin der Moose. Jahrb Wiss Botanik. 1928;69:762–818. [Google Scholar]
- 31••.Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010 doi: 10.1016/j.cell.2010.09.009. Using a two-state Hidden Markov Model to analyze DamID data for 53 different nuclear proteins in Kc167 Drosophila cells, the authors define five different types of chromatin. This contrasts with the simplistic binary picture provided by electron microscopy of euchromatic (light stained) and heterochromatic (dark stained) domains. This study is the basis for new ways of thinking how chromatin is defined based on its interacting factors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. Similar to the previous study, a Hidden Markov Model is used to define chromatin regions, based on previously published data on 38 different histone modifications in CD4+ human T cells. Surprisingly, a similar number of chromatin types is found, although slightly different, as a multivariate analysis is used, which leads to a high number of ‘states’ defining the different chromatin types. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vogel MJ, Pagie L, Talhout W, Nieuwland M, Kerkhoven RM, van Steensel B. High-resolution mapping of heterochromatin redistribution in a Drosophila position-effect variegation model. Epigenet Chromatin. 2009;2:1. doi: 10.1186/1756-8935-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 35••.Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. Molecular maps of the reorganization of genome nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. Using the DamID technology based on a lamin-dam fusion during the induction of astrocyte differentiation from mouse ESCs, the authors define Lamin-Associated Domains (LADs). About 90% of these domains are stable during differentiation. However, LADs containing neuronal genes activated during differentiation are released from the nuclear periphery upon activation. Surprisingly a number of LADs are released before transcriptional activation of the genes (‘unlocked’). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- 37.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 38.Towbin BD, Meister P, Gasser SM. The nuclear envelope — a scaffold for silencing? Curr Opin Genet Dev. 2009;19:180–186. doi: 10.1016/j.gde.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 39.Leung B, Hermann GJ, Priess JR. Organogenesis of the Caenorhabditis elegans intestine. Dev Biol. 1999;216:114–134. doi: 10.1006/dbio.1999.9471. [DOI] [PubMed] [Google Scholar]
- 40••.Yuzyuk T, Fakhouri TH, Kiefer J, Mango SE. The polycomb complex protein mes-2/E(z) promotes the transition from developmental plasticity to differentiation in C. elegans embryos. Dev Cell. 2009;16:699–710. doi: 10.1016/j.devcel.2009.03.008. Using staged C. elegans embryos to probe for cell fate plasticity upon ectopic expression of a ‘selector’ gene able to transform cell identity, the authors show that the Polycomb protein mes-2/E(z) is necessary to restrict plasticity, suggesting a role for H3K27 methylation in the restriction of cell fate choices. Moreover, they observe that cell fate restriction correlates with global compaction of the genome, partially dependent on mes-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-repressive complex 2. Genes Dev. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fukushige T, Krause M. The myogenic potency of HLH-1 reveals wide-spread developmental plasticity in early C. elegans embryos. Development. 2005;132:1795–1805. doi: 10.1242/dev.01774. [DOI] [PubMed] [Google Scholar]
- 43.Horner MA, Quintin S, Domeier ME, Kimble J, Labouesse M, Mango SE. pha-4, an HNF-3 homolog, specifies pharyngeal organ identity in Caenorhabditis elegans. Genes Dev. 1998;12:1947–1952. doi: 10.1101/gad.12.13.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiefer JC, Smith PA, Mango SE. PHA-4/FoxA cooperates with TAM-1/TRIM to regulate cell fate restriction in the C. elegans foregut. Dev Biol. 2007;303:611–624. doi: 10.1016/j.ydbio.2006.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gurdon JB, Laskey RA, Reeves OR. The developmental capacity of nuclei transplanted from keratinized skin cells of adult frogs. J Embryol Exp Morphol. 1975;34:93–112. [PubMed] [Google Scholar]
- 46.Cowan CA, Atienza J, Melton DA, Eggan K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science. 2005;309:1369–1373. doi: 10.1126/science.1116447. [DOI] [PubMed] [Google Scholar]
- 47.Rouquette J, Genoud C, Vazquez-Nin GH, Kraus B, Cremer T, Fakan S. Revealing the high-resolution three-dimensional network of chromatin and interchromatin space: a novel electron microscopic approach to reconstructing nuclear architecture. Chromosome Res. 2009;17:801–810. doi: 10.1007/s10577-009-9070-x. [DOI] [PubMed] [Google Scholar]
- 48.Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chambeyron S, Da Silva NR, Lawson KA, Bickmore WA. Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development. 2005;132:2215–2223. doi: 10.1242/dev.01813. [DOI] [PubMed] [Google Scholar]
- 50.Taddei A, Van Houwe G, Nagai S, Erb I, van Nimwegen E, Gasser SM. The functional importance of telomere clustering: global changes in gene expression result from SIR factor dispersion. Genome Res. 2009;19:611–625. doi: 10.1101/gr.083881.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most ‘dark matter’ transcripts are associated with known genes. PLoS Biol. 2010;8:e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee JH, Hart SR, Skalnik DG. Histone deacetylase activity is required for embryonic stem cell differentiation. Genesis. 2004;38:32–38. doi: 10.1002/gene.10250. [DOI] [PubMed] [Google Scholar]
- 53.Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 55.Wiblin AE, Cui W, Clark AJ, Bickmore WA. Distinctive nuclear organisation of centromeres and regions involved in pluripotency in human embryonic stem cells. J Cell Sci. 2005;118:3861–3868. doi: 10.1242/jcs.02500. [DOI] [PubMed] [Google Scholar]
- 56.Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Muller S, Eils R, Cremer C, Speicher MR, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 58.Yamanaka S. Induction of pluripotent stem cells from mouse fibroblasts by four transcription factors. Cell Prolif. 2008;41(Suppl 1):51–56. doi: 10.1111/j.1365-2184.2008.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ragoczy T, Bender MA, Telling A, Byron R, Groudine M. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 2006;20:1447–1457. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams RR, Azuara V, Perry P, Sauer S, Dvorkina M, Jorgensen H, Roix J, McQueen P, Misteli T, Merkenschlager M, et al. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J Cell Sci. 2006;119:132–140. doi: 10.1242/jcs.02727. [DOI] [PubMed] [Google Scholar]
- 61.Takizawa T, Gudla PR, Guo L, Lockett S, Misteli T. Allele-specific nuclear positioning of the monoallelically expressed astrocyte marker GFAP. Genes Dev. 2008;22:489–498. doi: 10.1101/gad.1634608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62••.Meister P, Towbin BD, Pike BL, Ponti A, Gasser SM. The spatial dynamics of tissue-specific promoters during C. elegans development. Genes Dev. 2010;24:766–782. doi: 10.1101/gad.559610. Using C. elegans, the authors show that copy number and developmentally regulated promoters are two critical parameters directing nuclear gene localization. While in undifferentiated embryonic cells, high copy number arrays are localized at the nuclear lamina, while the same sequences in lower repeats number is randomly localized. However, upon differentiation, in four different tissues of the three germ layers, transgenes are segregated either to the nuclear rim when inactive or the nuclear center when transcriptionally active, independent of their repeat number. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Towbin B, Meister P, Pike BL, Gasser SM. Repetitive transgenes in C. elegans accumulate heterochromatic marks and are sequestered at the nuclear envelope in a copy number- and lamin-dependent manner. Cold Spring Harb Symp Quant Biol. 2010 doi: 10.1101/sqb.2010.75.041. in press. [DOI] [PubMed] [Google Scholar]
- 64.Liu T, Rechtsteiner A, Egelhofer TA, Vielle A, Latorre I, Cheung MS, Ercan S, Ikegami K, Jensen M, Kolasinska-Zwierz P, et al. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. 2011;21:227–236. doi: 10.1101/gr.115519.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65•.Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. This modENCODE consortium generated a number of large-scale data-sets ranging from chromatin modifications, transcription factors and RNA polymerase binding sites, RNA and micro-RNA sequencing. The meta-analysis of the data is presented here. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66•.Ikegami K, Egelhofer T, Strome S, Lieb JD. Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol. 2010;11:R120. doi: 10.1186/gb-2010-11-12-r120. This companion paper to the modENCODE integrative analysis [60] describes in more detail the LEM-2 ChIP-chip and ChIP-seq experiments. It also provides experimental data on the boundary between LEM-2 bound domains at the nuclear periphery and the rest of the genome, as well as sequence characteristics of the peripherally located worm genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fakhouri TH, Stevenson J, Chisholm AD, Mango SE. Dynamic chromatin organization during foregut development mediated by the organ selector gene PHA-4/FoxA. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brown KE, Amoils S, Horn JM, Buckle VJ, Higgs DR, Merkenschlager M, Fisher AG. Expression of alpha- and beta-globin genes occurs within different nuclear domains in haemopoietic cells. Nat Cell Biol. 2001;3:602–606. doi: 10.1038/35078577. [DOI] [PubMed] [Google Scholar]
- 69.Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 70.Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mellor J. The dynamics of chromatin remodeling at promoters. Mol Cell. 2005;19:147–157. doi: 10.1016/j.molcel.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 72.Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci U S A. 2009;106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gasser SM. Visualizing chromatin dynamics in interphase nuclei. Science. 2002;296:1412–1416. doi: 10.1126/science.1067703. [DOI] [PubMed] [Google Scholar]
- 77.Heun P, Laroche T, Shimada K, Furrer P, Gasser SM. Chromosome dynamics in the yeast interphase nucleus. Science. 2001;294:2181–2186. doi: 10.1126/science.1065366. [DOI] [PubMed] [Google Scholar]
- 78.Gartenberg MR, Neumann FR, Laroche T, Blaszczyk M, Gasser SM. Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell. 2004;119:955–967. doi: 10.1016/j.cell.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 79.Chubb JR, Boyle S, Perry P, Bickmore WA. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr Biol. 2002;12:439–445. doi: 10.1016/s0960-9822(02)00695-4. [DOI] [PubMed] [Google Scholar]


