Abstract
Osterix (Osx) is an osteoblast-specific transcription factor required for osteoblast differentiation and bone formation. Osx knock-out mice lack bone completely. Recent findings that Osx inhibits Wnt signaling provide a feedback control mechanism involved in bone formation. Mechanisms of Osx inhibition on Wnt signaling are not fully understood. Our results in this study revealed that the expression of a Wnt antagonist Sclerostin (Sost) was downregulated in Osx-null calvaria. Overexpression of Osx in stable C2C12 mesenchymal cell line resulted in Sost upregulation. Transient transfection assay showed that Osx activated 1 kb Sost promoter reporter activity in a dose-dependent manner. To define Sost promoter activated by Osx, we made a series of deletion mutants of Sost constructs, and narrowed down the minimal region to the proximal 260 bp. Gel shift assay indicated that Osx bound to GC-rich site within this minimal region, and that point mutations of this binding site disrupted Osx binding. Moreover, the same point mutations in 260 bp Sost promoter reporter disrupted the promoter activation by Osx, suggesting that the GC-rich binding site was responsible for Sost promoter activation by Osx. To further examine physical association of Osx with Sost promoter in vivo, Chromatin immunoprecipitation (ChIP) assays were performed using primary osteoblasts from mouse calvaria. Osx was found to associate with endogenous Sost promoter. Taken together, these findings support our hypothesis that Sost is a direct target of Osx. This provides a new additional mechanism through which Osx inhibits Wnt signaling during bone formation.
Keywords: Osx, Sost, Osteoblast, Wnt signaling, Bone formation
1. Introduction
Bone formation takes place through two distinct processes: endochondral ossification involving a cartilage model and intramembranous ossification by which bones form directly from condensations of mesenchymal cells without a cartilage intermediate. Bone formation is a highly regulated process involving the differentiation from mesenchymal progenitor cells into preosteoblast, then into osteoblast lineage, and finally into osteocytes [1,2]. Osteoblast differentiation is regulated by different transcription factors and signaling proteins including Indian hedgehog (Ihh), Runx2, Osterix (Osx), and Wnt signaling pathway. Ihh is required for endochondral but not for intramembranous bone formation [3] and is needed for the establishment of the osteogenic portion of the perichondrium/periosteum and for the initial activation of the gene for Runx2. Runx2 is needed for bone formation since no endochondral and no membranous bones are formed in Runx2-null mice [4]. Runx2 is required for the differentiation of mesenchymal cells into preosteoblasts. As a downstream gene of Runx2, Osx is required for the differentiation of preosteoblasts into mature osteoblasts. Osx is specifically expressed in all osteoblasts. In Osx-null embryos, cartilage is formed normally, but the embryos completely lack bone formation [2]. Osx expression pattern in mice indicates that the presence of Osx transcript begins as early as the commitment time for mesenchymal cells to enter osteoblast lineage and its signal becomes stronger as osteoblast differentiation occurs. The C terminal region of Osx contains the DNA-binding domain which can bind to specific GC-rich sequences to control target gene expression, such as osteoblast differentiation markers type 1 collagen, bone sialoprotein (BSP), and osteocalcin.
Wnt signaling has been studied for its broad range of activities in cell proliferation, differentiation and cell death during both embryonic development, and the adult stage in a variety of tissue types including bone [5]. Wnts are secreted glycoproteins that bind to Frizzled family receptors and low-density lipoprotein receptorrelated proteins (LRP) 5/6 coreceptors. In the absence of Wnt, β-catenin forms a complex with the APC, Axin, and the kinases glycogen synthase kinase 3 (GSK3), which facilitates phosphorylation and proteosomal degradation of β-catenin. Stimulation of these receptors by Wnts leads to the intracellular molecule β-catenin to accumulate and translocate into the nucleus, where it interacts with TCF/Lef1 transcription factor to activate transcription of target genes. Wnt/β-catenin pathway has been known to play a crucial role in bone formation and bone metabolism [6]. Gain-of-function mutants of Lrp5 cause high bone mass syndrome in patients [7] and in mice [8]. Conditional inactivation of b-catenin in either skeletal progenitor cells or at a later stage of osteoblast development in mouse embryos blocks osteoblast differentiation [9–12]. Dickkopf (Dkk) is a Wnt antagonist. It binds to LRP5/6 receptor to form a complex with Kremen1 and 2 and inhibits Wnt signaling by reducing the availability of LRP5/6 [5]. Sclerostin (Sost), another extracellular Wnt antagonist, binds to LRP5/6 receptor to prevent Wnt binding to LRP5/6 and inhibits Wnt signaling [5,13–15]. It has been demonstrated that the Sost loss-of-function mutations are the cause for Sclerosteosis and Van Buchem disease with dramatically increased bone mass due to increased Wnt signal [5]. On the other hand, transgenic mice that overexpress Sost are osteopenic due to reduced bone formation, consistent with a model whereby Sost negatively regulates osteoblast activity [16].
Recent study has discovered that in addition to its essential role in osteoblast differentiation, the osteoblast-specific transcription factor Osx also inhibits osteoblast proliferation and negatively regulates Wnt/β-catenin signaling [17]. Further data have indicated that Osx controls Wnt signaling by two different mechanisms: (i) stimulates Wnt antagonist DKK1 expression and (ii) disrupts Tcf1 binding to DNA to inhibit the transcriptional activity of β-catenin/Tcf. Those findings cannot exclude the possibility that Osx might also target other components of Wnt signaling as part of its inhibitory effect. Osx inhibition of Wnt signaling provides a feedback control mechanism involved in bone formation. The mechanisms of Osx inhibition on Wnt signaling are not fully understood.
In this study, our results from quantitative real-time RT-PCR revealed that Sost expression was downregulated in the calvaria of Osx-null mouse embryos, suggesting Osx is essential for Sost expression. Furthermore, we provide evidences to demonstrate that Osx directly targets Sost. This is a new additional mechanism through which Osx inhibits Wnt signaling during bone formation.
2. Materials and methods
2.1. RNA isolation and real-time RT-PCR
Total RNA was isolated from the calvaria of E18.5 wild type and Osx-null mouse embryos with TRIzol reagent (Invitrogen) followed by RNeasy mini kit (Qiagen). Total RNA from C2C12 cells was isolated using QIAGEN RNeasy Mini Kit. RNA was subjected to quantitative RT-PCR, using the TaqMan One-Step RT-PCR Master Mix reagent. Relative transcript levels were measured by real-time PCR in 50 ll reaction volume on 96-well plates, using an ABI 7500 real-time PCR system (Applied Biosystem). Transcript levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels. All experiments were done in duplicates.
2.2. Plasmid constructs and subcloning
The fragments of Sost promoter region were generated by PCR using mouse genomic DNA as a template and subcloned into the XhoI and MluI sites of pGL-3 vector. Primers were obtained from Integrated DNA Technologies (IDT) (Coralville, IA), and the sequences were as follows: (1) SOST-Xho-3: 5′GCG CCT CGA GTG TCC AGC CTA GAT ACG GTT G; (2) SOST-Mlu-1K-5: 5′GCG CAC GCG TGA AAG ACA CCT CCT CAG GTC; (3) Sost-Mlu-540: 5′GCG CAC GCG TAA GGC ATC CTT CTG; (4) Sost-Mlu-260: 5′GCG CAC GCG TTG TGT CCC TGC CTC; and (5) Sost-Mlu-106: 5′GCG CAC GCG TTG AGG AGG AGG GTG A. Sost point mutants were made with the QuickChange site-directed mutagenesis kit (Stratagene) using Sost-260 as a template by the following primers: (1) SOSTM1-1: 5′CCT CGG GTC ACC TGA AAA ATA CCA GCA GCA ATT TGGAAG; (2) SOST-M1-2: 5′CTTCCA AAT TGC TGC TGG TAT TTT TCA GGT GAC CCG AGG; (3) SOST-M2-1: 5′CCC TGC CTC ACC AAA GAA AAA CCC CCC AAC ACA CAC; and (4) SOST-M2-2: 5′GTG TGT GTT GGG GGG TTT TTC TTT GGT GAG GCA GGG. All constructs including mutants were verified by DNA sequencing.
2.3. Cell culture and transient transfection assay
HEK293 cells were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen, CA) with 10% fetal bovine serum and 100 units/ml penicillin plus 100 lg/ml streptomycin at 37 °C. HEK293 cells were plated in 12-well plates, cultured to 60–80% confluence and transfected with FuGENE 6 (Roche) according to the manufacture’s instruction. Cells were cotransfected with 300 ng of Sost promoter reporter, Osx expression plasmid as indicated, and 25 ng of pSV2-beta-gal. After transfection, cells were incubated for 24 h before harvest. The reporter assays were analyzed with BD Monolight system (BD Biosciences). Luciferase activity was normalized by βgalactosidase activity. Every transfection experiment was done at least three times. Values were presented as the mean ± S.D.
2.4. Electrophoretic mobility shift assay (EMSA)
EMSA was performed according to protocols previously described [17]. Briefly, Sost promoter oligoes were ordered from IDT (Coralville, IA). Oligo sequences are as following: wild-type Sost-1: 5′-GGG A CCT GGG AGG TGC CAG CAG CAA TTT GGAAGTT-3′ and Sost-2: 5′-CCCAACTT CCA AAT TGC TGC TGG CAC CTC CCA GGT-3′; mutant Sost-M-1: 5′-GGG A CCT GAA AAA TAC CAG CAG CAA TTT GGAAGTT-3′ and Sost-M-2: 5′-CCC AACTT CCA AAT TGC TGC TGG TAT TTT TCA GGT-3′. Baculovirus-expressed Osx was used as the protein resource as previously described [18]. In the gel shift assays, P32-labeled oligo was incubated with Osx. Protein–DNA complexes were resolved on a non-denaturing polyacrylamide gel and visualized by autoradiography.
2.5. Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed as previously described [19] provided in Supplementary materials.
3. Results
3.1. Sost expression is downregulated in the absence of Osx
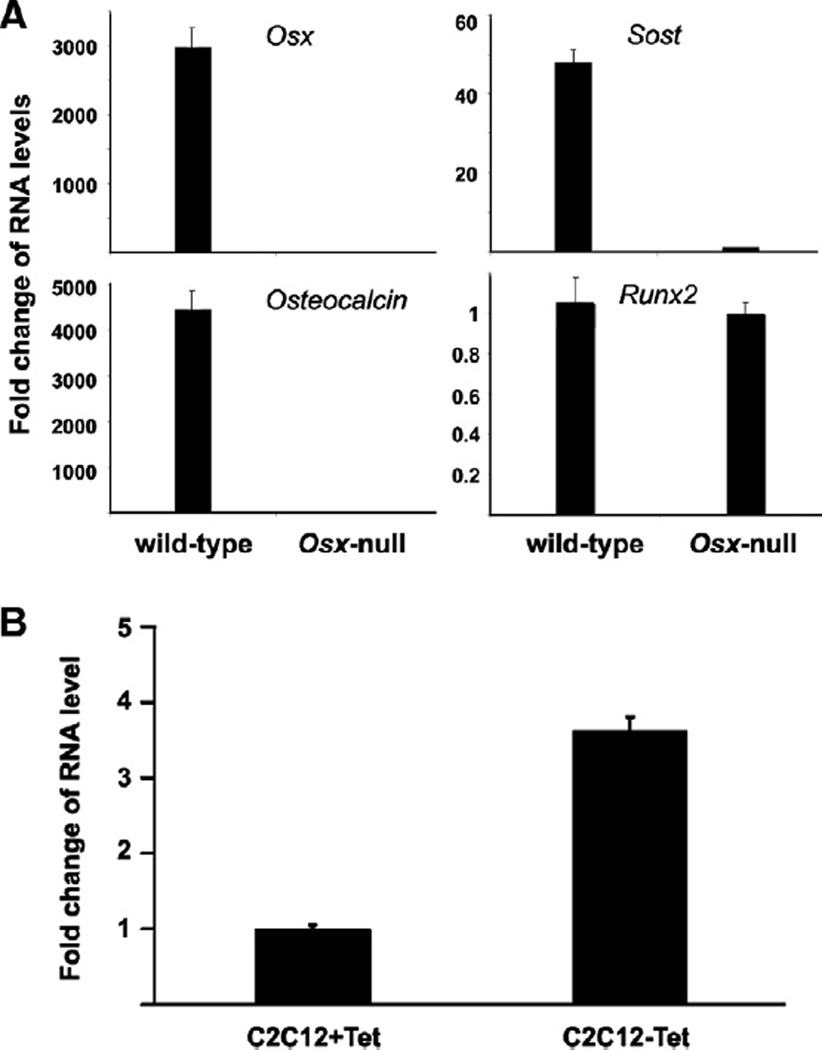
Our recent studies have demonstrated that Osx can inhibit Wnt signaling, a possible mechanism for Osx to inhibit osteoblast proliferation [17]. The mechanisms of Osx inhibition on Wnt signaling are not fully understood. We carried out quantitative real-time RT-PCR to compare RNA of some genes between Osx wild type and knock-out mice. RNA was isolated from calvaria of E18.5 mouse embryos. As shown in Fig. 1A, Osx expression was abolished in Osx-null calvaria. Osteoblast marker gene osteocalcin was at undetected level as expected. Expression of Runx2, which is upstream of Osx, was unchanged as a negative control. Interestingly, Wnt signaling antagonist Sost expression was found downregulated by about 47-folds in Osx-null calvaria compared with that in wild-type calvaria. The marked decrease in Sost RNA level in Osx knock-out mice suggests that Osx controls Sost gene expression.
Figure 1.
Effect of Osx on Sost expression. (A) Fold change in RNA levels from E18.5 wild-type and Osx-null mice embryos. Calvaria RNAs were isolated from E18.5 Osx wild-type and Osx-null embryos. RNA levels for Osx, Osteocalcin, Runx2, and Sost were analyzed by real-time RT-PCR. The level of each RNA from Osx-null calvaria was normalized to a value of 1. (B) Fold change of RNA levels for Sost gene expression in C2C12 stable cells using Tet-off system. RNA was isolated from C2C12 stable cells. In C2C12 stable cell line, Osx expression was induced in the absence of tetracycline. RNA levels were measured by real-time RT-PCR. The RNA level from Tet-present cells was normalized to a value of 1. Values were presented as the mean ± SD.
3.2. Overexpression of Osx activates Sost expression
Next we asked whether Osx could positively regulate Sost expression. To address this question, a C2C12 stable cell line was used in which the expression of Osx could be induced by using the tetracycline (Tet) system as previously described [17]. Osx expression was tuned on in the absence of Tet. Total RNA was isolated from C2C12 stable cell line in the presence or absence of Tet and measured by real-time RT-PCR for Sost expression. As shown in Fig. 1B, in the absence of Tet when Osx expression was induced, Sost expression was upregulated by about 4-folds. This observation indicates that Osx activates Sost expression.
3.3. Osx stimulates Sost promoter activity in a dose-dependent manner
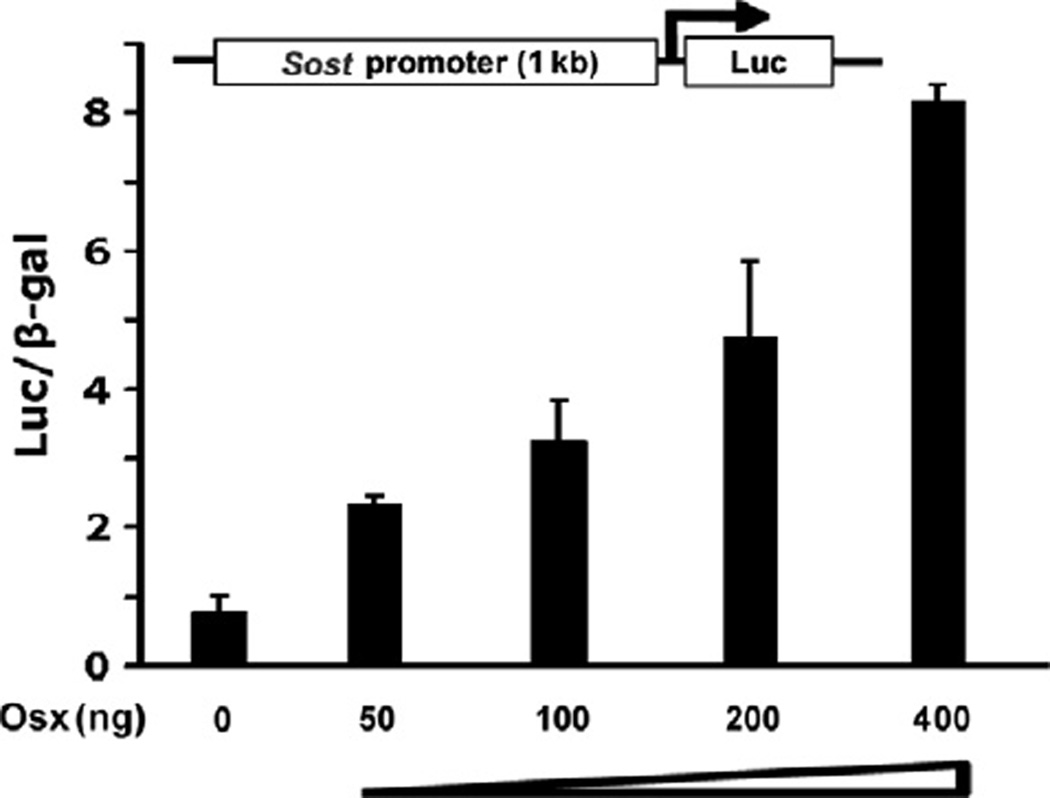
Real-time RT-PCR and C2C12 stable cell line results suggest that Osx activates Sost expression. To further confirm this observation, we generated luciferase reporter construct driven by 1 kb Sost promoter. Transient transfection assays were carried out in HEK293 cells. Sost promoter construct plasmid was cotransfected with increasing amounts of Osx expression plasmid. As shown in Fig. 2, the activation of Sost promoter reporter by Osx was detected when as low as 50 ng of Osx was transfected. Increasing amounts of Osx resulted in higher expression of Sost promoter reporter. This demonstrated that Osx stimulated 1 kb Sost promoter luciferase reporter in a dose-dependent manner, suggesting that Osx transcriptionally activated the Sost gene.
Figure 2.
Osx activates Sost promoter activity in a dose-dependent manner. HEK293 cells were transfected with 1 kb Sost promoter luciferase reporter without or with increasing amounts of Osx as indicated. Luciferase activity was normalized by β-galactosidase activity. Values were presented as the mean ± SD.
3.4. Identification of the Osx binding site in the promoter of Sost gene
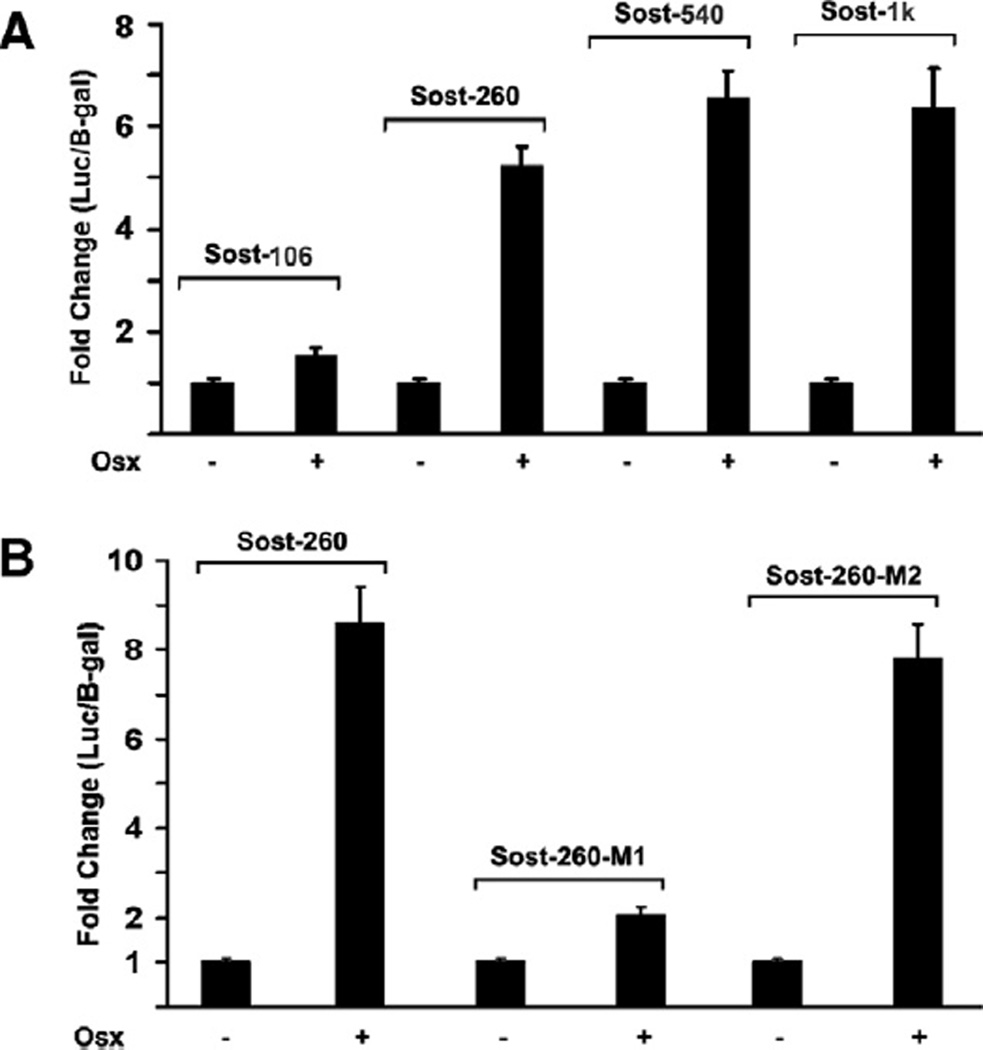
We have shown Osx can stimulate Sost promoter activity, however it is still not clear which region within Sost promoter is responsible for Osx activation. To address this question, the deletion analysis and transient transfection assay were carried out to narrow down responsible region within 1 kb Sost promoter for Osx activation. Sost luciferase reporter constructs driven by different lengths of Sost promoter region were generated. As shown in Fig. 3A, Osx was able to activate Sost promoter reporters of Sost-1 kb, Sost-540 bp, and Sost-260 bp in transient transfection assay. However Osx activation was dramatically reduced in Sost-106 bp reporter. These data suggest that the Sost promoter region between Sost-260 bp and Sost-106 bp contains critical binding site for Osx. Previous study has shown that Osx is a zinc finger-containing transcription factor and belongs to the Sp/XKLF family [2]. This family of transcription factors tends to bind to GC-rich sequence in target gene promoter region to control target gene expression. According to the sequence analysis of the region between 260 bp and 106 bp, two tentative binding sites for Osx were identified as GC-rich elements. To test which binding site is responsible for Sost promoter activation by Osx, we generated two point mutants of Sost-260 promoter reporter named Sost-260-M1 and Sost-260-M2, in which GC-rich element was replaced with A. In transient transfection assay, Sost-260-M1 mutant resulted in significantly reduced reporter activation by Osx as shown in Fig. 3B. On the other hand, Sost-260-M2 mutant reporter was activated by Osx in the similar level to the wild-type Sost-260 reporter. Thus, these results indicated that GC-rich sequence in Sost-260-M1 was responsible for Sost promoter activation by Osx.
Figure 3.
Osx binding site is located within 260 bp of Sost promoter. (A) Deletion analysis of Sost promoter reporter activated by Osx. Sost-1 kb, Sost-540 bp, Sost-260 bp, and Sost-106 bp Sost promoter reporter plasmids were constructed. Each plasmid was cotransfected with Osx expression plasmid in HEK293 cells. Luciferase activity was normalized by β-galactosidase activity. (B) The GC-rich element in Sost-260-M1 is responsible for Sost promoter reporter activation by Osx. Sost-260-M1 and Sost-260-M2 point mutants were constructed. Each plasmid was cotransfected with Osx expression plasmid in HEK293 cells. Luciferase activity was normalized by β-galactosidase activity.
3.5. Osx directly binds to GC-rich sequence within Sost promoter
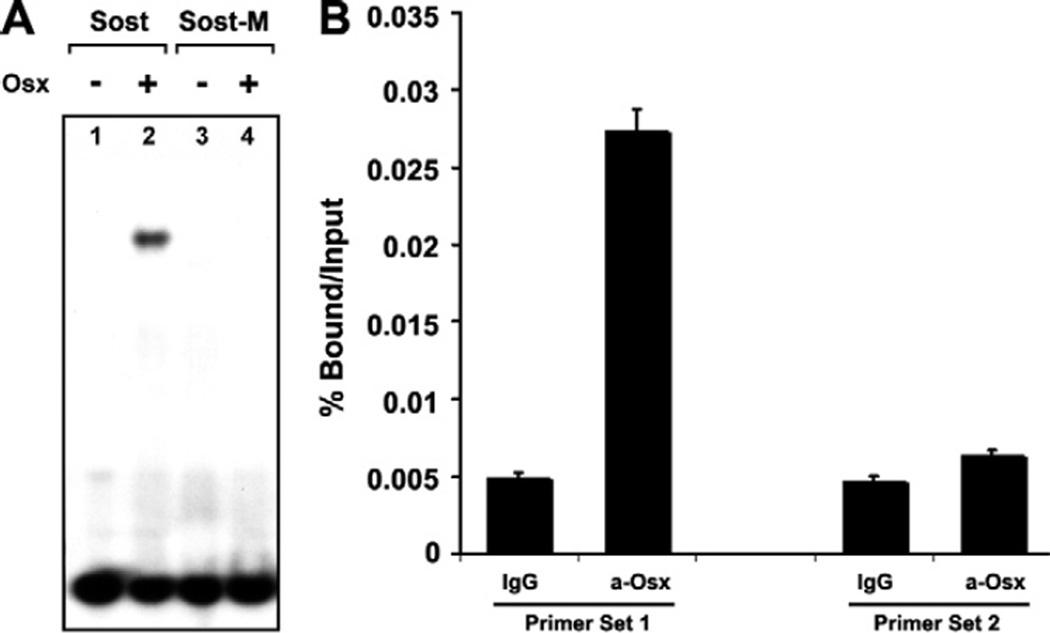
Having shown that a GC-rich sequence between 260 bp and 106 bp region of Sost promoter was critical for Sost promoter activation by Osx, we tested whether Osx could physically bind to this GC-rich element directly by Gel shift assay. The oligo containing the GC-rich sequence in Sost-260-M1 was radio-labeled and incubated with Osx protein. Protein–DNA complexes were resolved on a non-denaturing polyacrylamide gel and visualized by autoradiography. As shown in Fig. 4A, Osx bound to the GC-rich oligo in lane 2. No Osx was added in lane 1 as a negative control. To confirm that Osx binding to this oligo was specific, we designed point mutant oligo named by Sost-M replacing GC-rich with A, corresponding to point mutant sequences of Sost-260-M1 used in Fig. 3B. As shown in Fig. 4A, no Osx binding was observed in the Sost-M in lane 4. Thus, Sost-M mutant abolished Osx binding to this element. These data clearly indicated that Osx specifically bound to GC-rich sequence in Sost promoter.
Figure 4.
Osx associates with Sost promoter. (A) Osx directly binds to Sost GC-rich sequence in EMSA. In the gel shift assays, P32-labeled oligo corresponding to Sost GC-rich sequence identified within 260 bp of Sost promoter region was incubated with Osx. Baculovirus-expressed Osx was used as the protein resource. Protein–DNA complexes were resolved on a non-denaturing polyacrylamide gel and visualized by autoradiography. (B) Osx associates with native Sost promoter in ChIP assay. Calvarial cells were isolated and cultured from wild-type new born mice. Anti-Osx antibody was used for ChIP analysis, and IgG was used as a negative control. The precipitated chromatin was analyzed by quantitative real-time PCR. Primer set 1 corresponds to a segment of the Sost promoter containing GC-rich element between 106 bp and 260 bp. As a negative control, Primer set 2 covers a distal region of the Sost promoter, which does not contain GC-rich sequence.
3.6. Osx associates with native Sost promoter in vivo
The studies above indicate that Osx can activate Sost promoter activity in vitro and that Osx can specifically bind to the GC-rich region of Sost promoter. It is currently unknown whether endogenous Osx can bind to the native Sost promoter in vivo. Therefore, ChIP assay was performed to determine whether Osx could associate with Sost promoter in primary calvarial cells isolated from new born wild-type mice. Two sets of primers were designed. Primer set 1 covered GC-rich element identified within 260 bp Sost promoter in Figs. 3 and 4A. Primer set 2 covered a sequence without GC-rich region at the distal 3 kb within Sost promoter. Anti-Osx antibody was used as previously described for ChIP analysis [17]. The precipitated chromatin was analyzed by quantitative real-time PCR using Primer sets 1 and 2. As shown in Fig. 4B, Osx was found to associate with the Sost promoter region containing GC-rich sequence (Primer set 1) compared with IgG control group. Osx did not associate with the distal Sost promoter region without GC-rich sequence (Primer set 2). The data suggest that Osx associates with Sost promoter in vivo.
4. Discussion
We performed in vitro and in vivo experiments to study the effect of osteoblast-specific transcription factor Osx on the Wnt antagonist Sost. In the calvaria of Osx-null mice embryos, the Sost expression was downregulated, suggesting that Osx positively regulates the expression of Sost. This was supported by additional findings: Osx activated a 1 kb Sost promoter region in a dose-dependent manner, and bound directly to a GC-rich sequence between 260 bp and 106 bp within Sost promoter. More importantly, ChIP assay demonstrated that Osx was associated with Sost promoter in vivo in primary osteoblasts. Our data elucidated that Osx bound to and activated Sost promoter. A GC-rich element within the Sost promoter region was responsible for Sost activation by Osx. Taken together, these evidences strongly indicate that Osx directly targets Sost.
Wnt signaling is known to have the major impact at different stages of bone formation and bone metabolism [5,6]. Wnt signaling-mediated gene expression can promote osteoblast proliferation and differentiation. Some studies investigated the role of Wnt/β-catenin signaling in non-union and osteoporosis, suggesting Wnt signaling could possibly have potential to become a target of pharmacological intervention to increase bone formation [20,21]. Sost is one of the Wnt antagonists. The Sost loss-of-function mutations in human cause the autosomal recessive bone dysplasias Sclerosteosis and Van Buchem disease, which are characterized by progressive bone overgrowth throughout life, enlargement of the jaw and facial bones, and increased bone formation [14,22]. Sost was also shown to be able to mediate bone response to mechanical unloading by antagonizing Wnt/β-catenin signaling [23]. It suggests that Sost is probably a mechanical loading sensor, which is secreted by osteocytes and acts on osteoblast on bone surface.
Due to the important role of Wnt signaling during bone formation, it will be interesting to investigate regulations of Wnt signaling. The extracellular Wnt antagonist Sost shows more advantage over other agent as the therapeutic target due to the bone-specificity of its expression. Currently, it is not well understood how Sost is regulated during bone formation. Parathyroid hormone (PTH) was found to induce bone gain through inhibition of Sost [24]. PTH might affect Sost mainly through cAMP/PKA signaling pathway. Osx is required for osteoblast differentiation and bone formation [2]. Recent study has demonstrated that Osx inhibits Wnt signaling activity during bone formation, a possible mechanism for the inhibition by Osx of osteoblast proliferation [17]. Osx inhibition of Wnt signaling provides a feedback control balance in bone formation. The mechanisms of Osx inhibition on Wnt signal are not fully understood. One mechanism is through Osx activation of Dkk1 [17]. However, different from Dkk1, Sost is highly specifically expressed in osteoblast lineage cells [16,25]. Because of bonespecific expression pattern of both Osx and Sost, the observation in this study that Osx controls Sost expression may provide a clue to develop a potential strategy to manipulate bone anabolic pathway for therapeutic applications.
In summary, Sost is a direct target of Osx. This provides a new additional mechanism through which Osx inhibits Wnt signaling. Elucidation of Osx inhibition of Wnt signaling will help to better understand the molecular mechanism of Osx effect on bone formation.
Supplementary Material
Acknowledgments
Work in Bone Research Laboratory is supported by Research Grant from Arthritis Foundation (To Chi Zhang) and RAP01 Grant from Texas Scottish Rite Hospital for Children (To Chi Zhang).
Abbreviations
- Osx
Osterix
- Sost
Sclerostin
- ChIP
chromatin immunoprecipitation
- Ihh
Indian hedgehog
- LRP
low-density lipoprotein receptor-related proteins
- Dkk
Dickkopf
- Tet
tetracycline
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bbrc.2010.08.128.
References
- 1.Franceschi RT, Ge C, Xiao G, Roca H, Jiang D. Transcriptional regulation of osteoblasts. Cells Tissues Organs. 2009;189:144–152. doi: 10.1159/000151747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 3.St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 5.Piters E, Boudin E, Van Hul W. Wnt signaling: a win for bone. Arch. Biochem. Biophys. 2008;473:112–116. doi: 10.1016/j.abb.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Baron R, Rawadi G, Roman-Roman S. Wnt signaling: a key regulator of bone mass. Curr. Top. Dev. Biol. 2006;76:103–127. doi: 10.1016/S0070-2153(06)76004-5. [DOI] [PubMed] [Google Scholar]
- 7.Little RD, Carulli JP, Del Mastro RG, Dupuis J, Osborne M, Folz C, Manning SP, Swain PM, Zhao SC, Eustace B, Lappe MM, Spitzer L, Zweier S, Braunschweiger K, Benchekroun Y, Hu X, Adair R, Chee L, FitzGerald MG, Tulig C, Caruso A, Tzellas N, Bawa A, Franklin B, McGuire S, Nogues X, Gong G, Allen KM, Anisowicz A, Morales AJ, Lomedico PT, Recker SM, Van Eerdewegh P, Recker RR, Johnson ML. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am. J. Hum. Genet. 2002;70:11–19. doi: 10.1086/338450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Babij P, Zhao W, Small C, Kharode Y, Yaworsky PJ, Bouxsein ML, Reddy PS, Bodine PV, Robinson JA, Bhat B, Marzolf J, Moran RA, Bex F. High bone mass in mice expressing a mutant LRP5 gene. j. Bone Miner. Res. 2003;18:960–974. doi: 10.1359/jbmr.2003.18.6.960. [DOI] [PubMed] [Google Scholar]
- 9.Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev. Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 10.Hill TP, Spater D, Taketo MM, Birchmeier W, Hartmann C. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev. Cell. 2005;8:727–738. doi: 10.1016/j.devcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Hu H, Hilton MJ, Tu X, Yu K, Ornitz DM, Long F. Sequential roles of Hedgehog and Wnt signaling in osteoblast development. Development. 2005;132:49–60. doi: 10.1242/dev.01564. [DOI] [PubMed] [Google Scholar]
- 12.Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133:3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- 13.Balemans W, Piters E, Cleiren E, Ai M, Van Wesenbeeck L, Warman ML, Van Hul W. The binding between sclerostin and LRP5 is altered by DKK1 and by high-bone mass LRP5 mutations. Calcif. Tissue Int. 2008;82:445–453. doi: 10.1007/s00223-008-9130-9. [DOI] [PubMed] [Google Scholar]
- 14.Semenov M, Tamai K, He X. SOST is a ligand for LRP5/LRP6 and a Wnt signaling inhibitor. J. Biol. Chem. 2005;280:26770–26775. doi: 10.1074/jbc.M504308200. [DOI] [PubMed] [Google Scholar]
- 15.ten Dijke P, Krause C, de Gorter DJ, Lowik CW, van Bezooijen RL. Osteocytederived sclerostin inhibits bone formation: its role in bone morphogenetic protein and Wnt signaling. J. Bone Joint Surg. Am. 2008;90(Suppl. 1):31–35. doi: 10.2106/JBJS.G.01183. [DOI] [PubMed] [Google Scholar]
- 16.Winkler DG, Sutherland MK, Geoghegan JC, Yu C, Hayes T, Skonier JE, Shpektor D, Jonas M, Kovacevich BR, Staehling-Hampton K, Appleby M, Brunkow ME, Latham JA. Osteocyte control of bone formation via sclerostin, a novel BMP antagonist. EMBO J. 2003;22:6267–6276. doi: 10.1093/emboj/cdg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang C, Cho K, Huang Y, Lyons JP, Zhou X, Sinha K, McCrea PD, de Crombrugghe B. Inhibition of Wnt signaling by the osteoblast-specific transcription factor Osterix. Proc. Natl. Acad. Sci. USA. 2008;105:6936–6941. doi: 10.1073/pnas.0710831105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang C, Baudino TA, Dowd DR, Tokumaru H, Wang W, MacDonald PN. Ternary complexes and cooperative interplay between NCoA-62/Ski-interacting protein and steroid receptor coactivators in vitamin D receptor-mediated transcription. J. Biol. Chem. 2001;276:40614–40620. doi: 10.1074/jbc.M106263200. [DOI] [PubMed] [Google Scholar]
- 19.Zhang C, Dowd DR, Staal A, Gu C, Lian JB, van Wijnen AJ, Stein GS, MacDonald PN. Nuclear coactivator-62 kDa/Ski-interacting protein is a nuclear matrix-associated coactivator that may couple vitamin D receptor-mediated transcription and RNA splicing. J. Biol. Chem. 2003;278:35325–35336. doi: 10.1074/jbc.M305191200. [DOI] [PubMed] [Google Scholar]
- 20.Issack PS, Helfet DL, Lane JM. Role of Wnt signaling in bone remodeling and repair. HSS J. 2008;4:66–70. doi: 10.1007/s11420-007-9072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krishnan V, Bryant HU, Macdougald OA. Regulation of bone mass by Wnt signaling. J. Clin. Invest. 2006;116:1202–1209. doi: 10.1172/JCI28551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wergedal JE, Veskovic K, Hellan M, Nyght C, Balemans W, Libanati C, Vanhoenacker FM, Tan J, Baylink DJ, Van Hul W. Patients with Van Buchem disease, an osteosclerotic genetic disease, have elevated bone formation markers, higher bone density, and greater derived polar moment of inertia than normal. J. Clin. Endocrinol. Metab. 2003;88:5778–5783. doi: 10.1210/jc.2003-030201. [DOI] [PubMed] [Google Scholar]
- 23.Lin C, Jiang X, Dai Z, Guo X, Weng T, Wang J, Li Y, Feng G, Gao X, He L. Sclerostin mediates bone response to mechanical unloading through antagonizing Wnt/beta-catenin signaling. J. Bone Miner. Res. 2009;24:1651–1661. doi: 10.1359/jbmr.090411. [DOI] [PubMed] [Google Scholar]
- 24.Keller H, Kneissel M. SOST is a target gene for PTH in bone. Bone. 2005;37:148–158. doi: 10.1016/j.bone.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 25.van Bezooijen RL, Roelen BA, Visser A, van der Wee-Pals L, de Wilt E, Karperien M, Hamersma H, Papapoulos SE, ten Dijke P, Lowik CW. Sclerostin is an osteocyte-expressed negative regulator of bone formation, but not a classical BMP antagonist. J. Exp. Med. 2004;199:805–814. doi: 10.1084/jem.20031454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.