Summary
Glioblastomas (GBM) harbor subpopulations of therapy-resistant tumor initiating cells (TICs) that are self-renewing and multipotent. To understand the regulation of the TIC state, we performed an image-based screen for genes regulating GBM TIC maintenance and identified ZFHX4, a 397-kDa transcription factor. ZFHX4 is required to maintain TIC-associated and normal human neural precursor cell phenotypes in vitro, suggesting that ZFHX4 regulates differentiation, and its suppression increases glioma-free survival in intracranial xenografts. ZFHX4 interacts with CHD4, a core member of the NuRD (nucleosome remodeling and deacetylase) complex. ZFHX4 and CHD4 bind to overlapping sets of genomic loci and control similar gene expression programs. Using expression data derived from GBM patients, we found that ZFHX4 significantly affects CHD4-mediated gene expression perturbations, which defines ZFHX4 as a master regulator of CHD4. These observations define ZFHX4 as a regulatory factor that links the chromatin remodeling NuRD complex and the GBM TIC state.
Introduction
Glioblastoma (GBM) is the most common and aggressive primary brain tumor, with an average survival of 14.6 months despite surgery, radiation, and chemotherapy (Stupp et al., 2005). Glioblastoma (GBM) tumor initiating cells (TICs) are stem cell-like, multipotent cells that are more resistant to therapy than the majority of the tumor. Their persistence may explain the inevitable recurrence of the tumor and the poor prognosis of patients with GBM (Bao et al., 2006; Piccirillo et al., 2006; Singh et al., 2003; Singh et al., 2004; Stiles and Rowitch, 2008). Although some genes that regulate aspects of TIC behavior have been identified (Ding et al., 2013; Gargiulo et al., 2013; Goidts et al., 2012; Hubert et al., 2013; Piccirillo et al., 2006; Rheinbay et al., 2013; Wurdak et al., 2010; Zheng et al., 2010), the molecular pathways that maintain the TIC state and drive the behavior of GBM TICs remain incompletely understood.
To identify genetic networks that regulate GBM TIC self-renewal and multipotency, we performed an image-based RNAi screen for genes involved in TIC maintenance. Previous shRNA screens in GBM TICs (Ding et al., 2013; Gargiulo et al., 2013; Goidts et al., 2012; Hubert et al., 2013; Wurdak et al., 2010) have targeted human kinases and phosphatases, Bmi1 targets, or a subset of known nucleic acid-binding proteins and assessed cell survival, proliferation and clustering. Given the association of transcription factors such as SOX2 with TIC functions, we sought to ascertain key transcriptional regulatory networks of GBM TICs by targeting a distinct and larger gene set that included most known transcription and chromatin binding factors. We identified ZFHX4 as essential for TIC and normal human neural precursor cells. We demonstrate that ZFHX4 associates with CHD4, a core member of the nucleosome remodeling and deacetylase (NuRD) complex and affects CHD4-mediated gene expression perturbations, which defines ZFHX4 as a master regulator of CHD4.
Results and Discussion
An RNAi screen in GBM TICs identifies ZFHX4
To identify candidate genes needed for human GBM TIC maintenance, we performed a high-content imaging-based RNAi screen (Figure 1A). The 0308 patient-derived TIC line (Lee et al., 2006) forms neurospheres when cultured under stem cell conditions (NBE medium) but acquires a flat and elongated morphology upon growth factor withdrawal or serum exposure (Figure S1A). In serum-exposed 0308 cells, expression of the stem cell markers SOX2 and NG2 is reduced, expression of differentiated lineage markers increased (Figures S1B and S1C), and the ability of the cells to form invasive brain tumors inhibited (Lee et al., 2006). The transition from neurosphere formation to flat, elongated morphology also occurs upon suppression of SOX2, a known driver of stem cell functions (Figure S1D). Thus, the morphologic features of flattening and elongation correlate with a transition from the TIC to a state resembling some but not necessarily all of the characteristics of terminally differentiated cells.
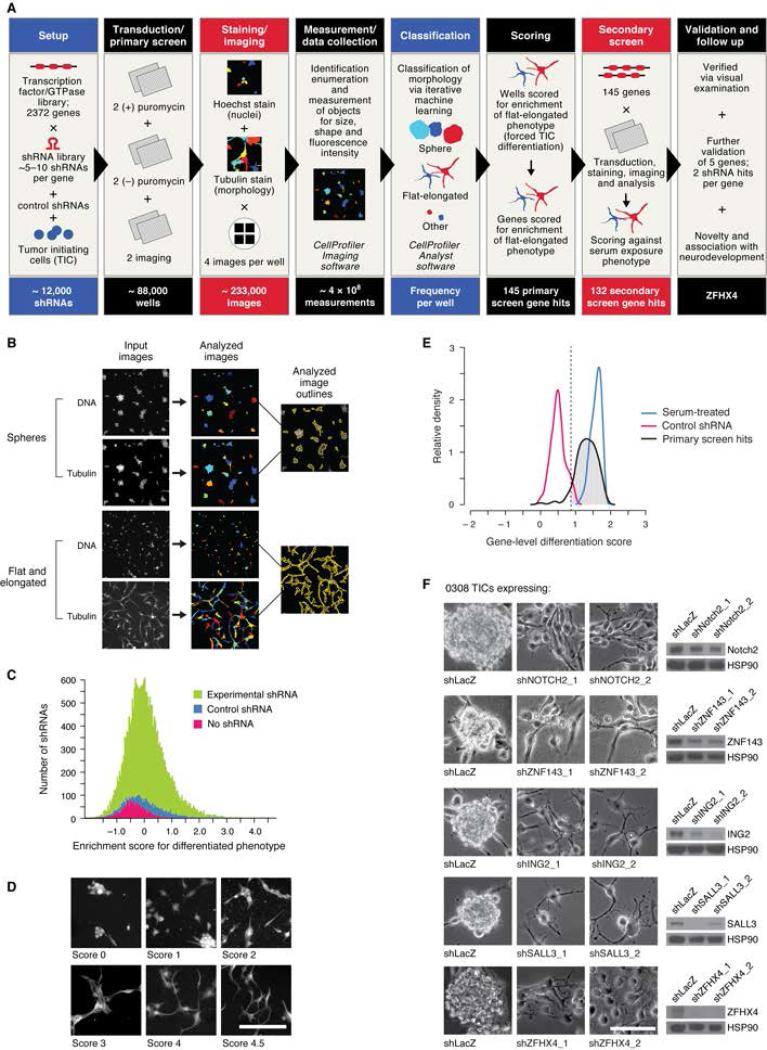
Figure 1.
Large-scale loss of function screen, confirmation screen, and validation experiments identify genes that regulate TIC morphology, including ZFHX4. (A) Schematic of high content imaging shRNA screen and follow up. (B) CellProfiler software identified individual cells and multicellular structures and measured 147 different parameters per object. (C) Distribution of enrichment scores of differentiated flat, elongated morphology by gene-targeting shRNA or control in primary screen. (D) Representative screen images with associated enrichment scores from the primary screen. (E) Confirmation screen results for 145 gene hits from the primary screen. Relative frequency histogram of gene-level differentiation scores. Serum-treated wells served as positive controls, and non-targeting shRNA wells served as negative controls. Dashed line indicates score cutoff for genes scoring at least as well as serum treatment; 132 genes (hash marks under the black curve, to the right of the dashed line) met this criteria. (F) Bright-field images (left) and immunoblots (right) of 0308 TICs transduced with a control shRNA or two different shRNAs targeting five validated gene hits from the confirmation screen. Scale bars: 100μm each. See also Figure S1, Table S1.
To screen for genes involved in TIC state maintenance, we transduced 0308 TICs in multiwell plates, under growth conditions that favor the stem cell-like state, using a library of 11,816 short hairpin RNAs (shRNAs) targeting 2372 human genes encoding most known and putative transcription factors, chromatin-binding proteins, GTPases, and members of the PI3KAKT-mTOR pathway (1 shRNA/well; median 5 shRNAs/gene) and 124 control shRNAs targeting non-mammalian genes (Figure 1A). To quantify cell morphologic changes associated with loss of the TIC state, we used high-content imaging and software-based image analysis. Assaying the cells at the time point at which we previously observed changes in morphology and marker expression after serum exposure or SOX2 suppression (Figure S1), we stained cells for DNA and tubulin and imaged in high-throughput, collecting over 200,000 images. We used CellProfiler and CellProfiler Analyst software (Carpenter et al., 2006; Jones et al., 2009) to identify individual cells and multicellular structures in each image (Figure 1B) and to collect size, shape, and fluorescence intensity data for each object. Using iterative machine learning, we classified objects into three morphologic categories: neurospheres; flat, elongated cells; and all other objects (Figure S1E). Each image from the screen was scored using these rules to obtain the per-well frequency of each of these phenotypes. This algorithm calculated enrichment scores for each shRNA, indicating the statistical significance of enrichment for each phenotype (Figures 1C, 1D, and S1F, see Extended Experimental Procedures).
Compared to uninfected wells or wells transduced with control shRNAs, the tail of the distribution of enrichment scores for the flat, elongated cell phenotype was shifted higher among gene-targeting shRNAs (Figure 1C), suggesting that a subset of the shRNAs altered the morphology of the TICs. We ranked genes by the strength of the second-best scoring shRNA for each gene and included the top 5% (116 genes) in a secondary screen. Ranking by the second-best shRNA prioritizes genes for which at least two shRNAs show strong and similar phenotypic effects, increasing confidence that the phenotype is due to on-target effects. To ensure re-evaluation of potentially important genes that failed to meet this criterion, we also included genes that for which a single shRNA exhibited a differentiated phenotype enrichment score in the top 1.5% of shRNAs (6 additional genes; Figure 1C), as well as genes that scored in the top 1.5% by the Kolmogorov–Smirnov-based method (23 additional genes) (Cheung et al., 2011; Luo et al., 2008) (see Extended Experimental Methods). In sum, we obtained 145 candidates whose suppression strongly alters TIC morphology.
In a morphology-based validation screen, we confirmed that 132 of these candidates (91%, Figure 1E, Table S1 Sheet 1) scored at least as strongly as the serum-differentiated controls, using the second-best scoring shRNA method. In a parallel screen of the same primary screen shRNA library in 0308 TICs, we identified 76 genes whose suppression by at least two different shRNAs significantly decreased relative cell number (Table S1 Sheet 2, Figure S1G). Notably, only 11 of the 132 confirmed genes whose suppression alters TIC morphology are found among those whose suppression significantly decreases relative cell number (Table S1), demonstrating that this imaging-based approach identified distinct pathways that drive glioma TIC functions. We note that suppression of several of the other 121 genes that scored in the morphology screen also decreased proliferation in the relative cell number screen; however, these candidates did not meet the stringent cutoff used to identify cell number hits (Figure S1H). We have made the entire primary image set available (http://science.wi.mit.edu/research/data/Glioma_TIC_Screen) to facilitate the use of these data with other analyses and screens, such as high-throughput immunofluorescent staining to assess marker expression (Kagey et al., 2010).
We focused on a small number of candidates, including NOTCH2, ZNF143, ING2, SALL3, and ZFHX4 (Figure S1H). NOTCH2, ZNF143, ING2, and SALL3 play known roles in stem cell functions and tumorigenesis. Specifically, Notch signaling drives neurosphere growth (Chen et al., 2010; Fan et al., 2010; Hu et al., 2011), and inhibiting the Notch signaling pathway is an active area of clinical development in GBM (National Cancer Institute, 2000a,b,c,d,e). The observation that NOTCH2 scored in our screen suggests our strategy can uncover potentially important TIC drivers. ZNF143 is essential for normal development (Halbig et al., 2012) and associated with cancer cell proliferation (Izumi et al., 2010), ING proteins bind to H3K4me2 and H3K4me3 marks after genotoxic stress (Ludwig et al., 2011), and SALL members are required for normal embryonic development (Sweetman and Munsterberg, 2006). We validated these five candidates: two different shRNAs targeting each of the genes caused a shift from neurosphere formation to flat, elongated morphology, and the phenotypic effect correlated with depletion of the target protein (Figure 1F). In addition, suppression of each of these candidates decreased the proliferation of GBM TICs (Figure S1H).
To discover novel transcriptional regulators of glioma, we focused on ZFHX4, a recently identified 397-kD putative transcription factor containing four homeodomains and 22 zinc fingers (Hemmi et al., 2006), because of its association with brain development and because 5 ZFHX4-specific shRNAs exhibited a correlation between ZFHX4 suppression and the strength of the observed phenotype. ZFHX4 may play a role in neural cell maturation and region-specific brain differentiation (Nogami et al., 2005). Zfhx4 expression peaks at the neural precursor stage along with that of the neural stem cell marker Nestin and is inversely correlated with expression of astrocyte marker GFAP (Hemmi et al., 2006) when murine embryonic carcinoma stem-like cells are differentiated with retinoic acid. ZFHX4 also plays a role in human neural development, as ZFHX4 disruption is associated with intellectual disability (Palomares et al., 2011) and congenital bilateral ptosis (McMullan et al., 2002; Nakashima et al., 2008; Palomares et al., 2011). When we examined ZFHX4 expression in previously published gene expression data for the four molecular subtypes of GBM (Verhaak et al., 2010), we found significantly higher expression of ZFHX4 in clinical samples from the Classical subtype as compared to the other subtypes (FDR=0, fold change=1.89; see Methods). However, potential roles of ZFHX4 in cancer have not previously been investigated.
To assess the role of ZFHX4 in maintaining the TIC state, we suppressed ZFHX4 in four additional patient-derived GBM TIC lines: BT112 (Mehta et al., 2011), BT145, BT147, and BT99 (Figure S2A). Although recent work has challenged the idea that TICs in culture maintain the molecular subtype of the GBM of origin (Bhat et al., 2013), the expression profiles of 0308, BT112, BT145, and BT147 cells were most similar to those of GBMs of the Classical subtype, whereas the BT99 expression profile resembled that of Mesenchymal GBMs. As in 0308 cells (Figure 1F), ZFHX4 suppression led to inhibition of neurosphere formation and induced a differentiated-like flat and elongated morphology in all four additional lines (Figure 2A). Thus, ZFHX4 is required for TIC morphology in all GBM TICs examined, and this requirement does not appear to be restricted to the Classical subtype.
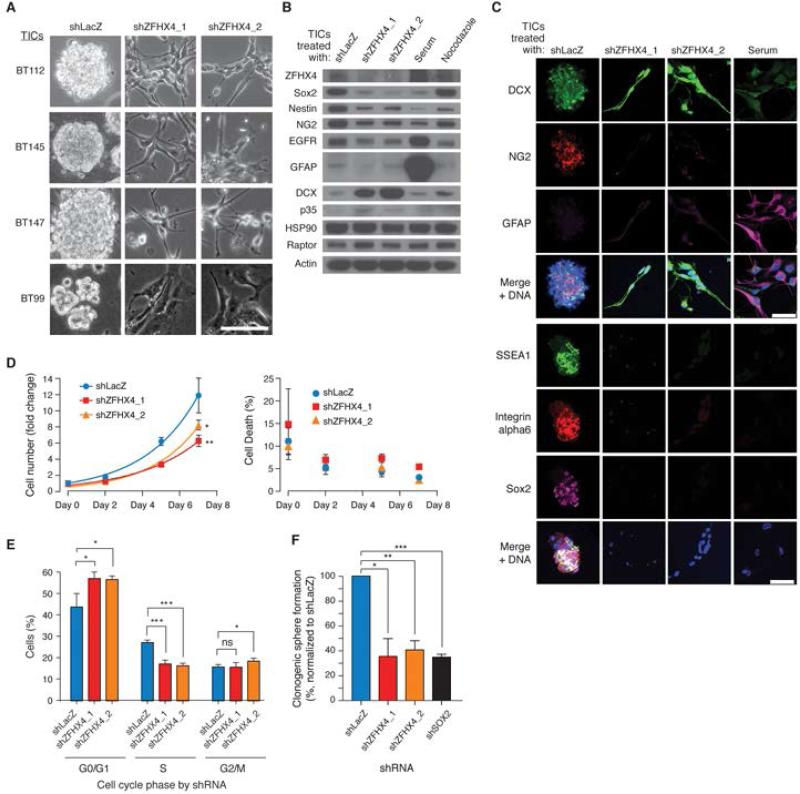
Figure 2.
ZFHX4 suppression causes loss of stem cell-like features and reduces proliferation of GBM TICs. (A) Bright-field images; ZFHX4 suppression induced a differentiated flat, elongated morphology in BT112, BT145, BT147, and BT99 glioma TIC lines. Scale bar: 100μm. (B) Decreased expression of stem cell markers Sox2, Nestin, and NG2 and the GBM oncogene EGFR, and increased expression of neuronal markers DCX and p35 in 0308 TICs in which ZFHX4 was suppressed. Serum treatment served as a positive control for loss of stem cell markers (Lee et al., 2006), whereas nocodazole treatment arrested cell growth. (C) Immunofluorescent images showing downregulation of the stem cell markers NG2, SSEA1, Integrin alpha6, and Sox2, and upregulation of the neuronal marker DCX in 0308 TICs upon ZFHX4 suppression. Scale bars: 50μm. (D) Effects of ZFHX4 suppression on cell growth and viability of 0308 TICs. For fold change at day 7, ** p=0.0025 shLacZ vs. shZFHX4_1, * p=0.0166 shLacZ vs. shZFHX4_2, two-tailed t-tests. Error bars reflect standard deviations; n=4. (E) Effects of ZFHX4 suppression on cell cycle of 0308 TICs. G0/G1: * p= 0.0258 shLacZ vs. shZFHX4_1, * p=0.0237 vs. shZFHX4_2; S: *** p=0.0007 shLacZ vs. shZFHX4_1, *** p=0.0001 shLacZ vs. shZFHX4_2; G2/M: * p=0.0343 shLacZ vs. shZFHX4_2, two-tailed t-tests. Error bars reflect standard deviations; n=3. (F) Effects of ZFHX4 suppression on clonogenic sphere formation by 0308 TICs. shSOX2 served as positive control. * p=0.0160 shLacZ vs. shZFHX4_1, ** p=0.0051 shLacZ vs. shZFHX4_2, and *** p=0.0004 shLacZ vs. shSOX2, two-tailed t-tests. Error bars reflect standard deviations; n=3. See also Figure S2.
Suppression of ZFHX4 reduces stem cell-like properties and tumorigenicity of GBM TICs
To determine whether ZFHX4 regulates the TIC state, we assessed the expression of TIC and differentiated cell markers after silencing ZFHX4. We found that ZFHX4 suppression downregulates the stem cell markers SOX2, NESTIN, NG2, SSEA-1, Integrin alpha6 and the GBM oncogene EGFR and induces the expression of neuronal markers DCX and p35 (Figures 2B and 2C). Unlike serum exposure, ZFHX4 suppression failed to increase GFAP expression significantly (Figure 2B). The downregulation of NG2, as well as either SSEA1 or Integrin alpha6, was confirmed in all GBM TIC lines tested (Figure S2B). Suppression of ZFHX4 decreased cell proliferation (Figure 2D) in association with a G0/G1 arrest (Figure 2E) and decreased clonogenic neurosphere formation in both 0308 (Figure 2F) and BT112 (Figure S2C) TICs, but it did not induce TIC death (Figure 2D). Unlike in TICs, ZFHX4 suppression did not affect proliferation or marker expression of non-stem-like GBM cell lines (Figures S2D, S2E). Together, these observations suggest that ZFHX4 plays a role in maintaining the undifferentiated, self-renewing state of GBM TICs. Others have reported a role for ZFHX4 in neural maturation (Nogami et al., 2005), suggesting that these observations may also be relevant to fate decisions by normal neural precursor cells. Indeed, ZFHX4 suppression in a human embryonic stem cell-derived neural precursor cell (NPC) line decreased expression of the stem cell marker SOX2 (Figure S2F) and increased expression of neuronal markers MAP2 and DCX (Figure S2G), suggesting that ZFHX4 also plays a role in maintaining the undifferentiated state of non-tumorigenic NPCs. The observations that ZHFX4 plays a role in the function of a subset of cells that constitute GBMs and NPCs are similar to what has been observed with the deletion of proto-oncogenes such as KRAS and MYC, which also affects normal development and cell survival (Koera et al., 1997; Laurenti et al., 2008; Nakhai et al., 2008; Stoelzle et al., 2009).
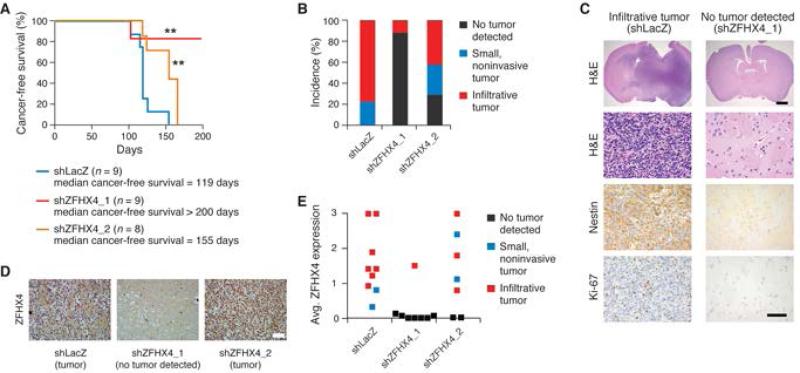
To examine whether ZFHX4-mediated maintenance of the TIC state is important for tumorigenesis in vivo, we assessed its role using a xenograft model in mice. We injected control BT112 cells or those in which we silenced ZFHX4 intracranially into NOD-SCID mice (Singh et al., 2004). ZFHX4 suppression significantly extended median cancer-free survival compared to control shRNA-transduced cells (p=0.0057 for shZFHX4_1; p=0.0098 for shZFHX4_2, Mantel-Cox Log-rank test), with one ZFHX4-targeting shRNA prolonging median survival beyond the experimental time frame (Figure 3A). All of the mice harboring the control cells developed tumors, the majority being high grade gliomas with a high mitotic index and expressing NESTIN, OLIG2, and SOX2; there were fewer infiltrative tumors in mice that received cells in which ZFHX4 was suppressed (Figures 3B, 3C). We note that we were unable to maintain long-term suppression of ZFHX4 in 0308 cells in vivo, as tumors derived from these TICs regained ZFHX4 expression. Indeed, because gliomas developed in some animals injected with cells that had been treated with anti-ZFHX4 shRNAs (particularly shZFHX4_2), we assessed ZFHX4 expression in brain tissue from all three groups using immunohistochemistry (Figures 3D, 3E, and S3). All tumors, including those arising from TICs transduced with ZFHX4-targeting shRNAs, expressed ZFHX4, while normal brain around the injection track exhibited little to no expression (Figures 3D and 3E). These observations indicate that tumors that formed in the mice receiving ZFHX4 shRNA-treated TICs originated from cells that had regained or failed to silence ZFHX4 expression. We found that the shZFHX4_2 shRNA was less effective in uniformly suppressing ZFHX4 over the long term compared to the shZFHX4_1 construct (Figure 3E), accounting for the shorter median survival and higher incidence of infiltrative tumors in mice injected with shZFHX4_2-treated cells than in those injected with shZFHX4_1-treated cells (Figures 3A, 3B). When ZFHX4 expression was extinguished, no tumors formed. We concluded that ZFHX4 expression is required for TIC-driven tumorigenesis in at least a subset of GBMs.
Figure 3.
ZFHX4 expression is required for TIC-driven tumorigenesis. (A) Kaplan-Meier cancer-free survival curves of mice intracranially injected with BT112 TICs transduced with indicated shRNAs. ** p=0.0057 shLacZ vs. shZFHX4_1 and p=0.0098 shLacZ vs. shZFHX4_2, Mantel-Cox log-rank test. (B) Percent incidence of animals with no tumor detected; small, noninvasive tumors; and infiltrative tumors in the three groups. (C) Representative immunohistochemical (IHC) images of H&E, NESTIN, and Ki-67 staining of infiltrative glioma tissue and tumor-free brain tissue. Scale bars: top row, 500μm; all others, 50μm. (D) Representative images of ZFHX4 IHC of brain tissue from the three groups. Scale bar: 50μm. (E) IHC-based scoring of ZFHX4 expression in all post-mortem tissue samples, on a scale of 0 to 3. A score of 0 indicates absence of 3,3’-diaminobenzidine (DAB) chromogen signal, while 3 indicates saturation of signal within the nuclei. See also Figure S3.
ZFHX4 interacts with NuRD core member CHD4
To investigate the mechanism by which ZFHX4 maintains the TIC state, we searched for ZFHX4-interacting proteins. Expression of FLAG-tagged ZFHX4 or a tagged control protein in HEK-293T cells followed by FLAG immunoprecipitation (IP) and mass spectrometry identified several candidate binding proteins (Table S2 Sheet 1), the most abundant of which was CHD4, one of the core catalytic subunits of the nucleosome remodeling and deacetylase (NuRD) complex. NuRD is a modular epigenetic regulatory complex that can activate or repress transcription of target genes via multiple processes, including nucleosome mobilization, histone deacetylation, and histone demethylation (Ramirez and Hagman, 2009). Components of the NuRD complex have been shown to play roles in maintenance of the stem cell state and in differentiation (Lai and Wade, 2011; Ramirez and Hagman, 2009; Whyte et al., 2012) and are suspected to be involved in oncogenesis (Lai and Wade, 2011; Ramirez and Hagman, 2009).
Immunoblot analysis of immune complexes confirmed that CHD4 binds to FLAG-tagged ZFHX4 (Figure 4A) and that endogenous CHD4 from TICs interacts with endogenous ZFHX4 (Figure 4B). When we suppressed ZFHX4 expression, ZFHX4 protein in CHD4 immune complexes was also reduced to undetectable levels (Figure 4C). In consonance with these findings, suppression of CHD4 (which was not targeted in our original screen) induced similar pro-differentiation effects to those caused by ZFHX4 silencing, including inhibition of neurosphere formation and increased frequency of flat, elongated cells, decreased expression of SOX2, NESTIN, NG2, and EGFR, and increased expression of p35 (Figure 4D).
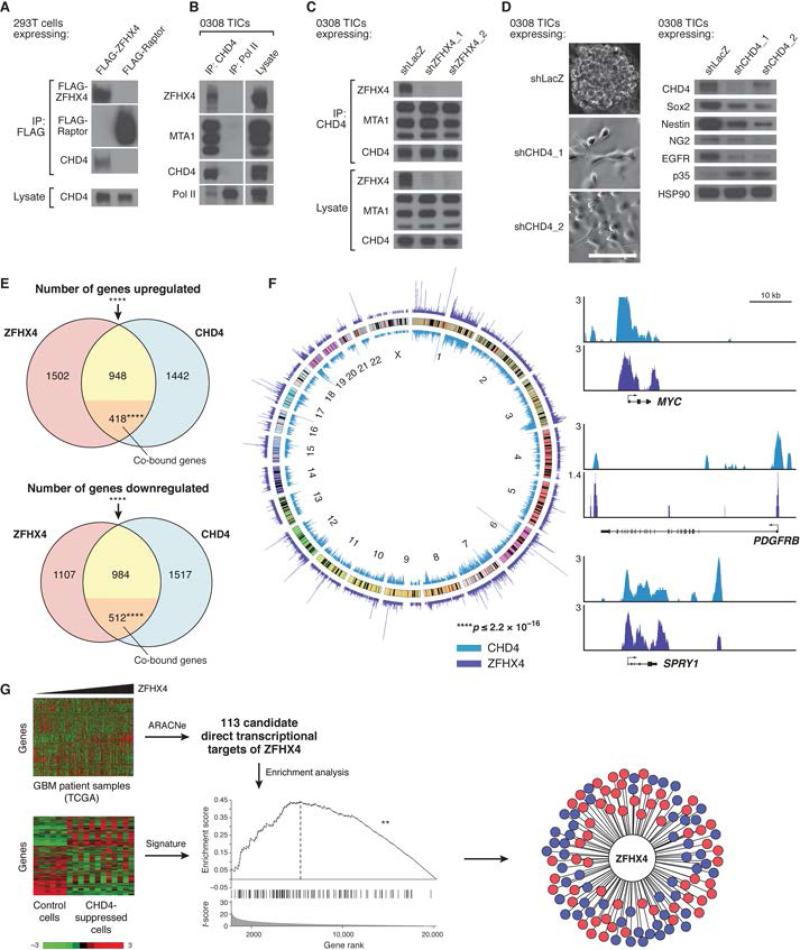
Figure 4.
ZFHX4 interacts with CHD4, CHD4 suppression phenocopies ZFHX4 suppression, ZFHX4 and CHD4 regulate overlapping gene sets and co-localize throughout the genome, and ZFHX4 is a master regulator of CHD4. (A) Immunoblots of FLAG immunoprecipitates (IPs) and lysates from HEK 293T cells transfected with FLAG tagged ZFHX4. FLAG tagged Raptor served as negative control. (B) Immunoblots of endogenous CHD4 and RNA Polymerase II (Pol II) IPs and lysate from 0308 TICs. MTA1, a known member of the NuRD complex (Lai and Wade, 2011; Ramirez and Hagman, 2009), was used as a positive control for co-IP with CHD4. (C) Immunoblots of endogenous CHD4 IPs and lysates from 0308 TICs transduced with control (shLacZ) or ZFHX4-targeting shRNAs. (D) Bright-field images in neurosphere culture (left) and immunoblots in adherent culture (right) of 0308 TICs transduced with control or CHD4-targeting shRNAs, showing induction of flat, elongated morphology, decreased expression of SOX2, NESTIN, NG2, and EGFR, and increased expression of p35 upon CHD4 silencing. Scale bar: 100μm. (E) Venn diagrams demonstrating the overlap of genes significantly upregulated or downregulated after ZFHX4 or CHD4 suppression in 0308 TICs, as well as enrichment of upregulated or downregulated intersecting genes for those genes that were shown in ChIP-Seq to be co-bound by ZFHX4 and CHD4 (orange sub-regions). **** p≤2.2×10−16 for significantly upregulated or downregulated intersecting genes, p=1.5×10−13 for enrichment of upregulated intersecting genes for co-bound genes, p≤2.2×10−16 for enrichment of downregulated intersecting genes for co-bound genes, chi-square tests. (F) ChIP-Seq results from 0308 TICs, showing significant overlap between ZFHX4-bound and CHD4-bound genomic regions. Left: whole-genome plot of mapped reads from CHD4 (blue) and ZFHX4 (purple) ChIP-Seq. Middle circle: chromosomes (labeled in innermost circle), with cytogenetic bands shown. Peaks show reads per million. For overlap of ZFHX4- and CHD4-bound regions, enrichment over expected = 30-fold, p≤2.2×10−16, two-tailed Fisher's exact test. Right: ChIP-Seq binding profiles (reads per million) for CHD4 and ZFHX4 at the MYC, PDGFRB, and SPRY1 loci, with the y-axis floor set to 1. (G) Identification of ZFHX4 as a likely master regulator of CHD4 using ARACNe and GSEA. Left: the ARACNe algorithm was used to identify candidate transcriptional targets of ZFHX4 from 273 microarray expression profiles (top) of primary glioblastoma patient samples. In parallel, microarray analysis (bottom) of control-treated 0308 TICs (expressing an anti-LacZ shRNA) and CHD4-suppressed TICs (both day 3 and day 5 post-transduction, both shRNAs) generated a signature consisting of a ranked list of genes, from most- to least-differentially-expressed, in CHD4-suppressed versus control cells. The heat maps were generated after z-score transforming the expression values for each gene across all samples and capping the values at −3 and 3. Middle: GSEA was used to compute enrichment of candidate ZFHX4 targets in the CHD4 suppression signature. Enrichment scores were computed using a Kolmogorov-Smirnov test. ** p=0.002, non-parametric test with sample shuffling. Right: wheel plot illustrating that ZFHX4 is likely a master regulator of CHD4, with 58 (red) out of the 113 (red and blue) candidate ZFHX4 targets enriched upon CHD4 suppression. See also Figure S4, Tables S2 and S3, and Data S1.
Notably, in the large-scale screen, suppression of another NuRD component, MTA1, also induced a shift to flat, elongated morphology (Table S1 Sheet 1). Furthermore, we found that endogenous MTA1 interacted with endogenous ZFHX4 in TICs (Figure S4A). These observations suggested that several members of the NuRD complex may play important roles in TIC functions.
Because ZFHX4 is a putative transcription factor and the NuRD complex is known to regulate gene expression via its interaction with transcription factors, we assessed potential direct transcriptional targets of ZFHX4 and CHD4. We obtained microarray expression profiles of TICs transduced with control, ZFHX4-targeting, and CHD4-targeting shRNAs at 3 days (prior to the overt shift from TIC to differentiated cell morphology) and 5 days after transduction. We identified 2603 genes whose expression significantly decreased and 2868 genes whose expression significantly increased in ZFHX4-depleted TICs [FDR≤0.05, Significance Analysis of Microarrays (SAM)]. Of these genes, 1496 and 1366, respectively, also significantly changed when we suppressed CHD4 (Figure 4E, Table S2 Sheets 2 and 3; gene intersection p≤ 2.2×10−16, chi-square test). Additionally, we observed that CHD4 and ZFHX4 expression are significantly correlated in 543 GBM samples in The Cancer Genome Atlas (TCGA) (TCGA, 2008) (Figure S4B; r=0.54, p≤2.2×10−16, Pearson correlation test). Together, this highly significant intersection of regulated genes and strong expression correspondence suggest ZFHX4 and CHD4 work in concert to regulate key effectors of TIC functions.
ZFHX4 co-localizes with CHD4 at genomic loci and is a master regulator of CHD4
To understand the consequences of the interaction between ZFHX4 and the NuRD complex on the regulation of target genes, we employed two orthogonal approaches. To test whether ZFHX4 and CHD4 co-localize to the same genomic regions, we performed chromatin immunoprecipitation, individually for endogenous ZFHX4 and CHD4 (Figure S4C), followed by high throughput DNA sequencing (ChIP-Seq; Table S2 Sheets 4-10, Data S1). CHD4 protein was detected in ZFHX4 immune complexes (Figure S4C), suggesting co-localization of CHD4 and ZFHX4 at genomic sites. Furthermore, 29% of ZFHX4-bound genomic regions overlap with those occupied by CHD4 (Figure 4F, Table S2 Sheets 4-6, Data S1; 30-fold enrichment over expected overlap, p≤2.2×10-16, two-tailed Fisher's exact test). 77.2% of the co-occupied regions fall into or near gene-coding regions (Figures S4D and S4E), including 59.6% in introns or near transcription start sites of genes, both being areas known to play important roles in the regulation of gene expression (Barrett et al., 2012). Based on the co-occupied regions, we identified 4118 target genes co-bound by ZFHX4 and CHD4 (Table S2 Sheet 10), and these targets include known drivers of tumorigenesis, such as MYC, PDGFRB, and ETS1, and tumor suppressors, such as SPRY1 (Figures 4F and S4E). Upon binding to regulatory regions of target genes, the NuRD complex can either activate or repress transcription (Hu and Wade, 2012; Ramirez and Hagman, 2009). Indeed, integrating the ChIP-Seq data with expression data, we found that 418 of the 1366 genes whose expression increased (30.6%; p=1.5×10−13, chi-square test) and 512 of the 1496 genes whose expression decreased (34.2%; p≤2.2×10−16, chi-square test) upon ZFHX4 suppression and upon CHD4 suppression are direct binding targets of ZFHX4 and CHD4 (Figure 4E, Table S2 Sheets 2 and 3). These observations suggest that the ZFHX4-CHD4 complex binds to regulatory regions of numerous target genes and directly regulates a large-scale gene expression program.
In hematopoietic cells, the NuRD complex localizes to genomic loci via various transcription factors (Ramirez and Hagman, 2009). To ascertain whether ZFHX4 may similarly regulate CHD4-mediated gene expression in GBM TICs, we integrated the results of gene suppression in these cells with GBM patient sample data. This analysis consisted of the following three steps.
First, we ran the Algorithm for the Reconstruction of Accurate Cellular Networks (ARACNe, (Margolin et al., 2006)) using 273 gene expression profiles of GBM patient samples from TCGA (TCGA, 2008) to assemble a GBM-specific genome-wide transcriptional network. The ARACNe method uses information-theoretic approaches to calculate the mutual information (MI) between every transcription factor and its putative targets, assesses the statistical significance of each MI value, and discards insignificant interactions as well as false positive interactions identified by the data processing inequality. This approach, using as few as 176 (Parsa, 2010) or 228 (Carro et al., 2010) patient samples to identify genome-wide transcriptional networks, has been used to identify C/EBPbeta and STAT3 as regulators of mesenchymal transformation in GBM (Carro et al., 2010). The GBM-specific ARACNe-inferred network contained approximately 270,000 interactions between 1173 transcription factors and their putative targets. In particular, it predicted 119 direct transcriptional targets of ZFHX4, of which 113 were represented in our transcriptional profiling studies (Figure 4G, Table S3). Second, we defined a CHD4 suppression signature in 0308 TICs, which consisted of a ranked list of genes, from most to least differentially expressed in CHD4-depleted TICs compared to control TICs (Figure 4G). Third, using gene set enrichment analysis (GSEA) with Kolmogorov-Smirnov (KS)-like statistics (Subramanian et al., 2005), we interrogated whether the inferred targets of ZFHX4 from patient GBM samples are enriched in the CHD4 suppression signature. We found that ZFHX4 targets were significantly enriched in the signature (Figure 4G, Table S3, p=0.002, non-parametric test), with 58 of the 113 targets in the GSEA leading edge (Subramanian et al., 2005). The observation that the candidate direct targets of ZFHX4 are enriched among genes that undergo the strongest expression changes upon CHD4 suppression in TICs suggests that the transcription-regulatory activity of ZFHX4 significantly affects the gene expression program downstream of CHD4, and that ZFHX4 is a master regulator of CHD4 in GBM. In this analysis, we define master regulators as genes that are sufficient and/or necessary to induce or describe a cellular state change, in this case CHD4-mediated gene expression perturbations (Ying et al., 2013). In summary, the ChIP-Seq, transcriptional profiling, and ARACNe/GSEA results suggest that a ZFHX4-driven, NuRD-mediated gene regulatory network serves to drive the TIC state.
Here, we identified ZFHX4 as a crucial molecular regulator of TIC stem cell-like functions and GBM pathogenesis in multiple patient-derived samples. Importantly, ZFHX4 silencing appeared to induce differentiation in GBM TICs, resulting in decreased tumorigenesis and prolonged cancer-free survival. We found that ZFHX4 interacts with CHD4, a core member of the NuRD complex, which is known to play roles in stem cell maintenance and differentiation within the hematologic system (Yoshida et al., 2008), as well as in cancer progression (Lai and Wade, 2011). The high concordance of gene expression signatures induced by ZFHX4 or CHD4 depletion, significant co-localization of ZFHX4 and CHD4 to target regions throughout the genome, and the finding that ZFHX4 is likely a master regulator of CHD4 are consistent with the notion that ZFHX4 acts as a transcription factor that modulates NuRD-mediated gene expression, possibly by localizing the NuRD complex to the regulatory regions of a specific subset of genes involved in TIC functions. Thus, not only ZFHX4 but also, more broadly, CHD4 and possibly the NuRD complex regulate genes important for stem cell-like functions and tumorigenicity in at least a subset of GBMs. Recent work implicates alterations in epigenetic regulation as a key step during gliomagenesis (Lewis et al., 2013; Schwartzentruber et al., 2012; Wu et al., 2012). Here we show that a transcription factor with previously unknown roles in cancer interacts with a core member of an epigenetic regulatory complex and thereby drives a specific gene expression program and regulates the GBM TIC state. Further studies of ZFHX4 and its downstream effectors, as well as its interaction with CHD4 and the NuRD complex, will enhance our understanding of glioma and may eventually lead to new targeted differentiation-based therapies for brain tumor patients.
Experimental Procedures
Materials, cell lines, and shRNA and expression constructs used, as well as detailed methods, are described in Extended Experimental Procedures.
RNAi Screen
0308 patient-derived TICs were seeded in 384-well plates (500 cells/well), transduced with experimental or control shRNAs, and either left unselected or selected with puromycin, to ensure efficient transduction. After 1 wk, 2 selected and 2 unselected replicates were assayed for relative cell number using CellTiter-Glo Luminescent Assay (Promega). Two additional unselected replicates were stained for DNA and Tubulin and imaged on an automated microscope. Cellular objects were identified, characterized, and classified by phenotype using CellProfiler and CellProfiler Analyst software (Carpenter et al., 2006; Jones et al., 2009), and gene hits were identified using GENE-E software (Cheung et al., 2011). The confirmation screen was performed as described in Extended Experimental Procedures.
GBM molecular subtype analysis on TIC lines
We determined the association of ZFHX4 expression with the 4 GBM molecular subtypes (Verhaak et al., 2010), and assigned a subtype to each TIC line as described in Extended Experimental Procedures.
Xenograft tumorigenesis studies
105 BT112 TICs, transduced and selected as above, were injected into the forebrain striata of 6-8 week-old NOD.CB17-Prkdcscid/NcrCrl female mice. Mice were sacrificed upon exhibiting signs of morbidity, and brains were excised and processed for histology and IHC. Studies were approved by the Dana-Farber Animal Care and Use Committee (institutional animal welfare assurance no. A-3023-01).
Chromatin immunoprecipitation followed by DNA sequencing (ChIP-Seq)
20-100 million target cells were cross-linked and processed through DNA elution steps following Agilent Technologies Mammalian ChIP-on-chip protocol (version 10.2, March 2011, http://www.chem.agilent.com/Library/usermanuals/Public/G4481-90010_MammalianProtocol_10.2.pdf), with IP performed on supernatants after sonication and centrifugation. After library preparation of eluted DNA, 7 barcoded libraries were run on 2 lanes of Hiseq 2000 in a 50bp/50bp paired end run, using the TruSeq SBS Kit v3 (Illumina). ChIP-Seq reads were aligned using Bowtie software (Langmead et al., 2009) and sequences uniquely mapping to the genome were used in further analysis for peak detection as described (Whyte et al., 2012). ZFHX4/CHD4 co-bound regions were defined as having overlap of at least 1bp between a ZFHX4-occupied region and a CHD4-occupied region. ChIP antibody and protocol details, ChIP-Seq library preparation, and details of ChIP-Seq data analysis, assessment of significance of ZFHX4 and CHD4 co-localization, and identification of bound and co-bound genes are described in Extended Experimental Procedures.
Identification of candidate transcriptional targets of ZFHX4 using ARACNe
Using the bootstrap version of the ARACNe (Margolin et al., 2006) algorithm, we generated a GBM-specific transcriptional network based on 273 gene expression profiles (HG-U133A GeneChip arrays, Affymetrix) from 262 primary glioblastoma patients, 10 control samples, and 1 cell line control obtained from TCGA (TCGA, 2008), resulting in approximately 270,000 interactions for 1173 total transcription factors. This network was parsed to identify 119 interactions for ZFHX4; 113 of these were represented in transcriptional profiles obtained after suppressing CHD4.
Gene set enrichment analysis of candidate ZFHX4 targets in CHD4 suppression signature
Gene Set Enrichment Analysis (GSEA (Subramanian et al., 2005)) with Kolmogorov-Smirnov (KS) test was used to determine whether the 113 candidate transcriptional targets of ZFHX4, identified using ARACNe) are over-represented in the ranked list of genes from most to least differentially expressed in CHD4-suppressed 0308 TICs compared to control TICs. Significance of enrichment was calculated using a non-parametric test with sample shuffling. Details of ARACNe analysis, GSEA, and significance assessment are described in Extended Experimental Procedures.
Accession Numbers and Image Set
Transcriptional profiling and ChIP-Seq data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO), under accession numbers GSE52363 and GSE52419, respectively. The entire image set from the primary RNAi screen is available at http://science.wi.mit.edu/research/data/Glioma_TIC_Screen.
Supplementary Material
Highlights.
An image-based screen identifies regulators of glioblastoma tumor initiating cells
ZFHX4 is required for the glioblastoma tumor initiating cell state
ZFHX4 interacts with and is a master regulator of CHD4, core member of NuRD complex
ZFHX4 and CHD4 co-localize throughout the genome and co-regulate gene expression
Acknowledgements
We thank members of the Sabatini, Hahn, and Mellinghoff labs for advice, members of the Fine, Kornblum and Ligon labs for advice on work with GBM TICs, C. Maire for technical assistance, L. Solomon, L. Gaffney, and T. DiCesare for extensive graphical help preparing manuscript figures, E. Spooner for mass spectrometric analysis, H. Le, S. Ponduru, M. Vokes, V. Ljosa, A. Papallo, and other members of the RNAi and Imaging Platforms at the Broad Institute for assistance with RNAi screening and analysis. This work was supported in part by an American Cancer Society postdoctoral fellowship and an American Brain Tumor Association Discovery Grant to Y.C.; grants from National Institutes of Health (NIH) (P30CA016672 and U24CA143883) to S.Z. and R.G.W.V.; a grant from the European Leukodystrophy Association to J.M.; a grant from MEXT/JSPS (KAKENHI 25670168) to N.S.; a grant from NIH (R01GM089652) and funding from the Broad Institute to A.E.C.; grants from NIH (U01CA168426 and U54CA121852) to A.C and M.B; grants from NIH (R01HG002668 and R01CA146455) to R.A.Y.; grants from NIH (R01CA170592, P01CA095616), Sontag Foundation, and Goldhirsh Foundation to K.L.L.; a grant from NIH (R01NS080944) to I.K.M.; a grant from NIH (R01CA129105) and awards from the Starr Foundation, Koch Institute Research Program, Goldhirsh Foundation, and National Brain Tumor Foundation to D.M.S.; grants from NIH (P01CA095616, P01CA142536, U01CA176058) to W.C.H.; career development grants from NIH (K08NS062907 and K12CA090354), American Brain Tumor Association Fellowship, American Association for Cancer Research/National Brain Tumor Foundation Fellowship, and Dana-Farber Cancer Institute Pediatric Low Grade Astrocytoma Program to M.G.C.; and a SPARC award from the Broad Institute to D.M.S., W.C.H., and D.E.R. D.M.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental Information
Supplemental Information includes Extended Experimental Procedures, four figures, three tables, one data file, and supplemental references.
References
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Barrett LW, Fletcher S, Wilton SD. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell Mol Life Sci. 2012;69:3613–3634. doi: 10.1007/s00018-012-0990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KP, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, Wani K, Heathcock L, James JD, Goodman LD, et al. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24:331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Kesari S, Rooney C, Strack PR, Shen H, Wu L, Griffin JD. Inhibition of notch signaling blocks growth of glioblastoma cell lines and tumor neurospheres. Genes Cancer. 2010;1:822–835. doi: 10.1177/1947601910383564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung HW, Cowley GS, Weir BA, Boehm JS, Rusin S, Scott JA, East A, Ali LD, Lizotte PH, Wong TC, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci U S A. 2011;108:12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzynkiewicz Z, Juan G. DNA Content Measurement for DNA Ploidy and Cell Cycle Analysis. In: Robinson JP, editor. Current Protocols in Cytometry. John Wiley & Sons, Inc.; Somerset, NJ: 2001. pp. 7.5.1–7.5.24. [DOI] [PubMed] [Google Scholar]
- Ding Y, Hubert CG, Herman J, Corrin P, Toledo CM, Skutt-Kakaria K, Vazquez J, Basom R, Zhang B, Risler JK, et al. Cancer-Specific requirement for BUB1B/BUBR1 in human brain tumor isolates and genetically transformed cells. Cancer Discov. 2013;3:198–211. doi: 10.1158/2159-8290.CD-12-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Khaki L, Zhu TS, Soules ME, Talsma CE, Gul N, Koh C, Zhang J, Li YM, Maciaczyk J, et al. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 2010;28:5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargiulo G, Cesaroni M, Serresi M, de Vries N, Hulsman D, Bruggeman SW, Lancini C, van Lohuizen M. In Vivo RNAi Screen for BMI1 Targets Identifies TGF-beta/BMP-ER Stress Pathways as Key Regulators of Neural- and Malignant Glioma-Stem Cell Homeostasis. Cancer Cell. 2013;23:660–676. doi: 10.1016/j.ccr.2013.03.030. [DOI] [PubMed] [Google Scholar]
- Goidts V, Bageritz J, Puccio L, Nakata S, Zapatka M, Barbus S, Toedt G, Campos B, Korshunov A, Momma S, et al. RNAi screening in glioma stem-like cells identifies PFKFB4 as a key molecule important for cancer cell survival. Oncogene. 2012;31:3235–3243. doi: 10.1038/onc.2011.490. [DOI] [PubMed] [Google Scholar]
- Halbig KM, Lekven AC, Kunkel GR. The transcriptional activator ZNF143 is essential for normal development in zebrafish. BMC Mol Biol. 2012;13:3. doi: 10.1186/1471-2199-13-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmi K, Ma D, Miura Y, Kawaguchi M, Sasahara M, Hashimoto-Tamaoki T, Tamaoki T, Sakata N, Tsuchiya K. A homeodomain-zinc finger protein, ZFHX4, is expressed in neuronal differentiation manner and suppressed in muscle differentiation manner. Biol Pharm Bull. 2006;29:1830–1835. doi: 10.1248/bpb.29.1830. [DOI] [PubMed] [Google Scholar]
- Hu G, Wade PA. NuRD and pluripotency: a complex balancing act. Cell Stem Cell. 2012;10:497–503. doi: 10.1016/j.stem.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YY, Zheng MH, Cheng G, Li L, Liang L, Gao F, Wei YN, Fu LA, Han H. Notch signaling contributes to the maintenance of both normal neural stem cells and patient-derived glioma stem cells. BMC Cancer. 2011;11:82. doi: 10.1186/1471-2407-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert CG, Bradley RK, Ding Y, Toledo CM, Herman J, Skutt-Kakaria K, Girard EJ, Davison J, Berndt J, Corrin P, et al. Genome-wide RNAi screens in human brain tumor isolates reveal a novel viability requirement for PHF5A. Genes Dev. 2013;27:1032–1045. doi: 10.1101/gad.212548.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi H, Wakasugi T, Shimajiri S, Tanimoto A, Sasaguri Y, Kashiwagi E, Yasuniwa Y, Akiyama M, Han B, Wu Y, et al. Role of ZNF143 in tumor growth through transcriptional regulation of DNA replication and cell-cycle-associated genes. Cancer Sci. 2010;101:2538–2545. doi: 10.1111/j.1349-7006.2010.01725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TR, Carpenter AE, Lamprecht MR, Moffat J, Silver SJ, Grenier JK, Castoreno AB, Eggert US, Root DE, Golland P, et al. Scoring diverse cellular morphologies in image-based screens with iterative feedback and machine learning. Proc Natl Acad Sci U S A. 2009;106:1826–1831. doi: 10.1073/pnas.0808843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koera K, Nakamura K, Nakao K, Miyoshi J, Toyoshima K, Hatta T, Otani H, Aiba A, Katsuki M. K-ras is essential for the development of the mouse embryo. Oncogene. 1997;15:1151–1159. doi: 10.1038/sj.onc.1201284. [DOI] [PubMed] [Google Scholar]
- Lai AY, Wade PA. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat Rev Cancer. 2011;11:588–596. doi: 10.1038/nrc3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti E, Varnum-Finney B, Wilson A, Ferrero I, Blanco-Bose WE, Ehninger A, Knoepfler PS, Cheng PF, MacDonald HR, Eisenman RN, et al. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 2008;3:611–624. doi: 10.1016/j.stem.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. Inhibition of PRC2 Activity by a Gain-of-Function H3 Mutation Found in Pediatric Glioblastoma. Science. 2013 doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig S, Klitzsch A, Baniahmad A. The ING tumor suppressors in cellular senescence and chromatin. Cell Biosci. 2011;1:25. doi: 10.1186/2045-3701-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin AA, Wang K, Lim WK, Kustagi M, Nemenman I, Califano A. Reverse engineering cellular networks. Nat Protoc. 2006;1:662–671. doi: 10.1038/nprot.2006.106. [DOI] [PubMed] [Google Scholar]
- McMullan TW, Crolla JA, Gregory SG, Carter NP, Cooper RA, Howell GR, Robinson DO. A candidate gene for congenital bilateral isolated ptosis identified by molecular analysis of a de novo balanced translocation. Hum Genet. 2002;110:244–250. doi: 10.1007/s00439-002-0679-5. [DOI] [PubMed] [Google Scholar]
- Mehta S, Huillard E, Kesari S, Maire CL, Golebiowski D, Harrington EP, Alberta JA, Kane MF, Theisen M, Ligon KL, et al. The central nervous system-restricted transcription factor Olig2 opposes p53 responses to genotoxic damage in neural progenitors and malignant glioma. Cancer Cell. 2011;19:359–371. doi: 10.1016/j.ccr.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima M, Nakano M, Hirano A, Kishino T, Kondoh S, Miwa N, Niikawa N, Yoshiura K. Genome-wide linkage analysis and mutation analysis of hereditary congenital blepharoptosis in a Japanese family. J Hum Genet. 2008;53:34–41. doi: 10.1007/s10038-007-0214-6. [DOI] [PubMed] [Google Scholar]
- Nakhai H, Siveke JT, Mendoza-Torres L, Schmid RM. Conditional inactivation of Myc impairs development of the exocrine pancreas. Development. 2008;135:3191–3196. doi: 10.1242/dev.017137. [DOI] [PubMed] [Google Scholar]
- National Cancer Institute . ClinicalTrials.gov [Internet] National Library of Medicine (US); Bethesda (MD): 2000. [2013 May 6]. Gamma-Secretase/Notch Signalling Pathway Inhibitor RO4929097 in Treating Patients With Recurrent or Progressive Glioblastoma. Available from: http://clinicaltrials.gov/ct2/show/NCT01122901 NLM Identifier: NCT01122901. [Google Scholar]
- National Cancer Institute . ClinicalTrials.gov [Internet] National Library of Medicine (US); Bethesda (MD): 2000. [2013 May 6]. RO4929097, Temozolomide, and Radiation Therapy in Treating Patients With Newly Diagnosed Malignant Glioma. Available from: http://clinicaltrials.gov/ct2/show/NCT01119599 NLM Identifier: NCT01119599. [Google Scholar]
- National Cancer Institute . ClinicalTrials.gov [Internet] National Library of Medicine (US); Bethesda (MD): 2000. [2013 May 6]. RO4929097 in Treating Patients With Recurrent Invasive Gliomas. Available from: http://clinicaltrials.gov/ct2/show/NCT01269411 NLM Identifier: NCT01269411. [Google Scholar]
- National Cancer Institute . ClinicalTrials.gov [Internet] National Library of Medicine (US); Bethesda (MD): 2000. [2013 May 6]. Gamma-Secretase Inhibitor RO4929097 and Cediranib Maleate in Treating Patients With Advanced Solid Tumors. Available from: http://clinicaltrials.gov/ct2/show/NCT01131234 NLM Identifier: NCT01131234. [Google Scholar]
- National Cancer Institute . ClinicalTrials.gov [Internet] National Library of Medicine (US); Bethesda (MD): 2000. [2013 May 6]. RO4929097 and Bevacizumab in Treating Patients With Progressive or Recurrent Malignant Glioma. Available from: http://clinicaltrials.gov/ct2/show/NCT01189240 NLM Identifier: NCT01189240. [Google Scholar]
- Nogami S, Ishii Y, Kawaguchi M, Sakata N, Oya T, Takagawa K, Kanamori M, Sabit H, Obata T, Kimura T, et al. ZFH4 protein is expressed in many neurons of developing rat brain. J Comp Neurol. 2005;482:33–49. doi: 10.1002/cne.20382. [DOI] [PubMed] [Google Scholar]
- Palomares M, Delicado A, Mansilla E, de Torres ML, Vallespin E, Fernandez L, Martinez-Glez V, Garcia-Minaur S, Nevado J, Simarro FS, et al. Characterization of a 8q21.11 microdeletion syndrome associated with intellectual disability and a recognizable phenotype. Am J Hum Genet. 2011;89:295–301. doi: 10.1016/j.ajhg.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsa AT. A newly identified transcriptional network for mesenchymal transformation of brain tumors: potential targets for therapeutic intervention. World Neurosurg. 2010;73:424. doi: 10.1016/j.wneu.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo SG, Reynolds BA, Zanetti N, Lamorte G, Binda E, Broggi G, Brem H, Olivi A, Dimeco F, Vescovi AL. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761–765. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- Ramirez J, Hagman J. The Mi-2/NuRD complex: a critical epigenetic regulator of hematopoietic development, differentiation and cancer. Epigenetics. 2009;4:532–536. doi: 10.4161/epi.4.8.10108. [DOI] [PubMed] [Google Scholar]
- Rheinbay E, Suva ML, Gillespie SM, Wakimoto H, Patel AP, Shahid M, Oksuz O, Rabkin SD, Martuza RL, Rivera MN, et al. An aberrant transcription factor network essential for wnt signaling and stem cell maintenance in glioblastoma. Cell Rep. 2013;3:1567–1579. doi: 10.1016/j.celrep.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Stiles CD, Rowitch DH. Glioma stem cells: a midterm exam. Neuron. 2008;58:832–846. doi: 10.1016/j.neuron.2008.05.031. [DOI] [PubMed] [Google Scholar]
- Stoelzle T, Schwarb P, Trumpp A, Hynes NE. c-Myc affects mRNA translation, cell proliferation and progenitor cell function in the mammary gland. BMC Biol. 2009;7:63. doi: 10.1186/1741-7007-7-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetman D, Munsterberg A. The vertebrate spalt genes in development and disease. Dev Biol. 2006;293:285–293. doi: 10.1016/j.ydbio.2006.02.009. [DOI] [PubMed] [Google Scholar]
- TCGA Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurdak H, Zhu S, Romero A, Lorger M, Watson J, Chiang CY, Zhang J, Natu VS, Lairson LL, Walker JR, et al. An RNAi screen identifies TRRAP as a regulator of brain tumor-initiating cell differentiation. Cell Stem Cell. 2010;6:37–47. doi: 10.1016/j.stem.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Ying CY, Dominguez-Sola D, Fabi M, Lorenz IC, Hussein S, Bansal M, Califano A, Pasqualucci L, Basso K, Dalla-Favera R. MEF2B mutations lead to deregulated expression of the oncogene BCL6 in diffuse large B cell lymphoma. Nat Immunol. 2013;14:1084–1092. doi: 10.1038/ni.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Hazan I, Zhang J, Ng SY, Naito T, Snippert HJ, Heller EJ, Qi X, Lawton LN, Williams CJ, et al. The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev. 2008;22:1174–1189. doi: 10.1101/gad.1642808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Ying H, Wiedemeyer R, Yan H, Quayle SN, Ivanova EV, Paik JH, Zhang H, Xiao Y, Perry SR, et al. PLAGL2 regulates Wnt signaling to impede differentiation in neural stem cells and gliomas. Cancer Cell. 2010;17:497–509. doi: 10.1016/j.ccr.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.