Abstract
The human thymine-DNA glycosylase (TDG) initiates the base excision repair (BER) pathway to remove spontaneous and induced DNA base damage. It was first biochemically characterized for its ability to remove T mispaired with G in CpG context. TDG is involved in the epigenetic regulation of gene expressions by protecting CpG-rich promoters from de novo DNA methylation. Here we demonstrate that TDG initiates aberrant repair by excising T when it is paired with a damaged adenine residue in DNA duplex. TDG targets the non-damaged DNA strand and efficiently excises T opposite of hypoxanthine (Hx), 1,N6-ethenoadenine, 7,8-dihydro-8-oxoadenine and abasic site in TpG/CpX context, where X is a modified residue. In vitro reconstitution of BER with duplex DNA containing Hx•T pair and TDG results in incorporation of cytosine across Hx. Furthermore, analysis of the mutation spectra inferred from single nucleotide polymorphisms in human population revealed a highly biased mutation pattern within CpG islands (CGIs), with enhanced mutation rate at CpA and TpG sites. These findings demonstrate that under experimental conditions used TDG catalyzes sequence context-dependent aberrant removal of thymine, which results in TpG, CpA→CpG mutations, thus providing a plausible mechanism for the putative evolutionary origin of the CGIs in mammalian genomes.
INTRODUCTION
In mammals, post-replicative methylation of cytosine at the 5-position (5mC) in DNA provides molecular basis of the epigenetic regulation of gene expression. DNA methylation is essential for the organism development, cell differentiation, genomic imprinting and suppression of repetitive elements. The drawback of this mode of regulation is that spontaneous deamination of 5mC generates thymine, resulting in G•T mismatch which, if not repaired, leads to C→T transition mutations at CpG dinucleotides. In fact, it was proposed that low CpG content in mammalian genomes is due to this high mutability of 5mC (1). In mammalian cells, both the mismatch-specific thymine-DNA glycosylase (TDG) and methyl-binding domain protein 4 (MBD4/MED1) prevent mutagenic impact of 5mC deamination by excising thymine from G•T mispairs in CpG context which is then replaced by cytosine completing base excision repair (BER) (2,3). In the BER pathway, a DNA glycosylase recognizes the abnormal base and catalyzes cleavage of the base-sugar bond, generating an abasic site, which in turn is repaired by an apurinic/apyrimidinic (AP) endonuclease (4,5). The human TDG and MBD4 were first biochemically characterized for their ability to remove T mispaired with G. A more detailed characterization showed that TDG exhibits a wide DNA substrate specificity: it excises 3,N4-ethenocytosine (ϵC) (6,7), thymine glycol (8), 5-hydroxycytosine (9), 7,8-dihydro-8-oxoadenine (8oxoA) (10), mismatched uracil (11) and its derivative with modifications at the C5 position (12). In contrast, MBD4 has a narrow DNA substrate specificity, in addition to T excising uracil, 5-fluorouracil and 5-hydroxymethyluracil when these bases are opposite to a guanine in duplex DNA (3,13,14). Importantly, TDG is highly conserved in vertebrates (Supplementary Figure S1).
The TDG and MBD4 proteins have a very low turnover rate because of a high affinity for the AP site, generated after base excision (15). Major human AP endonuclease 1 (APE1) can stimulate TDG-catalyzed DNA glycosylase activities by increasing the dissociation rate of TDG from AP site (16). A second factor facilitating TDG enzymatic turnover is the SUMOylation of TDG by SUMO-1 and SUMO-2/3, which reduces drastically TDG's affinity to AP sites and increases its enzymatic turnover toward DNA substrates (17). TDG is also implicated in the regulation of transcription of retinoic acid and estrogen receptors, c-jun and thyroid transcription factor 1 (18,19). The TDG protein interacts with the transcriptional co-activator CBP/p300 and the resulting TDG/CBP/p300 complex is competent for both BER and histone acetylation (20). The TDG protein enhances the CBP/p300 transcriptional activity and in turn CBP/p300 acetylates TDG. Acetylation of TDG regulates the recruitment of APE1. Thus TDG-catalyzed repair and transcriptional activities are coupled via post-translational modifications (SUMOylation and acetylation) and by protein–protein interactions. Importantly, TDG expression is strictly cell-cycle regulated: it is present in cells throughout the G2-M and G1 phases, but rapidly disappears in the S phase (21). The presence of ectopically expressed TDG hinders S-phase progression and cell proliferation.
Structural studies revealed that TDG binds to 23–28-mer DNA duplex in 2:1 ratio with a second molecule of protein generating the non-catalytic enzyme:substrate complex, this protein dimer complex cannot be formed on short 15-mer DNA duplex (22,23). In relation to these observations, it has been shown that human O6-alkylguanine-DNA alkyltransferase (AGT) and Escherichia coli mismatch-specific uracil-DNA glycosylase (MUG), a bacterial homolog of TDG, exhibit cooperative binding to DNA substrates and can form protein oligomers on DNA with 4–12 base periodicity (24,25). It was hypothesized that cooperative mode of binding could enable more efficient lesion search and/or protect DNA repair intermediates before holding them to downstream processing. However, the formation of dimeric complex on 28-mer DNA duplex had no measurable effect on the uracil-DNA glycosylase activity of TDG (22).
Recent advances in understanding of the mechanisms of active DNA demethylation in mammals have identified the Ten-eleven translocation family of proteins (TETs) as 5-methylcytosine (5mC) hydroxymethylases. TETs convert 5mC to 5-hydroxymethylcytosine and then further oxidize it to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), both in vitro and in vivo (26–29). TDG excises with high-efficiency 5fC and 5caC residues in CpG context suggesting a direct involvement of the TDG-initiated BER pathway in the active erasure of 5mC from the genome (28,30). Furthermore, it has been found that TDG knockout mice are embryonic lethal due to aberrant de novo DNA methylation of CpG islands (CGIs) promoters of developmental genes, which results in failure to establish and/or maintain cell-type-specific gene expression programs during embryonic development (31,32). Vertebrate CGIs are short interspersed CG-rich unmethylated genomic regions that are present near the transcription start sites (TSSs) of genes (33). In mammalian genomes, CGIs are typically 500–3000 base pairs in length and have been found in or near half of the promoters of mammalian genes (34). CGIs are key regulatory elements in transcription regulation, they are enriched in permissive histone modifications, poor in DNA cytosine methylation and contain multiple sites for transcription factors (35). Despite low CpG content of mammalian genomes, enrichment of CGIs at TSSs appeared early in evolution of vertebrates suggesting that association of CGIs with promoter regions was a consequence of warm-blooded vertebrate evolution (36). It was hypothesized that emergence and stabilization of CpG-rich context of CGIs could be due to hypodeamination regime associated with low level of DNA methylation and/or GC-biased gene conversion—a non-reciprocal copying of a DNA sequence from one homologous chromosome onto the other during meiotic recombination (37,38). Interestingly, the CGI-containing primate promoters exhibit the highest rate of divergence/mutation when compared with other distant mammalian species suggesting a heterotachy—accelerated evolution of primate promoters (39). At present, understanding the molecular mechanisms underlying the CpG enrichment at transcriptional regulatory regions and emergence of CGIs in evolution require further investigations.
In the BER pathway, DNA glycosylases specifically recognize and excise modified DNA bases among the vast majority of regular bases. It is generally agreed that the main function of BER is to thwart the genotoxic effects of spontaneous and oxidative DNA base damage (5). In this respect, mismatches between two regular bases owing to spontaneous deamination of 5mC to T and also to DNA polymerase errors during replication present a challenging puzzle to repair systems. To counteract these mutagenic threats to genome stability, cells evolved special mono-functional DNA glycosylases that can target non-damaged DNA strand to remove mismatched regular bases. The sequence-specific E. coli Vsr endonuclease, the mismatch-specific adenine-DNA glycosylases (E. coli MutY and human MutY homologue (MYH)) and thymine-DNA glycosylases TDG/MBD4 recognize and remove regular bases in mismatched DNA duplexes (40–42). Intriguingly, it was shown that E. coli MutY can act in a mutagenic manner during DNA replication by excising regular A in the non-damaged template DNA strand opposite to mis-incorporated 8-oxoguanine residue consequently leading to fixation of A•T→C•G mutation (43). This observation raises the possibility of existence of specific DNA repair mechanisms that can introduce bias in spontaneous mutation spectra. Remarkably, certain mutations in the active sites of classic human uracil-DNA glycosylase (UDG) and TDG, which might occur in vivo, can result in dramatic change of their DNA substrate specificity (44,45). UDG-Y147A and UDG-N204D mutants excise regular thymine and cytosine residues in DNA, respectively, whereas TDG-A145G, TDG-H151A and double TDG-A145G-H151Q mutants exhibit non-specific activity to thymine in A•T base pair. These artificially engineered DNA glycosylases with aberrant activities can be highly cytotoxic and mutagenic in vivo (44).
Here, we characterized the aberrant DNA glycosylase activities of wild-type MBD4 and TDG enzymes in vitro. Unexpectedly, we found that TDG can introduce T→C mutation in the sequence context-dependent and a DNA replication-independent manner. Moreover, data obtained from analysis of single-nucleotide polymorphisms (SNPs) from human genome revealed very biased spectrum of spontaneous mutation in the CGIs. The role of TDG-catalyzed DNA repair activities in the evolution of CpG-rich regions in mammalian genomes is discussed.
MATERIALS AND METHODS
Chemicals, reagents and proteins
Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs (Evry, France). The E. coli BL21(DE3) cells were purchased from Novagen-EMD4Biosciences (Merck Chemicals, Nottingham, UK). Collection of the purified DNA glycosylases and AP endonucleases was from the laboratory stock (46). The activities of various DNA repair proteins were tested using their principal substrates and were checked just prior to use.
Oligonucleotides
Sequences of the oligonucleotide duplexes used in the present work are shown in Table 1. All oligonucleotides containing modified bases and their complementary strands were purchased from Eurogentec (Seraing, Belgium) including the following: 40-mer d(AATTGCTATCTAGCTCCGCXCGCTGGTACCCATCTCATGA) where X is either hypoxanthine (Hx), 8oxoA, 1,N6-ethenoadenine (ϵA), tetrahydrofuran (THF, an abasic site analog), 7,8-dihydro-8-oxoguanine (8oxoG), ϵC, 5,6-dihydrouracil (DHU), alpha-2′-deoxyadenosine and complementary 40-mer d(TCATGAGATGGGTACCAGCGTGCGGAGCTAGATAGCAATT) where T is opposite to the lesion. The oligonucleotides were 5′-end labeled with [γ-32P]-adenosine triphosphate (ATP) (PerkinElmer, France) and then annealed with corresponding complementary strands as described previously (47). The resulting oligonucleotide duplexes are referred to in the text as X•Y (NYN), where X is a residue in the [32P]-labeled top strand, Y is a residue opposite to X in the complementary non-labeled bottom strand and N is a regular DNA base immediately neighboring the 5′ and 3′ sites of Y.
Table 1.
DNA sequence of the oligonucleotide duplexes used in the studya
| Name of duplex | DNA sequence of oligonucleotide substrates | |||||||||||||
| G-34 | C | TAT | CCA | CTA | CTA | TCC | TCA | TGA | TCT | ACT | TCA | ATC | ||
| G | ATA | GGT | GAT | GAT | AGG | AGT | ACT | AGA | TGA | AGT | TAG | |||
| T•X(NXN) | T | CAT | GAG | ATG | GGT | ACC | AGC | NTN | CGG | AGC | TAG | ATA | GCA | ATT |
| A | GTA | CTC | TAC | CCA | TGG | TCG | NXN | GCC | TCG | ATC | TAT | CGT | TAA | |
| T•Y(CXC) | T | CAT | GAG | ATG | GGT | ACC | AGC | GTG | CGG | AGC | TAG | ATA | GCA | ATT |
| A | GTA | CTC | TAC | CCA | TGG | TCG | CYC | GCC | TCG | ATC | TAT | CGT | TAA | |
| Hx•C | A | ATT | GCT | ATC | TAG | CTC | CGC | XCG | CTG | GTA | CCC | ATC | TCA | TGA |
| T | TAA | CGA | TAG | ATC | GAG | GCG | CGC | GAC | CAT | GGG | TAG | AGT | ACT | |
| G•N | A | ATT | GCT | ATC | TAG | CTC | CGC | GCG | CTG | GTA | CCC | ATC | TCA | TGA |
| T | TAA | CGA | TAG | ATC | GAG | GCG | NGC | GAC | CAT | GGG | TAG | AGT | ACT | |
| T•G | A | ATT | GCT | ATC | TAG | CTC | CGC | TGG | CTG | GTA | CCC | ATC | TCA | TGA |
| T | TAA | CGA | TAG | ATC | GAG | GCG | GCC | GAC | CAT | GGG | TAG | AGT | ACT | |
| T•ϵA-mHa-ras | G | CAT | GGC | ACT | ATA | CTC | TTC | TTG | ACC | TGC | TGT | GTC | TAA | GAT |
| C | GTA | CCG | TGA | TAT | GAG | AAG | AXC | TGG | ACG | ACA | CAG | ATT | CTA | |
| T•ϵA-hp53 | C | ACT | GGA | GTC | TTC | CAG | TGT | GAT | GCT | TGT | GAG | GAT | GGG | CCT |
| G | TGA | CCT | CAG | AAG | GTC | ACA | CTX | CGA | ACA | CTC | CTA | CCC | GGA | |
| T•Z 28 mer | G | TGT | CAC | CAC | CGC | TCA | TGT | ACA | GAG | CTG | ||||
| C | ACA | GTG | GTG | GCG | AGT | ZCA | TGT | CTC | GAC | |||||
| 8oxoA•N 28 mer | G | TGT | CAC | CAC | CGC | TCA | NGT | ACA | GAG | CTG | ||||
| C | ACA | GTG | GTG | GCG | AGT | XCA | TGT | CTC | GAC | |||||
| U•G 28 mer | G | TGT | CAC | CAC | CGC | TCA | UGT | ACA | GAG | CTG | ||||
| C | ACA | GTG | GTG | GCG | AGT | GCA | TGT | CTC | GAC | |||||
| T•Z 15 mer | T | CAT | GTA | CAG | AGC | TG | ||||||||
| A | GTZ | CAT | GTC | TCG | AC | |||||||||
| 8oxoA•N 15 mer | T | CAX | GTA | CAG | AGC | TG | ||||||||
| A | GTN | CAT | GTC | TCG | AC | |||||||||
| U•G 15 mer | T | CAU | GTA | CAG | AGC | TG | ||||||||
| AC | ||||||||||||||
| A | GTG | CAT | GTC | TCG | ||||||||||
aFollowing symbols are used to designate the modified and regular DNA bases: X is for 8oxoA and Hx; Y is for THF, ϵA, 2oxoA, 8oxoG, DHU, ϵC and αA; Z is for ϵA, Hx and G; N is for C, G, A and T.
Expression and purification of TDG and MBD4
The expression and purification of TDG, TDGcat and MBD4 proteins were performed as described previously (10,46). Briefly, Rosetta 2 (DE3) cells were transformed with the expression vectors pET28c-TDG (for full-length TDG protein), pET28c-TDGcat (for catalytic domain TDG protein) and pET6H-MBD4 (for full-length MBD4 protein) and then grown at 37°C in Luria Broth (LB) medium, supplemented with 50 μg•ml−1 of kanamycin or ampicillin, on an orbital shaker to OD600 nm = 0.6-0.8. Then temperature was reduced to 30°C and the proteins expression was induced by 0.2 mM isopropyl β-D-1-thiogalactopyranoside and the cells were further grown either for 3 h for TDG and MBD4 inductions or 15 h for TDGcat induction. Bacteria were harvested by centrifugation and cell pellets were lysed using a French press at 18 000 psi in buffer containing 20 mM HEPES-KOH pH 7.6, 50 mM KCl supplemented with CompleteTM Protease Inhibitor Cocktail (Roche Diagnostics, Switzerland). Lysates were cleared by centrifugation at 40 000 x g for 1 h at 4°C, the resulting supernatant was adjusted to 500 mM NaCl and 20 mM imidazole and loaded onto HiTrap Chelating HP column (Amersham Biosciences, GE Health). All purification procedures were carried out at 4°C. The column was washed with buffer A (20 mM HEPES, 500 mM NaCl, 20 mM imidazole) and the bound proteins were eluted with a linear gradient of 20-500 mM imidazole in buffer A. Eluted fractions were analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and fractions containing the pure His-tagged TDG, TDGcat and MBD4 proteins were stored at −80°C in 50% glycerol. The concentration of purified proteins was determined by the method of Bradford.
Mammalian cell culture and protein extracts preparation
Mouse embryonic fibroblasts (MEFs) WT and MEF Tdg−/− cell lines were obtained as previously described (48). MEF-WT and MEF-Tdg−/− cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum, 100 U•ml−1 penicillin and 100 μg•ml−1 streptomycin at 37°C in the presence of 5% CO2. Nuclear and cytosolic MEF extracts were prepared as previously described (49) with minor modifications. All manipulations were carried out at 4°C. The cell pellets were washed twice in cold phosphate-buffered saline (PBS), followed by a wash in the hypotonic buffer containing 250 mM sucrose supplemented with CompleteTM Protease Inhibitor Cocktail (Roche Diagnostics, Switzerland). After pellets were re-suspended in the hypotonic buffer without sucrose and let to swell on ice for 10 min, these pellets were lysed by 35 strokes of a tight-fitting Dounce homogenizer. The resulting lysates were centrifuged at 2000 x g for 5 min at 4°C (for collecting of nuclei) and the supernatants were further clarified by centrifugation at 15 000 x g for 20 min at 4°C. The supernatants (cytosolic extracts) were stored in aliquots at −80°C. For nuclear extracts, nuclei pellets previously collected were suspended (v/v) in buffer containing 0.5 M NaCl and 0.2% NP-40. The nuclear suspension was left for an additional 10 min at 4°C, after which the nuclei were spun down 20 min at 12 000 x g. The supernatants (nuclear extracts) were stored in aliquots at −80°C.
DNA repair activity
The standard reaction mixture (20 μl) for DNA repair assays contained 5 nM of 5′-[32P]-labeled duplex oligonucleotide, 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM DTT, 100 μg•ml−1 bovine serum albumin (BSA) and 50 nM of TDG, TDGcat (truncated catalytic domain TDG protein) or MBD4 for 1 h at 37°C, unless otherwise stated. The standard reaction mixture (50 μl) for repair assays in cell-free extracts contained 2.5 nM 5′-[32P]-labeled oligonucleotide duplex in 50 mM KCl, 20 mM HEPES-KOH (pH 7.6), 0.1 mg/ml BSA, 1 mM DTT, 1 mM EDTA and either 30 μg cytosolic or nuclear protein extracts from MEFs, unless otherwise stated. The reaction mixtures were incubated at 37°C for 4 h when measuring TDG-specific activities (T•G, T•Hx and 8oxoA•G duplexes in which T- and 8oxoA-containing strands were [32P]-labeled) or 100 min when measuring human alkyl-N-purine DNA glycosylase (ANPG) specific activities (Hx•T duplex in which Hx-containing strand was [32P]-labeled). After incubation, the samples were treated either with 0.1 M NaOH for 3 min at 99°C and then neutralized by 0.1 M HCl or with light piperidine (10% (v/v) piperidine at 37°C for 40 min) in order to cleave at AP sites left after excision of damaged bases. To analyze reaction products, the samples were desalted using Sephadex G25 column (Amersham Biosciences) equilibrated in 7.5 M urea and the cleavage fragments were separated by electrophoresis in denaturing 20% (w/v) polyacrylamide gels (7-M Urea, 0.5 x TBE, 42°C). The gels were exposed to a Fuji FLA-3000 Phosphor Screen, then scanned with Fuji FLA-3000 and/or Typhoon FLA 9500 and quantified using Image Gauge V4.0 software. The release of T residue was measured by the cleavage of the oligonucleotide containing a single T•X pair, where X is a residue opposite to T in the complementary strand.
Single turnover kinetics
Here we used single turnover kinetics under large excess of enzyme over substrate ([E]>> [S]>Kd) to obtain rate constants (kobs) that are not affected by enzyme-substrate association or by product inhibition, such that kobs reflects the maximal base excision rate (kobs ≈ kmax). The data were fitted by nonlinear regression to one-phase exponential association [Equation (1)] using GraphPad Prism 5 software,
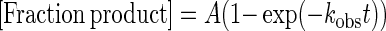 |
(1) |
where A is the amplitude, kobs is the rate constant and t is the reaction time (in minutes).
The enzymatic assays were performed in large volume reaction mixture with 500 nM TDG and 50 nM duplex oligonucleotide for varying periods of time at 37°C. At each time point, 20 μl of sample was withdrawn and treated by NaOH (0.1 M) for 3 min at 99°C, as previously described (10). Reaction products were analyzed by electrophoresis on denaturing 20% (w/v) polyacrylamide gels (7 M urea, 0.5 x TBE at 42°C) before quantification as described above.
In vitro reconstitution of Hx•T repair by TDG
5 nM [32P]-labeled Hx•T duplex was incubated in the presence of 5 nM APE1, 0.1 units Polβ and 2 units T4 DNA Ligase, 50 μM deoxyribonucleoside triphosphates (dNTPs), and either 300 nM TDG or 80 nM ANPG, in buffer containing 20 mM HEPES-KOH (pH 7.6), 100 mM NaCl, 0.1 mg•ml−1 BSA, 1 mM DTT, 2 mM ATP and 5 mM MgCl2 for 30 min at 37°C, and then reaction mixtures were incubated with 1 units of BstUI restriction enzyme for 40 min at 60°C. Reaction products were analyzed on denaturing PAGE as described above.
Construction of circular DNA plasmid containing T•Hx pair within stop codon
pT7Blue-3Rev-TGA and control pT7Blue-3Rev-CGA plasmids are derivatives of pT7Blue-3 AmpR, KamR (Novagen, EMD Millipore, MA, USA), they contain reversed f1 origin and either TGA stop codon or CGA-Arg codon, respectively, inserted after Met19 of kanamycin resistance gene. Note that, pT7Blue-3 and its derivatives do not have any mammalian replication origins and cannot replicate in MEFs. The plasmid vectors were obtained by a polymerase chain reaction-based site-directed mutagenesis. Circular heteroduplex DNA substrate pT7Blue-3Rev-TGA-Hx containing Hx opposite to T within TGA codon (TpCpHx/TpGpA context) was constructed by primer extension, using 5′-phosphorylated pHx-Kan29 d(pTCAGCATCTCHxCATGTTGGAATTTAATCG) oligonucleotide containing single Hx residue as a primer and single-stranded phagemid DNA as a template, as described previously (50,51). After synthesis of a second strand and ligation, the covalently closed circular heteroduplex plasmid DNA was agarose gel purified using Qiagen MinElute Gel Extraction Kit (Qiagen, France).
Transient pT7Blue-3Rev-TGA-Hx transfection assay of MEFs cells
For in vitro assay, 6 ng pT7Blue-3Rev-TGA-Hx plasmid DNA was incubated or not in the presence of 500 nM TDG or 50 nM ANPG proteins under standard reaction conditions (30 min and 15 min at 37°C, respectively) and then electroporated into E. coli XL1-Blue competent cells (Stratagene, CA, USA), the transformants were selected on LB agar plates containing either kanamycin (Kam) or ampicillin (Amp). The mutation rates to KamR were calculated as the ratio of KamR/AmpR colonies, individual KamR clones were isolated and sequenced by GATC-Biotech (Germany) to characterize the mutation spectra.
For ex vivo transient transfection assay, pT7Blue-3Rev-TGA-Hx plasmid DNA was transfected into MEF-WT and MEF-Tdg−/− cells grown to 80% confluence, using ExGen 500 reagent (Euromedex, Souffelweyersheim, France) according to the manufacturer's recommendations. Following 6 h after transfection, the cells were washed three times by PBS buffer and then treated with 1.75 U•μl−1 DNase I (New England Biolabs, France) in buffer containing 10 mM Tris-HCl, pH 7.6, 2.5 mM MgCl2, 0.5 mM CaCl2 for 30 min at 37°C. After DNAse I treatment cells were washed three times by PBS buffer and harvested by centrifugation at 5000 x g for 10 min at 4°C. The plasmid DNA was purified from cell pellets using QIAGEN Plasmid Mini Kit as recommended by the manufacturer (Qiagen, France). Purified DNA was then electroporated into E. coli XL1-Blue competent cells and plated on LB agar plates as described above. The mutation rates measurement and sequencing were performed as described above.
Bioinformatics
TDG sequence data were downloaded from NCBI site. Multiple sequence alignment was performed using ClustalX 2.1. Phylogenetic tree was created with PHYLIP 3.6. CGIs data and dbSNP build 127 were downloaded from the UCSC Genome Bioinfomatics site as plain text files. For SNP analysis, we developed special programs to parse input data and compute required statistics.
RESULTS
Human TDG excises thymine opposite to damaged adenine in oligonucleotide duplexes
Since TDG exhibits a wider substrate specificity compared to MBD4, we decided to further explore DNA substrate specificity of the former. For this, a 5′-[32P]-labeled 34-mer oligonucleotide containing a single G residue at position 21 was hybridized to a complementary strand and then treated by chloroacetaldehyde (CAA), followed by incubation with APE1, mono-functional DNA glycosylases 3-methyladenine-DNA glycosylase II (AlkA), MUG and MutY from E. coli and TDG and alkyl-N-purine-DNA glycosylase (ANPG) from human. After reaction with DNA glycosylases samples were treated with alkali to reveal the presence of AP sites (Figure 1A). It should be noted that we can observe cleavage of labeled DNA strand only, actions of DNA repair enzymes on non-labeled complementary strand cannot be detected by this approach. CAA is a carcinogenic compound that reacts with all DNA bases except thymine and generates ϵC, ϵA and N2,3-ethenoguanine (ϵG) adducts (52). As expected from the previous studies (53), the E. coli AlkA protein cleaves CAA-DNA at G21 position and generates 20-mer product indicating the presence of ϵG (lane 8), whereas incubations with E. coli MUG and human ANPG resulted in DNA cleavage at C and A residues, respectively (lanes 9 and 11) indicating the presence of ϵC and ϵA residues, respectively. Interestingly, the E. coli MutY protein excises G21 residue suggesting that it may recognize ϵG (lane 10). Unexpectedly, incubation of the CAA-DNA with TDG resulted in the formation of a single 19-mer cleavage fragment indicating the excision of T at position T20 which is located in TpG sequence context (lane 12). Importantly, no cleavage is observed at other thymine positions and also when untreated DNA was incubated with TDG (lane 4) suggesting that TDG recognizes T in the damaged DNA duplex, and only in the TpG context. Based on these observations, we hypothesized that CAA reacted with adenines in the non-labeled complementary strand of 34-mer duplex and generated T•ϵA base pairs in which regular T was recognized by TDG as a mismatched base.
Figure 1.

Action of mono-functional DNA glycosylases on oligonucleotide duplexes containing base lesions and mismatches. (A) Cleavage of duplex oligonucleotide containing ethenobases by various DNA glycosylases. 20 nM 5′-[32P]-labeled 34-mer oligonucleotide duplex was treated or not with 1% CAA and then incubated in the presence of 100 nM DNA glycosylase or 10 nM APE1 for 1 h at 37°C. Lane 1: control DNA; lane 2: DNA cleaved at G20 position; lanes 3–6: as 1 but with enzymes; lane 7: as 1 but 1% CAA; lanes 8–13: as 7 but with enzymes. (B) Excision of mismatched T in various oligonucleotide duplexes by full-length TDG, TDGcat and MBD4. 5 nM 5′-[32P]-labeled 40-mer oligonucleotide duplex containing T paired with G, ϵA, Hx and 8oxoA was incubated with 100 nM DNA glycosylase for 1 h at 30°C. After reaction, all samples were treated by light piperidine (10% (v/v) 40 min at 37°C) to cleave at AP sites. The reaction products were analyzed as described in the Materials and Methods section.
Next, we suggested that other modifications of adenine in DNA duplex in the specific sequence context may also expose the non-damaged complementary thymine residues to TDG action. To examine these, we constructed several 5′-[32P]-labeled 40-mer oligonucleotide duplexes containing single T•G, T•ϵA, T•Hx and T•8oxoA base pairs in TpG/CpX sequence context, where X is a modified adenine. The duplexes were incubated with the human TDG (full length), catalytic domain TDG (TDGcat, amino acids 111–308) and full-length MBD4 proteins. As expected, all three DNA glycosylases excise thymine in T•G duplex and generate a 20-mer cleavage product (Figure 1B, lanes 2–4). In addition, TDG excises thymine opposite to ϵA, Hx and 8oxoA residues, with the relative order of efficiency T•G ≥ T•Hx > T•ϵA >> T•8oxoA (lanes 3, 7, 11 and 15). This result confirms our finding that TDG excises T opposite to ϵA in CAA-treated DNA (Figure 1A, lane 12). TDGcat exhibits the same DNA substrate preference as TDG but excises T opposite to modified A very weakly (lanes 2, 6, 10 and 14) suggesting that the N- and C-terminal portions of TDG are required for the efficient recognition of damaged T•A pairs. Interestingly, MBD4 can excise T in T•ϵA duplex albeit with very low efficiency (lanes 4 and 8) but not in T•Hx and T•8oxoA duplexes (lanes 12 and 16). In an additional screen, we show that TDG can also excise T opposite to 8oxoG, ϵC, DHU, alpha-anomeric 2′-deoxyadenosine and THF, a synthetic analog of AP site (Supplementary Figure S2). Taken together, these results show that TDG can recognize T in TpG/CpA* sequence context when complementary A* is absent or undergoes chemical modifications.
Kinetic parameters for the excision of T paired with damaged adenine residues in duplex DNA by TDG
We further substantiated substrate specificity of TDG by measuring the cleavage rates of 40-mer T•G, T•Hx, T•ϵA, T•THF and T•8oxoA duplexes, where T is in TpG/CpX sequence context (where X is G or a damaged residue), under single-turnover conditions, using a molar excess of enzyme over DNA substrate, which provides the maximal rate of base excision (kobs) for a given substrate (Figure 2A and Table 2). Time course of the cleavage product generation shows that T•G is the most preferred substrate for TDG followed by T•Hx and then by T•ϵA, T•THF and T•8oxoA that are cleaved with lower efficiency (Figure 2A). Importantly, TDG cleaves only 60% of T•Hx, 15% of T•ϵA, 10% of T•THF and 3% of T•8oxoA after 90 min of incubation, and the kobs values of TDG-catalyzed cleavage of T•Hx, T•ϵA, T•THF and T•8oxoA are 2.5-, 4-, 6- and 28-fold lower than that of T•G (0.165 min−1) (Table 2), indicating that the human enzyme has lower affinity to the chemically modified, damaged DNA as compared to mismatched T•G duplex. Previously, it was shown that T•G-specific activity of TDG exhibits strong preference for 5′-TpG-3′/5′-CpG-3′ context (54). Here, we observed the same sequence context preference for TDG when it excises T in T•Hx and T•ϵA duplexes. TDG excises T in 5′-TpG-3′/5′-CpX-3′ (where X is Hx or ϵA) (Figure 2B, lanes 1, 3, 5, 7 and Figure 3A, lane 1) but fails or removes very weakly T in TpC, TpT and TpA contexts, respectively (Figure 2B, lanes 9, 11, 15, 17, 13, 19 and Figure 3A, lanes 3, 5 and 7). Interestingly, the nature of 5′-flanking base next to T has no significant influence on the cleavage efficiency of TDG (Figure 2B, lanes 1, 3, 5, 7 and Supplementary Table S1).
Figure 2.

Kinetics and sequence context dependences of TDG-catalyzed excision of mismatched T. (A) Graphic presentation of pre-steady-state single turnover kinetic of TDG-catalyzed cleavage of various oligonucleotide duplexes. Time kinetics were performed using 500 nM TDG and 50 nM 5′-[32P]-labeled 40-mer oligonucleotide duplex (⋄) T•G, (•) T•Hx, (▪) T•ϵA, (▾) T•THF and (Δ) T•8oxoA. Each bar represents the mean values of TDG activity ± SD of three independent experiments. (B) Separation of TDG-cleavage products on denaturing PAGE. 5 nM 5′-[32P]-labeled 40-mer T•Hx oligonucleotide duplexes, where T-containing strand is labeled and Hx placed in different sequence context: CXC, CXG, CXA, CXT, GXC, AXC, TXC, GXG, AXG and TXG, where X is Hx, were incubated with 50 nM TDG for 30 min at 37°C. Arrows 40 mer and 20 mer indicate substrate and cleavage products, respectively. The reaction products were analyzed as described in the Materials and Methods section.
Table 2.
Pre-steady-state kinetic parameters of TDG-catalyzed excision of T opposite to various DNA adducts
| Substratea | kobs (min−1)b |
|---|---|
| T•G (CXG) | 0.165 ± 0.014 |
| T•Hx (CXG) | 0.066 ± 0.008 |
| T•ϵA DL (CXC) | 0.042 ± 0.012 |
| T•THF (CXC) | 0.027 ± 0.006 |
| T•8oxoA (CXC) | 0.006 ± 0.001 |
aLetters within the parentheses represent nearest neighbor nucleotide sequence context where X is G, Hx, ϵA, THF or 8oxoA.
bConstants were calculated by one-phase exponential association equation using GraphPad Prism 5.
Figure 3.

Kinetics and sequence context dependence of TDG-catalyzed excision of T opposite to ϵA. (A) Separation of TDG-cleavage products on denaturing PAGE. 5 nM 5′-[32P]-labeled 40-mer T•ϵA oligonucleotide duplexes (T strand is labeled), where ϵA is placed in different sequence context: CXC, GXC, AXC, TXC, CXA and CXT where X is ϵA, were incubated with 50-nM TDG for 30 min at 37°C. Arrows 40 mer and 20 and 21 mer indicate substrate and cleavage products, respectively. The reaction products were analyzed as described in the Materials and Methods section. (B) Graphic presentation of pre-steady-state single turnover kinetic of TDG-catalyzed cleavage of various oligonucleotide duplexes. Time kinetics were performed using 500 nM TDG and 50 nM 5′-[32P]-labeled 40-mer oligonucleotide duplex (⋄) T•G DL, (•) T•ϵA DL, (○) T•ϵA-hp53 and (▪) T•ϵA-Ha. Each bar represents the mean values of TDG activity ± SD of three independent experiments.
Importantly, the aberrant activity of TDG and MBD4 on ϵA-DNA shown above (Figures 1B and 3A) may have implications in the observed hotspot mutations at adenine sites in the ras and p53 genes of tumors induced by chemical carcinogens (55). Indeed, in mouse liver tumors induced by the urethane- and vinyl carbamate exposure, the H-ras gene is activated by CAA→CTA transversion mutation at codon 61 with a higher frequency, compared to spontaneous tumors. In human cancers triggered by vinyl chloride exposure, the specific A→T transversions at codons 179 and 255 of the p53 gene also occur in the CpA context (56). To examine whether TDG is able to excise T opposite to ϵA in these mutational hotspot contexts, we constructed 40-mer duplex oligonucleotides T•ϵA-Ha and T•ϵA-p53 containing ϵA within the mouse H-ras codon 61 and human p53 codon 179 sequences (Table 3). As shown in Figure 3A, TDG excises T opposite to ϵA in both codons (lanes 9 and 11) suggesting that this aberrant repair may initiate error-prone translesion DNA synthesis across ϵA in mammalian cells. However, it should be stressed that the relative efficiency of TDG-catalyzed excision of T in T•ϵA-Ha and T•ϵA-p53 duplexes was much lower as compared to T•G duplex (Figure 3B), suggesting that this aberrant activity is minor relative to the other DNA repair functions of TDG, but nevertheless it may play a role under genotoxic stress.
Table 3.
The mutation spectrum in dinucleotide contexts in the human genome inferred from single nucleotide polymorphismsa
| SNPs within a whole genome | SNPs within CGIs | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. | XX-YYb | Counts | Fraction (%) | Probability of XX (%) | Probability of YY (%) | XX-YY | Counts | Fraction (%) | Probability of XX (%) | Probability of YY (%) |
| 1. | AT-GT | 4747845 | 5.97 | 7.73 | 5.05 | CA-CG | 44879 | 7.99 | 5.60 | 9.89 |
| 2. | AC-AT | 4728528 | 5.95 | 5.03 | 7.73 | CG-TG | 44439 | 7.91 | 9.89 | 5.60 |
| 3. | CG-TG | 4191791 | 5.27 | 0.99 | 7.27 | CC-CT | 34022 | 6.06 | 12.43 | 6.56 |
| 4. | CA-CG | 4189811 | 5.27 | 7.25 | 0.99 | AG-GG | 33929 | 6.04 | 6.59 | 12.46 |
| 5. | AA-AG | 3467707 | 4.36 | 9.78 | 6.99 | AC-GC | 27555 | 4.91 | 4.26 | 12.02 |
| 6. | CT-TT | 3466696 | 4.36 | 7.00 | 9.80 | GC-GT | 26737 | 4.76 | 12.02 | 4.26 |
| 7. | TA-TG | 3326902 | 4.19 | 6.57 | 7.27 | CC-TC | 22113 | 3.94 | 12.43 | 5.65 |
| 8. | CA-TA | 3310846 | 4.17 | 7.25 | 6.57 | GA-GG | 22110 | 3.94 | 5.68 | 12.46 |
| 9. | CC-CT | 3036893 | 3.82 | 5.21 | 7.00 | CG-GG | 17020 | 3.03 | 9.89 | 12.46 |
| 10. | AG-GG | 3033929 | 3.82 | 6.99 | 5.21 | CC-CG | 16751 | 2.98 | 12.43 | 9.89 |
aCounts are shown in descending order and only for the first 10 SNPs. Full data set of all possible dinucleotide contexts can be found in Supplementary Table S3.
bXX and YY represent dinucleotide sequence context.
Role of cooperative DNA binding in TDG-catalyzed DNA glycosylase activities
Previously, it has been shown that TDG and its bacterial homolog MUG bind to DNA substrates in a cooperative manner with a 2:1 stoichiometry. To examine a possible role of the TDG-dimer complex on its DNA glycosylase activities, we constructed 15-mer and 28-mer DNA duplexes containing T•ϵA, T•Hx, T•G, U•G and T•8oxoA base pairs positioned within 5′-TpG-3′/5′-CpX-3′ (where X is G or damaged A) context. It should be noted that according to the crystallographic studies the 28-mer but not 15-mer duplex can accommodate 2:1 TDG-binding. As shown in Figure 4, TDG excises T opposite to ϵA, Hx and G in 28-mer duplex with good efficiency (lanes 1, 3 and 5) but it fails to excise T from the corresponding 15-mer duplexes (lanes 7, 9 and 11). At the same time, TDG was able to excise U opposite to G in both 28- and 15-mer duplexes (Figure 4, lanes 13 and 15). Taken together these results suggest that formation of dimeric 2:1 TDG-DNA complex is necessary for the excision of mismatched T in duplex DNA but not for the removal of U residues.
Figure 4.

Effect of DNA substrate length on TDG-catalyzed cleavage of T•G, T•Hx, T•ϵA and U•G oligonucleotide duplexes. 5 nM 3′-[α-32P]cordycepin-labeled 28- and 15-mer duplexes were incubated with 50 nM TDG for 30 min at 37°C. After reaction, all samples were treated by 0.1 M NaOH for 3 min at 99°C to cleave at AP sites. The cleavage products were analyzed as described in the Materials and Methods section. Arrows indicate substrate (29 and 16 mer) and cleavage products (12 mer).
Next, we have examined whether TDG-catalyzed excision of 8oxoA residues depends on the size of DNA duplex. The full-length TDG protein excised 8oxoA opposite to T in 40- and 28-mer duplexes but not in 15-mer duplex (Supplementary Figure S3A). At the same time, TDG and TDGcat were able to remove 8oxoA paired with C and G in all three 40-, 28- and 15-mer duplexes (Supplementary Figure S3A and B). Taken together, these results suggest that DNA substrate specificity of TDG toward 8oxoA and mismatched T varies strongly depending not only on the opposite base but also on the length of DNA duplex. The ability of TDG to excise both 8oxoA and T in 8oxoA•T duplex (Figure 1B, lane 15 and Supplementary Figure S3A) is quite puzzling since in the absence of other BER enzymes it may result in the formation of bi-stranded AP site cluster.
In vitro reconstitution of the DNA glycosylases-initiated BER pathway of T•Hx pair in oligonucleotides and plasmid DNA
One of the outcomes of the TDG-catalyzed removal of T opposite to a damaged A in duplex DNA would be a DNA polymerase synthesis across a damaged DNA template in the downstream step of BER pathway. Consequently, one would expect that the removal of T in T•Hx and T•8oxoA would direct DNA polymerase-catalyzed misincorporation of C across Hx or 8oxoA, followed by ligation and restoration of duplex DNA. Altogether this would result in the persistence of a lesion and appearance of T→C mutation in the non-damaged DNA strand. To examine this, we have reconstituted in vitro the BER pathway for T•Hx pair positioned in 5′-TpG-3′/5′-CpHx-3′ context using the purified TDG, APE1, DNA polymerase β (POLβ) and T4 DNA ligase proteins, dNTPs and 40-mer Hx•T duplex in which the Hx-containing strand was labeled with [32P] at 5′ end (Figure 5, lanes 2–6). To detect Hx•T→Hx•C mutation, after reconstitution reaction we probed DNA duplex with restriction endonuclease BstUI that recognizes the sequence CGC∧G. As expected, BstUI cleaves Hx•T duplex only after reconstitution assay in the presence of BER enzymes and generates 18-mer cleavage fragment indicating that the TDG-initiated aberrant BER leads to CGTG→CGCG mutation (lane 4). In control experiment, in vitro reconstitution of BER with the ANPG protein, a human DNA glycosylase that removes Hx, did not create BstUI restriction site (Supplementary Figure S4). Taken together, these results suggest that in vivo upon spontaneous deamination of A, TDG could introduce A•T→G•C mutation predominantly in CpA and TpG contexts in the absence of Hx repair and DNA replication.
Figure 5.

In vitro reconstitution of the aberrant BER pathway using TDG and Hx•T duplex. 5 nM 5′-[32P]-labeled oligonucleotide duplex was incubated with 300-nM TDG, 5 nM APE1, 0.1 unit Polβ, 2 units T4 DNA Ligase and 50 μM of dNTPs for 30 min at 37°C. BstUI digestion was carried out at 60°C for 40 min. Lane 1: T•Hx duplex in which T-containing strand is 5′-[32P]-labeled incubated with TDG and then treated by 0.1 M NaOH to cleave at AP sites; lanes 2–8: Hx•T and Hx•C duplexes in which Hx-containing strand is 5′-[32P]-labeled; lanes 9 and 10: G•T duplex in which G-containing strand is 5′-[32P]-labeled. The reaction products were analyzed as described in the Materials and Methods section. Arrows indicate TDG- and BstUI-catalyzed cleavage products.
To further examine the repair of T•Hx duplex we developed a plasmid DNA vector carrying selectable antibiotic-resistance markers for phenotypic selection in E. coli that can be used for in vitro and ex vivo mammalian cell-based transient transfection assays. The pT7Blue-3Rev-TGA-Hx vector, carrying T•Hx pair within an artificial stop codon TGA (5′-TpGpA-3′/5′-TpCpHx-3′) inserted into the kanamycin-resistance (KmR)-encoding gene 57 bp downstream from the start codon, was used to detect T→C mutation by phenotypic screening of E. coli transformants. The TDG-initiated repair of T•Hx pair in pT7Blue-3Rev-TGA-Hx should result in an increase of the ratio of KmR clones to total ampicillin-resistance (AmpR) transformants due to mutation TGA→CGA in the stop codon, whereas ANPG action should result in a decrease of the ratio of KmR clones due to removal of Hx and restoration of the stop codon. For in vitro testing, the pT7Blue-3Rev-TGA-Hx vector was transformed into E. coli either directly or after pre-treatment with the purified TDG or ANPG proteins. As shown in Figure 6, pre-treatment of the plasmid DNA with TDG and ANPG resulted in 12.6-fold increase and 43-fold decrease, respectively, in the relative frequency of KmR E. coli transformants as compared to that of the control non-treated plasmid DNA (see also Supplementary Table S2). These results are in agreement with data obtained in the in vitro reconstitution assay using T•Hx duplex oligonucleotide (Figure 5) and indicate that TDG induces T→C mutations whereas ANPG prevents the mutagenic effect of Hx in E. coli. For ex vivo testing, we transfected the pT7Blue-3Rev-TGA-Hx vector into MEF cell lines either proficient (MEF-WT) or deficient (MEF-Tdg−/−) for TDG. After 6 h incubation, transfected plasmid DNA was recovered from MEFs and transformed into E. coli, and the ratio of KmR/AmpR transformants was determined. The results showed that the plasmid DNA from MEF-WT cells yields 1.7-fold higher frequency of KmR clones as compared to that of the plasmid from MEF-Tdg−/− cells (Figure 6 and Supplementary Table S3). DNA sequencing of the plasmids from KmR clones from in vitro and ex vivo experiments confirmed the presence of TGA→CGA mutation. However, when the plasmid DNA isolated from MEF-WT cells was treated with the purified ANPG protein, the yield of KmR is decreased 7-fold as compared to that of the plasmid DNA not treated with ANPG (Supplementary Table S3). These results suggest that the pT7Blue-3Rev-TGA-Hx plasmid DNA recovered 6 h after transfection into MEF-WT cells still contains unrepaired Hx residues and that a small difference in the relative frequencies of KmR clones between MEF-WT and MEF-Tdg−/− cells could also be attributed to the presence of unrepaired plasmid molecules. Taken together, the results obtained using the transient transfection assay are not conclusive and other approaches are required to address a possible role of the aberrant repair function of TDG in vivo.
Figure 6.

The relative reversion KamS→KamR rates of the plasmid DNA treated with the purified DNA glycosylases and/or transiently transfected to MEFs. (A) The fold change in the KamS→KamR reversion rates between pT7Blue-3Rev-TGA-Hx plasmid DNA treated and non-treated with ANPG. (B) The fold change in the KamS→KamR reversion rates between pT7Blue-3Rev-TGA-Hx plasmid DNA treated and non-treated with TDG. (C) The fold change in the KamS→KamR reversion rates between pT7Blue-3Rev-TGA-Hx plasmid DNA transiently transfected to MEF-WT and MEF-Tdg−/− cells. For details see Supplementary Tables S2 and S3.
Repair activities on oligonucleotide duplexes containing T•G, T•Hx and 8oxoA•G pairs in mouse cell-free extracts
Based on the above observations, we conclude that T•Hx pair in 5′-TpG-3′/5′-CpHx-3′ context will be a target for at least two human enzymes TDG and ANPG. Therefore, in vivo the order of action of these two enzymes on T•Hx duplex will determine the mutagenic outcome of the BER pathway. To examine this, we measured thymine- and Hx-DNA glycosylase activities in nuclear and cytosolic cell-free extracts prepared from WT and Tdg−/− MEFs using 5′-[32P]-labeled 40-mer T•G, T•Hx, Hx•T and 8oxoA•G oligonucleotide duplexes as DNA substrates. It should be noted that in these duplexes only the upper T-, T-, Hx-, and 8oxoA-containing DNA strands were labeled, respectively. In addition, the 40-mer T•Hx and Hx•T duplexes have the same sequence and contain a single Hx residue placed either in CXC or GXC sequence context, where X is Hx. As shown in Figure 7A, incubation of 40-mer T•G duplex in cytosolic and nuclear extracts from MEF-WT generated 19-mer cleavage fragment indicating the presence of a robust mismatch-specific thymine-DNA glycosylase activity (lanes 2 and 3). To note, the nuclear extracts from MEF-WT exhibited higher cleavage efficiency (lane 2) as compared to that of the cytosolic extracts from the same cells (lane 3) suggesting preferential nuclear distribution of TDG in MEFs. As expected, both cytosolic and nuclear extracts from MEF-Tdg−/− completely lack thymine-DNA glycosylase activity (lanes 4 and 5) indicating the absence of TDG in these cells. Incubation of the 40-mer T•Hx duplex, in which T-containing strand is 5′-[32P]-labeled, with extracts from MEF-WT generated 20-mer cleavage fragment (lanes 7 and 8), with much lower efficiency as compared to T•G duplex, indicating excision of T at the position 21 opposite to Hx. Interestingly, the cytosolic extracts from MEF-WT exhibited higher activity on T•Hx duplex (lane 7) as compared to nuclear extracts (lane 8), which is opposite to the cleavage pattern observed on T•G duplex (lanes 2 and 3). Importantly, no cleavage of the 40-mer T•Hx duplex was observed in the both cytosolic and nuclear extracts from MEF-Tdg−/− (lanes 9 and10) indicating that TDG is a major DNA glycosylase that excises T opposite to Hx in mammalian cells. Next, we examined Hx-DNA glycosylase activity in MEF extracts using the 40-mer Hx•T duplex that has the same sequence as T•Hx one and in which the Hx-containing strand is 5′-[32P]-labeled. Nuclear extracts from both WT and Tdg−/− MEF exhibited highly efficient Hx-DNA glycosylase activity that cleaves most of the Hx•T duplex and generates 19-mer product (lanes 13 and 15). In contrast, cytosolic extracts from MEF exhibited much weaker Hx-DNA glycosylase activity (lanes 13 and 15) suggesting that the mouse ANPG protein is mainly localized in nucleus of MEFs, with very little cytoplasmic distribution. Taken together, these results indicate that mammalian TDG can catalyze aberrant removal of T in T•Hx duplex in the cell-free extracts although to a much lesser extent as compared to its damage-specific DNA glycosylase activities on T•G and 8oxoA•G substrates. Furthermore, TDG and ANPG exhibit antagonistic DNA glycosylase activities on T•Hx duplex: in the nuclear extracts highly efficient excision of Hx by ANPG inhibits thymine-DNA glycosylase activity of TDG (lanes 13 and 8) whereas in the cytosolic extracts weak Hx-activity of ANPG stimulates TDG-catalyzed aberrant removal of T opposite to Hx (lanes 12 and 7).
Figure 7.
DNA glycosylase activities in cytosolic and nuclear protein extracts from MEF-WT and MEF-Tdg−/− cells. Denaturing PAGE analysis of the cleavage products after incubation of 5′-[32P]-labeled 40-mer oligonucleotide duplexes containing T•G, T•Hx, Hx•T and 8oxoA•G base pairs with cell-free extracts. The DNA repair assay (50 μl) was performed in BER+EDTA buffer containing 2.5 nM 5′-[32P]-labeled duplex, 50 mM KCl, 20 mM HEPES-KOH (pH 7.6), 0.1 mg•ml−1 BSA, 1 mM DTT, 1 mM EDTA and either 30 μg of cytosolic or nuclear proteins extracts from MEFs. Reactions mixtures were incubated 4 h or 100 min at 37°C to measure TDG or ANPG-catalyzed activities, respectively. (A) DNA glycosylase activities on Hx and mismatched T residues in the cell-free extracts. Lanes 1–5: T•G duplex in which T-containing strand is 5′-[32P]-labeled and G opposite to T in complementary strand is in the sequence context CGG; Lanes 6–10: T•Hx duplex in which T-strand is labeled and Hx opposite to T is in the context CXC where X is Hx; Lanes 11–15: Hx•T duplex in which Hx strand is labeled and T opposite to Hx is in the context GTG. (B) DNA glycosylase activities on T, Hx and 8oxoA residues in the cell-free extracts. Lanes 1–5: T•Hx duplex in which T-containing strand is labeled and Hx opposite to T is in the context GXC where X is Hx; Lanes 6–10: Hx•T duplex in which Hx strand is labeled and T opposite to Hx is in the context GTC; Lanes 11–15: 8oxoA•G duplex in which 8oxoA strand is labeled and G opposite to 8oxoA is in the context GGG. Arrows denote the 40-mer DNA substrate and 20- and 19-mer cleavage products. For details see the Materials and Methods section.
Next, we have examined whether TDG also exhibits sequence context preference in the cell-free extracts. As shown in Figure 7B, no thymine-DNA glycosylase activity was observed in extracts from MEF-WT on T•Hx (GXC) duplex, in which T is in GpTpC/GpXpC context where X is Hx (lanes 7 and 8) indicating that TDG in cell-free extracts also exhibits strong preference for 5′-TpG-3′/5′-CpX-3′ context where X is a damaged adenine residue. This result is in agreement with data obtained with the purified TDG protein (Figure 2B, lane 9). Importantly, the nuclear extracts from MEF exhibited efficient Hx-DNA glycosylase activity on Hx•T (GXC) duplex, in which the Hx-containing strand is 5′-[32P]-labeled (Figure 7B, lanes 13 and 15), indicating that activity of the mouse ANPG is not dependent on sequence context. In addition, we measured TDG-catalyzed 8oxoA-DNA glycosylase activity in extracts from MEFs using 8oxoA•G duplex, in which the 8oxoA-containing strand is 5′-[32P]-labeled (Figure 7B, lanes 11–15). Again, the nuclear extracts exhibited higher activity as compared to the cytosolic ones (lane 13 versus 12) from MEF-WT and no 8oxoA activity was observed in the extracts from MEF-Tdg−/−. These results confirm that TDG is a major T- and 8oxoA-DNA glycosylase in mammalian cells and a preferential nuclear distribution of TDG in MEFs.
Comparison of SNPs in whole genome and CGIs
Based on our biochemical data, we propose that in vivo TDG may promote mutagenic conversion of both CpA and TpG dinucleotides to CpG ones. In chromosomes TDG is mainly localized in CGIs of promoter regions to protect them from cytosine methylation by DNMT3a and DNMT3b de novo methylases, therefore CGIs may exhibit mutational bias for TpG, CpA→CpG mutations. To examine this we analyzed the mutation spectra in human genome inferred from the SNP data (NCBI dbSNP build 127 for human). For this, we measured the relative frequencies of base substitutions within dinucleotide contexts. Remarkably, the relative frequencies of CpA↔CpG (7.99% of total SNPs) and CpG↔TpG (7.91%) in CGIs of human genome were the highest among other types of SNPs suggesting that TpG and CpA dinucleotides are mutation hotspots in CpG-rich regions (Table 3 and Supplementary Table S4). At the same time, the highest frequencies of SNPs for the whole genome were ApT↔GpT (5.97% of total SNPs) and ApC↔ApT (5.95%), followed by TpG and CpA (5.27% each), indicating that the genome-wide mutation spectra are different from that of CGIs (Table 3 and Supplementary Table S4). Importantly, the probability of occurrence of CpA and CpG dinucleotides in CGIs is 5.6% and 9.89%, whereas in a whole genome it is the opposite, 7.25% and 0.99%, respectively. These dramatic imbalances in the frequencies of dinucleotides occurrence between whole genome and CGIs might be due to directionality of spontaneous mutagenesis in vivo: at the genome-wide level CpG→TpG/CpA mutations are more frequent than the reverse mutations because of the specific pattern of DNA methylation, whereas in the CGIs, this is in fact the opposite, as TpG and CpA mutate more frequently to CpG, possibly due to TDG-catalyzed aberrant BER.
DISCUSSION
Here, we report that the TDG protein is able to excise T opposite to various adenine lesions with a good efficiency when it placed in the specific 5′-TpG-3′/5′-CpX-3′ (where X is Hx, ϵA, AP site or 8oxoA) context. Importantly, under our experimental conditions we do not observe TDG activity toward non-damaged A•T duplex. Also TDG-catalyzed excision of T strongly depends on the TpG dinucleotide context, as the enzyme was not at all or only weakly active in the TpA, TpC and TpT contexts (Figures 2B and 3A). However, in the present study, we did not perform an exhaustive search for all possible sequence contexts and DNA lesions, hence one cannot exclude a possibility of the presence of a weak TDG activity on certain non-TpG contexts. Interestingly, MBD4, a functional human counterpart of TDG, is also able to remove T opposite to ϵA residue but not across other types of adenine damage (Figure 1B and Supplementary Figure S2). This aberrant excision of non-damaged T could lead to translesion repair synthesis across ϵA adduct and introduce A•T→T•A transversions that were typically observed previously in the plasmid transfection experiments and in human liver angiosarcomas associated with exposure to vinyl chloride (55,57). Intriguingly, the carcinogen-induced A→T hotspot mutations occurred at CpA sites in codon 61 of the c-Ha-ras gene and codons 179 and 255 of the p53 gene suggesting a possible involvement of MBD4 and TDG in the vinyl chloride-induced tumors (55). The aberrant BER repair initiated by thymine-DNA glycosylases on ϵA•T and 8oxoA•T duplexes may explain the higher mutagenic potentials of exocyclic DNA adduct and oxidized adenine residues in mammalian cells, compared to E. coli, as described in the previous studies (57,58).
The ability of TDG to excise T opposite to 8oxoA, ϵA and/or AP site with significant efficiency is quite unexpected because of the requirement of G in complementary strand for excision of both U and T residues by TDG. Indeed, crystal structures of TDGcat in complex with various DNA substrates containing either G or A opposite to the lesion showed that G, but not A, is contacted by the backbone oxygens of amino acids Ala274 and Pro280 (14,23,45). These specific interactions explain the preference of TDG for G over non-damaged A in the complementary strand. However, it is not clear whether TDG could interact with modified adenine residues such as Hx, 8oxoA and ϵA in mismatches with T. Moreover, the capacity of TDG to excise ϵC, 5caC and 5fC without a requirement for opposite base (7,23) suggests that the nature of the target base and of the specific interactions between the base and protein residues within the active site of the enzyme play a primordial role in the DNA substrate recognition by TDG. Crystallographic studies of MBD4 in complex with DNA revealed that, in contrast to TDG, MBD4 has very limited DNA substrate preference due to conformation of its active site pocket and due to specific interactions between the orphan G and Arg468 that helps to stabilize the flipped-out target base for efficient catalysis (46). Still, MBD4 can excise with very low efficiency T opposite to ϵA, but not opposite to Hx or 8oxoA, suggesting that Arg468 may interact with unpaired ϵA in enzyme-substrate complex. Overall, the available crystallographic data on DNA glycosylases:substrate complexes do not provide sufficient insight (i) into the structural basis of the strong sequence context dependence of excision of mismatched T by TDG and MBD4 and (ii) for whether the DNA glycosylases are able to make specific contacts with the orphan damaged A residue during DNA substrate recognition.
The phenomenon of cooperative DNA binding by repair enzymes has been studied previously (24,25). It was proposed that this mode of binding may protect DNA and help to coordinate downstream repair steps (25). Here, we demonstrated that long 28–40-mer DNA duplexes are required for efficient excision of mismatched T and 8oxoA, but not U residues, by full-length TDG (Figure 4 and Supplementary Figure S3) suggesting that cooperative binding of TDG to DNA is required for efficient processing of a number of DNA substrates. The molecular mechanism and structural basis for this molecular recognition phenomenon remain unclear. We may propose that formation of oligomeric TDG filaments along the length of DNA promoter regions enables both the protection from de novo DNA methylation and the efficient repair of mismatched thymine residues.
In vitro reconstitution of the TDG-initiated BER pathway of T•Hx duplex and the plasmid mutagenesis studies (Figures 5 and 6 and Supplementary Tables S2 and S3) demonstrated that the aberrant excision of T by TDG can introduce TpG, CpA→CpG mutations in the absence of DNA replication. Data obtained in cell-free extracts from MEF-WT and MEF- Tdg−/− cells indicate that mammalian TDG is the main DNA glycosylase involved in the aberrant repair of T•Hx and 8oxoA•G duplexes in mouse cells (Figure 7). Importantly, in cell-free extracts, ANPG-catalyzed Hx and ϵA removal can efficiently prevents TDG-mediated aberrant removal of T in the complementary strand in 5′-TpG-3′/5′-CpX-3′ sequence context, however not to absolute extent (Figure 7A). Therefore, in vivo minor fraction of T•Hx and T•ϵA base pairs in DNA could be repaired by TDG in aberrant error-prone manner. Interestingly, the nuclear fraction of TDG associates tightly with euchromatin and binds to the CpG-rich promoters of transcribed genes including the pluripotency and developmental genes (31,59). It was suggested that TDG scans CpG sites for mismatches and regulates gene expressions, either by protecting CpG from de novo DNA methylation or via interaction with chromatin and transcription factors. Comparison of the spontaneous genome-wide mutation spectra versus those at the CGIs using human SNP database revealed that CGIs, in contrast to whole genome, exhibit strong mutational bias for TpG, CpA→CpG mutations (Table 3 and Supplementary Table S4). The observed mutation spectra suggest that the TDG-catalyzed aberrant BER might be involved in the stabilization and extension of CG content in CpG-rich promoters.
Based on our results, it is tempting to speculate that the increased rate of TpG, CpA→CpG mutations in CGIs would depend on their transcriptional activity given the association of TDG with the CpG-rich promoter regions to protect them from de novo methylation. Furthermore, emergence of CGIs at TSSs of genes during evolution of warm-blooded vertebrates (36) may be a direct consequence of the TDG-catalyzed aberrant BER pathway of spontaneous and oxidative damage to adenine residues occurred within regulatory regions of a genome. Here, we propose a model in which the non-methylated regulatory DNA regions associated with TDG would be driven toward an increased CpG dinucleotide content by conversion of CpA and TpG to CpG dinucleotides, whereas the reverse processes would occur in methylated genome area due to spontaneous deamination of 5mC residues (Figure 8). In line with our hypothetical mechanism, it has been shown that the relative rate of CpG-rich promoter evolution is substantially accelerated in primates and/or remains neutral in mammals (39). Furthermore, the presence of multiple 5mC-DNA glycosylases in plants that contain mismatch-specific thymine-DNA glycosylase activity may provide a rational for the increased CG content of the plant genomes despite dense methylation of cellular DNA (60). Finally, the aberrant BER pathway described here could be considered as an evolutionary capacitor mechanism that accelerates mutation rate at the regulatory DNA sequences which enables environmentally induced gene expression to become a developmentally programmed expression pattern, which in turn may play a role in the formation of the placenta and exponential growth in taxonomic diversity of mammals (61).
Figure 8.

Dynamics of mutational spectra in the mammalian genomes. (A) Mammalian DNA methyltransferases (DNMTs) transfer methyl group to cytosines at position 5 in CpG dinucleotide contexts resulting in 5mC residues in DNA. The spontaneous deamination of 5mC induces the transition mutations CpG→TpG and CpG→CpA. (B) TDG localizes in CGIs promoter regions to protect them from de novo DNA methylation either in passive or active TET-mediated manner. Spontaneous damage to adenines in TpG and CpA dinucleotide contexts in non-methylated DNA promote the TDG-catalyzed aberrant BER and TpG→CpG and CpA→CpG transition mutations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We wish to thank Dr Andrei Kuzminov for critical reading of the manuscript and thoughtful discussions and also to A. Bellacosa, A. Drohat and P. Schar for the MBD4 and TDG protein expression constructs and MEF cell lines, respectively.
FUNDING
Agence Nationale pour la Recherche [ANR Blanc 2010 Projet ANR-10-BLAN-1617 to M.K.S.]; Fondation pour la Recherche Médicale [EQUIPES FRM 2007 DEQ20071210556 to M.K.S.]; Electricité de France [RB 2014-26 to M.K.S.]; Fondation de France [#2012 00029161 to A.A.I.]; Science Committee of the Ministry of Education and Science of the Republic of Kazakhstan to B.M.; I.T, S.C. and L.G. were supported by postdoctoral fellowships funded by the grants from Fondation ARC [PDF20110603195], FRM [DEQ20071210556] and European Community [FP6 Euroatom Grant RISC-RAD FI6R-CT-2003-508842], respectively. Funding for open access charge: Grant from Nazarbayev University Research and Innovation System [RB 2014-26].
Conflict of interest statement. None declared.
REFERENCES
- 1.Bird A.P. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8:1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neddermann P., Jiricny J. The purification of a mismatch-specific thymine-DNA glycosylase from HeLa cells. J. Biol. Chem. 1993;268:21218–21224. [PubMed] [Google Scholar]
- 3.Hendrich B., Hardeland U., Ng H.H., Jiricny J., Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 4.Hitomi K., Iwai S., Tainer J.A. The intricate structural chemistry of base excision repair machinery: implications for DNA damage recognition, removal, and repair. DNA Repair (Amst) 2007;6:410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Krokan H.E., Bjoras M. Base excision repair. Cold Spring Harb. Perspect. Biol. 2013;5:a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hang B., Medina M., Fraenkel-Conrat H., Singer B. A 55-kDa protein isolated from human cells shows DNA glycosylase activity toward 3,N4-ethenocytosine and the G/T mismatch. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13561–13566. doi: 10.1073/pnas.95.23.13561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saparbaev M., Laval J. 3,N4-ethenocytosine, a highly mutagenic adduct, is a primary substrate for Escherichia coli double-stranded uracil-DNA glycosylase and human mismatch-specific thymine-DNA glycosylase. Proc. Natl. Acad. Sci. U.S.A. 1998;95:8508–8513. doi: 10.1073/pnas.95.15.8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoon J.H., Iwai S., O’Connor T.R., Pfeifer G.P. Human thymine DNA glycosylase (TDG) and methyl-CpG-binding protein 4 (MBD4) excise thymine glycol (Tg) from a Tg:G mispair. Nucleic Acids Res. 2003;31:5399–5404. doi: 10.1093/nar/gkg730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett M.T., Rodgers M.T., Hebert A.S., Ruslander L.E., Eisele L., Drohat A.C. Specificity of human thymine DNA glycosylase depends on N-glycosidic bond stability. J. Am. Chem. Soc. 2006;128:12510–12519. doi: 10.1021/ja0634829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Talhaoui I., Couve S., Ishchenko A.A., Kunz C., Schar P., Saparbaev M. 7,8-Dihydro-8-oxoadenine, a highly mutagenic adduct, is repaired by Escherichia coli and human mismatch-specific uracil/thymine-DNA glycosylases. Nucleic Acids Res. 2013;41:912–923. doi: 10.1093/nar/gks1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neddermann P., Jiricny J. Efficient removal of uracil from G.U mispairs by the mismatch-specific thymine DNA glycosylase from HeLa cells. Proc. Natl. Acad. Sci. U.S.A. 1994;91:1642–1646. doi: 10.1073/pnas.91.5.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardeland U., Bentele M., Jiricny J., Schar P. The versatile thymine DNA-glycosylase: a comparative characterization of the human, Drosophila and fission yeast orthologs. Nucleic Acids Res. 2003;31:2261–2271. doi: 10.1093/nar/gkg344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petronzelli F., Riccio A., Markham G.D., Seeholzer S.H., Genuardi M., Karbowski M., Yeung A.T., Matsumoto Y., Bellacosa A. Investigation of the substrate spectrum of the human mismatch-specific DNA N-glycosylase MED1 (MBD4): fundamental role of the catalytic domain. J. Cell. Physiol. 2000;185:473–480. doi: 10.1002/1097-4652(200012)185:3<473::AID-JCP19>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 14.Hashimoto H., Liu Y., Upadhyay A.K., Chang Y., Howerton S.B., Vertino P.M., Zhang X., Cheng X. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012;40:4841–4849. doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waters T.R., Swann P.F. Kinetics of the action of thymine DNA glycosylase. J. Biol. Chem. 1998;273:20007–20014. doi: 10.1074/jbc.273.32.20007. [DOI] [PubMed] [Google Scholar]
- 16.Waters T.R., Gallinari P., Jiricny J., Swann P.F. Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J. Biol. Chem. 1999;274:67–74. doi: 10.1074/jbc.274.1.67. [DOI] [PubMed] [Google Scholar]
- 17.Hardeland U., Steinacher R., Jiricny J., Schar P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 2002;21:1456–1464. doi: 10.1093/emboj/21.6.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Um S., Harbers M., Benecke A., Pierrat B., Losson R., Chambon P. Retinoic acid receptors interact physically and functionally with the T:G mismatch-specific thymine-DNA glycosylase. J. Biol. Chem. 1998;273:20728–20736. doi: 10.1074/jbc.273.33.20728. [DOI] [PubMed] [Google Scholar]
- 19.Cortazar D., Kunz C., Saito Y., Steinacher R., Schar P. The enigmatic thymine DNA glycosylase. DNA Repair (Amst) 2007;6:489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 20.Tini M., Benecke A., Um S.J., Torchia J., Evans R.M., Chambon P. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol. Cell. 2002;9:265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- 21.Hardeland U., Kunz C., Focke F., Szadkowski M., Schar P. Cell cycle regulation as a mechanism for functional separation of the apparently redundant uracil DNA glycosylases TDG and UNG2. Nucleic Acids Res. 2007;35:3859–3867. doi: 10.1093/nar/gkm337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maiti A., Morgan M.T., Pozharski E., Drohat A.C. Crystal structure of human thymine DNA glycosylase bound to DNA elucidates sequence-specific mismatch recognition. Proc. Natl. Acad. Sci. U.S.A. 2008;105:8890–8895. doi: 10.1073/pnas.0711061105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L., Lu X., Lu J., Liang H., Dai Q., Xu G.L., Luo C., Jiang H., He C. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat. Chem. Biol. 2012;8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rasimas J.J., Kar S.R., Pegg A.E., Fried M.G. Interactions of human O6-alkylguanine-DNA alkyltransferase (AGT) with short single-stranded DNAs. J. Biol. Chem. 2007;282:3357–3366. doi: 10.1074/jbc.M608876200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grippon S., Zhao Q., Robinson T., Marshall J.J., O’Neill R.J., Manning H., Kennedy G., Dunsby C., Neil M., Halford S.E., et al. Differential modes of DNA binding by mismatch uracil DNA glycosylase from Escherichia coli: implications for abasic lesion processing and enzyme communication in the base excision repair pathway. Nucleic Acids Res. 2011;39:2593–2603. doi: 10.1093/nar/gkq913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito S., D’Alessio A.C., Taranova O.V., Hong K., Sowers L.C., Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maiti A., Drohat A.C. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cortazar D., Kunz C., Selfridge J., Lettieri T., Saito Y., MacDougall E., Wirz A., Schuermann D., Jacobs A.L., Siegrist F., et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 32.Cortellino S., Xu J., Sannai M., Moore R., Caretti E., Cigliano A., Le Coz M., Devarajan K., Wessels A., Soprano D., et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bird A., Taggart M., Frommer M., Miller O.J., Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985;40:91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- 34.Akan P., Deloukas P. DNA sequence and structural properties as predictors of human and mouse promoters. Gene. 2008;410:165–176. doi: 10.1016/j.gene.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deaton A.M., Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharif J., Endo T.A., Toyoda T., Koseki H. Divergence of CpG island promoters: a consequence or cause of evolution. Dev. Growth Differ. 2010;52:545–554. doi: 10.1111/j.1440-169X.2010.01193.x. [DOI] [PubMed] [Google Scholar]
- 37.Duret L., Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu. Rev. Genomics Hum. Genet. 2009;10:285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- 38.Cohen N.M., Kenigsberg E., Tanay A. Primate CpG islands are maintained by heterogeneous evolutionary regimes involving minimal selection. Cell. 2011;145:773–786. doi: 10.1016/j.cell.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 39.Taylor M.S., Kai C., Kawai J., Carninci P., Hayashizaki Y., Semple C.A. Heterotachy in mammalian promoter evolution. PLoS Genet. 2006;2 doi: 10.1371/journal.pgen.0020030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neddermann P., Gallinari P., Lettieri T., Schmid D., Truong O., Hsuan J.J., Wiebauer K., Jiricny J. Cloning and expression of human G/T mismatch-specific thymine-DNA glycosylase. J. Biol. Chem. 1996;271:12767–12774. doi: 10.1074/jbc.271.22.12767. [DOI] [PubMed] [Google Scholar]
- 41.Hennecke F., Kolmar H., Brundl K., Fritz H.J. The vsr gene product of E. coli K-12 is a strand- and sequence-specific DNA mismatch endonuclease. Nature. 1991;353:776–778. doi: 10.1038/353776a0. [DOI] [PubMed] [Google Scholar]
- 42.Tsai-Wu J.J., Radicella J.P., Lu A.L. Nucleotide sequence of the Escherichia coli micA gene required for A/G-specific mismatch repair: identity of micA and mutY. J. Bacteriol. 1991;173:1902–1910. doi: 10.1128/jb.173.6.1902-1910.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fowler R.G., White S.J., Koyama C., Moore S.C., Dunn R.L., Schaaper R.M. Interactions among the Escherichia coli mutT, mutM, and mutY damage prevention pathways. DNA Repair (Amst) 2003;2:159–173. doi: 10.1016/s1568-7864(02)00193-3. [DOI] [PubMed] [Google Scholar]
- 44.Kavli B., Slupphaug G., Mol C.D., Arvai A.S., Peterson S.B., Tainer J.A., Krokan H.E. Excision of cytosine and thymine from DNA by mutants of human uracil-DNA glycosylase. EMBO J. 1996;15:3442–3447. [PMC free article] [PubMed] [Google Scholar]
- 45.Maiti A., Noon M.S., MacKerell A.D., Jr, Pozharski E., Drohat A.C. Lesion processing by a repair enzyme is severely curtailed by residues needed to prevent aberrant activity on undamaged DNA. Proc. Natl. Acad. Sci. U.S.A. 2012;109:8091–8096. doi: 10.1073/pnas.1201010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morera S., Grin I., Vigouroux A., Couve S., Henriot V., Saparbaev M., Ishchenko A.A. Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA. Nucleic Acids Res. 2012;40:9917–9926. doi: 10.1093/nar/gks714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gelin A., Redrejo-Rodriguez M., Laval J., Fedorova O.S., Saparbaev M., Ishchenko A.A. Genetic and biochemical characterization of human AP endonuclease 1 mutants deficient in nucleotide incision repair activity. PLoS ONE. 2010;5:e12241. doi: 10.1371/journal.pone.0012241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kunz C., Focke F., Saito Y., Schuermann D., Lettieri T., Selfridge J., Schar P. Base excision by thymine DNA glycosylase mediates DNA-directed cytotoxicity of 5-fluorouracil. PLoS Biol. 2009;7:e91. doi: 10.1371/journal.pbio.1000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Challberg M.D., Kelly T.J., Jr Adenovirus DNA replication in vitro. Proc. Natl. Acad. Sci. U.S.A. 1979;76:655–659. doi: 10.1073/pnas.76.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baerenfaller K., Fischer F., Jiricny J. Characterization of the “mismatch repairosome” and its role in the processing of modified nucleosides in vitro. Methods Enzymol. 2006;408:285–303. doi: 10.1016/S0076-6879(06)08018-9. [DOI] [PubMed] [Google Scholar]
- 51.Zlatanou A., Despras E., Braz-Petta T., Boubakour-Azzouz I., Pouvelle C., Stewart G.S., Nakajima S., Yasui A., Ishchenko A.A., Kannouche P.L. The hMsh2-hMsh6 complex acts in concert with monoubiquitinated PCNA and Pol eta in response to oxidative DNA damage in human cells. Mol. Cell. 2011;43:649–662. doi: 10.1016/j.molcel.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 52.Choi J.H., Pfeifer G.P. DNA damage and mutations produced by chloroacetaldehyde in a CpG-methylated target gene. Mutat. Res. 2004;568:245–256. doi: 10.1016/j.mrfmmm.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 53.Gros L., Ishchenko A.A., Saparbaev M. Enzymology of repair of etheno-adducts. Mutat. Res. 2003;531:219–229. doi: 10.1016/j.mrfmmm.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 54.Sibghat U., Gallinari P., Xu Y.Z., Goodman M.F., Bloom L.B., Jiricny J., Day R.S., III Base analog and neighboring base effects on substrate specificity of recombinant human G:T mismatch-specific thymine DNA-glycosylase. Biochemistry. 1996;35:12926–12932. doi: 10.1021/bi961022u. [DOI] [PubMed] [Google Scholar]
- 55.Barbin A. Etheno-adduct-forming chemicals: from mutagenicity testing to tumor mutation spectra. Mutat. Res. 2000;462:55–69. doi: 10.1016/s1383-5742(00)00014-4. [DOI] [PubMed] [Google Scholar]
- 56.Trivers G.E., Cawley H.L., DeBenedetti V.M., Hollstein M., Marion M.J., Bennett W.P., Hoover M.L., Prives C.C., Tamburro C.C., Harris C.C. Anti-p53 antibodies in sera of workers occupationally exposed to vinyl chloride. J. Natl. Cancer Inst. 1995;87:1400–1407. doi: 10.1093/jnci/87.18.1400. [DOI] [PubMed] [Google Scholar]
- 57.Levine R.L., Yang I.Y., Hossain M., Pandya G.A., Grollman A.P., Moriya M. Mutagenesis induced by a single 1,N6-ethenodeoxyadenosine adduct in human cells. Cancer Res. 2000;60:4098–4104. [PubMed] [Google Scholar]
- 58.Kamiya H., Miura K., Ishikawa H., Inoue H., Nishimura S., Ohtsuka E. c-Ha-ras containing 8-hydroxyguanine at codon 12 induces point mutations at the modified and adjacent positions. Cancer Res. 1992;52:3483–3485. [PubMed] [Google Scholar]
- 59.Tini M., Benecke A., Um S.J., Torchia J., Evans R.M., Chambon P. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol. Cell. 2002;9:265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- 60.Morales-Ruiz T., Ortega-Galisteo A.P., Ponferrada-Marin M.I., Martinez-Macias M.I., Ariza R.R., Roldan-Arjona T. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6853–6858. doi: 10.1073/pnas.0601109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sahney S., Benton M.J., Ferry P.A. Links between global taxonomic diversity, ecological diversity and the expansion of vertebrates on land. Biol. Lett. 2010;6:544–547. doi: 10.1098/rsbl.2009.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



