Abstract
In Neurospora crassa, the methionine synthase gene met-8 plays a key role in methionine synthesis. In this study, we found that MET-8 protein levels were compromised in several mutants defective in proper heterochromatin formation. Bioinformatics analysis revealed a 50-kb AT-rich region adjacent to the met-8 promoter. ChIP assays confirmed that trimethylated H3K9 was enriched in this region, indicating that heterochromatin may form upstream of met-8. In an H3K9R mutant strain, the output of met-8 was dramatically reduced, similar to what we observed in mutant strains that had defective heterochromatin formation. Furthermore, the production of ectopically expressed met-8 at the his-3 locus in the absence of a normal heterochromatin environment was inefficient, whereas ectopic expression of met-8 downstream of two other heterochromatin domains was efficient. In addition, our data show that the expression of mig-6 was also controlled by an upstream 4.2-kb AT-rich region similar to that of the met-8 gene, and we demonstrate that the AT-rich regions adjacent to met-8 or mig-6 are required for their peak expression. Our study indicates that met-8 and mig-6 may represent a novel type of gene, whose expression relies on the proper formation of a nearby heterochromatin region.
INTRODUCTION
Large regions in higher eukaryotic genomes are composed of repetitive deoxyribonucleic acid (DNA) elements, which are preferential targets for heterochromatin assembly. In multicellular organisms, constitutive heterochromatin is located at pericentromeric and telomeric regions. An additional form of facultative heterochromatin, found in the inactive mammalian X chromosome, is characterized by the presence of trimethylated H3K27 (1,2). Heterochromatic regions are highly condensed in structure and are characterized by their transcriptionally repressed state, as exemplified by the phenomenon of position-effect variegation in Drosophila melanogaster, in which the expression of a euchromatic gene is compromised when it is relocated near or within a block of heterochromatin (3). In animals and fungi, histone H3 lysine 9 trimethylation (H3K9me3) is a distinct feature of heterochromatic loci, whereas in the plant Arabidopsis thaliana, histone H3 monomethylation and dimethylation at lysine 9 are the predominant markers for silenced heterochromatin. Heterochromatin protein 1 (HP1) recognizes and binds to H3K9 methylation through its chromodomain for heterochromatin assembly in most well-studied model systems (4).
In fission yeast, the protein Rik1 interacts with Cul4 to target the histone methyltransferase Clr4, forming the ClrC complex for H3K9 methylation (5–7). In plants and metazoans, DDB1 is the only Rik1-like protein (8,9) and is a component of Cul4-based E3 ubiquitin ligases in eukaryotes (10–12). Knockdown of the ddb1 or cul4 genes in mammalian cells decreases trimethylation of H3K4, H3K9 and H3K27 (13). Moreover, DDB1 and Cul4 proteins appear to associate with trimethylated H3K9 and H3K27 (13), the principal markers of heterochromatic domains and transcriptionally silenced loci. These findings suggest that DDB1 and Cul4 may be important for gene silencing and heterochromatin formation in mammals, in addition to their role in the DNA damage repair pathway. In Neurospora crassa, the histone methyltransferase DIM-5 is required for H3K9 methylation and it functions with HP1 in DNA methylation and heterochromatin formation at most regions (14–16). Recent studies demonstrated that in N. crassa, DDB1 and Cul4 interact with DIM-5 via a bridge comprising DCAF26 (DDB1-Cul4-Associated Factor 26) and DIM-7 (Defective In Methylation 7). These proteins form a complex that is essential for histone H3K9 trimethylation and DNA methylation at AT-rich DNA regions, where heterochromatin may potentially form (17–20). These results suggest that the Cul4-based histone methyltransferase complex may be involved in the regulation of heterochromatin formation and gene expression in N. crassa and other higher eukaryotes.
Many genes from diverse organisms are known to be silenced by the nearby heterochromatin (3), but the expression of certain genes is known to be dependent on heterochromatin (21–24). In D. melanogaster, these heterochromatic genes are immune to the silencing effects of heterochromatin and are known to depend on heterochromatic locations and SU(VAR) proteins for normal expression (25–27). The notion that repetitive elements can create heterochromatic repressive environments by recruiting silencing components is well accepted, but it is unknown how this environment might facilitate active expression of heterochromatic genes. The expression of some essential heterochromatic genes within these repressive regions presents a paradox, and the understanding of which could provide key insights into the effects of heterochromatin structure on gene expression (24).
To further understand the mechanism of heterochromatin for gene regulation in eukaryotes, we sought to identify pathways that selectively regulate the expression of heterochromatic genes. Here we report that normal, high-efficiency expression of the methionine synthase gene (met-8) requires the presence of a proximal 50-kb AT-rich region of heterochromatin in N. crassa. met-8 expression appears to be compromised when proper heterochromatin formation was bothered, which is in contrast with the well-accepted notion that repetitive elements create heterochromatic repressive environments by recruiting the silencing components for gene silencing (28,29). In addition, we found that the expression of menadione-induced gene-6 (mig-6) was positively controlled by its upstream 4.2-kb AT-rich region, in a similar way to the met-8 gene. Our findings not only strongly implicate Cul4-based complex activity in gene expression via its effects on the proper formation of heterochromatin but also provide evidence of a clear function for heterochromatin in maintaining expression of the met-8 and mig-6 genes.
MATERIALS AND METHODS
Strains and culture conditions
In this study, N. crassa 87–3 (bd, a) (30) was used as the wild-type strain. The H3K9R knock-in mutant was newly generated from the ku70RIP (bd, a) genetic background strain. cul4KO, ddb1KO, dcaf26KO, dim-7KO, dim-5KO, hpoKO and dim-2KO strains, generated previously (17,18), were also included in this study. The 301–6 (bd, his-3, A) and dim-7KO (bd, his-3) strains were used as the host strains for his-3 targeting constructs.
Liquid cultures were grown at 25°C with shaking in minimal medium (1× Vogel's and 2% glucose) for ∼22 h in constant light (LL).
Protein extraction for 2D gel
Tissues were harvested by filtration and ground in liquid nitrogen. The fine powder was precipitated overnight at −20°C with 10% (w/v) trichloroacetic acid in cold acetone containing 0.07% (v/v) 2-mercaptoethanol. The mixture was centrifuged at 40 000 g at 4°C for 1 h, and the precipitates were washed with cold acetone containing 0.07% (v/v) 2-mercaptoethanol, 1-mM phenylmethylsulfonyl fluoride (PMSF), 1-μg/ml pepstatin A and 1-μg/ml leupeptin. Dried pellets were then dissolved in a 7-M urea solution with 2-M thiourea, 20-mM dithiothreitol, 1-mM PMSF, 1-μg/ml pepstatin A and 1-μg/ml leupeptin before centrifugation at 100 000 g at 4°C for 1 h. The supernatant was collected and quantified with a 2-D Quant kit (GE Healthcare).
2D gel experiments and data analysis
A series of 2D gels were produced essentially as reported previously (31). Isoelectric focusing was performed with the Ettan IPGphor 3 Isoelectric Focusing System. Immobiline nonlinear pH 3–10 DryStrips (GE Healthcare) were run using rehydration buffer (8-M urea, 2% CHAPS and 20-mM Dithiothreitol, DTT) containing 0.5% v/v IPG Buffer (GE Healthcare). Sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using 12.5% polyacrylamide gels without a stacking gel in the Ettan Dalt Six Elect Unit 230 (GE Healthcare). Gels were stained with 0.04% w/v PhastGel Blue R (CBB R-350; GE Healthcare) in 10% acetic acid. In order to obtain reliable results from 2D images, the experiments were performed independently four times. The protein spots were excised from 2D gels and subjected to tryptic digestion and liquid chromatography–tandem mass spectrometry (LC–MS/MS).
Generation of antiserum against MET-8
GST-MET-8 (containing MET-8 amino acids 575–689) fusion protein was expressed in BL21 cells, and the soluble recombinant protein was purified and used as the antigen to generate rabbit polyclonal antiserum, as described previously (17,18).
Protein analyses
Protein extraction, quantification, western blot analysis and protein degradation assays were performed as described previously (32). Equal amounts of total protein (10 μg) were loaded into each protein lane. After electrophoresis, proteins were transferred onto PVDF membrane. Western blot analyses were performed by using antibodies against the proteins of interest.
Northern blot analysis
Ribonucleic acid (RNA) was extracted as described previously (33), and equal amounts of total RNA (20 μg) were loaded into each lane of 1.3% agarose gels for electrophoresis. The RNAs were transferred from gels to nylon membrane. The membranes were probed with RNA probes specific to met-8, NCU06171, NCU06511 and mig-6 mRNA.
Chromatin immunoprecipitation analysis
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (17). Briefly, N. crassa tissues harvested under the experimental conditions were fixed with 1% formaldehyde for 15 min at 25°C with shaking and then stopped with glycine at a final concentration of 125 mM. Cross-linked tissues were ground and resuspended at 1 g/ml in lysis buffer containing 1-mM PMSF, 1-μg/ml pepstatin A and 1-μg/ml leupeptin. Chromatin was sheared by sonication to ∼500–1000-bp fragments. One milliliter of protein (2-mg protein/ml) was used per immunoprecipitation, and 10 μl was kept as the input DNA. ChIP was carried out with 10 μl of antibody to trimethylated H3K9 (07–442; Millipore), 5 μl of antibody to H3 (2650; CST), 2 μl of antibody to trimethylated H3K4 (07–473; Millipore) or 10 μl of antibody 8WG16 to Pol II carboxyl-terminal domain (CTD) repeats (ab817; Abcam). Immunoprecipitated DNA was quantified using real-time polymerase chain reaction (PCR) (CFX96; Bio-Rad) with primer pairs (see Supplementary Table S1). ChIP-quantitative PCR (qPCR) data were normalized by the input and presented as a percentage of input DNA. Each experiment was independently performed at least three times.
Western blot analyses of histone H3 and trimethylated H3K9
Approximately 0.1 g of dried frozen tissue was resuspended in 1 ml of extraction buffer containing protease inhibitors. The mixture was vortexed and sonicated briefly. The homogenate was then aliquoted (100 μl) and centrifuged at 10 000 g for 15 min at 4°C. The precipitate was resuspended in 100 μl of loading buffer (50-mM Tris pH 6.8, 2% SDS, 1-mM ethylenediaminetetraacetic acid, 1% glycerol, 3-M urea, 100-mM DTT and 0.25% bromophenol blue) and boiled for 30 min. The crude extracts were centrifuged at 10 000 g for 10 min, and the supernatants were loaded onto 12% polyacrylamide gels for electrophoresis. After electrophoresis, proteins were transferred onto PVDF membranes and western blot analysis was performed using antibodies against trimethylated H3K9 (07–442; Millipore) and H3 (2650, CST), respectively.
RESULTS
MET-8 protein expression is decreased in strains with deletions in the Cul4-based histone methyltransferase complex
Previous studies showed that the DCDC (the DIM-5/-7/-9, CUL4/DDB1 complex) (19) or Cul4-DDB1-DCAF26-DIM-7-DIM-5 complex is essential for histone H3K9me3 and DNA methylation in N. crassa (17,18). To investigate how the complex affects gene expression, we examined changes in the proteome of cul4KO and ddb1KO strains during vegetative growth in liquid medium under constant light conditions. After 2D gel electrophoresis of soluble protein extracts, LC–MS/MS analysis of excised gel spots led to the identification of 172 spots representing 170 unique proteins with differences in intensity in the cul4KO and ddb1KO strains compared to the wild-type strain (Supplementary Figure S1). Among the spots that differed in intensity between wild-type and mutant strains, MET-8 (NCU06512) was quite prominent, because its expression was significantly decreased in the mutant strains (Figure 1A and B). MET-8 is responsible for transfer of the methyl group of 5-methyltetrahydrofolate to homocysteine, resulting in methionine for the SAM cycle in N. crassa (34). Three protein spots, labeled in Figure 1A and B, were identified by LC–MS/MS as the same MET-8 protein (Figure 1C). To further confirm that these three protein spots were MET-8, we generated a MET-8 polyclonal antibody and performed western blotting to probe the PVDF membrane that held proteins transferred from the 2D gel of the wild-type sample. As shown in Figure 1D, the MET-8 antibody signal fits with the three protein spots identified as MET-8 by LC–MS/MS, which suggests that the protein spots may represent post-translational modifications or other isoforms of MET-8.
Figure 1.
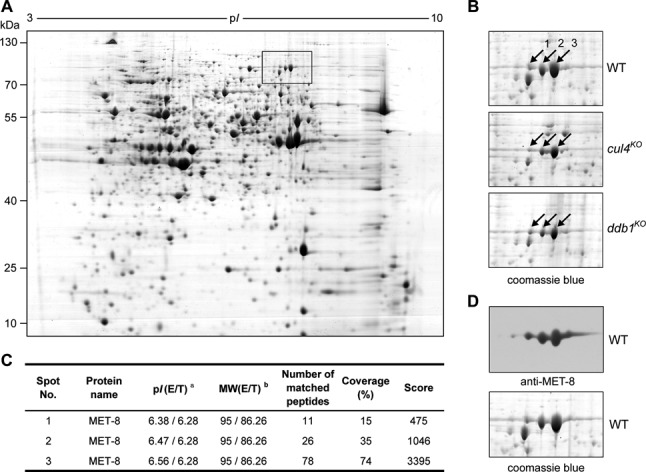
Expression of methionine synthase (MET-8) is decreased in cul4KO and ddb1KO strains. (A) Total protein extracts from N. crassa wild-type (WT) cells were separated by 2D gel electrophoresis. The rectangle shows the location of MET-8 proteins in the 2D map. (B) Expression pattern of MET-8 proteins in WT, cul4KO and ddb1KO strains. The arrows indicate the spots of MET-8 proteins identified by LC–MS/MS. (C) Identification of MET-8 proteins in N. crassa using LS-MS/MS. E: experimental values obtained directly from gels; T: theoretical values obtained from ExPASy search for full-length precursor proteins; MW: molecular weight (kDa). (D) Protein spots of MET-8 in 2D gels were confirmed by western blot analysis using antiserum against MET-8. Five protein spots were found on a PVDF membrane (top panel) containing proteins transferred from a 2D gel with WT sample (bottom panel) stained by CBB R-350.
Since the Cul4-based histone methyltransferase complex is essential for H3K9me3 and heterochromatin formation in N. crassa, we next tested whether the other members of this complex were involved in regulation of MET-8. To compare levels of MET-8 proteins among the wild-type, cul4KO and ddb1KO strains, we performed side-by-side comparisons of cultures harvested after growing under the same conditions. As shown in Figure 2A, protein levels of MET-8 were lower in each of the mutants compared to the wild-type strain, indicating that the Cul4-based histone methyltransferase complex was a positive regulator of MET-8 expression. The low MET-8 levels in the mutants suggest that either MET-8 protein was less stable or transcription of met-8 was decreased in the absence of each subunit. To test these possibilities, we compared the stability of MET-8 in wild-type, cul4KO and ddb1KO strains after addition of cycloheximide (CHX). MET-8 was very stable in the wild-type and mutant strains (Figure 2B and C), indicating that the low levels of MET-8 in mutants were not due to an increase in the degradation of MET-8 protein. We next measured met-8 mRNA in the mutant and wild-type strains by northern blot analysis. As shown in Figure 2D and Supplementary Figure S2, northern blot analyses revealed that levels of met-8 mRNA were reduced in each of the deletion strains compared to the wild-type strain, indicating that the expression of met-8 was affected in these mutants.
Figure 2.
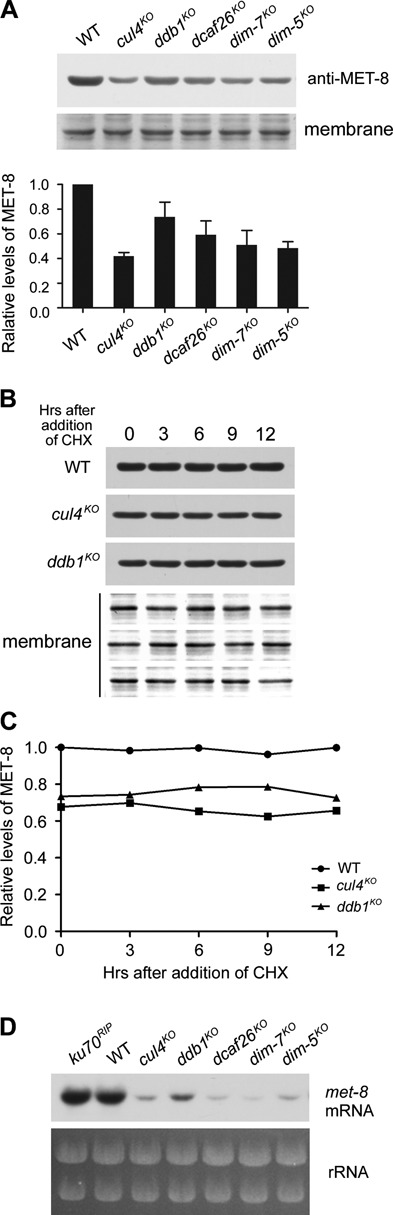
Highest expression of met-8 requires Cul4-based histone methyltransferase complex. (A) Representative results of western blot analyses of MET-8 in wild-type, cul4KO, ddb1KO, dcaf26KO, dim-7KO and dim-5KO strains. Stained PVDF membranes with Amido Black showing that the lanes were loaded with equal amounts of protein (top panel). Densitometric analyses from four independent experiments are shown in the bottom panel. (B) Western blot analyses showing stability of MET-8 protein in WT, cul4KO and ddb1KO strains after the addition of CHX (10 μg/ml). (C) Densitometric analyses from four independent experiments showing degradation of MET-8. (D) Northern blot analysis showing levels of met-8 mRNA in WT, ku70RIP, cul4KO, ddb1KO, dcaf26KO, dim-7KO and dim-5KO strains. The northern blot analysis was repeated five times.
Since the Cul4-based histone methyltransferase complex generally plays an important role in regulating histone H3K9 trimethylation at AT-rich DNA regions that are potentially packaged into heterochromatin (5,6,17–19), these results suggest that defects in heterochromatin formation may affect the proper expression of met-8.
Proper formation of heterochromatin is required for high expression of met-8
A crucial step in heterochromatin assembly is the binding of HP1 to the trimethylated H3K9 produced by DIM-5. Several studies have shown that HP1 proteins play important roles in heterochromatin formation and regulation of gene expression, but they contribute little to H3K9me3 (14,35). We compared levels of MET-8 between the wild-type strain and the hpo deletion mutant, which is deficient for HP1. MET-8 levels were decreased in the hpoKO strain compared to the wild-type and ku70RIP strains (Figure 3A). To further investigate whether proper formation of heterochromatin is required for high expression of met-8 in N. crassa, we measured the levels of met-8 mRNA in the hpoKO, wild-type, and ku70RIP stains. As in the cul4KO strain, levels of met-8 mRNA in the hpoKO strain were dramatically decreased compared to the wild-type and ku70RIP strains (Figure 3B and Supplementary Figure S2). In N. crassa, HP1 is known to be involved in DNA methylation by recruiting the DNA methyltransferase DIM-2 to chromatin (14). To examine whether proper DNA methylation is required for high expression of met-8 in N. crassa, we measured the levels of MET-8 protein and met-8 mRNA in dim-2KO and wild-type strains. Both MET-8 protein levels (Figure 3C) and met-8 mRNA levels (Figure 3D) in the dim-2KO strain were comparable to those in the wild-type strain. Together, these results indicate that proper formation of heterochromatin, not proper DNA methylation, is required for expression of met-8 in N. crassa.
Figure 3.
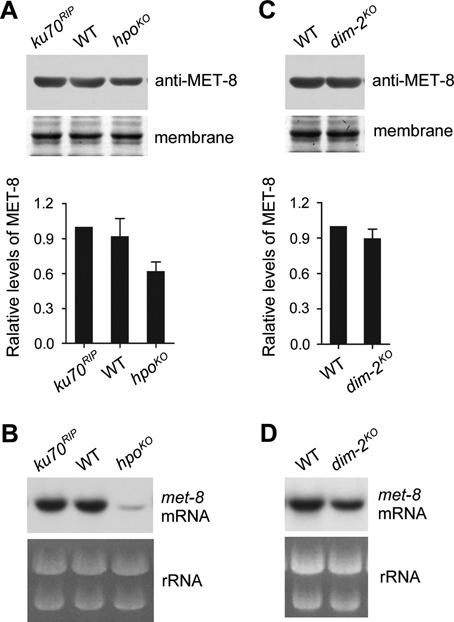
Proper formation of heterochromatin is required for the highest expression of met-8. (A) Results of western blot analysis of MET-8 (top panel) and the densitometric analyses from four independent experiments (bottom panel) in WT, ku70RIP and hpoKO strains. (B) Northern blot analysis showing met-8 mRNA in WT, ku70RIP and hpoKO strains. (C) Results of western blot analysis of MET-8 (top panel) and the densitometric analyses from four independent experiments (bottom panel). (D) Northern blot analysis showing met-8 mRNA in WT and dim-2KO strains.
Proper establishment of H3K9me3 is required for the highly efficient expression of met-8
Previous studies showed that N. crassa DIM-5 is essential for histone H3K9me3 and heterochromatin formation (15,19,35). To determine whether this modification was involved in maintaining high levels of met-8 expression, we generated a mutant strain with an amino acid substitution (lysine 9 to arginine; hereafter referred to as the H3K9R mutant). This mutation mimics loss of trimethylation on H3K9. The N. crassa genome has a single histone H3 gene (NCU01635) (36), which facilitates testing of mutations in this gene. To obtain an H3K9R mutant, we made a knock-in cassette containing the mutated histone H3 gene and a hygromycin resistance gene (hph) inserted downstream of the H3 gene 3′ untranslated region (UTR) (Figure 4A). This cassette was then transformed into a ku70RIP genetic background strain, and hph-resistant transformants were selected on a plate with hygromycin B. PCR analysis confirmed the integration of the knock-in cassette at the endogenous H3 gene locus, and the homokaryotic nature of the H3K9R strains was ensured by microconidia purification and germination of ascospores from crossing H3K9R mutant (bd, a) to 301–6 strain (bd, A) (Supplementary Figure S3). DNA sequencing of the endogenous H3 gene further confirmed the mutated H3K9R and the presence of a nonsense mutation for the introduction of a SalI site in these strains (Figure 4B and Supplementary Figure S3A). We tested the levels of histone H3 or trimethylated H3K9 using western blot analysis in wild-type, ku70RIP, cul4KO, dim-5KO and H3K9R strains. As shown in Figure 4C, all of the strains had similar levels of histone H3. In contrast to the robust H3K9 trimethylation in the wild-type and ku70RIP strains, no trimethylated H3K9 was detected in the H3K9R mutant or in the cul4KO and dim-5KO strains (Figure 4C). This confirmed that the H3K9R mutant strains had lost H3K9me3. Moreover, the H3K9R mutants exhibited dense, cauliflower-like growth patterns with abnormal hyphae and asexual spores on plates, similar to the phenotypes of the cul4KO and dim-5KO strains (Figure 4D and Supplementary Figure S3B) (17,18). The H3K9R strain exhibited slow growth rates on race tubes compared to the wild-type strain (Figure 4E), the growth rate being similar to that of the cul4KO and dim-5KO strains. These results suggest that the lost H3K9 trimethylation contributed to most of the defective phenotypes we observed in strains with mutations in the Cul4-based histone methyltransferase complex in N. crassa. Next, we measured levels of met-8 mRNA in the H3K9R, ku70RIP and wild-type strains. Northern blot analyses showed that the level of met-8 mRNA in the H3K9R mutants was dramatically decreased compared to those in the ku70RIP and wild-type strains (Figure 4F and Supplementary Figure S3C), demonstrating that H3K9 trimethylation is required for the high expression of met-8 in N. crassa.
Figure 4.
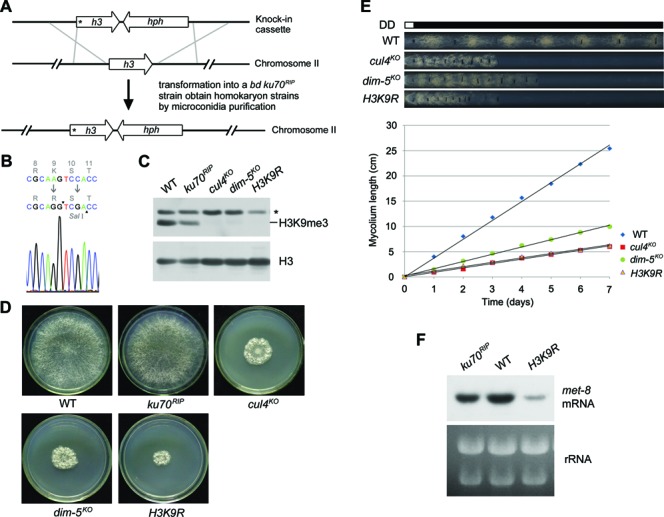
Mutation of histone H3 results in decreased expression of met-8. (A) Diagram depicting procedures used to create the homokaryotic H3K9R knock-in strain by homologous recombination. The asterisk in the H3K9R ORF indicates the location of the Lys9-to-Arg mutation at the N terminus of H3. (B) Diagram showing sequencing of the point mutation (from codon AAG to AGG) and a nonsense mutation (from codon TCC to TCG) in serine10 to introduce a SalI site in the H3K9R mutant. Sequencing result from an H3K9R strain showing that it is a homokaryotic H3K9R knock-in strain. (C) Western blot analysis of global H3 and H3K9 trimethylation in the WT, ku70RIP, cul4KO, dim-5KO and H3K9R strains. (D) Phenotypes of aerial hyphae and conidia of wild-type, ku70RIP, cul4KO, dim-5KO and H3K9R strains on plates (30°C, 32 h). (E) Growth rates of wild-type, cul4KO, dim-5KO and H3K9R strains, measured at 25°C by race tube assays in constant darkness after 1 day of light treatment. (F) Northern blot analysis showing met-8 mRNA levels in wild-type, ku70RIP and H3K9R strains.
Location and organization of the met-8 gene in linkage group III of N. crassa
To determine why the proper formation of heterochromatin is required for the high-efficiency expression of met-8 in N. crassa, we explored its chromosomal location and nearby DNA sequence features in the N. crassa database (http://www.broadinstitute.org/annotation/genome/neurospora/). We found that the met-8 (NCU06512) gene was located in the left arm of linkage group III (located in Supercontig 3: 1698305–1701771) between the NCU06511 and NCU06171 genes. Unexpectedly, immediately upstream of the met-8 promoter, there was an ∼50-kb AT-rich DNA region located between met-8 and NCU06171 (Figure 5A and B). In N. crassa, histone H3K9 is always trimethylated at AT-rich DNA regions (35), thus histone H3K9 at this 50-kb AT-rich region upstream of met-8 is likely to be trimethylated. We carried out a series of ChIP assays using antisera against trimethylated H3K9 and oligonucleotide primer pairs (1–5; see Figure 5B) designed to target the area from the met-8 promoter region to the middle of the proximal 50-kb AT-rich region in the wild-type strain and each mutant strain. qPCR with primer pairs 1 or 2 and immunoprecipitated DNA from the wild-type or any of the mutant strains did not generate product signals, indicating that there was no H3K9me3 at the met-8 promoter region (Figure 5C and D). However, as revealed by primer pairs 3–5, H3K9me3 was dramatically enriched at the 50-kb AT-rich region in the wild-type and hpoKO strains, but not in the other mutants (Figure 5E–G). This result indicates that histone H3K9 trimethylation of the 50-kb AT-rich region began at the region between primer pairs 2 and 3. Our ChIP data demonstrate that the 50-kb AT-rich DNA region was modified by histone H3K9 trimethylation and that this region may form heterochromatin.
Figure 5.
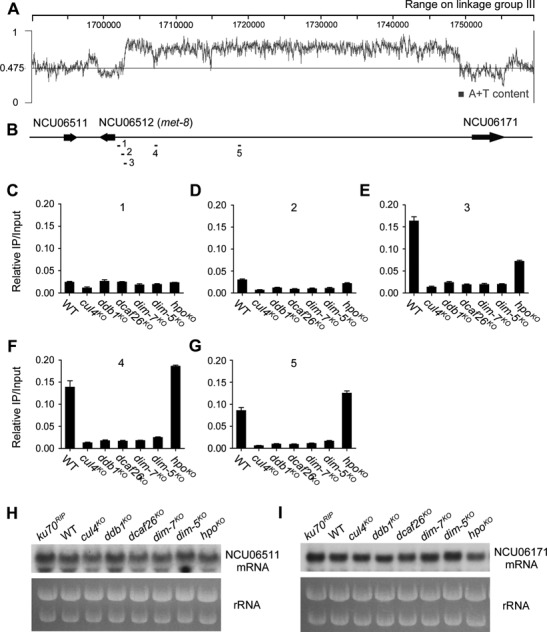
Localization and organization of the met-8 gene on linkage group III of N. crassa. (A) and (B) Schematic depicting a 50-kb AT-rich DNA region located between met-8 and NCU06171 on linkage group III of N. crassa genome. Bars (primer pairs 1–5) above the schematic indicate the regions tested by qPCR in ChIP assays. (C)–(G) ChIP analyses showing enrichment of H3K9 trimethylation at different regions upstream of met-8 in wild-type, cul4KO, ddb1KO, dcaf26KO, dim-7KO, dim-5KO and hpoKO strains. The loci detected by ChIP are indicated in (B). (H) and (I) Northern blot analysis showing levels of NCU06171 and NCU06511 mRNA in wild-type, ku70RIP, cul4KO, ddb1KO, dcaf26KO, dim-7KO, dim-5KO and hpoKO strains.
To confirm whether proper heterochromatin formation in the 50-kb AT-rich region is required for high met-8 expression specifically, we measured the expression of NCU06511 and NCU06171, which are located in the vicinity of the 50-kb AT-rich region. Northern blot analyses showed that the levels of NCU06511 and NCU06171 mRNA expressed in the mutants were comparable to those in the wild-type strain (Figure 5H and I), indicating that proper formation of the 50-kb AT-rich heterochromatin region only affects met-8 expression. Taken together, these results strongly suggest that the 50-kb AT-rich heterochromatin region may play an important role in regulating met-8 expression.
Heterochromatin positively regulates mig-6 gene expression
We tried to delete the 50-kb AT-rich DNA region at the endogenous location by replacement with the hph gene to see whether it was indeed responsible for activating met-8 expression. However, we could only obtain heterokaryon strains, which showed no visible change in met-8 mRNA levels. Next, we wondered whether other heterochromatins can activate proximal gene expression other than the met-8 gene in the Neurospora genome. After screening a set of heterochromatin-nearby genes, we found the mig-6 gene (NCU09285) located nearby a 4.2-kb AT-rich region on linkage group VII (Figure 6A). A previous study showed that this 4.2-kb AT-rich region (referred to as peak3) enriches peaks of H3K9me3 and DNA methylation (35). Northern blot analyses revealed that levels of mig-6 mRNA were reduced in each of the heterochromatin-defective strains compared to wild-type (Figure 6B). Moreover, deletion of its upstream heterochromatin region led to a decrease of mig-6 mRNA levels compared to that in the wild-type strain (Figure 6C). Consistent with the regulation of met-8 expression, these results confirm that the 4.2-kb AT-rich heterochromatin region positively regulates mig-6 gene expression.
Figure 6.
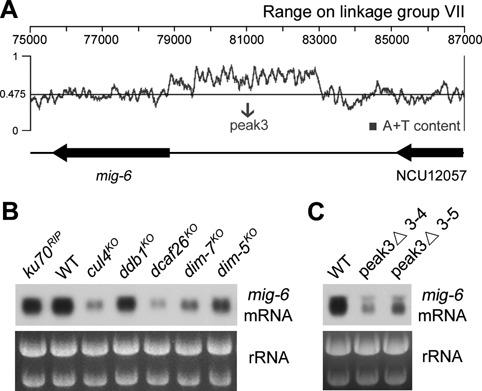
High expression of the mig-6 gene depends on its proximal 4.2-kb AT-rich heterochromatin region. (A) Schematic depicting a 4.2-kb AT-rich DNA region (peak3) located between mig-6 and NCU12057 on linkage group VII of N. crassa genome. (B) Northern blot analysis showing levels of mig-6 mRNA in WT, ku70RIP, cul4KO, ddb1KO, dcaf26KO, dim-7KO and dim-5KO strains. (C) Northern blot analysis showing levels of mig-6 mRNA in the WT and AT-rich region peak3 deletion strains.
Production of ectopically expressed met-8 is inefficient at the his-3 locus but efficient at downstream of heterochromatic regions
To investigate a direct correlation between met-8 expression and its proximal 50-kb AT-rich heterochromatin region, we checked met-8 expression in an environment lacking this region by cloning the entire met-8 gene, integrating it into the his-3 locus and detecting the output in different mutant strains. Bioinformatics analysis of the met-8 promoter by TESS (37) showed that its core region was about 1.0 kb from the start codon to the boundary (between primer pairs 2 and 3) of the proximal 50-kb AT-rich region (Figure 7A). To confirm this prediction, ChIP assays with antibody 8WG16 against the CTD repeats of the Pol II Rbp1 subunit were performed to measure the enrichment of Pol II on the met-8 promoter. As shown in Figure 7B, enrichment of Pol II was dramatically increased at the primer pair 1 region, but not at the other regions. Consequently, the 1.0-kb core region and the 1.0-kb core region plus 500 bp of AT-rich sequences were cloned as promoters for ectopic met-8 expression. Full-length open reading frames and the 3′ UTR of met-8 were amplified from genomic DNA and cloned together into both the pmet-8(1.0-kb)-5Myc-6His and pmet-8(1.5-kb)-5Myc-6His vectors (Figure 7C). We then introduced pmet-8(1.0-kb)- or pmet-8(1.5-kb)-driven Myc-tagged MET-8 constructs into the his-3 locus of 301–6 (bd, his-3, A) and dim-7KO (bd, his-3) strains. As shown in Figure 7D, the expression levels of Myc-MET-8 at the his-3 locus of wild-type or dim-7KO strains driven by met-8 promoters of different lengths were comparable, indicating that the defect in heterochromatin formation had no effect on ectopic expression of met-8. Next, we performed western blot analyses using antiserum against MET-8 proteins in these transformants to compare expression between the ectopic met-8 gene and endogenous met-8 gene in wild-type and dim-7KO strains. As shown in Figure 7E, the outputs of ectopic MET-8 were much lower than endogenous MET-8. Densitometry analyses revealed that the difference in protein expression between ectopic and endogenous MET-8 was at least 3-fold in the wild-type background (Figure 7F). Interestingly, when we introduced the pmet-8(1.0-kb)-driven Myc-tagged MET-8 construct with a 4-kb AT-rich sequence from its upstream 50-kb AT-rich region into the his-3 locus of 301–6 (bd, his-3, A) strain, the outputs of ectopic myc-met-8 gene in transformants were higher than those of the pmet-8(1.0-kb)-driven Myc-MET-8 construct (Supplementary Figure S4), suggesting that the ectopic expression of met-8 genes was positively regulated by this 4-kb AT-rich region.
Figure 7.
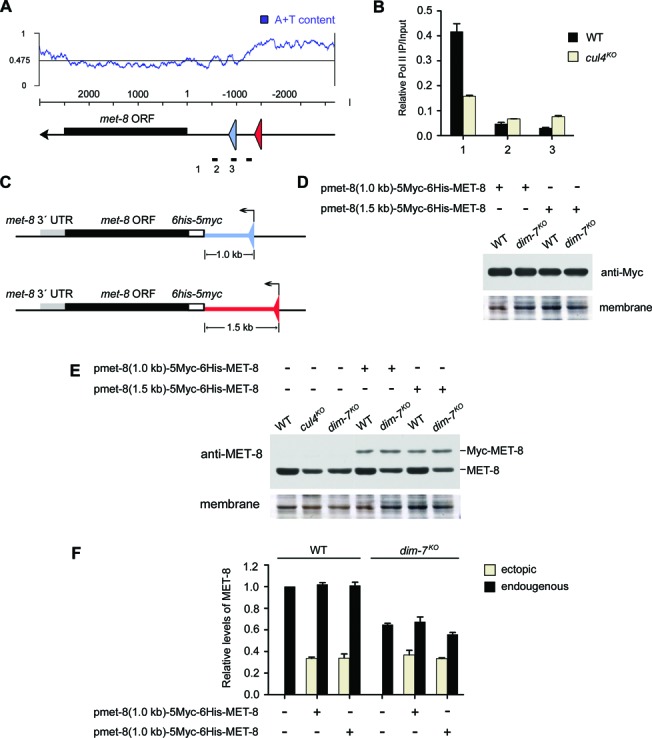
The highest expression of the met-8 gene requires its proximal 50-kb AT-rich heterochromatin region. (A) Schematic representation of the promoter region of met-8. The proximal triangle indicates the start site of the 1.0-kb-length promoter; the distal triangle indicates the start site of the 1.5-kb-length promoter, which includes an ∼500-bp AT-rich DNA fragment. (B) ChIP analysis showing recruitment of RNA polymerase II at different regions of the met-8 promoter in WT and cul4KO strains. (C) Schematic representation of two constructs with different promoter lengths for ectopic met-8 expression. The black box is the MET-8 coding region, the gray box is its 3′ untranslated region, the blank box is the 5Myc-6His tag sequence, the upstream bar is the 1.0-kb-length promoter or the 1.5-kb-length promoter. (D) Western blot analyses were performed using antibodies against c-Myc to show MET-8 expression of pmet-8(1.0-kb)-5Myc-6His-MET-8 or pmet-8(1.5-kb)-5Myc-6His-MET-8 at the his-3 locus of 301–6 (bd, his-3, A) or dim7KO (bd, his-3) transformants, respectively. (E) Western blot analyses were performed using antiserum against MET-8 to show protein expression levels of ectopic and original met-8 genes in 301–6 (bd, his-3, A) or dim7KO (bd, his-3) transformants. (F) Densitometric analyses from four independent experiments showing the western blot analysis of (E).
We also wondered whether the expression of the met-8 gene can be stimulated by other heterochromatin regions. To test this possibility, we examined the Neurospora genome and focused on two AT-rich DNA regions located in linkage group III (Figure 8A, top panel) and linkage group I (Figure 8A, bottom panel), respectively. ChIP assays confirmed the enrichment of trimethylated H3K9 in these regions (Figure 8B), indicating that they are potential heterochromatin domains.
Figure 8.
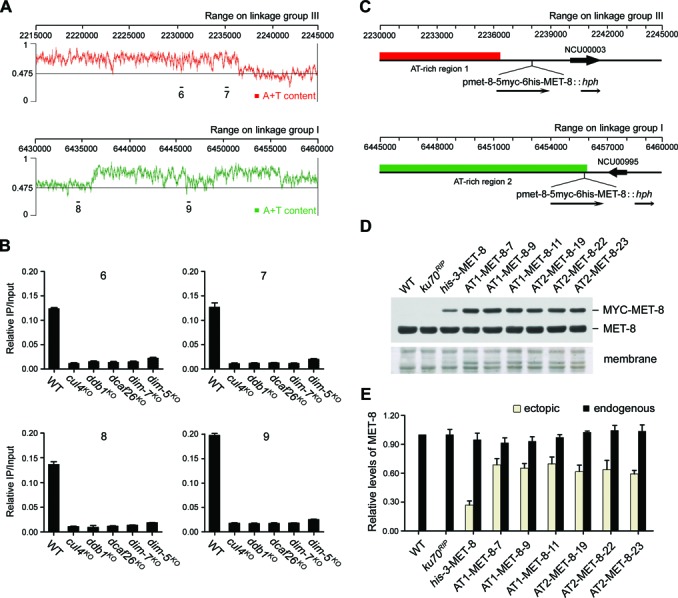
Ectopically expressed met-8 can be stimulated by its upstream heterochromatic domains. (A) Schematic depicting two AT-rich DNA regions located on linkage group III (top panel) and linkage group I (bottom panel) of N. crassa genome, respectively. Bars (primer pairs 6–9) above the schematic indicate the regions used by qPCR in ChIP assays. (B) ChIP analyses showing enrichment of H3K9 trimethylation at the AT-rich region 1 or region 2 in wild-type, cul4KO, ddb1KO, dcaf26KO, dim-7KO and dim-5KO strains. (C) Schematic showing the loci where the pmet-8(1.0-kb)-5Myc-6His-MET-8::hph cassette integrated into the downstream of AT-rich region 1 or AT-rich region 2. (D) Western blot analyses using antiserum against MET-8 showing protein levels of ectopic myc-met-8 genes integrated into the his-3 locus (his-3-MET-8) and the nearby AT-rich region 1 (AT1–MET-8) or AT-rich region 2 (AT2–MET-8), and endogenous MET-8 levels, respectively. (E) Densitometric analyses from four independent experiments showing the western blot analysis of (D).
We next generated two knock-in cassettes containing pmet-8(1.0-kb)-5Myc-6His-MET-8 and the selectable marker hph targeting downstream of each previously mentioned region. These cassettes were then transformed into a ku70RIP genetic background strain, respectively. The homokaryon pmet-8(1.0-kb)-5Myc-6His-MET-8::hph strains integrated downstream of AT-rich region 1 or region 2 were obtained by microconidia purification (hereafter referred to as AT1-MET-8 and AT2-MET-8, respectively) (Figure 8C). As shown in Figure 8D and E, the outputs of ectopic met-8 gene downstream of AT-rich region 1 or region 2 were much higher than those at the his-3 locus, indicating that the ectopic expression of met-8 genes was positively regulated by these two proximal heterochromatin domains. Interestingly, the outputs of ectopic myc-met-8 gene downstream of AT-rich region 1 or region 2 in ddb1KO strains were lower than those in ku70RIP genetic background strains (Supplementary Figure S5), further confirming that the ectopic expression of met-8 genes was positively regulated by the proper formation of its proximal heterochromatin domains.
Taken together, our experiments indicate that peak production of protein encoded by the met-8 gene is dependent on its proximal AT-rich heterochromatin regions.
Defects in heterochromatin formation affect H3K4me3 and recruitment of RNA Pol II at the met-8 5′ UTR
To provide a possible mechanistic explanation for how met-8 expression is regulated by its proximal 50-kb AT-rich heterochromatin, we first checked the density of histone H3 at the met-8 promoter region in wild-type and mutant strains. ChIP assays showed that the density of H3 at the met-8 promoter region was comparable in wild-type and mutant strains (Figure 9A), indicating that the defect in heterochromatin formation had no effect on nucleosome assembly at the met-8 5′ UTR.
Figure 9.
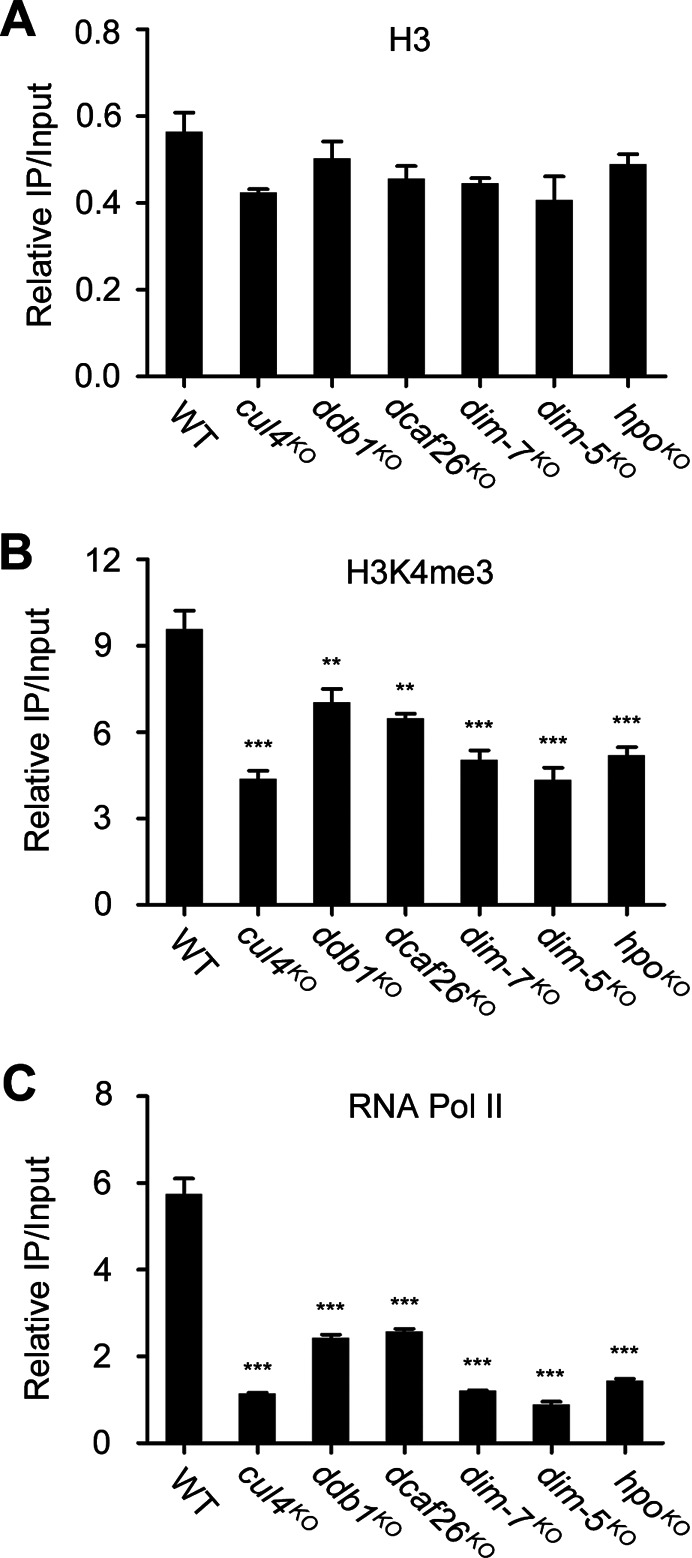
H3K4me3 and recruitment of RNA Pol II at met-8 5′ UTR are decreased in heterochromatin-defective strains. ChIP analysis shows the nucleosome density (A), H3K4 trimethylation (B) and recruitment of Rpb1 (C) at the met-8 5′ untranslated region in wild-type, cul4KO, ddb1KO, dcaf26KO, dim-7KO, dim-5KO and hpoKO strains. Significance was assessed compared with wild-type strains using a two-tailed t-test. **P < 0.01 and ***P < 0.001. Error bars indicate the SD (n = 3).
Histone H3 lysine 4 trimethylation (H3K4me3) occurs in the promoter regions and 5′ UTR region of actively transcribed genes, and it is associated with transcription initiation. We next carried out ChIP assays to compare H3K4me3 at the met-8 5′ UTR region in wild-type and mutant strains. As shown in Figure 9B, levels of H3K4me3 were enriched at the 5′ UTR region of met-8 in the wild-type strain, whereas levels of this modification were decreased at this region in mutant strains, indicating that histone modifications at met-8 were altered in mutant strains. These results suggest that the defect in met-8 proximal heterochromatin affected histone modifications of the met-8 gene. Considering the distribution of H3K4me3, which serves as a marker of active genes, these results may explain the downregulation of met-8 in mutants. ChIP assays with antibody 8WG16 were carried out in wild-type and mutant strains to examine the enrichment of RNA Pol II at the 5′ UTR of met-8. In the absence of Cul4 or each of the other subunits, enrichment of RNA Pol II at met-8 promoter regions was lower compared to wild-type strains (Figure 9C), suggesting that the defect in heterochromatin formation affected the recruitment of RNA Pol II to the met-8 promoter in mutants. Consistent with the decreased expression of met-8 in the mutant strains, these results indicate that proper formation of heterochromatin is required for proper histone modifications of met-8 and recruitment of RNA Pol II to the met-8 promoter.
DISCUSSION
N. crassa, a filamentous fungus, is a powerful, genetically tractable system for studies of heterochromatin formation and gene expression. In this study, we found that a heterochromatic region was necessary for the high-efficiency expression of met-8, contradicting the well-accepted notion that heterochromatic regions create repressive environments for gene silencing by recruiting certain components (38). The met-8 gene has an ∼50-kb AT-rich DNA region contiguous with its promoter, which forms heterochromatin and positively regulates met-8 expression. In the absence of the proximal 50-kb AT-rich DNA region, expression of met-8 was dramatically decreased. Furthermore, we showed that mig-6 expression was directly regulated by its proximal 4.2-kb heterochromatin region. To the best of our knowledge, this is the first systematic functional analysis of heterochromatin positively regulating proximal gene expression in filamentous fungi.
In D. melanogaster, Howe et al. demonstrated that the level of light gene expression was correlated with the size of the heterochromatin block adjacent to that gene (23). In fact, it is well known that heterochromatic genes in D. melanogaster are repressed when moved to distal euchromatin by chromosomal rearrangements (39). This indicates that proximity to heterochromatin is an important regulatory requirement for the normal functioning of heterochromatic genes (22,25). Consistent with the observations in D. melanogaster, our results demonstrate that expression of the met-8 or mig-6 gene relies on their proximal heterochromatic region in N. crassa.
In D. melanogaster, the expression of several well-studied heterochromatic genes, including light and rolled, is compromised in a genetic background in which the HP1 (named SU(VAR)205 in D. melanogaster) has been reduced (26,40). Here, we show that peak expression of met-8 was compromised in a genetic background deficient for HP1 (hpoKO strain) in N. crassa. A major challenge is to understand how the molecular functions of SU(VAR) proteins that affect heterochromatic gene expression can be reconciled with the current models of SU(VAR)- and heterochromatin-mediated euchromatic gene silencing. A large-scale mapping analysis in Drosophila embryonic cells showed that HP1 protein is distributed throughout the concertina, light and rolled heterochromatic gene regions and that it binds both unique and repetitive sequences in intronic and exonic portions of the genes, respectively (41,42). These data suggest that genes around heterochromatin may have evolved a transcriptional dependence on factors that are known to silence gene expression. However, most Su(var) mutations are pleiotropic and most SU(VAR) proteins are multifunctional, making it more difficult to determine a clear-cut explanation for these phenomena.
To determine if the trimethylation at lysine 9 of H3 plays an important role in met-8 transcription, we created a H3K9R substitution strain. As in the cul4KO strain, expression of met-8 was dramatically decreased in the H3K9R mutants, indicating that the proper formation of heterochromatin is required for the highest-efficiency expression of met-8 in N. crassa. Consistent with the idea that SU(VAR) proteins affect expression of these genes by direct binding, our results clearly show the effects of these modifiers and that H3K9me3 directly targets heterochromatic regions, which in turn influences met-8 expression.
Another interesting question is how might the peak expression of met-8 be compatible with the known silencing properties of heterochromatin, and what factors account for the difference between functional and silent heterochromatin?
As we have mentioned, methionine synthase is a key enzyme for methionine synthesis in N. crassa, and highly expressed met-8 is required throughout all phases of the cell cycle. One explanation, therefore, is that the requirement for the continuous activity of MET-8 and similar gene products resulted in a robust adaptation strategy to promote continued expression in spite of repressive heterochromatic microenvironments. Our ChIP data revealed a sharp enrichment of H3K9me3 just beyond the met-8 promoter at the boundary of its proximal 50-kb AT-rich DNA region (between primers 2 and 3) in the wild-type strain, suggesting that there is a clear boundary between the met-8 promoter and its proximal heterochromatin. Therefore, we would expect that strong boundaries are required both to maintain the integrity of the heterochromatin domains that facilitate heritable gene silencing and to protect neighboring gene loci from pervasive spreading of heterochromatin.
In addition, in our study, although levels of ectopic expression of met-8 were dramatically lower than those of the original met-8 gene, western blot analyses with c-Myc antibody revealed high levels of Myc-MET-8 under normal growth conditions, indicating that the met-8 gene has a strong promoter, which is necessary to ensure that enough methionine synthase will be produced to generate sufficient methionine and SAM for cell viability. Several studies in yeast, fruit fly and human cells have shown that artificial recruitment of specific transcription activators, such as Reb1 and Gal4, to DNA elements is sufficient to block the spread of heterochromatin (43). Moreover, strong RNA polymerase II promoters can also insulate genes in S. cerevisiae and S. pombe, again without requiring transcription (43,44). Therefore, another plausible explanation is that met-8 gene is insulated from repressive heterochromatin by its strong promoter, essentially maintaining a microenvironment that is compatible with transcription.
Remarkably, our ChIP data revealed a sharp enrichment of RNA polymerase II at the met-8 promoter side of the met-8/50-kb heterochromatin boundary, coincident with the increased enrichment of H3K4me3 at this region. Consistent with our observations, a truncated human CD2 gene lacking part of its locus control region (LCR) showed variegated expression in the thymus when the transgene was integrated into or close to pericentromeric heterochromatin in mice (45). Surprisingly, a CD2 gene that contained the complete CD2 LCR yielded full nonvariegated expression when integrated into heterochromatin. Although this is a particularly striking result, since it contradicts the long-held view that heterochromatin has a dominant silencing effect on euchromatic genes that become associated with it, these results strongly indicate that insulation at these sites is a consequence of direct competition between the spreading heterochromatin and transcription factor binding coupled with chromatin remodeling at the barrier (46).
CONCLUSION
The findings of heterochromatin-mediated stimulation of gene expression expand our understanding of the function of heterochromatin and demonstrate that heterochromatic genes are evolutionarily conserved in eukaryotes. They also emphasize the importance of the chromosomal context in complex eukaryotic genomes. We predict that the mechanisms of heterochromatin-mediated stimulation of gene expression in eukaryotes will soon be revealed in future studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGMENTS
We thank Zhuo Shen for excellent technical assistance.
FUNDING
National Natural Science Foundation of China [31171208 to Q.H.]. Funding for open access charge: National Natural Science Foundation of China [31171208 to Q.H.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Plath K., Fang J., Mlynarczyk-Evans S.K., Cao R., Worringer K.A., Wang H., de la Cruz C.C., Otte A.P., Panning B., Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 2.Silva J., Mak W., Zvetkova I., Appanah R., Nesterova T.B., Webster Z., Peters A.H., Jenuwein T., Otte A.P., Brockdorff N. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev. Cell. 2003;4:481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 3.Grewal S.I., Elgin S.C. Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 2002;12:178–187. doi: 10.1016/s0959-437x(02)00284-8. [DOI] [PubMed] [Google Scholar]
- 4.Maison C., Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 2004;5:296–304. doi: 10.1038/nrm1355. [DOI] [PubMed] [Google Scholar]
- 5.Horn P.J., Bastie J.N., Peterson C.L. A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev. 2005;19:1705–1714. doi: 10.1101/gad.1328005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia S., Kobayashi R., Grewal S.I. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat. Cell Biol. 2005;7:1007–1013. doi: 10.1038/ncb1300. [DOI] [PubMed] [Google Scholar]
- 7.Horn P.J., Peterson C.L. Heterochromatin assembly: a new twist on an old model. Chromosome Res. 2006;14:83–94. doi: 10.1007/s10577-005-1018-1. [DOI] [PubMed] [Google Scholar]
- 8.Angers S., Li T., Yi X., MacCoss M.J., Moon R.T., Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- 9.Pickart C.M. Back to the future with ubiquitin. Cell. 2004;116:181–190. doi: 10.1016/s0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- 10.Higa L.A., Zhang H. Stealing the spotlight: CUL4-DDB1 ubiquitin ligase docks WD40-repeat proteins to destroy. Cell Div. 2007;2:5. doi: 10.1186/1747-1028-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson S., Xiong Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem. Sci. 2009;34:562–570. doi: 10.1016/j.tibs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He Y.J., McCall C.M., Hu J., Zeng Y., Xiong Y. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev. 2006;20:2949–2954. doi: 10.1101/gad.1483206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higa L.A., Wu M., Ye T., Kobayashi R., Sun H., Zhang H. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat. Cell Biol. 2006;8:1277–1283. doi: 10.1038/ncb1490. [DOI] [PubMed] [Google Scholar]
- 14.Honda S., Selker E.U. Direct interaction between DNA methyltransferase DIM-2 and HP1 is required for DNA methylation in Neurospora crassa. Mol. Cell. Biol. 2008;28:6044–6055. doi: 10.1128/MCB.00823-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamaru H., Selker E.U. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- 16.Freitag M., Hickey P.C., Khlafallah T.K., Read N.D., Selker E.U. HP1 is essential for DNA methylation in neurospora. Mol. Cell. 2004;13:427–434. doi: 10.1016/s1097-2765(04)00024-3. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Y., Shen Y., Yang S., Wang J., Hu Q., Wang Y., He Q. Ubiquitin ligase components Cullin4 and DDB1 are essential for DNA methylation in Neurospora crassa. J. Biol. Chem. 2010;285:4355–4365. doi: 10.1074/jbc.M109.034710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu H., Wang J., Hu Q., Quan Y., Chen H., Cao Y., Li C., Wang Y., He Q. DCAF26, an adaptor protein of Cul4-based E3, is essential for DNA methylation in Neurospora crassa. PLoS Genet. 2010;6, e1001132 doi: 10.1371/journal.pgen.1001132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis Z.A., Adhvaryu K.K., Honda S., Shiver A.L., Knip M., Sack R., Selker E.U. DNA methylation and normal chromosome behavior in Neurospora depend on five components of a histone methyltransferase complex, DCDC. PLoS Genet. 2010;6:e1001196. doi: 10.1371/journal.pgen.1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis Z.A., Adhvaryu K.K., Honda S., Shiver A.L., Selker E.U. Identification of DIM-7, a protein required to target the DIM-5 H3 methyltransferase to chromatin. Proc. Natl. Acad. Sci. U.S.A. 2010;107:8310–8315. doi: 10.1073/pnas.1000328107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devlin R.H., Bingham B., Wakimoto B.T. The organization and expression of the light gene, a heterochromatic gene of Drosophila melanogaster. Genetics. 1990;125:129–140. doi: 10.1093/genetics/125.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eberl D.F., Duyf B.J., Hilliker A.J. The role of heterochromatin in the expression of a heterochromatic gene, the rolled locus of Drosophila melanogaster. Genetics. 1993;134:277–292. doi: 10.1093/genetics/134.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howe M., Dimitri P., Berloco M., Wakimoto B.T. Cis-effects of heterochromatin on heterochromatic and euchromatic gene activity in Drosophila melanogaster. Genetics. 1995;140:1033–1045. doi: 10.1093/genetics/140.3.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schulze S.R., Sinclair D.A., Fitzpatrick K.A., Honda B.M. A genetic and molecular characterization of two proximal heterochromatic genes on chromosome 3 of Drosophila melanogaster. Genetics. 2005;169:2165–2177. doi: 10.1534/genetics.103.023341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wakimoto B.T., Hearn M.G. The effects of chromosome rearrangements on the expression of heterochromatic genes in chromosome 2L of Drosophila melanogaster. Genetics. 1990;125:141–154. doi: 10.1093/genetics/125.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hearn M.G., Hedrick A., Grigliatti T.A., Wakimoto B.T. The effect of modifiers of position-effect variegation on the variegation of heterochromatic genes of Drosophila melanogaster. Genetics. 1991;128:785–797. doi: 10.1093/genetics/128.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yasuhara J.C., Wakimoto B.T. Oxymoron no more: the expanding world of heterochromatic genes. Trends Genet. 2006;22:330–338. doi: 10.1016/j.tig.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 28.Verdel A., Jia S., Gerber S., Sugiyama T., Gygi S., Grewal S.I., Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dorer D.R., Henikoff S. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell. 1994;77:993–1002. doi: 10.1016/0092-8674(94)90439-1. [DOI] [PubMed] [Google Scholar]
- 30.Belden W.J., Larrondo L.F., Froehlich A.C., Shi M., Chen C.H., Loros J.J., Dunlap J.C. The band mutation in Neurospora crassa is a dominant allele of ras-1 implicating RAS signaling in circadian output. Genes Dev. 2007;21:1494–1505. doi: 10.1101/gad.1551707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y., Lam K.S., Chan L., Chan K.W., Lam J.B., Lam M.C., Hoo R.C., Mak W.W., Cooper G.J., Xu A. Post-translational modifications of the four conserved lysine residues within the collagenous domain of adiponectin are required for the formation of its high molecular weight oligomeric complex. J. Biol. Chem. 2006;281:16391–16400. doi: 10.1074/jbc.M513907200. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Z., Wang Y., Cai G., He Q. Neurospora COP9 signalosome integrity plays major roles for hyphal growth, conidial development, and circadian function. PLoS Genet. 2012;8:e1002712. doi: 10.1371/journal.pgen.1002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aronson B.D., Johnson K.A., Loros J.J., Dunlap J.C. Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science. 1994;263:1578–1584. doi: 10.1126/science.8128244. [DOI] [PubMed] [Google Scholar]
- 34.Borkovich K.A., Alex L.A., Yarden O., Freitag M., Turner G.E., Read N.D., Seiler S., Bell-Pedersen D., Paietta J., Plesofsky N., et al. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 2004;68:1–108. doi: 10.1128/MMBR.68.1.1-108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis Z.A., Honda S., Khlafallah T.K., Jeffress J.K., Freitag M., Mohn F., Schubeler D., Selker E.U. Relics of repeat-induced point mutation direct heterochromatin formation in Neurospora crassa. Genome Res. 2009;19:427–437. doi: 10.1101/gr.086231.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hays S.M., Swanson J., Selker E.U. Identification and characterization of the genes encoding the core histones and histone variants of Neurospora crassa. Genetics. 2002;160:961–973. doi: 10.1093/genetics/160.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schug J. Using TESS to predict transcription factor binding sites in DNA sequence. Curr. Protoc. Bioinform. 2008;Chapter 2:2.6.1–2.6.15. doi: 10.1002/0471250953.bi0206s21. [DOI] [PubMed] [Google Scholar]
- 38.Weiler K.S., Wakimoto B.T. Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- 39.Schultz J., Dobzhansky T. The relation of a dominant eye color in Drosophila melanogaster to the associated chromosome rearrangement. Genetics. 1934;19:344–364. doi: 10.1093/genetics/19.4.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu B.Y., Emtage P.C., Duyf B.J., Hilliker A.J., Eissenberg J.C. Heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in Drosophila. Genetics. 2000;155:699–708. doi: 10.1093/genetics/155.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Wit E., Greil F., van Steensel B. Genome-wide HP1 binding in Drosophila: developmental plasticity and genomic targeting signals. Genome Res. 2005;15:1265–1273. doi: 10.1101/gr.3198905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Wit E., Greil F., van Steensel B. High-resolution mapping reveals links of HP1 with active and inactive chromatin components. PLoS Genet. 2007;3:e38. doi: 10.1371/journal.pgen.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fourel G., Boscheron C., Revardel E., Lebrun E., Hu Y.F., Simmen K.C., Muller K., Li R., Mermod N., Gilson E. An activation-independent role of transcription factors in insulator function. EMBO Rep. 2001;2:124–132. doi: 10.1093/embo-reports/kve024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valenzuela L., Kamakaka R.T. Chromatin insulators. Annu. Rev. Genet. 2006;40:107–138. doi: 10.1146/annurev.genet.39.073003.113546. [DOI] [PubMed] [Google Scholar]
- 45.Festenstein R., Tolaini M., Corbella P., Mamalaki C., Parrington J., Fox M., Miliou A., Jones M., Kioussis D. Locus control region function and heterochromatin-induced position effect variegation. Science. 1996;271:1123–1125. doi: 10.1126/science.271.5252.1123. [DOI] [PubMed] [Google Scholar]
- 46.Valenzuela L., Dhillon N., Kamakaka R.T. Transcription independent insulation at TFIIIC-dependent insulators. Genetics. 2009;183:131–148. doi: 10.1534/genetics.109.106203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


