Abstract
For toxin/antitoxin (TA) systems, no toxin has been identified that functions by cleaving DNA. Here, we demonstrate that RalR and RalA of the cryptic prophage rac form a type I TA pair in which the antitoxin RNA is a trans-encoded small RNA with 16 nucleotides of complementarity to the toxin mRNA. We suggest the newly discovered antitoxin gene be named ralA for RalR antitoxin. Toxin RalR functions as a non-specific endonuclease that cleaves methylated and unmethylated DNA. The RNA chaperone Hfq is required for RalA antitoxin activity and appears to stabilize RalA. Also, RalR/RalA is beneficial to the Escherichia coli host for responding to the antibiotic fosfomycin. Hence, our results indicate that cryptic prophage genes can be functionally divergent from their active phage counterparts after integration into the host genome.
INTRODUCTION
Toxin/antitoxin (TA) systems are widespread among prokaryotes (1). Five different types of TA systems have been characterized, depending on the interaction of the TA and the nature of the antitoxin (2,3). For type I systems, an RNA antitoxin interacts with the toxin transcript and inhibits translation of the toxic protein (4). The toxins and antitoxins of type II systems interact through direct protein–protein binding (4). Type III systems rely upon the direct interaction of an RNA antitoxin with the toxin protein (5). A type IV designation has been proposed for a TA system in which the protein antitoxin does not interact with the toxin directly but suppresses the toxicity of the toxin by stabilizing its target (6), and a type V designation has been proposed in which the proteic antitoxin cleaves specifically the mRNA of the toxin to prevent the translation of the toxin (7,8).
In Escherichia coli, most well-studied TA systems belong to type II TA systems, in which the labile proteic antitoxin binds to the more stable toxin and inhibits its activity. Type I and type III systems are less well studied probably due to the delay in the identification of small RNAs (sRNAs) (9). However, in recent years, bioinformatic searches for putative type I TA systems based on tandem copies of the full loci and the presence of transmembrane domains has led to the identification of multiple copies of potential type I loci within new hosts across 774 bacterial genomes (10).
The role of TA systems in cell physiology, specifically in biofilm formation (11,12), persister cell formation (13,14), the general stress response (15,16), phage inhibition (17,18), and differential mRNA decay (19,20) is becoming more clear. Type II toxins with mRNA endoribonuclease activity usually alter gene regulation by cleaving specific mRNAs (21). Type I toxin TisB and type V toxin GhoT cause membrane damage when overproduced and are stress-response elements that are actively involved in persistence (7,8,22). Type I toxin Hok utilizes post-segregational killing to stabilize the R1 plasmid (23), but most of the Hok-like toxins in the E. coli K-12 chromosome appear inactive (24). Another major group of plasmid-based toxins function as gyrase inhibitors, and homologues of the plasmid RK2 ParE/ParD TA system in the Vibrio cholerae genome help maintain the integrity of the two chromosomes (25). These results indicate that chromosomal toxins are functionally divergent from plasmid loci.
Prophages or prophage remnants carrying toxic genes have been found to harbor TA systems, and five pairs have been reported in E. coli K12: RelE/RelB (26), RnlA/RnlB (27), YpjF/YfjZ (28), YkfI/YafW (28) and CbtA/YeeU (28). Furthermore, two interesting protein toxins have been described in cryptic prophage rac in E. coli K 12. KilR is a toxic peptide that inhibits cell division by inhibiting FtsZ (29). It has been suggested that RalR alleviates restriction modification possibly to protect the bacterial chromosome when recombination generates unmodified products by the same mechanism as Ral of phage lambda (30,31). However, the amino acid identity between the Ral protein in phage lambda and the RalR protein in rac prophage is very low (24%) (30) and the function of RalR remains unclear. Lambdoid prophage rac has lost about 60% of its original DNA (32), and ralR is differentially regulated in the development of E. coli biofilms (33). Also, ydaC, downstream of ralR, has been named as rcbA for its ability to reduce the frequency of double-strand chromosome breaks (34). However, it remains unclear whether rcbA encodes RNA or protein (34), or whether ydaC is transcribed.
In this study, unexpectedly, we found that RalR does not inhibit restriction modification but instead is part of a toxin/antitoxin system and functions as a non-specific DNase. Furthermore, we found that the adjacent gene product, a trans-encoded sRNA RalA (for RalR antitoxin), functions as an antitoxin for RalR. The ralR and ralA genes are adjacent but in the opposite orientation, and RalA RNA has 16 nucleotides of complementarity to the coding region of RalR mRNA. We show that RalA RNA interacts with the mRNA of RalR via base-pairing, thus preventing the translation of RalR. The activity of antitoxin RalA requires RNA chaperone Hfq. Thus, RalR/RalA belongs to a type I TA system where the antitoxin sRNA interferes with translation of the toxin mRNA via the 16-nt base-pairing.
MATERIALS AND METHODS
Bacterial strains, plasmids and growth conditions
The isogenic E. coli BW25113 K-12 strains and plasmids used in this study are listed in Table 1. For deleting and overexpressing single genes, the Keio collection (35) and the ASKA library (36) were used. The ΔralRA mutant strain was created using the λ Red method (37) using primers DralRA-f and DralRA-r (Supplementary Table S1). The kanamycin resistance cassette from the Keio and ΔralRA mutants was removed as previously described to ensure that only the impact of the deleted gene was studied (38). The ΔralR and ΔralRA mutations were verified by amplifying chromosomal DNA using the primers CralRA-f and CralRA-r, or CralRA-f2 and CralRA-r2, respectively, and sequencing the resulting fragments using CralRA-f (Supplementary Table S1) to also verify that genes upstream and downstream of the mutation (recT and ralR or ydaQ, respectively) were left intact. Removal of hfq was verified using the same method using primer pairs hfq-up and hfq-r, hfq-up and hfq-down, hfq-f and hfq-r, or hfq-f and hfq-down as described previously (35). All experiments were conducted at 37°C in Luria-Bertani medium (LB) (39) unless specified otherwise. Chloramphenicol (30 μg/ml) was used for maintaining pCA24N-based plasmids, and ampicillin (50 μg/ml) was used to maintain plasmid pBAD-ralA.
Table 1.
Bacterial strains and plasmids used in this study
| Bacterial strains/Plasmids | Description | Source |
|---|---|---|
| E. coli K12 BW25113 strains | ||
| Wild-type | rrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1 | (35) |
| Δrac | Whole prophage rac removed | (38) |
| Δhfq | Δhfq Δkan | This study |
| ΔralR | ΔralR Δkan | This study |
| ΔralRA | ΔralRA Δkan | This study |
| Plasmids | ||
| pCA24N | CmR; lacIq | (36) |
| pCA24N-ralR | CmR; lacIq, PT5-lac::ralR | (36) |
| pCA24N-ralA | CmR; lacIq, PT5-lac::ydac | (36) |
| pCA24N-ralR-ralA | CmR; lacIq, PT5-lac::ralR-ydaC | This study |
| pBAD-ralA | AmpR; PBAD::ralA+ | This study |
| pCA24N-hfq | CmR; lacIq, PT5-lac::hfq | (36) |
| Mutated pCA24N-ralR-ralA plasmids | This study | |
| pCA24N-ralR-ralA-G81CG83A | G to C at position 81 in ralA; G to A at position 83 in ralA | |
| pCA24N-ralR-ralA-G83A | G to A at position 83 in ralA | |
| pCA24N-ralR-ralA-G289del | Sequences removed after G at position 289 in ralR-ralA | |
| pCA24N-ralR-T189C-ralA | T to C at position 189 in ralR, changed putative ydaC start aa from Met to Thr, no change in RalR aa sequence | |
| pCA24N-ralR-ralA-C161A | C to A at position 161 in ralA, introduced a stop codon TAA at the ninth aa of putative YdaC | |
| pCA24N-ralR-ralA-G64C | G to C at position 64 in ralA | |
| pCA24N-ralR-ralA-T63C | T to C at position 63, no change in aa sequence | |
| pCA24N-ralR-ralA-M1 | Modified the 16-nt repeat in ralA to AGCCTCCTTCTTACCT, no change in aa of putative YdaC | |
| pCA24N-ralR-M2-ralA | Modified the 16-nt repeat in ralR, change aa from Gly-Ser-Glu-Lys-Glu-Ala to Gly-Ser-Ala-Ile-Ala-Glu | |
| pCA24N-ralR-M1-ralA-M1 | Modified the 16-nt repeat both in ralR and ralA to AAGTGAGAAGGAGGCT and AGCCTCCTTCTTACCT | |
| Mutated pCA24N-ralR plasmids | This study | |
| pCA24N-ralR-A154G | Encodes RalR-mutant RalR K52G | |
| pCA24N-ralR-M1 | Modified the 16-nt repeat in ralR to AAGTGAGAAGGAGGCT, no change in aa | |
| pCA24N-ralR-M2 | Modified the 16-nt repeat in ralR, change aa from Gly-Ser-Glu-Lys-Glu-Ala to Gly-Ser-Ala-Ile-Ala-Glu | |
| Note. Cm R and Amp R indicate chloramphenicol and ampicillin resistance, respectively, and aa indicates amino acid. | ||
Cloning of pCA24N-ralR-ralA, pCA24N-ralR-ralA-G289del and pBAD-ralA
The full-length gene region ralR-ralA and partial gene region ralR-ralA-G289 were cloned as described previously (36). They were polymerase chain reaction (PCR) amplified from E. coli K12 BW25113 chromosomal DNA using one front primer (p-ralR-f) and two different rear primers (p-ralA-r and p-ralR-ralA-G289del-r) (Supplementary Table S1). PCR products were phosphorylated using a kination kit (Takara, Dalian, China) and purified using a PCR product purification kit (Qiagen, Hilden, Germany). The purified products were then ligated into vector pCA24N that was digested with the StuI restriction enzyme (New England Biolabs, Beverly, MA, USA). The ligation mixture was transformed into BW25113 and the Δrac mutant. The constructs were verified by DNA sequencing using primer pair of p-f and p-r (Supplementary Table S1). Plasmid pBAD-ralA was constructed with the full-length gene ralA, and 124 bp upstream of ralA was controlled by the arabinose promoter using primer pair pBAD-ralA-f and -r (Supplementary Table S1) and cloning into NcoI–HindIII-digested pBAD/Myc-HisA (Invitrogen, Carlsbad, CA, USA).
Mutant library creation by epPCR
Error-prone PCR (epPCR) was conducted on plasmid pCA24N-ralR and pCA24N-ralR-ralA using primers epPCR-f and epPCR-r (Supplementary Table S1) as described earlier (40). The epPCR program was as follows: 94°C for 5 min, 30 cycles of 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C, followed by 10 min at 72°C final extension. The PCR products were gel-purified and digested using BseRI and HindIII before being ligated into pCA24N-ralR-ralA. The ligation mixture was transformed into BW25113 Δrac.
Single and multiple site-directed mutagenesis
Single site-directed mutagenesis (16) was used to mutate the putative start codon of ydaC from ATG to ACG using primer pair p-ralR-T189C-ralA-f and -r, and to introduce a stop codon TAA into the ninth putative coding region of ydaC in pCA24N-ralR-ralA using primer pair p-ralR-ralA-C161A-f and -r (Supplementary Table S1). For the 16-nt repeat in RalA, positions 63 (T to C) and 64 (G to C) were mutated using corresponding primer pairs. In addition, the AGC site in ralA was mutated to AAC using p-ralR-ralA-G83A-f and -r. Site-directed mutagenesis at multiple sites was applied to mutate the GAGC sequence into CAAC using p-ralR-ralA-G81CG83A-f and -r. Moreover, p-ralR-ralA-M1-f and -r were used to mutate the 16-nt repeat sequence of ralA on pCA24N-ralR-ralA without changing the potential amino acid sequence. p-ralR-M1-f and -r were used to mutate the 16-nt repeat in ralR with no amino acid changes in pCA24N-ralR and pCA24N-ralR-ralA, and p-ralR-M2-f and -r were used to mutate the 16-nt repeat in ralR in pCA24N-ralR and pCA24N-ralR-ralA with amino acid changes (Table 1). The correct mutations were verified by DNA sequencing.
Protein expression and purification
RalR, RalR-mutant, and Hfq each with six histidines and 10 additional cloning artifacts at the N-terminus were purified via BW25113 with pCA24N-ralR, pCA24N-ralR-A154G (encodes inactive RalR K52Q) and pCA24N-hfq (36). Strains were grown in LB with chloramphenicol and were induced with 1 mM IPTG at a turbidity of 0.1 for 5 h. Cells were collected and resuspended in 25 ml lysis buffer [50 mM potassium phosphate buffer (pH 8.0), 300 mM NaCl, and protease inhibitor cocktail (Sigma-Aldrich, USA)]. Then samples were sonicated using a Sonic Dismembrator (Ningbodongzhi, China) at level 2 for 5 min twice with 2 s sonication and 4 s break while being cooled in ice. Ni-NTA resin (Qiagen) was used according to the manufacturer's protocol. Purified proteins were desalted by passage on disposable Sephadex G-25 pre-packed PD-10 columns pre-equilibrated in 20 mM Tris-HCl buffer (pH 8.0), and the protein concentration was measured by using a Bi Yuntian BCA assay kit (Haimen, China).
DNA cleavage assay
The DNA cleavage assay was performed as described earlier (41,42) with some modifications. Purified RalR and RalR-mutant (150 pmol each) were incubated separately with different substrates at 37°C in 250 mM NaCl, 100 mM Tris-HCl (pH 7.5), 10 mM CaCl2 and 10 mM MgCl2 for 30 min and 120 min. Plasmid pBR322 isolated from BW25113, genomic DNA of BW25113, as well as methylated and unmethylated lambda DNA (dam−, dcm−) (1 μg) were used as substrates, respectively. The reaction was stopped by the addition of a stop solution (25% glycerol, 0.5% SDS, 0.05% bromophenol blue and 50 mM EDTA) and was analyzed by electrophoresis on 1% agarose gels stained with SYBR safe (Invitrogen). The equal amount of deoxyribonuclease I (DNase I; New England Biolabs, Beverly, MA, USA) was used as a positive control, and excess EDTA (20 mM) was added in the reaction to inhibit the nuclease activity. To study whether Ca2+ and Mg2+ are cofactors for RalR, 250 mM NaCl and 100 mM Tris-HCl (pH 7.5) buffer with or without the addition of 10 mM Ca2+ or 10 mM Mg2+ were used to conduct the DNA cleavage assay. The RalR-mutant was used as a negative control.
In vitro transcription and RNA labeling
For in vitro synthesis of T7-RalA sRNA and T7-ompA mRNA, PCR products were obtained from genomic DNA using primer pairs T7-ompA-f and -r and T7-ralA-f and -r, respectively (Supplementary Table S1). The PCR-amplified products were used as templates for in vitro transcription with T7 RNA polymerase. The T7 RNA polymerase promoter sequence was included in the forward primers. About 1 μg gel-purified PCR products were used as the templates for in vitro RNA reactions with the T7 High Yield RNA Synthesis Kit (New England Biolabs, Beverly, MA, USA). About 50 pmol of the in vitro transcribed RNA was labeled using the PierceTM RNA 3′ end biotinylation kit (Thermo Scientific, Hudson, NH, USA) according to the manufacturer's protocol.
RNA cleavage assay
The RNA cleavage assay was performed as described earlier (7). The reaction mixture for the endoribonuclease cleavage assay (10 μl) contained 2 μg in vitro synthesized T7-ompA mRNA, 50 mM Tris-HCl (pH 8.5), 100 mM KCl, 2.5 mM MgCl2 and 30 μg of purified RalR or RalR-mutant protein. The reaction mixture was incubated at 37°C for 10, 20 and 30 min and quenched by the addition of an equal volume of 2 × TBE-urea sample buffer (Invitrogen). Inactive RalR-mutant was used as a negative control. The reaction products were resolved by 15% TBE-urea gels. RNA was stained with SYBR safe.
Tricine-SDS-PAGE and western blot analysis
To investigate RalR protein levels, Tricine-SDS-PAGE and a western blot were performed. BW25113 Δrac strains containing pCA24N, pCA24N-ralR, pCA24N-ralA and pCA24N-ralR-ralA were grown to a turbidity of 0.2 in LB with chloramphenicol, then 1 mM IPTG was added to produce RalR for 5 h, and cells were washed with TE buffer. Samples were sonicated and total protein was quantified as mentioned above. Protein was denatured at 95°C for 5 min. The Tricine-SDS-PAGE was performed as described earlier (43). Total protein (25 μg) of each sample was loaded for SDS-PAGE, and a western blot was performed using 2.5 μg protein of each sample with primary antibodies raised against a His-tag (Cell Signaling Technology, Danvers, MA, USA) and horseradish-peroxidase-conjugated goat anti-mouse secondary antibodies (Bio-Rad, Richmond, CA, USA).
RNA–protein electrophoretic mobility shift assay (EMSA)
Biotinylated RalA sRNA (4 × 10−3 pmol) was mixed with different concentrations of Hfq protein in 20 μl buffer containing 10 mM HEPES (pH 7.3), 20 mM KCl, 1 mM MgCl2, 1 mM DTT and 5% glycerol at 37°C for 30 min. Samples from the reactions were loaded onto a pre-run 6% native polyacrylamide gel, and electrophoretic transfer was used to bind the proteins to nylon membranes (400 mA for 30 min) after the separation. After crosslinking for 5 min using UV-light, biotin-labeled RNA was detected using a LightShift® Chemiluminescent RNA EMSA kit (Thermo Scientific, Hudson, NH, USA) according to the manufacturer's protocol. In this assay, in vitro synthesized ompA mRNA was used as a negative RNA control and inactive Hfq protein was used as a negative protein control.
Quantitative real-time reverse-transcription PCR (qRT-PCR)
Total RNAs were isolated as described previously (11) and was used as the template for the qRT-PCR reaction using the SuperScriptTM III Platinum SYBR® Green One-Step qRT-PCR Kit (Invitrogen). Primers for qRT-PCR of ralR-, ralA- and ralA-related fragments are listed in Supplementary Table S1. The level of rrsG transcript was used to normalize the gene expression data.
Primer extension
5′ end FAM dye (6-carboxyfluorescein) labeled primers FAM-ralA-r2 and FAM-ralA-r3 (Supplementary Table S1) were ordered from Invitrogen. A total of 30 μg RNA was added to 2 × 10−4 pmol of 5′ end labeled primer, and the mixture was added to 3 μl of 10× first-strand and 37.5 U AMV reverse transcriptase (Promega, Madison, WI, USA). The RNA mix was annealed to the primers by incubating at 37°C for 1 h. To enhance the concentrations of FAM-labeled reverse transcriptions, the incubated mixture was re-transcribed by adding 6 μl of 10× first-strand buffer and 75U AMV reverse transcriptase and re-incubated at 37°C for another 1 h. The products were screened by an ABI3730 DNA Analyzer (Applied Biosystems, Foster City, California, USA) and the results were analyzed using Genemapper (Version 4.1). In this study, only RNA isolated from BW25113 Δrac/pCA24N-ralR-ralA induced with 1 mM IPTG for 20 min was used due to the low expression level of ralA in BW25113.
Prediction of the ralA Rho-independent terminator
To determine whether RalA sRNA contains a Rho-independent structure, we folded its sequence with RNASTRUCTURE 4.6 (44). A Rho-independent terminator was predicted based on properties characterized previously (45).
Fosfomycin resistance assay
Metabolic activities in the presence of fosfomycin and glucose 6-phosphate (to enhance the activity of fosfomycin) were measured using reagents from Biolog, Inc. (Hayward, CA, USA). Cells were grown to OD600 1.0, then diluted to a turbidity of 0.07 in IF-10a (Cat. No. 72264), and further diluted 200-fold into a solution containing IF-10a, BioLog Redox Dye D (Cat. No. 74224), and rich medium (2.0 g/l tryptone, 1.0 g/l yeast extract and 1.0 g/l NaCl) to a final OD600 of 0.0003. Volumes of 100 μl of this cell suspension (with 50 mg/ml glucose 6-phosphate and 0 or 0.2 μg/ml fosfomycin) were transferred into 96-well microtiter plates and incubated at 37°C. Metabolic activity was measured via the absorbance at 590 nm, which indicates the intracellular reduced state due to formazan (purple) formed from a tetrazolium dye. Growth in the presence of fosfomycin was determined in minimal medium; cells were washed once with M9 minimal medium with 0.4% glucose (M9-Glu) (46), then diluted to OD600 0.01 in M9-Glu with or without 2 μg/ml fosfomycin. Turbidities were measured at 600 nm.
RESULTS
RalR is a proteic toxin with endonuclease activity
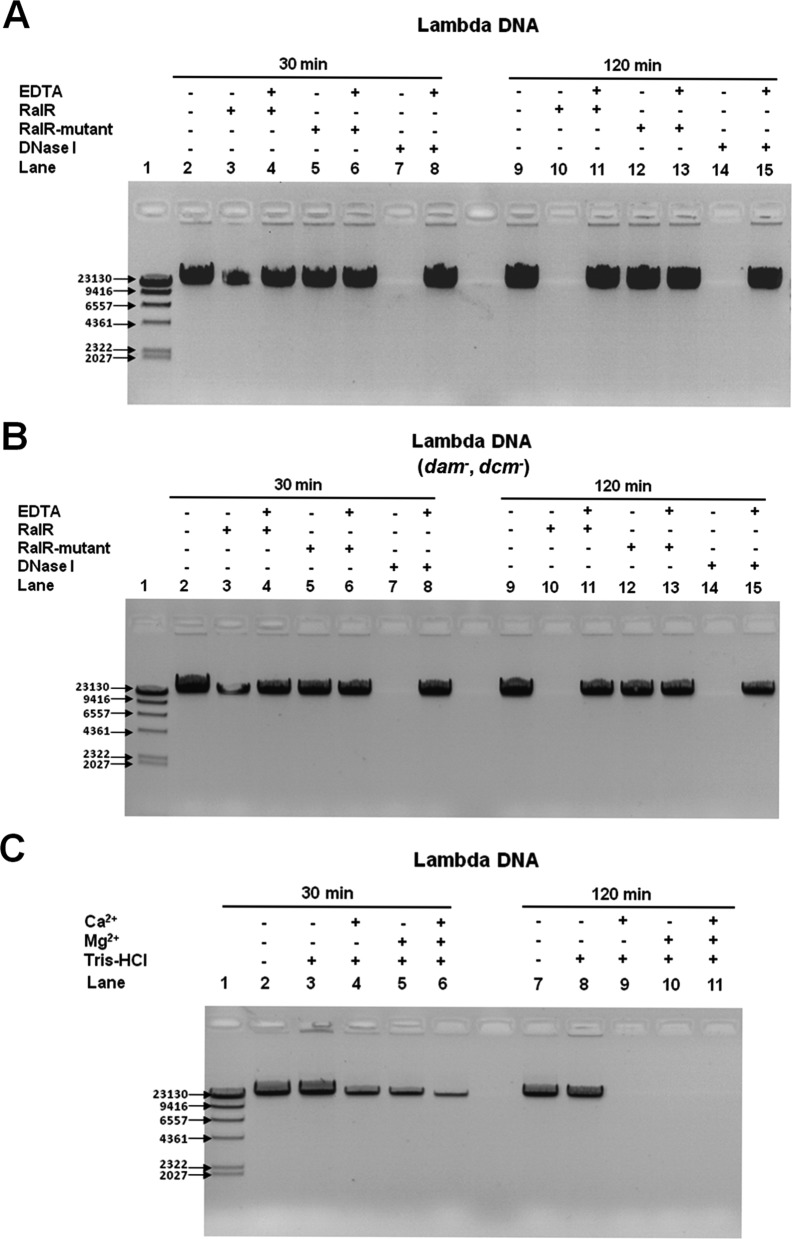
RASTA-bacteria has predicted RalR of prophage rac to be a small toxin (64 aa) (Figure 1A, Supplementary Figure S1) (47). Indeed, induction of ralR via pCA24N-ralR in E. coli results in growth inhibition (Figure 1B), along with a 103-fold reduction in colony-forming units (CFU) per milliliter in the wild-type host (Figure 1C). For the strain with the complete rac prophage of 23 kb removed by natural excision, Δrac, which is devoid of all rac prophage genes (38), overproducing RalR was also very toxic (Figure 1B and C). Hence, RalR is a potent toxin.
Figure 1.
RalR is toxic and RalA reduces the toxicity of RalR. (A) The chromosomal region of ralR-ralA in E. coli is shown in the upper panel. ralR is shown by the blue arrow, ydaC is shown by the gray arrow, ralA is shown by the red arrow, and the ORFs in the neighborhood of ralR/ralA are shown as gray arrows. The direction and the specific region cloned into each of the plasmids used in this study are shown in the lower panel. The numbers indicate the position of the related nucleotides (position 1 on the sense strand indicates the first base A of the ralR ORF, and position 1 on the anti-sense strand indicates the first base G of RalA sRNA). (B) Cell growth in LB plates supplemented with chloramphenicol (30 μg/ml) with and without 0.5 mM IPTG for cells producing RalR and RalA in the wild-type host and in the Δrac host. (C) CFU test in LB medium supplemented with chloramphenicol (30 μg/ml) with 1 mM IPTG (added at OD600 0.1) in the wild-type host and in the Δrac host. Three independent cultures of each strain were evaluated.
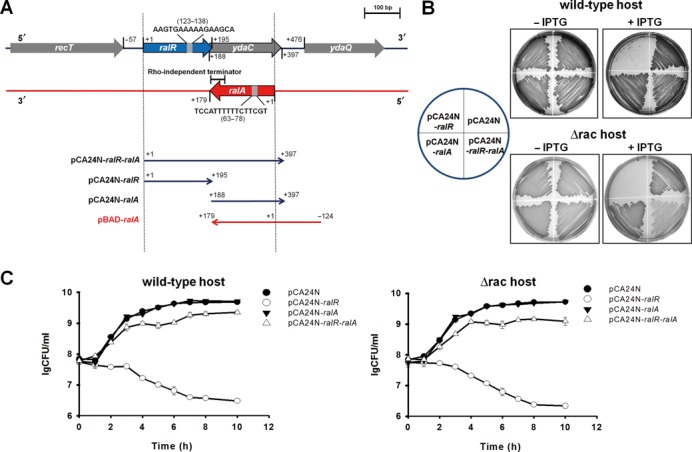
We investigated the cause of the RalR toxicity and found it was due to DNase activity. As shown by an in vitro DNA cleavage assay, purified RalR cleaves lambda DNA in a pattern similar to DNase I (Figure 2A). DNase I is an endonuclease that non-specifically cleaves DNA to release di-, tri- and oligonucleotides with 5′-phosphorylated and 3′-hydroxylated ends (48). After 30 min, ∼70% of the lambda DNA was cleaved by RalR, and all of the lambda DNA was cleaved by DNase I. After 120 min, all of the lambda DNA was cleaved by RalR (Figure 2A and B). Moreover, RalR also cleaved native genomic DNA of E. coli K-12 BW25113 and plasmid DNA pBR3222 (Supplementary Figure S2A).
Figure 2.
RalR cleaves DNA. (A) RalR cleaves lambda DNA after 30 min and 120 min. (B) RalR cleaves unmethylated lambda DNA (dam−, dcm−) after 30 min and 120 min. In (A) and (B), the positive control is degradation of DNA by DNase I, and the negative control is the inactive RalR-mutant (RalR K52Q). EDTA blocks RalR and DNase I activity. (C) RalR activity requires co-factor Mg2+ and/or Ca2+.
To test whether the endonuclease activity of RalR depends on modification of substrates by methylation, unmethylated lambda DNA was also tested, and the results show that RalR cleaves unmethylated DNA equally well, indicating that methylation is not required (Figure 2B). Moreover, Ca2+ and/or Mg2+ were essential for RalR endonuclease activity (Figure 2C), and as expected, the addition of excess EDTA inhibited the activity of both RalR and DNase I (Figure 2A and B).
Some nucleases degrade all nucleic acids, i.e. both RNA and DNA (49). Thus, the endoribonuclease activity of RalR was also tested, but no activity was detected in the in vitro RNA cleavage assay (Supplementary Figure S2B). Therefore, RalR is a non-specific endonuclease which cleaves DNA but not RNA. Furthermore, we found that overproduction of RalR after 5 h leads to filamentous growth as a result of the SOS response induced by degrading DNA (Supplementary Figure S2C).
We further determined the key residues in RalR using epPCR using pCA24N-ralR as the template. After screening ∼400 colonies for toxicity, 39 of them with reduced toxicity were sequenced, and changes at amino acid position 14, 37, 39, 49, 51 and 52 reduced RalR toxicity (Supplementary Figure S3). One of the mutated proteins (at aa 52) was also purified (RalR-mutant, RalR K52Q) (Supplementary Figure S4), and since it did not cleave DNA under the same conditions (Figure 2A and B), the DNase activity of RalR is not due to contamination.
DNA associated with ydaC functions as an antitoxin
Next, we tested whether previously annotated YdaC works as the antitoxin for RalR. YdaC is annotated as a rac-encoded protein (69 aa), and the start codon of ydaC overlaps with the stop codon of ralR (http://ecogene.org) (Supplementary Figure S1). The chromosomal region from the translational start of ralR to the translational stop of ydaC was used to construct a new plasmid pCA24N-ralR-ralA (Figure 1A). Results showed that when co-expressed with ralR, ralA, via the pCA24N-ralR-ralA construct, reduces the toxicity of RalR, demonstrating that the DNA associated with ydaC is necessary for repression of RalR toxicity (Figure 1B and C). However, no ydaC-encoded protein was detectable via SDS-PAGE or western blot when ydaC was tagged with an N-terminal hexahistidine sequence (Figure 3A).
Figure 3.
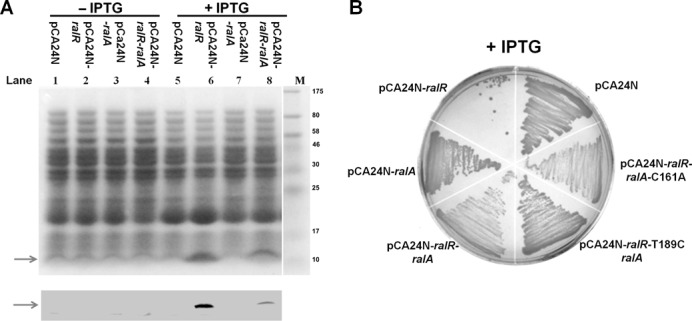
RalR is a protein and RalA functions as RNA. (A) Upper panel (SDS-PAGE) and lower panel (western blot) show that there is less RalR protein in the presence of RalA. His-tagged RalR (marked with arrows) was produced from pCA24N-ralR and pCA24N-ralR-ralA and has six histidine residues and 10 other amino acids attached at the N-terminus (expected size of ∼9.05 kDa). Production of RalR and RalA was induced via 0.2 mM IPTG at a turbidity of 0.1 for 5 h, and no IPTG treatment was used for the negative control. (B) Cell growth on LB plates with 0.5 mM IPTG in the Δrac host. T189C refers to the ydaC mutation in which the putative start codon ATG at position 189 was mutated into ACG, and C161A refers to the ralA mutation containing a stop codon instead of the ninth codon of putative protein YdaC.
To further test whether YdaC protein is the antitoxin, we introduced a stop codon by a single nucleotide change into ydaC at the putative amino acid position 9 (nucleotide position 212 from G to T) (pCA24N-ralR-ralA-C161A) and tested its impact on cell growth. However, the G212T substitution did not affect the ability of the ydaC-associated DNA to block the toxicity of RalR in cell growth (Figure 3B). In addition, when the putative start codon ATG at position 189 relative to the first nucleotide of start codon of ralR (lies inside the coding region of RalR) was mutated into a non-start codon ACG (pCA24N-ralR-T189C-ralA), which would prohibit the translation of the hypothetical YdaC, the DNA associated with ydaC still functioned as an antitoxin (Figure 3B). Thus, the DNA associated with ydaC inhibits the toxicity of RalR, but ydaC does not encode a protein. When we aligned ralR with the region associated with ydaC, we found two 16-nt repeats inside ralR and inside ydaC, respectively (Figure 1A). One of the 16-nt repeats lies in the coding region of the ralR, and the other, in the opposite orientation, lies 105-nt downstream of the end of the coding region of ralR on the complementary strand. In light of the recent discoveries that non-coding regulatory RNAs could act as antitoxins by base-pairing with the toxin mRNAs using limited complementarity (10–25 nt) (10,50–52), we hypothesized that the DNA strand complementary to ydaC encodes an RNA antitoxin for RalR and propose the name RalA for RalR antitoxin.
RalA is a trans-encoded sRNA
To further study the antitoxin activity of RalA, we constructed a dual plasmid system to allow expression of ralR and ralA independently. Plasmid pCA24N-ralR was induced by IPTG to express ralR and pBAD-ralA was induced by L-arabinose to express ralA using the complementary strand of the ydaC gene (from 99-nt downstream of the stop codon to the start codon of ydaC) (Figure 1A). pBAD-ralA was constructed to have a stop codon (TAA) immediately after the ATG start codon so that only RNA was formed. Toxicity test results indicated that there was a 100-fold reduction in viability when ralR is induced by IPTG for 4 h; however, when L-arabinose was also added to induce ralA, the toxicity of RalR was completely inhibited (Figure 4A). These results show that the RalA sRNA provides the antitoxin activity for RalR.
Figure 4.
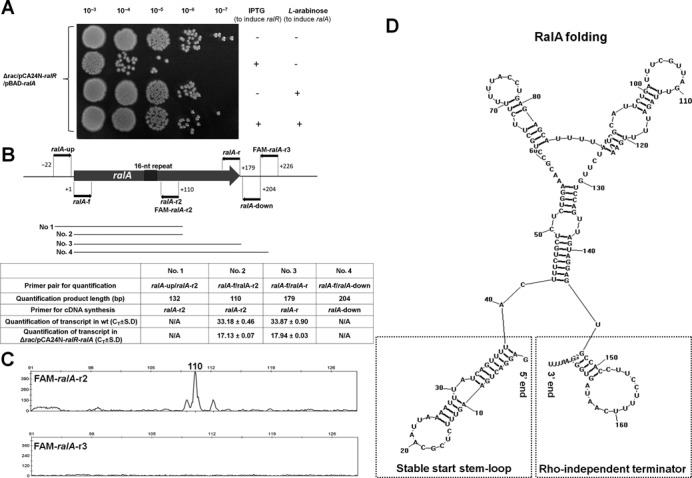
RalA is a trans-encoded sRNA. (A) The dual plasmid system pCA24N-ralR and pBAD-ralA was used to produce RalR protein and RalA sRNA by 1 mM IPTG and 0.1% L-arabinose in the Δrac host, respectively. The two inducers were added simultaneously at OD600 0.1 and the results of 4 h induction with or without the inducers are shown here. Two independent cultures of each strain were evaluated. (B) The position and direction of the primers used for primer extension and qRT-PCR to identify the start of transcription of ralA. The number 1 indicates the first base G of RalA sRNA. The length of the four fragments (Nos. 1–4) used for qRT-PCR are indicated. The Ct values of each fragment detected in the wild-type strain (OD600 1.0) or in the Δrac/pCA24N-ralR-ralA strain are indicated after 1 mM IPTG for 20 min once the OD600 reached ∼1.0, and the lower Ct indicates higher expression level. N/A indicates no signal was detected. (C) Results of the primer extension using FAM-ralA-r2 or FAM-ralA-r3, respectively. (D) RNA structure of RalA predicted by RNASTRUCTURE 4.6, with a stable start stem-loop at 5′ end and a Rho-independent terminator at 3′ end.
To map the 5′ end of RalA, we carried out primer extension experiments with oligonucleotide FAM-ralA-r2 with complementarity to RalA at regions 34-nt downstream from the 16-nt repeat region (Figure 4B). The results revealed a major extension product that is 110 nt in size, which indicates that the start of the transcript is 62-nt upstream of the 16-nt repeat region (Figure 4C, upper panel). No signal was detected using another primer that binds further downstream (FAM-ralA-r3) for primer extension (Figure 4C, lower panel).
Since non-coding sRNAs frequently terminate transcription via Rho-independent transcription terminators (53), we first searched for a potential Rho-independent termination signal in ralA. After folding RalA RNA by RNASTRUCTURE 4.6 (44), we identified a Rho-independent termination signal, with a stem-loop structure composed of 5 base pairs (GC rich) and a loop of 14 bases followed by four stretches of uridine residues (Figure 4D). This stretch is likely a Rho-independent terminator since (i) it is predicted to form a stem-loop structure, (ii) the formed stem has 5–10 base pairs in length and is GC rich with at least 60% GC base pairs, (iii) the formed loop generally contains 3–8 bases, (iv) in most structures, the average free energy calculated for a Rho-independent terminator is around −7 kcal/mol and here it was found to be −6.8 kcal/mol, and (v) the stem structure usually is followed by a stretch of Us (the average number is 4) and here it was found to have 4 (45). The Rho-independent termination signal thus gives an RNA product of 179 nucleotides (Figure 4D), which indicates that RalA sRNA is wholly contained within what was previously annotated as ydaC (on the opposite DNA strand). This explains why the ydaC DNA fragment was found to reduce RalR toxicity (Figure 1B and C). Moreover, the 5′ end of RalA also formed a stable stem-loop at the transcription start site determined by primer extension, which helps to stabilize the sRNA molecule (Figure 4D). Furthermore, both transcripts were detected in the exponential phase (OD600 0.8) and stationary phase ( OD600 6.2) of the wild-type strain using qRT-PCR with primer pair ralR-f and ralR-r (for ralR) and primer pair ralA-f2 and ralA-r3 (for ralA) (Supplementary Table S1), which shows that both native promoters function in vivo.
We further confirmed the length of the RalA transcripts using qRT-PCR and using cDNA templates synthesized from strain-specific primers in exponentially growing cells of BW25113 and BW25113 Δrac carrying pCA24N-ralR-ralA (Figure 4B). To test further whether RalA is an untranslated RNA, we searched for open reading frames in which a putative initiation codon (ATG, TTG or GTG) was preceded at an appropriate distance by a ribosome-binding site. This transcript does not contain a potential initiation codon preceded by a potential ribosome-binding site, thus it is unlikely that RalA encodes a protein.
RalA functions through base-pairing with RalR
We next investigated whether RalA RNA functions through a base-pairing mechanism. epPCR was employed to screen for the mutations in ralR and ralA that result in reduced antitoxin activity using pCA24N-ralR-ralA as template. After screening ∼400 colonies for toxicity, 20 of them with toxicity were sequenced, and the results showed that half of them had mutations within the two 16-nt repeats (Figure 5A) in the ralR gene and in the ralA gene. These results suggest that the 16-nt repeat is important for RalA RNA to inhibit RalR mRNA.
Figure 5.
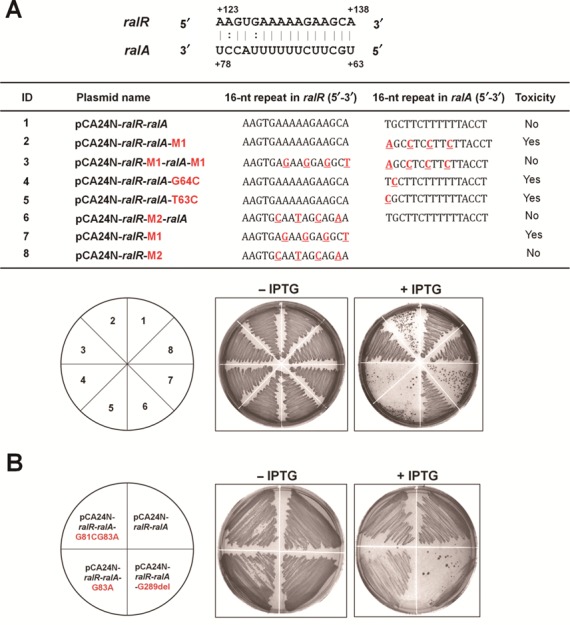
RalA interacts with RalR mRNA through 16-nt repeats. (A) The 16-nt repeat sequences of both RalR and RalA are shown in the upper schematic, and the numbers indicate the positions of the related bases. The table indicates the 16 nt in each plasmid, and the mutated nucleotides are marked in red font. ‘Toxicity’ indicates whether the toxin was toxic under the conditions indicated. Lower figures indicate growth on LB plates with cells producing both RalR and RalA via 0.5 mM IPTG in the Δrac host. (B) Cell growth on LB plates producing both RalR and RalA via 0.5 mM IPTG in the Δrac host. Two independent cultures of each strain were evaluated, and no IPTG treatment was used as the negative control.
To further investigate the involvement of the 16 nt in the base-pairing of the toxin and antitoxin RNAs, we performed site-directed mutagenesis on the 16-nt repeats as well as other neighboring regions. Our results showed that single mutations in the 16-nt repeat of ralA reduced its antitoxin function (pCA24N-ralR-ralA-G64C and pCA24N-ralR-ralA-T63C), and four mutations in the 16-nt repeat of ralA completely abolished the antitoxin activity of RalA (pCA24N-ralR-ralA-M1). In contrast, a single point mutation (pCA24N-ralR-ralA-G83A) and two point mutations (pCA24N-ralR-ralA-G81CG83A) that changed one or two nucleotides in regions outside of the 16-nt repeat of ralA did not affect the antitoxin activity of the RalA RNA (Figure 5B). In addition, to determine the minimal length of RalA RNA required for maintaining the function as an antitoxin, we removed all of the sequence after position 289, which eliminated the 16-nt repeat in RalA (pCA24N-ralR-ralA-G289del). As a result, the antitoxin function of RalA was completely abolished (Figure 5B). These results confirm the importance of the 16-nt repeat in the RalR/RalA TA pair in regard to the interaction of the toxin and antitoxin.
Moreover, when the 16 nt in ralR was altered to cause a synonymous mutation and non-synonymous mutation using pCA24N-ralR, the synonymous mutation retained the RalR toxicity (pCA24N-ralR-M1) while the non-synonymous mutation led to a RalR variant (pCA24N-ralR-M2, pCA24N-ralR-M2-ralA) that was no longer toxic (Figure 5A). These results confirm that RalR functions as a protein and, moreover, that amino acid changes at the 16-nt repeat coding region affect RalR toxicity.
To further test whether changes in the ralA 16-nt repeat can neutralize the toxic effect of RalR when the corresponding mutation is encoded by ralR, we introduced two complementary mutations and constructed pCA24N-ralR-M1-ralA-M1, which contains two identically mutated 16-nt repeats. The results show that the mutated ralA retains its antitoxin function when the mutation matches that of ralR, in contrast to the result that mutated ralA could not attenuate the toxic effect of the unmutated ralR (pCA24N-ralR-ralA-M1) (Figure 5A). Therefore, we provide several lines of evidence that RalA is a novel RNA antitoxin in which its activity to block the toxicity of RalR relies on a 16-nt repeat.
RalA likely inhibits RalR mRNA translation
To determine how RalA RNA attenuates the toxic effects of RalR, we checked the RalR protein levels with and without RalA. Through SDS-PAGE and western blot analysis, we found that the protein levels of RalR are reduced in the presence of RalA (Figure 3A), which indicates that RalA interferes with the accumulation of RalR protein. Base-pairing between sRNA and its target mRNA usually leads to repression of protein levels through translational inhibition and/or mRNA degradation (51). Our qRT-PCR results showed that in the presence of more RalA, the levels of RalR mRNA were not reduced substantially (Table 2), thus it is unlikely that RalA RNA promotes RalR mRNA degradation. Translational inhibition of sRNA with a target gene often occurs when sRNA base-pairs with the 5′-UTR of target mRNA; however, other locations for base-pairing and consequent mechanisms of regulation are possible (51).
Table 2.
Levels of RalR and RalA transcripts (Ct values ± S.D.) quantified by qRT-PCR in BW25113/pBAD-ralA
| Conditions | Ct (target gene) | Ct (control) | Fold change |
|---|---|---|---|
| ralR | rrsG | ||
| 20 min − ara | 28 .0 ± 0.1 | 8.9 ± 0.2 | 1.7 ± 0.2 (ralR) |
| 20 min + ara | 27.0 ± 0.2 | 8.7 ± 0.1 | |
| 8 h − ara | 29.8 ± 0.2 | 8.9 ± 0.0 | 1.5 ± 0.4 (ralR) |
| 8 h + ara | 29.0 ± 0.4 | 8.7 ± 0.1 | |
| ralA | rrsG | ||
| 20 min − ara | 19.7 ± 0.6 | 8.9 ± 0.2 | 22.6 ± 0.2 (ralA) |
| 20 min + ara | 15.0 ± 0.4 | 8.7 ± 0.1 | |
| 8 h − ara | 20.3 ± 0.1 | 8.9 ± 0.0 | 11.3 ± 0.1 (ralA) |
| 8 h + ara | 16.6 ± 0.1 | 8.7 ± 0.1 |
Exponentially growing cells (OD600 0.8) were induced with 1% L-arabinose (+ ara) or without L-arabinose (− ara) for 20 min and 8 h. Lower Ct values indicate higher expression levels, and rrsG was used to normalize total RNA levels. Two independent cultures were used for the assay. Fold changes in the transcription of various targets with or without L-arabinose were calculated as 2∧−(Cttarget(+ ara)−Ct rrsG(+ ara))/2∧−(Cttarget(− ara)−CtrrsG(− ara)). Means and standard errors are indicated.
Hfq is required for RalA activity
To facilitate limited base-pairing for the trans-encoded sRNAs and the target mRNAs, in many cases the RNA chaperone Hfq is required for trans-encoded sRNA-mediated regulation (51). The antitoxin RalA was tested in an hfq deletion mutant strain and, as expected, the hfq mutation abolished antitoxin activity of RalA (Figure 6A and B). Hence, Hfq is required for RalA RNA activity.
Figure 6.
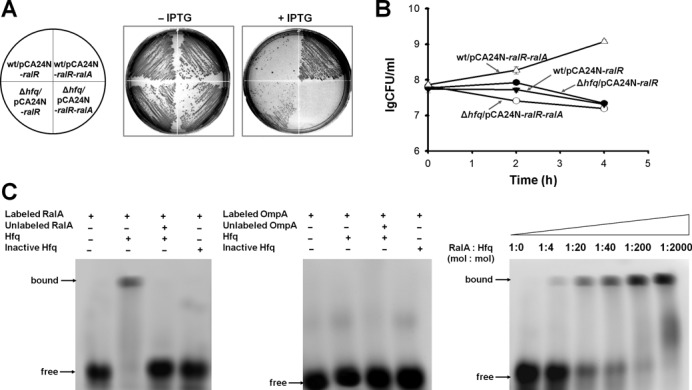
Hfq is required for RalA antitoxin activity. (A) Cell growth on LB plates supplemented with chloramphenicol (30 μg/ml) with and without 0.5 mM IPTG for cells producing RalR and RalR/RalA in the wild-type host and in the Δhfq host, respectively. (B) CFU test over time for cells producing RalR and RalR/RalA in the wild-type host and in the Δhfq host, respectively. Overnight cultures were diluted to OD600 0.1, and 1 mM IPTG was added initially. This assay was performed twice with two independent cultures, and one standard deviation is shown. (C) EMSA shows that Hfq binds to RalA sRNA (left panel) but not to the 3′ end of the coding region of ompA mRNA (middle panel). The binding of Hfq to RalA increases with increasing Hfq (right panel).
To provide additional evidence for the requirement of Hfq for RalA RNA activity, we investigated direct binding of Hfq to RalA RNA in vitro. Hfq interacts strongly with small non-coding RNAs at single-stranded AU-rich regions (54), and RalA is AU rich (63.1%) and has four stretches of more than seven continuous U/As. RNA–protein binding was performed, and Hfq was found to bind RalA RNA (Figure 6C). In contrast, Hfq did not bind to the negative control (3′ end of the coding region of OmpA mRNA, Figure 6C). Moreover, it seems that Hfq helps to stabilize the RalA transcript as RalA RNA was degraded faster in the absence of Hfq as shown by qRT-PCR, while both RalR and RalA RNAs appeared stable in the wild-type cells with half-times of more than 20 min (Table 3).
Table 3.
Levels of RalR and RalA transcripts (Ct values ± S.D.) quantified by qRT-PCR in BW25113 and BW25113 Δhfq after the addition of rifampicin
| Strains | Conditions | Ct (target gene) | Ct (control) | Fold change | ||
|---|---|---|---|---|---|---|
| ralR | ralA | rrsG | ralR | ralA | ||
| BW25113 | Rifampicin 0 min | 26.3 ± 0.3 | 27.5 ± 0.6 | 9.2 ± 0.3 | ||
| Rifampicin 5 min | 26.5 ± 0.1 | 27.7 ± 0.1 | 9.6 ± 0.2 | 1.1 ± 0.2 | 1.1 ± 0.2 | |
| Rifampicin 10 min | 26.8 ± 0.1 | 28.4 ± 0.2 | 10.1 ± 0.1 | 1.3 ± 0.2 | 1.0 ± 0.2 | |
| Rifampicin 20 min | 25.9 ± 0.5 | 27.6 ± 0.2 | 9.6 ± 0.2 | 1.7 ± 0.5 | 1.2 ± 0.3 | |
| ralR | ralA | rrsG | ralR | ralA | ||
| Δhfq | Rifampicin 0 min | 26.1 ± 0.9 | 26.5 ± 0.8 | 10.0 ± 0.2 | ||
| Rifampicin 5 min | 27.2 ± 0.2 | 28.8 ± 0.6 | 10.0 ± 0.1 | −2.1 ± 0.2 | −4.9 ± 0.6 | |
| Rifampicin 10 min | 26.6 ± 0.2 | 28.5 ± 0.4 | 10.0 ± 0.2 | −1.4 ± 0.3 | −4.0 ± 0.4 | |
| Rifampicin 20 min | 25.3 ± 0.1 | 28.2 ± 0.5 | 9.6 ± 0.3 | 1.3 ± 0.3 | −4.3 ± 0.6 | |
Overnight cultures were diluted to OD600 0.1, and re-grown in LB till OD600 1. Rifampicin (50 μg/ml) was added and samples were taken after 5 min, 10 min and 20 min. Fold changes in the concentrations of the targets at different times points were calculated as 2∧−(Cttarget_T = 5 or 10 or 20−CtrrsG_ T = 5 or 10 or 20)/2∧−(Cttarget_T = 0−CtrrsG_T = 0). Values less than one are indicated as negative fold changes (i.e. the amount of RNA that is reduced). Means and standard errors are indicated.
RalR increases resistance to fosfomycin
Large-scale phenotypic screening of all Keio mutants from PortEco showed that the ΔralR knockout strain has a negative fitness score when exposed to fosfomycin (55). Fosfomycin is a broad-spectrum antibacterial agent that targets bacterial mucopeptide synthesis by inhibiting phosphoenolpyruvate transferase, the first enzyme involved in the synthesis of peptidoglycan (56). Our results show that both the ΔralR and the ΔralRA strain are more sensitive to fosfomycin (0.2 μg/ml) compared to the wild-type strain based on their reduced metabolic activity (Figure 7A and B). Corroborating these results, the ΔralRA strain is more sensitive to fosfomycin (2 μg/ml) compared to the wild-type in terms of growth as well (Figure 7C and D). Hence, the RalR/RalA TA locus has an impact on cell physiology and is beneficial for resistance to an antibiotic.
Figure 7.
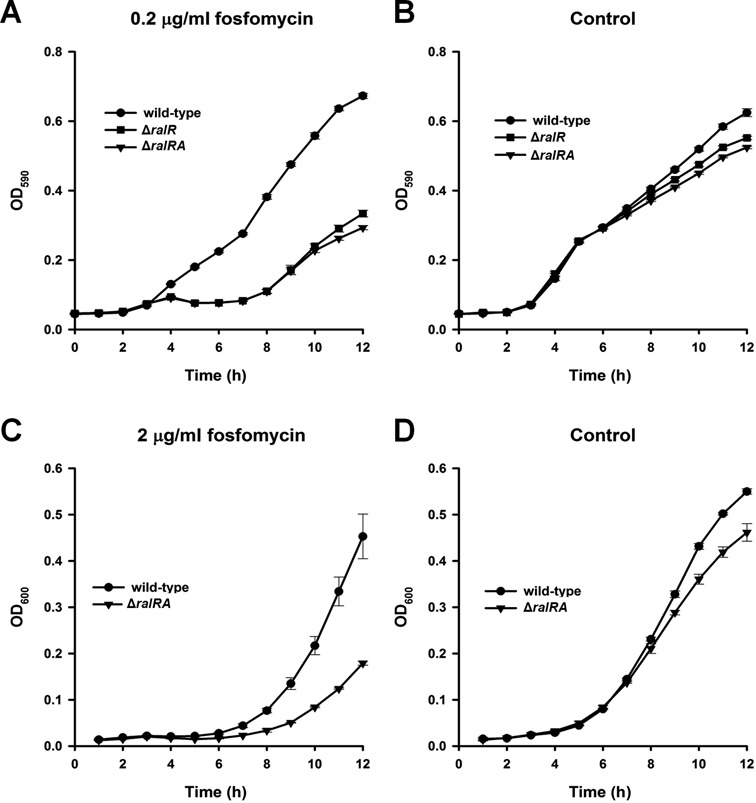
The RalR/RalA TA system increases resistance to fosfomycin. (A) Metabolic activity of the wild-type, ΔralR and the ΔralRA strains subjected to 0.2 μg/ml fosfomycin with 50 μg/ml glucose-6-phosphate to enhance fosfomycin activity and (B) metabolic activity with 50 μg/ml glucose-6-phosphate alone (negative control). This assay was performed twice with two independent cultures, and one standard deviation is shown. Growth of the wild-type and ΔralRA strains (C) with 2 μg/ml fosfomycin and (D) without fosfomycin. Two independent cultures were evaluated, and one standard deviation is shown.
DISCUSSION
Collectively, our results support that RalR/RalA forms a type I TA pair. These results are: (i) both genes are small and are adjacent, (ii) RalR functions as a toxin that inhibits growth, (iii) RalA functions as an antitoxin and is a non-coding RNA, (iv) RalA blocks RalR-mediated toxicity by the base-pairing of 16 key nucleotides and prevents the translation of RalR mRNA, and (v) Hfq is required for RalA antitoxin activity. As a novel type I TA system, RalR is the first toxin that functions as an endonuclease in E. coli. A recent study showed that when compared with a wild-type strain, a ydaC deletion mutant has an increased level of double-strand breaks that is remedied by expression of ydaC from a plasmid, suggesting that ydaC helps to maintain the integrity of the bacterial chromosome (34). These results fit well with our results here in that deletion of ralA (on the opposite strand as ydaC) leads to activation of toxin RalR that should result in double-stranded DNA breaks. Although overproduction of RalR leads to filamentous growth due to RalR-mediated DNA damage that activates the SOS response, it is not clear whether RalR can trigger cell death under physiological conditions.
Another novel feature of this TA system is that RalA is a trans-encoded sRNA antitoxin that neutralizes the protein toxin by base-pairing within the toxin mRNA coding region. Traditional type I toxin and antitoxin genes are usually located at the same locus, and sRNA antitoxins can have greater than 60-nt base-pairing with the 5′ UTR (e.g. Ldr of the Ldr/Rdl family in E. coli (57)) or the 3′ UTR of the toxin mRNA (e.g. TxpA/RatA in Bacillus subtilis (58)), and the two transcripts can also overlap (e.g. Ibs/Sib in E. coli) (50,52). Recently, four unconventional type I TA systems have been reported where the antitoxins are transcribed divergently from the toxin gene and have limited complementation regions to the toxin RNA (often 18–21 nucleotides), including TisB/IstR-1 (59), ShoB/OhsC (50) and ZorO/OrzO loci (50). Here with RalR/RalA, we provided evidence that RalA reduces RalR toxicity via the complementarity of 16 nt. For the trans-encoded base-pairing sRNAs, the potential base-pairing with target mRNA typically is 10–25 nt. Studies on ZorO/OrzO showed that for the 18-nt complementarity region, not all of the base-pairing interaction is necessary, and it is dependent on whether the complementary region is contiguous or not (52). Our results confirmed this and showed that 16 nt of complementarity (with 11 nt of contiguous perfect complement) is sufficient for base-pairing interaction between toxin and antitoxin.
Cryptic prophage rac is a DNA fossil since it was acquired over 4.5 million years ago (60), yet it has become a stable resident in the E. coli chromosome. Ral in active phage lambda and RalR in prophage rac share a low sequence identity of 24%, indicating that the two proteins are not related. In contrast, a high identity of 77% exists for P22 Ral and lambda Ral with highly conserved N- and C-termini (Supplementary Figure S5). Moreover, an even higher identity of 94% is found for recombinant Enterobacteria phage phi21 Ral and lambda Ral with highly conserved N- and less conserved C-termini (Supplementary Figure S5). Therefore, Ral is more conserved in phages while RalR is more conserved in E. coli strains. Ral of lambda phage acts by modulating the restriction and modification (RM) activities of the type I restriction systems in E. coli (61), and RM systems and TA systems are related (3). In general, they originate from phages and plasmids and are used to invade hosts or to keep them in the host by post-segregational killing. However, after integration into the chromosome, the function of these two systems has diverged. We found that RalR in E. coli functions differently, acting as a toxin by cleaving DNA, and that it belongs to a type I toxin–antitoxin system. We show here that removal of ralRA reduces the resistance of the cell to the antibiotic fosfomycin; hence, there is a clear benefit to the cell for harboring the RalR/RalA TA system. Therefore, our results provide further evidence that the genes of cryptic prophage impact cell physiology and that these genes may be used to increase resistance to stress (38).
Besides RalR/RalA, five other TA systems located in the E. coli K12 cryptic prophages have been reported, including the well-studied RelE/RelB (in Qin prophage) (26) and four recently identified TA pairs YpjF/YfjZ (in CP4-57) (28), RnlA/RnlB (in CP4-57) (27), YkfI/YafW (in CP4-6) (28) and CbtA/YeeU (in CP4-44) (6). These TA systems are located in prophages that are not inducible by SOS responses (38), suggesting a relatively stable residence in the host genome. Type I TA pair TxpA/RatA in B. subtilis is also in a phage-like element, and the phage-like element is retained in the host chromosome for spore formation (58). TA systems have been linked to persistence, biofilm formation and the general stress responses, and ralR was found to be highly induced in the later stage of E. coli biofilm development (24 h biofilms compared to 4 h biofilms) (33). Indubitably, other uncharacterized TA systems in cryptic prophages might also contribute to the stress response of the host, and there are probably additional intricate regulation mechanisms that need to be investigated.
Unconventional type I TA systems are more limited in distribution than typical type I TA systems, and are found mainly in Escherichia, Shigella and Salmonella species. By comparing the phylogenetic tree of those identified type I loci with the host taxonomy, earlier studies suggest that type I loci have not been freely disseminated by horizontal gene transfer but instead may have a common ancient ancestor (10). This agrees with what we found for TA pair RalR/RalA in rac prophage. The discovery of a high occurrence of TA systems in E. coli prophages will help to search for new TA systems in other prokaryotes. Since TA genes from bacterial prophages are actively expressed in the human gastrointestinal tract, TA systems may play a dynamic role in more ecosystems than expected (62). The mechanism of harboring TA systems in prophages, the origins of these TA systems, and the regulation of these loci in response to various stress conditions need to be investigated further.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We are grateful for the Keio and ASKA strains provided by the Genome Analysis Project in Japan. X.W. is the 1000-Youth Elite Program member in China; T.W. is the Biotechnology Endowed Professor at the Pennsylvania State University.
FUNDING
National Basic Research Program of China [2013CB955701 to X.W.]; National Natural Science Foundation of China [NFSC31270214, NFSC31290233 to X.W.]; National Institutes of Health [NIH R01 GM089999 to T.W.]; 1000-Youth Elite Program (the Recruitment Program of Global Experts in China) (X.W.). Source of open access funding: China [2013CB955701 to X.W.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Gerdes K., Christensen S.K., Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Micro. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 2.Brantl S. Bacterial type I toxin-antitoxin systems. RNA Biol. 2012;9:1488–1490. doi: 10.4161/rna.23045. [DOI] [PubMed] [Google Scholar]
- 3.Mruk I., Kobayashi I. To be or not to be: regulation of restriction–modification systems and other toxin–antitoxin systems. Nucleic Acids Res. 2014;42:70–86. doi: 10.1093/nar/gkt711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayes F., Van Melderen L. Toxins-antitoxins: diversity, evolution and function. Crit. Rev. Biochem. Mol. Biol. 2011;46:386–408. doi: 10.3109/10409238.2011.600437. [DOI] [PubMed] [Google Scholar]
- 5.Fineran P., Blower T., Foulds I., Humphreys D., Lilley K., Salmond G. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. U.S.A. 2009;106:894–899. doi: 10.1073/pnas.0808832106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masuda H., Tan Q., Awano N., Wu K., Inouye M. YeeU enhances the bundling of cytoskeletal polymers of MreB and FtsZ, antagonizing the CbtA (YeeV) toxicity in Escherichia coli. Mol. Microbiol. 2012;84:979–989. doi: 10.1111/j.1365-2958.2012.08068.x. [DOI] [PubMed] [Google Scholar]
- 7.Wang X., Lord D.M., Cheng H.-Y., Osbourne D.O., Hong S.H., Sanchez-Torres V., Quiroga C., Zheng K., Herrmann T., Peti W., et al. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat. Chem. Biol. 2012;8:858–861. doi: 10.1038/nchembio.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X., Lord D.M., Hong S.H., Peti W., Benedik M.J., Page R., Wood T.K. Type II toxin/antitoxin MqsR/MqsA controls type V toxin/antitoxin GhoT/GhoS. Environ. Microbiol. 2013;15:1734–1744. doi: 10.1111/1462-2920.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fozo E.M., Hemm M.R., Storz G. Small toxic proteins and the antisense RNAs that repress them. Microbiol. Mol. Biol. Rev. 2008;72:579–589. doi: 10.1128/MMBR.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fozo E.M., Makarova K.S., Shabalina S.A., Yutin N., Koonin E.V., Storz G. Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010;38:3743–3759. doi: 10.1093/nar/gkq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren D., Bedzyk L.A., Thomas S.M., Ye R.W., Wood T.K. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 2004;64:515–524. doi: 10.1007/s00253-003-1517-y. [DOI] [PubMed] [Google Scholar]
- 12.Kim Y., Wang X., Qun M., Zhang X.-S., Wood T.K. Toxin-antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae. J. Bacteriol. 2008;191:1258–1267. doi: 10.1128/JB.01465-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim Y., Wood T.K. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem. Biophys. Res. Commun. 2010;391:209–213. doi: 10.1016/j.bbrc.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dörr T., Vulić M., Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010;8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Y., Benedik M.J., Wood T.K. Antitoxin DinJ influences the general stress response through transcript stabilizer CspE. Environ. Microbiol. 2012;14:669–679. doi: 10.1111/j.1462-2920.2011.02618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X., Kim Y., Hong S.H., Ma Q., Brown B.L., Pu M., Tarone A.M., Benedik M.J., Peti W., Page R., et al. Antitoxin MqsA helps mediate the bacterial general stress response. Nat. Chem. Biol. 2011;7:359–366. doi: 10.1038/nchembio.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pecota D.C., Wood T.K. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J. Bacteriol. 1996;178:2044–2050. doi: 10.1128/jb.178.7.2044-2050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hazan R., Engelberg-Kulka H. Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol. Genet. Genomics. 2004;272:227–234. doi: 10.1007/s00438-004-1048-y. [DOI] [PubMed] [Google Scholar]
- 19.González Barrios A.F., Zuo R., Hashimoto Y., Yang L., Bentley W.E., Wood T.K. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022) J. Bacteriol. 2006;188:305–316. doi: 10.1128/JB.188.1.305-316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belitsky M., Avshalom H., Erental A., Yelin I., Kumar S., London N., Sperber M., Schueler-Furman O., Engelberg-Kulka H. The Escherichia coli Extracellular Death Factor EDF induces the endoribonucleolytic activities of the toxins MazF and ChpBK. Mol. cell. 2011;41:625–635. doi: 10.1016/j.molcel.2011.02.023. [DOI] [PubMed] [Google Scholar]
- 21.Wang X., Wood T.K. Toxin/antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl. Environ. Microbiol. 2011;77:5577–5583. doi: 10.1128/AEM.05068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng H.-Y., Soo V.W.C., Islam S., McAnulty M.J., Benedik M.J., Wood T.K. Toxin GhoT of the GhoT/GhoS toxin/antitoxin system damages the cell membrane to reduce adenosine triphosphate and to reduce growth under stress. Environ. Microbiol. 2014 doi: 10.1111/1462-2920.12373. doi:10.1111/1462-2920.12373. [DOI] [PubMed] [Google Scholar]
- 23.Gerdes K. The parB (hok/sok) locus of plasmid R1: a general purpose plasmid stabilization system. Nat. Biotechnol. 1988;6:1402–1405. [Google Scholar]
- 24.Pedersen K., Gerdes K. Multiple hok genes on the chromosome of Escherichia coli. Mol. Microbiol. 1999;32:1090–1102. doi: 10.1046/j.1365-2958.1999.01431.x. [DOI] [PubMed] [Google Scholar]
- 25.Yuan J., Yamaichi Y., Waldor M.K. The three Vibrio cholerae chromosome II-encoded ParE toxins degrade chromosome I following loss of chromosome II. J. Bacteriol. 2011;193:611–619. doi: 10.1128/JB.01185-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pedersen K., Zavialov A.V., Pavlov M.Y., Elf J., Gerdes K., Ehrenberg M. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell. 2003;112:131–140. doi: 10.1016/s0092-8674(02)01248-5. [DOI] [PubMed] [Google Scholar]
- 27.Koga M., Otsuka Y., Lemire S.b., Yonesaki T. Escherichia coli rnlA and rnlB compose a novel toxin-antitoxin system. Genetics. 2011;187:123–130. doi: 10.1534/genetics.110.121798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown J.M., Shaw K.J. A novel family of Escherichia coli toxin-antitoxin gene pairs. J. Bacteriol. 2003;185:6600–6608. doi: 10.1128/JB.185.22.6600-6608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conter A., Bouché J.P., Dassain M. Identification of a new inhibitor of essential division gene ftsZ as the kil gene of defective prophage Rac. J. Bacteriol. 1996;178:5100–5104. doi: 10.1128/jb.178.17.5100-5104.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King G., Murray N.E. Restriction alleviation and modification enhancement by the Rac prophage of Escherichia coli K-12. Mol. Microbiol. 1995;16:769–777. doi: 10.1111/j.1365-2958.1995.tb02438.x. [DOI] [PubMed] [Google Scholar]
- 31.Toothman P. Restriction alleviation by bacteriophages lambda and lambda reverse. J. Virol. 1981;38:621–631. doi: 10.1128/jvi.38.2.621-631.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blattner F.R., Guy Plunkett I., Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 33.Domka J., Lee J., Bansal T., Wood T.K. Temporal gene-expression in Escherichia coli K-12 biofilms. Environ. Microbiol. 2007;9:332–346. doi: 10.1111/j.1462-2920.2006.01143.x. [DOI] [PubMed] [Google Scholar]
- 34.Felczak M.M., Kaguni J.M. The rcbA gene product reduces spontaneous and induced chromosome breaks in Escherichia coli. J. Bacteriol. 2012;194:2152–2164. doi: 10.1128/JB.06390-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitagawa M., Ara T., Arifuzzaman M., Ioka-Nakamichi T., Inamoto E., Toyonaga H., Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 37.Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X., Kim Y., Ma Q., Hong S.H., Pokusaeva K., Sturino J.M., Wood T.K. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 2010;1:147. doi: 10.1038/ncomms1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning, A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Fishman A., Tao Y., Bentley W.E., Wood T.K. Protein engineering of toluene 4-monooxygenase of Pseudomonas mendocina KR1 for synthesizing 4-nitrocatechol from nitrobenzene. Biotechnol. Bioeng. 2004;87:779–790. doi: 10.1002/bit.20185. [DOI] [PubMed] [Google Scholar]
- 41.Bagnéris C., Briggs L.C., Savva R., Ebrahimi B., Barrett T.E. Crystal structure of a KSHV-SOX-DNA complex: insights into the molecular mechanisms underlying DNase activity and host shutoff. Nucleic Acids Res. 2011;39:5744–5756. doi: 10.1093/nar/gkr111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buisson M., Géoui T., Flot D., Tarbouriech N., Ressing M.E., Wiertz E.J., Burmeister W.P. A bridge crosses the active-site canyon of the Epstein-Barr virus nuclease with DNase and RNase activities. J. Mol. Biol. 2009;391:717–728. doi: 10.1016/j.jmb.2009.06.034. [DOI] [PubMed] [Google Scholar]
- 43.Schägger H. Tricine-SDS-PAGE. Nat. Protoc. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- 44.Mathews D.H., Disney M.D., Childs J.L., Schroeder S.J., Zuker M., Turner D.H. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. U.S.A. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Argaman L., Hershberg R., Vogel J., Bejerano G., Wagner E.G.H., Margalit H., Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 46.Rodriguez R.L., Tait R.C. Recombinant DNA Techniques: An Introduction. Menlo Park, CA: The Benjamin/Cummings Publishing Company, Inc.; 1983. [Google Scholar]
- 47.Sevin E., Barloy-Hubler F. RASTA-Bacteria: a web-based tool for identifying toxin-antitoxin loci in prokaryotes. Genome Biol. 2007;8:R155. doi: 10.1186/gb-2007-8-8-r155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vanecko S., Laskowski M. Studies of the specificity of Deoxyribonuclease I: III. Hydrolysis of chains of carrying a monoesterified phosphate on carbon 5′. J. Biol. Chem. 1961;236:3312–3316. [PubMed] [Google Scholar]
- 49.Suh Y., Alpaugh M., Krause K.L., Benedik M.J. Differential secretion of isoforms of Serratia marcescens extracellular nuclease. Appl. Environ. Microbiol. 1995;61:4083–4088. doi: 10.1128/aem.61.11.4083-4088.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fozo E.M. New type I toxin-antitoxin families from “wild” and laboratory strains of E. coli: Ibs-Sib, ShoB-OhsC and Zor-Orz. RNA Biol. 2012;9:1504–1512. doi: 10.4161/rna.22568. [DOI] [PubMed] [Google Scholar]
- 51.Waters L.S., Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wen J., Won D., Fozo E.M. The ZorO-OrzO type I toxin–antitoxin locus: repression by the OrzO antitoxin. Nucl. Acids Res. 2014;42:1930–1946. doi: 10.1093/nar/gkt1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 54.Valentin-Hansen P., Eriksen M., Udesen C. MicroReview: the bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- 55.Nichols R.J., Sen S., Choo Y.J., Beltrao P., Zietek M., Chaba R., Lee S., Kazmierczak K.M., Lee K.J., Wong A., et al. Phenotypic landscape of a bacterial cell. Cell. 2011;144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raz R. Fosfomycin: an old–new antibiotic. Clin. Microbiol. Infect. 2012;18:4–7. doi: 10.1111/j.1469-0691.2011.03636.x. [DOI] [PubMed] [Google Scholar]
- 57.Kawano M. Divergently overlapping cis-encoded antisense RNA regulating toxin-antitoxin systems from E. coli: hok/sok, ldr/rdl, symE/symR. RNA Biol. 2012;9:1520–1527. doi: 10.4161/rna.22757. [DOI] [PubMed] [Google Scholar]
- 58.Silvaggi J.M., Perkins J.B., Losick R. Small untranslated RNA antitoxin in Bacillus subtilis. J. Bacteriol. 2005;187:6641–6650. doi: 10.1128/JB.187.19.6641-6650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Unoson C., Wagner E.G.H. A small SOS-induced toxin is targeted against the inner membrane in Escherichia coli. Mol. Microbiol. 2008;70:258–270. doi: 10.1111/j.1365-2958.2008.06416.x. [DOI] [PubMed] [Google Scholar]
- 60.Perna N.T., Plunkett G., Burland V., Mau B., Glasner J.D., Rose D.J., Mayhew G.F., Evans P.S., Gregor J., Kirkpatrick H.A., et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 61.Zabeau M., Friedman S., Van Montagu M., J.S. The ral gene of phage lambda. I. Identification of a non-essential gene that modulates restriction and modification in E. coli. Mol. Gen. Genet. 1980;179:63–73. doi: 10.1007/BF00268447. [DOI] [PubMed] [Google Scholar]
- 62.Reyes A., Haynes M., Hanson N., Angly F.E., Heath A.C., Rohwer F., Gordon J.I. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.