Abstract
Post-translational modification of proteins is an evolutionarily conserved mechanism for regulating activity, binding affinities and stability. Compared with established post-translational modifications such as phosphorylation or uniquitination, post-translational modification by protons within physiological pH ranges is a less recognized mechanism for regulating protein function. By changing the charge of amino acid side chains, post-translational modification by protons can drive dynamical changes in protein conformation and function. Addition and removal of a proton is rapid and reversible and in contrast to most other post-translational modifications does not require an enzyme. Signaling specificity is achieved by only a minority of sites in proteins titrating within the physiological pH range. Here, we examine the structural mechanisms and functional consequences of proton post-translational modification of pH-sensing proteins regulating different cellular processes.
Keywords: pH sensor, protonation, intracellular pH, post-translational modification, coincidence detection, conformational change, ionization, charge, histidine
INTRODUCTION
Intracellular pH (pHi) dynamics is conventionally viewed as a homeostatic mechanism to protect cells from alkaline and acidic loads. Consistent with this view, cytosolic pH is tightly regulated at near neutral (11). For example, increased generation of metabolic acids is generally accompanied by increased H+ efflux by plasma membrane electroneutral ion transport proteins, such as the monocarboxylate lactate-H+ exchangers MCT1 and MCT4, the Na-H exchanger NHE1, and the Na+-dependent Cl-HCO3 transporter NBCn2. However, an emerging view is that pHi dynamics also functions as a signaling mechanism to regulate a number of cell processes. In mammalian cells a seemingly small increase in pHi of 0.2 to 0.3 units promotes cell proliferation and cell cycle progression (111; 121) and is now recognized to be necessary for directional cell migration, including cell polarity, actin filament assemblies, and focal adhesion remodeling (34; 35; 128; 130). Decreased pHi contributes to apoptosis, in part by activation of the pro-apoptotic protein BAX (65) and cytochrome C activation of caspases (85). Dysregulated pHi dynamics is also a hallmark of diseases, including a constitutive increase in pHi in cancers of different tissue origins and genetic backgrounds (18; 147), and a constitutive decrease in pHi in a number of neurodegenerative disorders (48; 133). In this review we present recent evidence supporting the view of pHi dynamics as a signaling mechanism and propose that this is best understood at the molecular level by considering protonation as a reversible post-translational modification.
Central to the view of signaling by pHi is an understanding of how physiological changes in pH regulate the function of selective proteins, termed pH sensors, by changing activities and binding affinities (127; 139). There is abundant structure-based evidence on how pH dynamics regulates activities of ion channels (56; 78; 117), ion transport proteins and pumps (58; 96) and enzymes (43; 61; 91). The protonation state of titratable residues also regulates the electrostatic energy of interactions to change binding affinities, including protein-protein, protein-phospholipid and macromolecular assemblies. Protein-protein binding can also be accompanied by proton uptake or release, leading to changes in the protonation states of ionizable residues (69; 84; 89). Also, pH regulation of binding affinities is a signaling mechanism inducing dynamic changes in protein localization, including the recruitment of cytosolic proteins to the plasma membrane or to intracellular membranes by pH-dependent binding to membrane-specific phospholipids (35; 54; 72). In turn, localization puts a constraint on the pH dependence of proteins to match subcellular differences in pHi (20). In mammalian cells, the cytoplasm, nucleus and endoplasmic reticulum have a near neutral pH, mitochondria are more basic, and lysosomes, endosomes and the Golgi network are more acidic (19). As recently noted (37), the plausible adaptation of protein-protein interactions to the pH in subcellular compartments has generally escaped the attention of researchers. Although there are limited structural analyses of pH-dependent protein-DNA binding (79) but given the importance of electrostatics in protein-DNA binding, the protonation state of titratable residues could markedly affect interactions with the phosphate backbone of DNA. We propose that the molecular basis for how physiological changes in pH regulate protein activities and binding affinities can be viewed as protonation being a post-translational modification.
PROTONS AS POST-TRANSLATIONAL MODIFICATIONS
As a mechanism for protein regulation, protons have much in common with small chemical post-translational modifications, particularly those that change protein charge in a site-specific manner, including phosphorylation and Lys acetylation. Although the proton is a particularly small chemical modification, its addition nonetheless increases the charge by one unit, and changes a hydrogen-bond acceptor to a group that can serve as a hydrogen-bond donor. Like phosphorylation, acetylation and ubiquitination, post-translational modification by protons can directly modulate binding to other macromolecules or small molecules, or can drive changes in conformation and dynamics that then modulate some aspect of function (99).
Moreover, changes in protonation state are rapid and reversible. One major difference compared with other post-translational modifications is that protonation or deprotonation does not need to be enzymatically catalyzed, such as transferases for adding methyl groups or kinases and phosphatases for adding and removing phosphate groups.1 Accordingly, an enzyme-mediated transfer “cascade” analogous to mitogen-activated protein kinases (MAPK) modules likely does not occur with pH-driven signaling.
The protonation state of any titratable residue is not in general uniquely defined. Rather, the pH and pKa together determine the probability of being in a particular protonation state, or the fraction of a population of protein molecules in a particular state. This property is similar to other post-translational modifications like phosphorylation, in that the fraction of proteins modified on a site can vary dramatically at any given time. However, single titratable groups can also rapidly change protonation state over time, with the fractional occupancy determined by the pH and pKa.
Virtually all proteins have titratable groups, and one major challenge in studying their regulation by pH is determining which sites could potentially serve as functionally relevant pH sensors in vivo. A similar challenge exists for other post-translational modifications; identifying a site of modification by mass spectroscopy does not guarantee that it has any significant functional role. Recent analyses of post-translational modifications by phosphorylation or acetylation suggest that only a fraction of identified modification sites likely have a significant biological role (9; 70). The challenge is magnified for protons because they are labile and cannot be detected directly by mass spectroscopy or by antibodies. Hence, large-scale proteomics analyses that have been used to identify phosphorylation, methylation or ubiquitination are not feasible for protonation state. Although bioinformatics searches based on isoelectric points (pI) have attempted to identify pH sensors, they have not detected meaningful correlations (37). However, unbiased systematic analyses of protein-protein complexes using Protein-Protein Docking Benchmark database (http://zlab.bu.edu/zdock/benchmark.html) (84; 89) have generated more global predictions on binding affinities dependent on physiological pH.
Although any titratable site is in principle a candidate pH sensor, among the many titratable sites in proteins only a minority titrates significantly within the normal cytosolic pH range. Histidine side chains are the most obvious candidates, with a nominal pKa of ~6.5, which in many cases can be modestly up-shifted due to the protein environment. Phosphate groups have a similar pKa, but the pKa’s of other common side chains are mostly far from 7, meaning that they are potentially relevant to cytoplasmic regulation only when the protein environment dramatically shifts the pKa, due to desolvation or the local electrostatic potential.
Computational methods for predicting pKa’s have been developed over several decades (1), including Multi Conformation Continuum Electrostatics (MCCE), Constant pH Molecular Dynamics (CpHMD) (145), and PROPKA (http://propka.ki.ku.dk). Although still imperfect, these methods are nonetheless powerful tools for helping to identify potential pH-sensing residues. Other computational methods such as molecular dynamics can make predictions about how pH-driven changes in protonation state may affect the structure and dynamics of a protein (90), as has also been done for post-translational modifications such as phosphorylation and acetylation (99). The most powerful biophysical methods for studying these effects in vitro are based on nuclear magnetic resonance (NMR), which can directly monitor titrating protons, and provide information about resulting changes in dynamics and conformation. However, advances in neutron diffraction crystallography may facilitate structural biology studies of pH sensors (156). In contrast to x-ray diffraction, which detects electron density, neutron diffraction can provide direct information on the positions of nuclei including protons and especially deuterons, which have a larger scattering cross-section.
As with other post-translational modifications, the most straightforward route to establishing the biological role of a particular titrating site is to mutate it to a non-titrating side chain. For example, Asn or Gln are reasonable isosteres for neutral His, in that the size is approximately the same, and they contain one hydrogen bond donor and one acceptor in similar positions (figure 1). Mimicking the positively charged form of His is more problematic in that Lys and Arg are substantially longer. In a similar way, Glu is an imperfect isostere for phosphorylated side chains such as pSer (82), and thus the results of such mutational experiments must be interpreted cautiously.
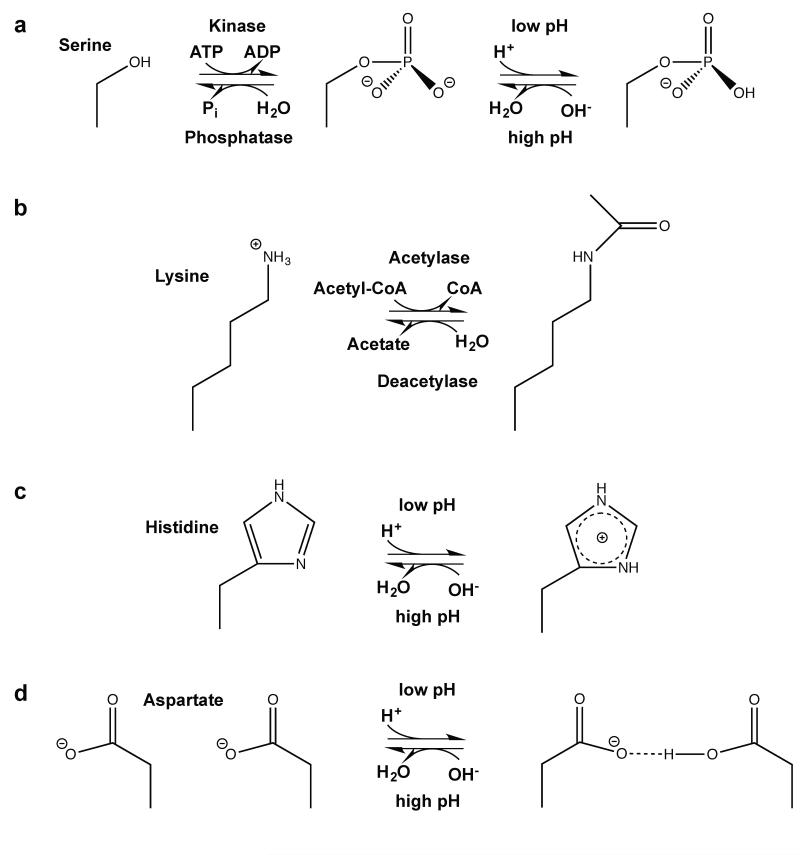
Figure 1.
Examples of amino acid post-translational modifications associated with changes in charge. (a): Phosphorylation of e.g. serines (shown), threonines, tryptophans and histidines leads to addition of a negative charge at weakly basic conditions (151). (b): Lysine acetylation shields the lysine amino group e.g. to decrease the affinity to DNA (126), (c): Histidines can quickly abstract protons to shuttle protons or to function as a pH sensor site, (d): When pKa values are upshifted, protonation of carboxyl groups of glutamate or aspartate (shown) can lead to formation of new hydrogen bonds important for conformational changes of proteins.
There remain important conceptual challenges to progress in understanding the molecular basis for pH regulation. A very fundamental one is the meaning of pHi in cells with small volumes. For example, in a typical prokaryotic cell with a volume of approximately 1 fL (10−15 L) and a pH of 7 (H+ concentration 10-7 M), there are only ~60 free protons, and at pH 7.5 only ~19. Such low numbers raise the question of whether the pH can be precisely defined in such a small volume, either theoretically or practically, for example by using pH-sensitive fluorescent dyes (BCECF, SNARFL) or genetically encoded biosensors (pHluorin, pHTomato). However, it should be noted that the number of labile protons - those on titratable site of macromolecules or metabolites - is many orders of magnitude larger, and these will exchange rapidly with those in the bulk. Thus, a pH reporter or a pH sensing protein is responding to the chemical potential of the overall pool of labile protons, and not just those that are free in solution.
Eukaryotic cells of course typically have dramatically larger volumes, although similar issues arise for defining pH in small sub-cellular compartments. Somewhat more problematic is the concept of pH gradients within a cellular compartment, such as local pH near a membrane, which is postulated to be modulated by electrostatic effects from lipid headgroups, or pH gradients due to large localized fluxes of protons by transporters (86). Although these are intriguing concepts, there are significant practical challenges associated with measuring such effects. With respect to pH gradients, a key parameter is the effective diffusion rate of protons. In pure water, protons diffuse exceptionally rapidly, aided by the Grotthuss mechanism (27). In water containing large concentrations of titratable sites (buffer), Grotthuss-based diffusion likely still occurs, but diffusion is highly hindered in that the effective path length for a free proton is small, and the effective diffusion rate of protons is limited by the rate of diffusion of the buffering sites (132). In cells, this effective rate of diffusion would likely remain rather high (higher than the diffusion rate of macromolecules) due to relatively high concentrations of metabolites with titrating groups, such as carnosine. As such, protons rapidly equilibrate between different portions of a cell not segregated by membranes, and maintaining a stable pH gradient requires large fluxes of protons. Hence, hydrogen ions are not generally “free” in the cytosol but rather complexed as hydronium ions or with proteins and metabolites.
MODES OF PROTEIN REGULATION BY PROTON POST-TRANSLATIONAL MODIFICATION
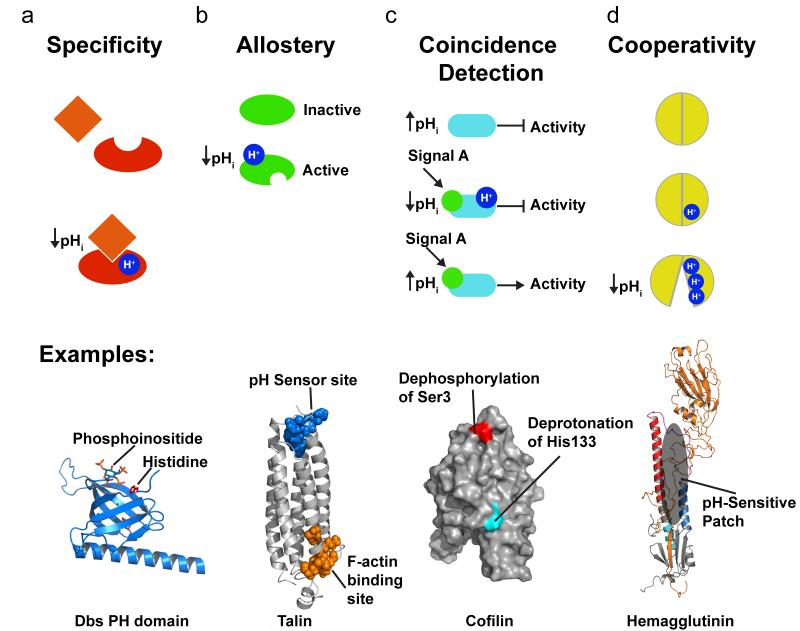
Although changes in pHi are pleiotropic, specificity of pH sensing is achieved by many of the mechanisms described above, most notably that among the many titratable sites in proteins only a small minority titrates significantly within the physiological pH range. Comparing structurally related proteins or protein domains reveals a number of examples of specificity in pH sensing (figure 2a). The pleckstrin homology (PH) domain of several guanine nucleotide exchange factors (GEFs), including GRP1 (general receptor for 3-phosphoinositides 1) (50) and Dbs (Dbl’s big sister) (35), contain pH-sensitive His switches for phosphoinoside binding. As discussed in the section below on pH sensing by actin regulatory proteins, specificity of pH sensitive PH domains is determined by critical His resides at phosphoinositide-binding sites that confer pH-dependent electrostatic interactions with negatively charged phospholipids. There are also examples of members of similar protein families having opposite regulatory responses to physiological changes in pH. The related actin severing proteins cofilin and twinfilin are respectively activated by increased (34; 109) and decreased pH (93). The focal adhesion kinase FAK and the related family member proline-rich tyrosine kinase 2 Pyk2 are structurally similar pH sensors; however, autophosphorylation of FAK increases at pHi > 7.2 (64; 137) and autophosphorylation of Pyk2 increases at pHi < 7.2 (75; 110).
Figure 2.
Signaling modes regulated by pHi. (a) Protonation can regulate specificity as in protonation of a histidine in some but not all Rho GEFs that is required for stereospecific interaction with phosphoinositides. (b) An allosteric regulation mode occurs when protonation of a distinct site induces a conformational change a remote, as in talin that has an actin binding side ~40 Å away from the pH sensor (pdb code 2JSW). (c) In coincidence detection two distinctly and generally unrelated input signals as necessary for the output of protein function, as shown for cofilin that requires dephosphorylation of an N-terminal serine and deprotonation of a C-terminal histidine for increased activity. (d) Cooperativity occurs when several protonation sites act together with electrostatic coupling affecting titration and sometimes pKa shifts, as occurs with disrupted interactions of hemagglutinin HA1 domain with the HA2 domain.
Post-translational modification by protons can also allosterically regulate protein function (figure 2b). The Bohr effect of pH-driven changes in the affinity of hemoglobin for oxygen binding is the classic example of proton-induced allosteric regulation (39). The charge interaction of a His-Asp salt bridge (His146 and Asp94 in human hemoglobin) is disrupted when His146 is deprotonated with increased blood pH, which induces conformational changes that increase oxygen-binding affinity at a distant site. In sections below we describe how protonation of pH-sensing residues allosterically regulates talin binding to actin filaments (127) (figure 2b) and unmasking of a myristoyl moiety in the human immunodeficiency virus gag/MA protein (33). Although not included in examples described below, the structural basis for allosteric regulation by pH has been determined for a number of functionally distinct membrane ion transport proteins and channels. Examples include System A (SNAT2) and System N (SNAT5) amino acid transporters (7); many K+ channels, including but not limited to Kir1.1 (112; 117), KcsA (134; 136; 161) and Kv1 (78); the Ca2+-ATPase (SERCA) (96); and NhaA, a Na-H exchanger in Escherichia coli. The pHi regulation of NhaA, which in turn maintains pHi homeostasis, is a feedback mechanism that is achieved by clusters of electrostatically coupled amino acids with shifted pKa’s near a negatively charged ion funnel that regulate long-range conformational changes affecting a distinct H+ exchange site (58; 101). Similar pH-dependent allosteric regulation of mammalian Na-H exchangers and other ion transport proteins controlling pHi homeostasis such as the proton-pumping V-ATPase is predicted although not structurally confirmed.
Another regulatory mode that is becoming increasingly apparent is the role of protonation in coincidence detection with other posttranslational modifications, including phosphorylation or binding of membrane phospholipids (figure 2c). Coincidence detectors require multiple inputs for a regulated output. In the section on pH-sensing actin regulatory proteins we describe the structural mechanisms for coincidence regulation of cofilin by phosphorylation of an N-terminal serine and protonation of a C-terminal histidine (figure 2c). Other examples include activity and trafficking of the gap junction protein connexin-43, which require changes in protonation state and ubiquitination (73) and dimer dissociation of dynein light chain LC8, which is regulated by a phosphorylation-dependent increase in the pKa of a critical histidine (154). Coincidence regulation by protonation allows integration of regulatory circuits, and viewed from the perspective of pHi dynamics being pleiotropic provides spatial discrimination by a locally restricted second regulator, like a kinase or ubiquitinating enzyme.
In the sections below we describe examples with structural details for distinct modes of protein regulation by proton post-translational modification. Several of these examples highlight two additional properties of regulation by protons. First is cooperativity involving electrostatic coupling of multiple proton or ligand binding sites. For example, protonation of one amino acid can induce changes in the pKa or cation binding of nearby residues, as described below for the influenza virus protein hemagglutinin (figure 2d). Second is the ability to regulate multiple proteins in unison to control a complex cell behavior. This latter principle is achieved by the pleiotropic nature of pHi dynamics, independence from enzymes, signaling specificity and coincidence regulation – and is best exemplified by pH sensors that collectively mediate pHi-dependent actin cytoskeleton remodeling during cell migration.
pH-SENSING ACTIN REGULATORY PROTEINS
The assembly of globular (G) actin to filamentous (F) actin and higher order filament structures and the reverse process of filament disassembly drive many cell processes, such as migration, contraction, vesicle trafficking, motility of pathogens in host cells, and cancer cell invasion and metastasis. Although intrinsic actin assembly rates in vitro are faster at acidic pH (146; 162), likely due to electrostatic effects, de novo actin assembly in mammalian cells requires pHi > 7.2 (147). Hence, how pH regulates purified actin is overridden in cells by the action of selective pH sensing actin regulatory proteins that respond to small changes in pHi of 0.3-0.4 units to induce dramatic differences in actin filament assemblies and architectures.
ADF/Cofilin
Members of the actin depolymerization factor (ADF)/cofilin (AC) family, including actin-depolymerizing factor (ADF), non-muscle cofilin-1 and muscle cofilin-2, are pH sensors that sever and nucleate actin filaments (3). In migrating cells, cofilin severing activity increases filament disassembly at the rear of actin networks and generates new filament free barbed ends for nucleation and assembly at the plasma membrane (10) (figure 3a). Cofilin severing activity requires a coincidence activation mechanism of dephosphorylation of a serine residue in the N-terminus and deprotonation of a C-terminal histidine residue (figure 3b). These two distinct mechanisms are independently regulated and are not cooperative (34).
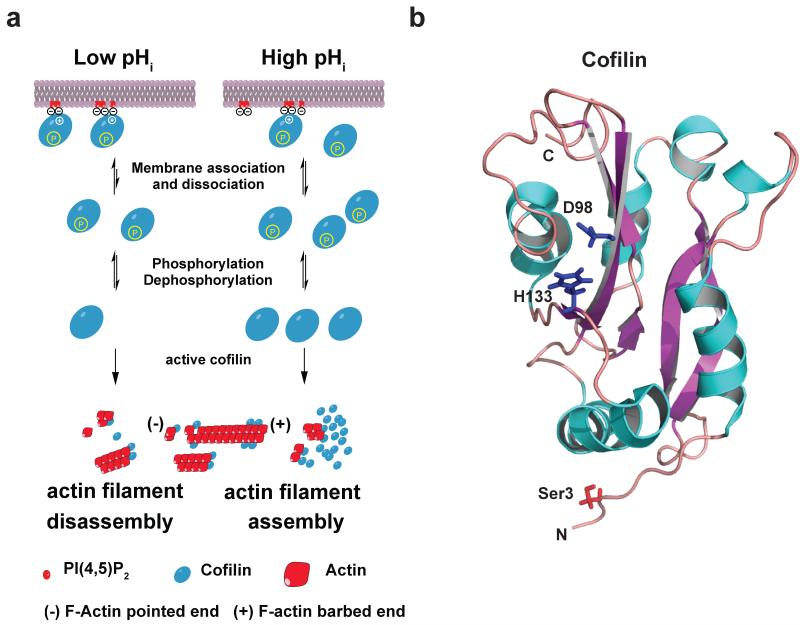
Figure 3.
Coincidence regulation of cofilin: (a) Model for coincidence detection of cofilin near the plasma membrane. At lower pHi < 7.2, the affinity of cofilin is higher, increasing binding to plasma membrane PI(4,5)P2 (red). At higher pHi > 7,2, less cofilin is bound to PI(4,5)P2, increasing the cytosolic pool of active cofilin if Ser3 is dephosphorylated. At higher pHi actin assembly is increased partly due to higher cofilin concentration. (b) Cartoon representation structure of human cofilin structure (pdb code: 1q8x) (109). The five α-helices are colored blue and the six β-sheets are colored purple. Side chains of Ser3, Asp98 and His133 are shown. N-terminal Ser3 is modified by phosphorylation. A salt bridge between Asp98 and His133 is formed under slight acidic conditions. His133 closely interacts with PI(4,5)P2 when doubly protonated (34).
For severing activity, two sites in A/C proteins bind to actin filaments. A G site includes an N-terminal region from the β1-strand, an N-terminal portion of the α1 helix and a central region from helices α4 to α5. An F site comprises an N-terminal portion of β5 and a region spanning from the C-terminal half of α6 to β7 (Figure 3b). For cofilins, a phosphate group on Ser3, which is added by LIM or TES kinase and removed by chronopHin or slingshot 1L phosphatases, prevents actin binding at the G site (10). It was previously speculated that cofilin phosphorylation causes extensive perturbations affecting critical residues in the G site (42; 109). Subsequently, molecular dynamics simulations suggested a plausible mechanism (34) whereby an electrostatic interaction between a phosphate group at Ser3 and Lys126 and Lys127 in the α4 helix sterically blocks actin filament binding at the G site. Dephosphorylation disrupts the electrostatic network and allows direct interaction between charged Lys126 and Lys127 with actin.
Severing activity also requires an alkaline pH (10). However, pH dependence differs between family members and acts through different mechanisms. Of the three mammalian isoforms, ADF is the most pH sensitive (143) and severing activity of yeast cofilin and the Acanthamoeba cofilin homologue actophorin are relatively pH-insensitive (80). A structural basis for these differences is indicated by an NMR study of human cofilin (109), which suggests a pH-dependent salt bridge in the F site between His133 and Asp98 in the β4-strand (figure 3a). The salt bridge is stable at slightly acidic conditions but likely disrupted at higher pH values leading to partial unfolding of the F site, and presumably increased actin binding. The relatively pH-independent actophorin and yeast cofilin lack this conserved histidine. Differences in pH sensitivity between ADF and cofilin are likely due to cofilin being more stable because of tighter packing of its C terminus to the central β-sheet (49; 109). Hence, because cofilin has more non-covalent interactions than ADF, disruption of the salt bridge by increasing pH has a more destabilizing effect on the ADF structure.
However, NMR analysis of cofilin at pH 7.5 compared with pH 6.5 shows minimal chemical shift differences in residues within the F site (34), suggesting an alternative mechanism independent of conformational changes for pH-dependent activity. Two recently reported alternative mechanisms include binding to PI(4,5)P2 in the plasma membrane (34) and to cortactin in invadopodia – invasive plasma membrane protrusions of cancer cells (81). Binding to PI(4,5)P2 maintains cofilin inactive at the plasma membrane but locally available for the rapid assembly of actin filaments (74; 141) (figure 3a). How cofilin is inactive when bound to PI(4,5)P2 was resolved by an NMR study (42) showing that the PI(4,5)P2-binding site includes His133 and basic residues in the F site. Hence PI(4,5)P2 and actin binding are mutually exclusive. We showed that cofilin binding to PI(4,5)P2 is pH-dependent, with increased binding at lower pH (34). Computational docking simulations suggest that doubly protonated His133 but not neutral His133 interacts with terminal phosphates of PI(4,5)P2. However, both phosphates in PI(4,5)P2 have pKa’s close to neutral (68), and cofilin includes alternative PI(4,5)P2-binding sites at basic residues near His133 (34; 160), which could contribute to fine-tuning binding avidity and maintaining a membrane-sequestered pool of cofilin (figure 3a). Binding to cortactin also sequesters inactive cofilin at the distal membrane of invadopodia. The binding affinity of cofilin for cortactin decreases at higher pH (81). Whether a pHi gradient occurs, higher at the distal margin of membrane protrusions as suggested (81), to locally disrupt cortactin-cofilin binding and increase cofilin activity is unclear; however, we propose that with coincidence activation by locally-regulated de-phosphorylation of Ser3, a global increase in cytosolic pH may be sufficient.
Talin
Changes in pHi also regulate dynamic remodeling of actin filaments at cell-substrate (focal) adhesion sites. This regulatory mechanism is particularly critical at the leading edge of migrating cells where increased pHi is necessary for cycles of focal adhesion disassembly (128; 130). The effect of pHi on focal adhesion remodeling is in part mediated by talin binding to F-actin, which tethers actin filaments to focal adhesions. The N-terminal FERM domain of talin binds the cytosolic domain of β-subunits of integrin receptors and contains a pH-insensitive F-actin-binding site (26). The C-terminal I/LWEQ module or THACTCH domain, which is conserved in huntingtin interacting protein-1 (Hip1), the Hip1-related protein (Hip1R/Hip12) and the yeast protein Sla2, also binds F-actin but with established pH dependence (26) (figure 4a).
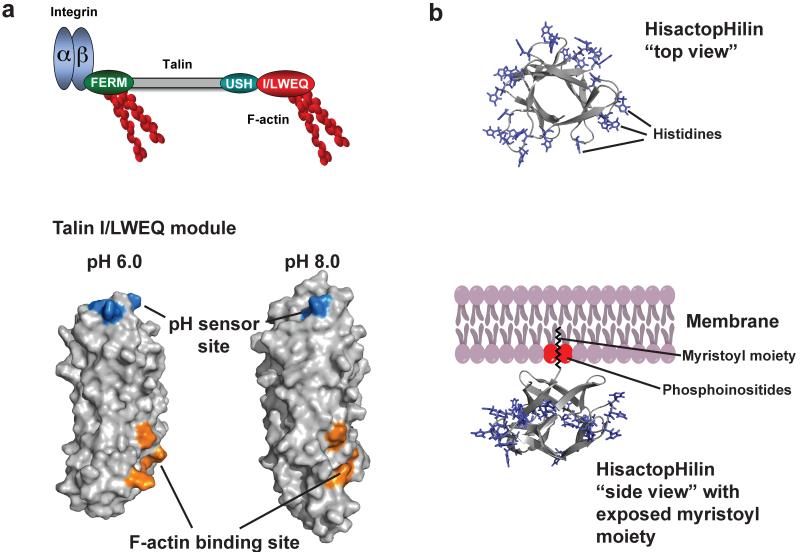
Figure 4.
Allosteric regulation of talin-actin binding by pH. (a) Domain organization of talin (upper panel), including an N-terminal FERM (domain that binds the β-subunit of integrin receptors and F-actin, a central rod domain (gray) and a C-terminal I/LWEQ (red) actin binding module that binding F-actin. F-actin binding by the I/LWEQ module but not by the FERM domain is pH sensitive with more binding at lower pH. Lower panel shows cartoon representation of C terminal actin-binding domain of talin. Residues E2337, E2342, H2418, D2482 form a pH sensor that induces pH-dependent conformations that allow actin binding only at lower pH (b) Cartoon representation of the histactopHilin structure and model of membrane attachment of hisactopHilin (pdb code: 1hcd). “Top view” shows that all histidines in loops in turns are equally distributed around the protein. However, viewed from the “side” these histidines point towards the cytoplasm, while the β-sheets point towards the membrane when the N-terminal myristoyl moiety is exposed partly by PI(4,5)P2.
As a pH sensor, talin highlights three of the properties we describe above on signaling modes, including cooperativity, allostery, and the ability of pHi dynamics to regulate multiple proteins in unison to control a complex cell behavior. As an example of cooperativity, the I/LWEQ module includes a five-helix bundle (40) that contains a cluster of residues - Glu2337, Glu2342, His2418, Glu2481, and Asp2482 - that have markedly upshifted pKa values (128) (Figure 4a). As an example of allostery, NMR and CpHMD simulations indicate that protonation of residues in the pH sensor induces significant changes in the structure and dynamics of a remote actin-binding site to increase actin binding (128) (figure 4a). A mutant talin-H2418F has reduced and pH-insensitive actin binding, indicating a critical role of His2418 in pH-dependent allosteric regulation (128). As an example of coordinated actions of pH sensors, in addition to talin-actin binding dynamics, focal adhesion remodeling also requires FAK activity, which requires pH > 7.2 (64; 137).
HisactopHilin
The slime mold Dictyostelium discoideum expresses the unique, remarkably pH-sensitive actin-binding proteins hisactopHilin I and hisactopHilin II. Both isoforms contain 118 amino acids and nearly a third of the residues are histidines. HistactopHilins bind to actin only at pH values below 7.2, suggesting that protonation of histidines promotes binding to actin in a switch like manner (119). HisactopHilins function in osmoprotection. In response to hyperosmolarity, the D. dicoideum cytosol acidifies and acid-increased hisactopHilin-actin binding generates a rigid osmoprotective actin cytoskeletal network (108). The structure of hisactopHilin has a β-trefoil fold consisting of 12 β-strands connected by turns and loops (figure 4b) (44). The β-trefoil fold is shared by a number of diverse proteins with unrelated amino acid sequence, including interleukin-1β (32) and the mammalian actin-bundling protein fascin, which consists of four β-trefoil domains (60). In hisactopHilin almost all of the histidines are located in loops and are at one side of the asymmetric protein, while the other side of the protein forms a tight β-barrel structure that is inserted into the plasma membrane by myristoylation (8), a common post-translational modification with a C14 fatty acyl chain at the N-terminal glycine residue. Myristoylation is required but energetically not sufficient for membrane targeting (104). HisactopHilin oscillates between a cytosolic form at pH 7.5 and a membrane-bound form at pH 6.5 (46), regulated by a cluster of charged amino acids adjacent to the myristoyl moiety (47). These two forms differ in the pH-dependent orientation of the myristoyl moiety, which can either be bound to a hydrophobic pocket or exposed to bind membranes (125). A similar pH-dependent myristoyl switch regulates multimerization and membrane targeting of the HIV gag/MA protein (33), and is described in more detail in the section below on pH Sensors in Pathogens.
Guanine Nucleotide Exchange Factors (GEFs)
Several GEFs show specificity of pH-dependent binding of their PH domains to membrane phosphoinositides. One example is Dbs, a Dbl family Rho GEF that activates the GTPase Cdc42 at the leading edge of migrating cells to control polarized movement. Dbs contains a PH domain that binds PI(4,5)P2 with higher affinity at lower pH (35). However, other Dbl family Rho GEFs, such as intersectin, contain a similar PH domain that has pH-independent binding to PI(4,5)P2. The PH domain in Dbl family Rho GEFs is adjacent and C-terminal to the catalytic Dbl homology (DH) domain (115) and PI(4,5)P2 binding can allosterically inhibit GEF activity (116). Dbs but not intersectin has a histidine (His843) at the PI(4,5)P2-binding site of the PH domain, which determines pH-sensitive binding (35), similar to pH-dependent binding of cofilin to PI(4,5)P2 as described above. Although pH-dependent phosphoinositide binding has been shown for only a limited number of Rho GEFs, the shared feature of a binding-site histidine could be used to predict pH sensing and hence pH-dependent GEF localization and activity.
The PH domain of Grp1, a GEF for Arf GTPases, also has pH-dependent binding to phosphoinositides. In contrast to Dbs, Grp1 binds PI(3,4,5)P3 at endosomal membranes, with higher affinity binding at lower pH (50), and phosphoinositide binding increases Grp1 activity (29). Increased binding affinity is directly determined by protonation of a histidine (His355) located in a 20-residue insertion within the β6/β7 loop of Grp1 that contacts the phosphate group 4 of inositol 1,3,4,5-tetrakisphosphate (IP4), a soluble analogue of PI(3,4,5)P3 (50; 77). Arf GTPases localize at acidic endosomes and regulate endosomal trafficking and actin dynamics (30). A predicted lower pH at the cytoplasmic side of endosomes (100) possibly regulates Grb1 localization and activity by increasing affinity for PI(3,4,5)P3.
Recent findings indicate physiological pH sensing by a number of other common phosphoinositide-binding domains. The FYVE (Fab1p, YOTB, Vac1p, and EEA1) domain (72), the ENTH and ANTH (Epsin and AP180 N-terminal) domain (54), and the PH domain of FAPP1 (four-phosphate-adaptor protein 1) (51) have increased phosphoinositide binding affinities at physiological pH values below neutral. A common feature of these domains is at least one histidine essential for stereospecific phosphoinositide recognition. Although not experimentally confirmed, proteins containing these domains likely have pH-dependent membrane localization and activity, and if associated with the actin cytoskeleton could coordinately regulate the pH-sensitive actin filament dynamics at membranes.
AMYLOIDOSIS
Although extreme changes in pH affect the folding and stability of most proteins, selective proteins show changes in electrostatic interactions by physiological pH changes that induce partial or full unfolding. Protein misfolding plays a role in pathological conditions such as amyloidosis, a group of more than 20 disparate human diseases including Alzheimer’s disease, Parkinson’s disease and type II diabetes (45; 105). Amyloid fibril formation is due to the folding of the secondary structure of an endogenous protein or protein fragment into a cross-β-sheet quaternary structure that oligomerizes into unbranched filaments. As folding is dependent on the sequence of the protein, misfolding induced by physiological changes in pH is highly specific. For example, low pH can enhance, in the case of prion protein, or inhibit, in the case of Islet Amyloid Polypeptide Protein (IAPP), amyloid formation through the protonation of specific histidine residues. In contrast to pathological amyloid formation, organisms also use highly regulated amyloid assembly for a diverse set of normal biological functions, including proteins involved in the production of mammalian melanosomes, E. coli biofilms, malarial coat proteins, and silk fibrils from some spider species (45).
Prion Protein and Spongiform Encephalopathies
Spongiform encephalopathies, including scrapie, “mad cow”, Kuru, and Creutzfeld-Jacob disease (CJD), are characterized by aggregation of the misfolded prion protein (PrP), a 209 amino acid protein on the surface of neuronal cells (for review, see (94)). The normal PrPC form of the protein misfolds into an aggregation-prone, protease-resistant and infectious PrPSc form that accumulates in the brain as amyloid plaques. The structure of PrPC includes a disordered, unfolded amino-terminus and a well-structured C terminus, which is comprised of 3 α-helices (A-C) and a short antiparallel β-sheet (113; 159). Although the atomic structure of PrPSc has not been resolved, low resolution structural analyses indicate a significant change in folding between PrPC and PrPSc, specifically a marked increase in β-sheet and decrease in α-helix content (87; 102). What triggers misfolding of PrPC is not known; however, because 1) low pH induces the formation of alternatively folded variants of PrP and 2) both PrPC and PrPSc are cycled through the endocytic pathway, it has been hypothesized that the conversion of PrPC into PrPSc occurs in the acidic environment of endosomes (17; 28; 144). Molecular dynamics simulations indicate that protonation of a critical His residue, His187, is important for the stability of PrPC; deprotonation at neutral pH increases β-sheet content and conformational mobility within PrPC (55; 140). His187, positioned between the 3 helixes, is partially buried and has a significantly downshifted pKa value of ~4 (6; 55). At neutral pH, His187 is hydrogen bonded to the backbone carbonyl of Arg156 (4; 66; 140). This salt bridge is not seen upon protonation of His187 or in a mutant PrP-H187R found in familial CJD. The positively charged Arg substitution results in a β-sheet-rich, aggregation-prone molecule. Because stabilizing the His187-Arg156 salt bridge at low pH would prevent misfolding of PrPC, it could be an effective therapeutic strategy to decrease infectivity of PrPSC.
IAPP and Type II Diabetes
Islet Amyloid Polypeptide (IAPP or amylin), a protein secreted from pancreatic β-cells, is the predominant component of amyloid flibrils found in a majority of patients with type II diabetes (150). Human IAPP (hIAPP) is a 37 amino acid peptide that is stored in the insulin secretory granule (pH ~6) and is co-secreted into the extracellular space (pH ~7.4) with insulin (24; 53; 150). The progression of soluble, monomeric hIAPP to a misfolded amyloid fibril is pH dependent – inhibited at the acidic pH of the secretory granule and enhanced at the higher extracellular pH (13; 67; 88). Although the precise mechanism of pH-dependent aggregation is unknown, one hint comes from rat IAPP (rIAPP), which in contrast to hIAPP does not form fibrils in solution. However, a single point mutant in rIAPP, Arg18His, (Figure 5a) is necessary and sufficient to induce fibrillogenesis (153). Above the pKa of His18, peptides encoding residues 10 to 19 of hIAPP aggregate to form amyloid whereas aggregation is slower at lower pH. Substitution of His18 to Ala abolishes pH-dependent aggregation (138). Further, insulin binding, which maintains an α-helical conformation of hIAPP and inhibits fibril formation, is dependent on the charged state of the amino acid residue at position 18. Introducing a charge either by protonation of His18 in hIAPP or an Arg18 substitution in rIAPP induces the formation of hydrogen bonds and salt bridges with the insulin β-chain (62; 148). Deprotonation of His18 at neutral pH reduces the polar interactions, destabilizes the α-helical motif of insulin-bound hIAPP, and decreases the affinity of hIAPP for insulin (148). Similarly, the hIAPP-membrane interaction, which facilitates hIAPP aggregation and amyloid toxicity, is regulated by pH. Using a truncated hIAPP construct (residues 1-19), it has been demonstrated that introduction of a charge at His18 by either protonation or mutation to Arg changes the hIAPP-membrane topology from buried to a surface associated conformation (12; 98). The structure of membrane-bound hIAPP has a kinked helix motif, with a neutral pH having a much more pronounced inter-helical angle (30° pH ~4.6, compared to 85° pH 7.3) (figure 5b) (97; 103). The change in membrane topology and conformation is linked to a reduced ability to disrupt phospholipid vesicles and cell membranes (12; 98). Taken together, these data suggest that when hIAPP is released from the acidic insulin secretory granule into the neutral pH of the cytoplasm or extracellular space, His18 becomes de-protonated, which decreases affinity for insulin, and enhances misfolding to promote amyloid fibril formation and cytotoxicity.
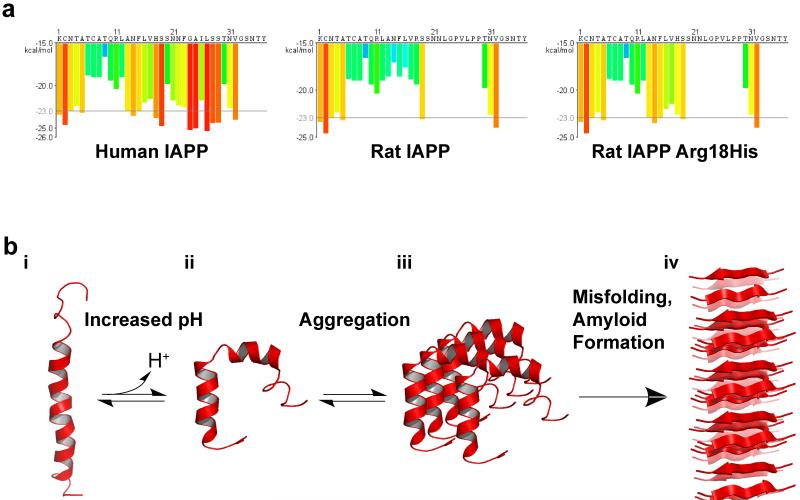
Figure 5.
Structural Regulation of hIAPP by pH. (a) IAPP sequences and segment propensity for fibril formation. The predicted energy for fibrillation of every six-residue segment of human IAPP, rat IAPP, and rat His18Arg IAPP are shown. Warmer colors represent a greater propensity for fibrillation, with red histogram bars represent hexapeptides that are predicted to form fibrils. Due to variations in the sequence, human IAPP has a much higher propensity to form fibrils than mouse IAPP. Mutation of Arg18 to His increases the propensity of rat IAPP to form fibrils. Graphs were generated using ZipperDB (http://services.mbi.ucla.edu/zipperdb/)(41) and modified from (153). (b) Potential model of pH dependent amyloid formation by IAPP. i) hIAPP, located in the insulin secretory granules, is bound to insulin b-chain (not shown) or to the membrane surface in an extended kinked helix conformation. ii) As IAPP is released from the acidic environment of the vesicle to the neurtral pH of the cytoplasma or extracellular space, His18 becomes deprotonated, weakening the insulin-hIAPP interaction and promoting insertion of hIAPP in the membrane by inducig a change in conformation. iii) Membrane associate hIAPP aggregates and, iv) undergoes further conformational changes leading to the formation of β-sheet-rich amyloid fibrils. Low pH structure: 2KB8 (103). Neutral pH structure: 2L86 (97). In both structures, the N terminus is located at the bottom of the figure. Figure adapted from (97; 150)
pH-SENSORS IN PATHOGENS
The pH-dependent dynamical structure of many bacterial and viral proteins is well characterized and examples highlight many principles of post-translational modification by protons, including allostery, specificity, and cooperativity. A mechanism shared by bacterial toxins and enveloped and non-enveloped viruses is spatial and temporal disassembly to transit host cells membranes. Most common is disassembly at endosomal membranes and is hence triggered by acidic pH. One example is the anthrax toxin component PA63 that undergoes a dramatic acidic pH-induced conformational change in the endosomal compartment. Protonation in loop β2-β3 region of PA63 causes structural reorganization and formation of a pore required for translocation of anthrax toxin subunits into the cytosol (157). Other toxins with confirmed pH-induced conformational changes include diphteria toxin (106; 114), botulinum toxin (124), and cholera toxin B (31). Here, we describe well-characterized pH-regulated structural changes necessary for the function of three viral proteins, hemagglutinin and the M2 pump in influenza virus, and the gag/matrix protein in human immunodeficiency virus (HIV).
Hemagglutinin (HA)
The influenza glycoprotein HA mediates receptor binding and membrane fusion and is an example of cooperativity in pH sensing. Pathogenesis of the enveloped influenza virus that causes seasonal flu requires dramatic structural changes of HA driven by transition from a neutral to an acidic pH (15). The precursor form HA0 contains protease cleavage sites to generate the subunits HA1 and HA2 that forms protein complexes on the surface of the influenza virus. HA1 is important for recognition of sialic acid receptors but also keeps HA2 in a prestressed conformation until exposure to acidic pH that induces a large conformational change in HA2 to drive membrane fusion (21).
Native HA is a trimer, with HA1 and HA2 linked by a disulfite bond. The C-terminus of HA2 is anchored in the viral membrane, and a triple coiled-coil formation of the helical segments C and D confers formation of a homotrimer of HA2. At neutral pH, a helix A packs against C and D connected by a 20 residue loop B (149) and the hydrophobic N-terminus of helix A, which contains the fusion peptide, is buried within the all-overall structure (figure 6a). However, at acidic pH, loop B adopts a helical structure that induces a change in the orientation of helix A from antiparallel to the end of a now elongated helix comprised of A, B and C (15). With this structural rearrangement, the fusion peptide moves 100 Å and drives insertion of the fusion peptide into the host cell membrane (figure 6a). Another consequence is that helical segment D now packs against the triple stranded coiled-coil, producing a new hydrophobic core that stabilizes this conformation. The HA2 C-terminal helix H close to the viral membrane transitions becomes unstructured, which gives the HA complex the necessary flexibility for membrane fusion (15). These conformational changes require cooperativity in pH regulation by histidine-rich patches throughout HA1 and HA2 (patch 1 includes HA1 residues His18 and His38; HA2 residues: His111, patch 2 includes: HA1 residues His47, His275, His/Lys285, His298, Lys46 and Lys50) that are highly conserved despite a high viral mutagenesis rate (129). Protonation of these histidines disrupts a number of contacts between HA1 and HA2, and an interaction of HA1 histidine residues with a basic region at the base of the globular domain. Although the “fusion” conformation of HA2 is thermodynamically favored, interactions with HA1 maintain HA2 in a metastable prefusion conformation that is relieved at low pH. In the absence of HA1, the HA2 domain spontaneously adopts the fusion conformation at neutral pH (22; 23).
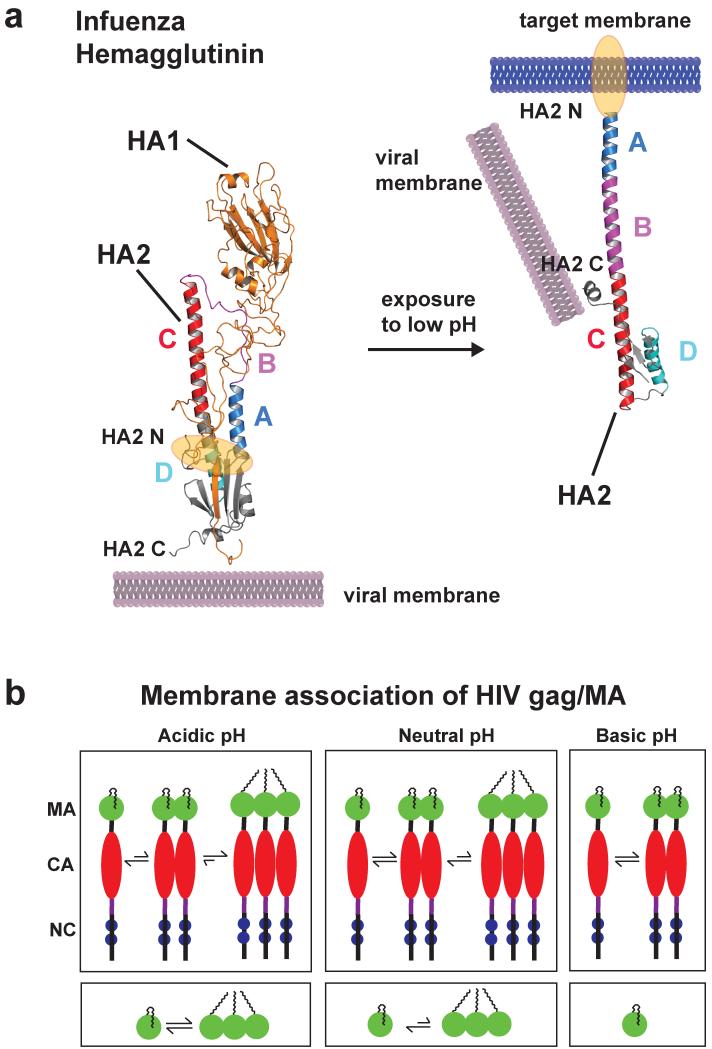
Figure 6.
Low pH induces a large conformational change of the influenza HA2 protein (only one monomer is shown). (a) The structure of prefusion complex (left) of HA1 (gold, pdb code: 2hmg) and HA2 (multicolor, pdb code: 1htm) and the HA2 at the fusion pH (right) structure are shown as cartoon representations. In the prefusion complex the HA2 has a metastable conformation, and HA1 acts as a clamp to keep HA2 in this conformation. The hydrophobic fusion peptide (highlighted in orange) is buried in a hydrophobic core distant from the tip of the protein complex. Low pH leads to protonation of several charged residues throughout HA1 and HA2, and HA1 dissociates partly. This leads to a spontaneous conformational change of region B (magenta) that becomes an α-helix to form a new continuous helix A (blue), B and C (red). The fusion peptide moves ~100 Å from the core to the tip for insertion into the target membrane. Helix D (cyan) now packs against helix A, and a new hydrophobic core is formed. The C-terminus moves more towards the new N terminus and has more flexibility important for membrane fusion. (b) Schematic representation showing multimerization events of HIV gag and MA proteins as a function of pH (MA: matrix, CA: capsid, NC: nucleocapsid). Decreasing pH promotes myristoyl exposure membrane targeting and formation of multimers of both gag and MA protein, critical mediators for HI virus assembly. Structure figure reprinted with permisson from Jamil S. Saad (33).
M2 pump
The influenza virus also expresses a pH-sensitive proton channel - the M2 pump that is a splice variant of mRNA encoding the matrix (M) protein. Formation of a homotetramer of the 96 amino acid polypeptide M2 generates a minimalistic proton-selective channel that is activated by cooperative proton post-translational modifications in the host cell endosome to acidify the virion for virus release (120; 131). Assembly and activity are regulated by the protonation state of a single histidine residue (His37) in the transmembrane (TM) domain of each M2 monomer (16). When the M2 pump is assembled in a lipid bilayer, the pKa’s of these four histidines change to 8.2, 8.2, 6.3, and <5.0, with the third imidazole ring crucial for channel activation (57). Recent structural studies using solid-state NMR spectroscopy identified a low pH (pH 4.5) and high pH (pH 8.5) state of the M2 pump (56). At high pH the four histidines are neutral (57) and their imidazole rings pack in an edge-face stacked fashion, creating an electron-rich region that prevents formation of an H-bonded water chain and disrupts Grotthuss hopping (56). At low pH, three of the four histidines are protonated and imidazolium rings repel each other, causing backbone conformational changes that result in a wider pore. In this model, water molecules in the C-terminal part of the pore are protonated after proper alignment with charged imidazoliums, which then flip back to bind another proton. Microsecond reorientations of the histidine actively transport protons into the viral interior. Here, cooperative protonation of histidines lead to activation of the M2 pump by requiring a proton threshold determined by pKa’s of the third of the four histidines present in the assembled pump.
Gag/matrix protein
HIV expresses a gag protein that has protonation-induced allosteric unmasking of an N-terminal myristoyl moiety (33). Assembly of the polyprotein HIV gag at punctate sites on the plasma or endosomal membrane is necessary for formation of immature virions. Gag is cleaved by viral protease to generate the proteins matrix (MA), capsid (CA), nucleocapsid (NC), spacer peptide 1 and 2, and P6 during or soon after budding. Subsequently, these proteins reorganize, and intrinsically form virus-like particles. Myristoylation is required but not sufficient for membrane of Gag/MA targeting (14; 25), and requires a cluster of conserved basic residues (158). Gag/MA proteins can expose or sequester the myristoyl moiety, similar to hisactopHilin as described above. The pH-dependent myristoyl switch requires a conserved histidine residue His89 that forms a salt bridge with Glu12 when protonated. Deprotonation destabilizes the salt bridge, increases sequestration of the myristoyl moiety by MA, and leads to monomer formation of MA (figure 6b). Importantly, mutation of His89 impairs correct targeting of gag/MA and significantly reduces virus production significantly (36). As a critical mediator of HIV-1 virus assembly (52), gag/MA could be considered a coincidence detector because in addition to pH-dependent myristoyl exposure its function also regulated by binding to plasma membrane PI(4,5)P2, oligomerization of MA, the presence of CA and calmodulin binding (38).
ENGINEERING pH-SENSITIVE SWITCHES
Structural design principles that we learn from understanding the biology of endogenous pH sensors can be applied to engineering pH sensitive switches. Engineered protein or small molecule switches can use environmental triggers to generate a predicted behavior (95), including ligand binding, membrane permeability, and changes in protein stability and lifetime. One important application is drug delivery. A strategy currently under intensive investigation is exploiting pH changes during the process of endocytosis to facilitate the ‘escape’ of biological therapeutics from the endosome into the cytosol (142).
To exploit the trigger of pHi dynamics or changes in pH between subcellular compartments, engineering changes in the ionization state of surface residues is a feasible approach that has been used for many applications. More difficult is designing changes in the pKa and ionization state of buried residues, although recent evidence with the E. coli protein thioredoxin (107) shows this can also be achieved. However, because the pKa of residues ishighly dependent on the structural environment, predicting the sensitivity of ionizable groups to physiological pH is challenging, even with sufficient structural or simulation data. One solution to this challenge is using a combinatorial histidine library, as recently described for sampling every possible combination of histidine and wild-type residue in a model anti-RNase A single domain VHH antibody (95).
The majority of validated pH-sensitive engineered switches involve histidines, although targeting modification of phosphorylated residues such as Tyr (83; 92; 135) can be achieved, particularly because the pKa of phosphorylated serine, threonine and tyrosine is generally near neutral. Switches using residues that are normally uncharged at physiological pH have also been generated, such as amphiphilic peptides rich in Glu and Leu designed to confer pH-dependent transit in different membrane compartments (71). A few of many examples of generated histidine switches include a modified cytokine granulocyte colony-stimulating factor (GCSF) to alter its endosomal trafficking and increase its half-life and efficacy (118); an interleukin receptor antibody to retain antigen binding in acidic compartments (59); and a Lys>His iso-1-cytochrome c mutant to increase the electron transfer gate (5).
Switches engineered to respond to the dysregulated pH in disease have been developed for direct therapeutic efficacy or for therapeutic targeting and stability. Most common are switches designed for the more acidic extracellular pH and more alkaline pHi of cancer cells. A number of peptides have been developed to exploit the low extracellular pH in tumors for site-specific drug delivery. Examples include soluble pH (Low) Insertion Peptides (pHLIPs) that fold and insert across a membrane to form a stable transmembrane α-helix under acidic conditions (2; 152) and GALA, a pH-responsive peptide that converts from a random coil to an amphipathic a-helix at acid pH to bind bilayer membranes (76). Engineered pH-responsive substrates such as polymers (122; 123; 155) and lipid micelles (63) have also been used as a strategy to release carried drugs at sites of tumor or intracellular vesicle acidity. Although therapeutic approaches to alter pH sensing by endogenous proteins have not been reported, one example would be a strategy to inhibit the pH-dependent activation of cofilin in alkaline cancer cells to limit metastatic progression. Albeit technically challenging, approaches such as this may be feasible as we attain an increased understanding of how protonation acts as a post-translational modification to regulate protein function.
SUMMARY POINTS.
-
-
A proton can be added to charged residues that have a pka near neutral
-
-
Protonation occurs rapid and reversible in absence of enzymes
-
-
Protonation can change the conformation and activity in a specific manner
-
-
Proteins that specific protonation sites that change activity or conformation upon protonation/deprotonation are termed pH sensors
-
-
Protonation can impair regulation modes including specificity, allostery, coincidence detection, and cooperativity
Acknowledgments
We thank members of the Diane Barber and the Torsten Wittmann groups for helpful suggestions; Bree Grillo-Hill for help creating figure 2; Jamil S. Saad for providing the HIV gag/matrix protein graphic. Work on the structure and function of actin-binding pH sensors was supported by a Research Fellowship of the Deutsche Forschungsgemeinschaft (SCHO 1410/1-1) to A.S. and a Canadian Institutes of Health Research Postdoctoral Fellowship to B.A.W. and National Institutes of Health grant GM58642 to D.L.B. MPJ is a consultant to Schrodinger LLC.
Footnotes
Although we know of no such examples, it is intriguing to speculate that protonation state changes could effectively be catalyzed if, e.g., binding of a partner protein caused conformational changes that caused the pKa of a titratable group to change dramatically, and the resulting change in protonation state then regulates other aspects of function.
LITERATURE CITED
We apologize to those in the field whose work we were unable to cite because of space limitations
- 1.Alexov E, Mehler EL, Baker N, Baptista AM, Huang Y, et al. Progress in the prediction of pKa values in proteins. Proteins. 2011;79:3260–75. doi: 10.1002/prot.23189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreev OA, Engelman DM, Reshetnyak YK. pH-sensitive membrane peptides (pHLIPs) as a novel class of delivery agents. Mol Membr Biol. 2010;27:341–52. doi: 10.3109/09687688.2010.509285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Molecular cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Antonyuk SV, Trevitt CR, Strange RW, Jackson GS, Sangar D, et al. Crystal structure of human prion protein bound to a therapeutic antibody. Proc Natl Acad Sci U S A. 2009;106:2554–8. doi: 10.1073/pnas.0809170106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baddam S, Bowler BE. Conformationally gated electron transfer in iso-1-cytochrome c: engineering the rate of a conformational switch. J Am Chem Soc. 2005;127:9702–3. doi: 10.1021/ja0527368. [DOI] [PubMed] [Google Scholar]
- 6.Bae SH, Legname G, Serban A, Prusiner SB, Wright PE, Dyson HJ. Prion proteins with pathogenic and protective mutations show similar structure and dynamics. Biochemistry. 2009;48:8120–8. doi: 10.1021/bi900923b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baird FE, Pinilla-Tenas JJ, Ogilvie WL, Ganapathy V, Hundal HS, Taylor PM. Evidence for allosteric regulation of pH-sensitive System A (SNAT2) and System N (SNAT5) amino acid transporter activity involving a conserved histidine residue. Biochem J. 2006;397:369–75. doi: 10.1042/BJ20060026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behrisch A, Dietrich C, Noegel AA, Schleicher M, Sackmann E. The actin-binding protein hisactophilin binds in vitro to partially charged membranes and mediates actin coupling to membranes. Biochemistry. 1995;34:15182–90. doi: 10.1021/bi00046a026. [DOI] [PubMed] [Google Scholar]
- 9.Beltrao P, Albanese V, Kenner LR, Swaney DL, Burlingame A, et al. Systematic functional prioritization of protein posttranslational modifications. Cell. 2012;150:413–25. doi: 10.1016/j.cell.2012.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernstein BW, Bamburg JR. ADF/cofilin: a functional node in cell biology. Trends in cell biology. 2010;20:187–95. doi: 10.1016/j.tcb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boron WF. Regulation of intracellular pH. Adv Physiol Educ. 2004;28:160–79. doi: 10.1152/advan.00045.2004. [DOI] [PubMed] [Google Scholar]
- 12.Brender JR, Hartman K, Reid KR, Kennedy RT, Ramamoorthy A. A single mutation in the nonamyloidogenic region of islet amyloid polypeptide greatly reduces toxicity. Biochemistry. 2008;47:12680–8. doi: 10.1021/bi801427c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brender JR, Lee EL, Hartman K, Wong PT, Ramamoorthy A, et al. Biphasic effects of insulin on islet amyloid polypeptide membrane disruption. Biophysical journal. 2011;100:685–92. doi: 10.1016/j.bpj.2010.09.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci U S A. 1990;87:523–7. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 16.Cady SD, Luo W, Hu F, Hong M. Structure and function of the influenza A M2 proton channel. Biochemistry. 2009;48:7356–64. doi: 10.1021/bi9008837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campos SR, Machuqueiro M, Baptista AM. Constant-pH molecular dynamics simulations reveal a beta-rich form of the human prion protein. The journal of physical chemistry. B. 2010;114:12692–700. doi: 10.1021/jp104753t. [DOI] [PubMed] [Google Scholar]
- 18.Cardone RA, Casavola V, Reshkin SJ. The role of disturbed pH dynamics and the Na+/H+ exchanger in metastasis. Nat Rev Cancer. 2005;5:786–95. doi: 10.1038/nrc1713. [DOI] [PubMed] [Google Scholar]
- 19.Casey J, Grinstein S, Orlowski J. Sensors and regulators of intracellular pH. Nat Rev Mol Cell Biol. 2010;11:50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- 20.Chan P, Warwicker J. Evidence for the adaptation of protein pH-dependence to subcellular pH. BMC Biol. 2009;7:69. doi: 10.1186/1741-7007-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J, Lee KH, Steinhauer DA, Stevens DJ, Skehel JJ, Wiley DC. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell. 1998;95:409–17. doi: 10.1016/s0092-8674(00)81771-7. [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Skehel JJ, Wiley DC. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc Natl Acad Sci U S A. 1999;96:8967–72. doi: 10.1073/pnas.96.16.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Wharton SA, Weissenhorn W, Calder LJ, Hughson FM, et al. A soluble domain of the membrane-anchoring chain of influenza virus hemagglutinin (HA2) folds in Escherichia coli into the low-pH-induced conformation. Proc Natl Acad Sci U S A. 1995;92:12205–9. doi: 10.1073/pnas.92.26.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooper GJ, Willis AC, Clark A, Turner RC, Sim RB, Reid KB. Purification and characterization of a peptide from amyloid-rich pancreases of type 2 diabetic patients. Proc Natl Acad Sci U S A. 1987;84:8628–32. doi: 10.1073/pnas.84.23.8628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Copeland NG, Jenkins NA, Nexo B, Schultz AM, Rein A, et al. Poorly expressed endogenous ecotropic provirus of DBA/2 mice encodes a mutant Pr65gag protein that is not myristylated. Journal of virology. 1988;62:479–87. doi: 10.1128/jvi.62.2.479-487.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Critchley DR. Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu Rev Biophys. 2009;38:235–54. doi: 10.1146/annurev.biophys.050708.133744. [DOI] [PubMed] [Google Scholar]
- 27.Cukierman S. Et tu, Grotthuss! and other unfinished stories. Biochim Biophys Acta. 2006;1757:876–85. doi: 10.1016/j.bbabio.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 28.DeMarco ML, Daggett V. Local environmental effects on the structure of the prion protein. Comptes rendus biologies. 2005;328:847–62. doi: 10.1016/j.crvi.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 29.DiNitto J, Delprato A, Gabe Lee M, Cronin T, Huang S, et al. Structural basis and mechanism of autoregulation in 3-phosphoinositide-dependent Grp1 family Arf GTPase exchange factors. Mol Cell. 2007;28:569–83. doi: 10.1016/j.molcel.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nature reviews. Molecular cell biology. 2011;12:362–75. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Falnes PO, Sandvig K. Penetration of protein toxins into cells. Current opinion in cell biology. 2000;12:407–13. doi: 10.1016/s0955-0674(00)00109-5. [DOI] [PubMed] [Google Scholar]
- 32.Finzel BC, Clancy LL, Holland DR, Muchmore SW, Watenpaugh KD, Einspahr HM. Crystal structure of recombinant human interleukin-1 beta at 2.0 A resolution. J Mol Biol. 1989;209:779–91. doi: 10.1016/0022-2836(89)90606-2. [DOI] [PubMed] [Google Scholar]
- 33.Fledderman EL, Fujii K, Ghanam RH, Waki K, Prevelige PE, et al. Myristate exposure in the human immunodeficiency virus type 1 matrix protein is modulated by pH. Biochemistry. 2010;49:9551–62. doi: 10.1021/bi101245j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frantz C, Barreiro G, Dominguez L, Chen X, Eddy R, et al. Cofilin is a pH sensor for actin free barbed end formation: role of phosphoinositide binding. J Cell Biol. 2008;183:865–79. doi: 10.1083/jcb.200804161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frantz C, Karydis A, Nalbant P, Hahn KM, Barber DL. Positive feedback between Cdc42 activity and H+ efflux by the Na-H exchanger NHE1 for polarity of migrating cells. J Cell Biol. 2007;179:403–10. doi: 10.1083/jcb.200704169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freed EO, Orenstein JM, Buckler-White AJ, Martin MA. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. Journal of virology. 1994;68:5311–20. doi: 10.1128/jvi.68.8.5311-5320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Moreno B. Adaptations of proteins to cellular and subcellular pH. J Biol. 2009;8:98. doi: 10.1186/jbiol199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghanam RH, Samal AB, Fernandez TF, Saad JS. Role of the HIV-1 Matrix Protein in Gag Intracellular Trafficking and Targeting to the Plasma Membrane for Virus Assembly. Front Microbiol. 2012;3:55. doi: 10.3389/fmicb.2012.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giardina B, Mosca D, De Rosa MC. The Bohr effect of haemoglobin in vertebrates: an example of molecular adaptation to different physiological requirements. Acta Physiol Scand. 2004;182:229–44. doi: 10.1111/j.1365-201X.2004.01360.x. [DOI] [PubMed] [Google Scholar]
- 40.Gingras AR, Bate N, Goult BT, Hazelwood L, Canestrelli I, et al. The structure of the C-terminal actin-binding domain of talin. The EMBO journal. 2008;27:458–69. doi: 10.1038/sj.emboj.7601965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldschmidt L, Teng PK, Riek R, Eisenberg D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc Natl Acad Sci U S A. 2010;107:3487–92. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorbatyuk VY, Nosworthy NJ, Robson SA, Bains NP, Maciejewski MW, et al. Mapping the phosphoinositide-binding site on chick cofilin explains how PIP2 regulates the cofilin-actin interaction. Molecular cell. 2006;24:511–22. doi: 10.1016/j.molcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 43.Gorfe AA, Caflisch A. Functional plasticity in the substrate binding site of beta-secretase. Structure. 2005;13:1487–98. doi: 10.1016/j.str.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 44.Habazettl J, Gondol D, Wiltscheck R, Otlewski J, Schleicher M, Holak TA. Structure of hisactophilin is similar to interleukin-1 beta and fibroblast growth factor. Nature. 1992;359:855–8. doi: 10.1038/359855a0. [DOI] [PubMed] [Google Scholar]
- 45.Hammer ND, Wang X, McGuffie BA, Chapman MR. Amyloids: friend or foe? Journal of Alzheimer’s disease: JAD. 2008;13:407–19. doi: 10.3233/jad-2008-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hanakam F, Albrecht R, Eckerskorn C, Matzner M, Gerisch G. Myristoylated and non-myristoylated forms of the pH sensor protein hisactophilin II: intracellular shuttling to plasma membrane and nucleus monitored in real time by a fusion with green fluorescent protein. The EMBO journal. 1996;15:2935–43. [PMC free article] [PubMed] [Google Scholar]
- 47.Hanakam F, Gerisch G, Lotz S, Alt T, Seelig A. Binding of hisactophilin I and II to lipid membranes is controlled by a pH-dependent myristoyl-histidine switch. Biochemistry. 1996;35:11036–44. doi: 10.1021/bi960789j. [DOI] [PubMed] [Google Scholar]
- 48.Harguindey S, Reshkin SJ, Orive G, Arranz JL, Anitua E. Growth and trophic factors, pH and the Na+/H+ exchanger in Alzheimer’s disease, other neurodegenerative diseases and cancer: new therapeutic possibilities and potential dangers. Curr Alzheimer Res. 2007;4:53–65. doi: 10.2174/156720507779939841. [DOI] [PubMed] [Google Scholar]
- 49.Hatanaka H, Ogura K, Moriyama K, Ichikawa S, Yahara I, Inagaki F. Tertiary structure of destrin and structural similarity between two actin-regulating protein families. Cell. 1996;85:1047–55. doi: 10.1016/s0092-8674(00)81305-7. [DOI] [PubMed] [Google Scholar]
- 50.He J, Haney RM, Vora M, Verkhusha VV, Stahelin RV, Kutateladze TG. Molecular mechanism of membrane targeting by the GRP1 PH domain. J Lipid Res. 2008;49:1807–15. doi: 10.1194/jlr.M800150-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He J, Scott JL, Heroux A, Roy S, Lenoir M, et al. Molecular basis of phosphatidylinositol 4-phosphate and ARF1 GTPase recognition by the FAPP1 pleckstrin homology (PH) domain. J Biol Chem. 2011;286:18650–7. doi: 10.1074/jbc.M111.233015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hearps AC, Jans DA. Regulating the functions of the HIV-1 matrix protein. AIDS research and human retroviruses. 2007;23:341–6. doi: 10.1089/aid.2006.0108. [DOI] [PubMed] [Google Scholar]
- 53.Higham CE, Jaikaran ET, Fraser PE, Gross M, Clark A. Preparation of synthetic human islet amyloid polypeptide (IAPP) in a stable conformation to enable study of conversion to amyloid-like fibrils. FEBS Lett. 2000;470:55–60. doi: 10.1016/s0014-5793(00)01287-4. [DOI] [PubMed] [Google Scholar]
- 54.Hom RA, Vora M, Regner M, Subach OM, Cho W, et al. pH-dependent binding of the Epsin ENTH domain and the AP180 ANTH domain to PI(4,5)P2-containing bilayers. J Mol Biol. 2007;373:412–23. doi: 10.1016/j.jmb.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hosszu LL, Tattum MH, Jones S, Trevitt CR, Wells MA, et al. The H187R mutation of the human prion protein induces conversion of recombinant prion protein to the PrP(Sc)-like form. Biochemistry. 2010;49:8729–38. doi: 10.1021/bi100572j. [DOI] [PubMed] [Google Scholar]
- 56.Hu F, Luo W, Hong M. Mechanisms of proton conduction and gating in influenza M2 proton channels from solid-state NMR. Science. 2010;330:505–8. doi: 10.1126/science.1191714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu J, Fu R, Nishimura K, Zhang L, Zhou HX, et al. Histidines, heart of the hydrogen ion channel from influenza A virus: toward an understanding of conductance and proton selectivity. Proc Natl Acad Sci U S A. 2006;103:6865–70. doi: 10.1073/pnas.0601944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hunte C, Screpanti E, Venturi M, Rimon A, Padan E, Michel H. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature. 2005;435:1197–202. doi: 10.1038/nature03692. [DOI] [PubMed] [Google Scholar]
- 59.Igawa T, Ishii S, Tachibana T, Maeda A, Higuchi Y, et al. Antibody recycling by engineered pH-dependent antigen binding improves the duration of antigen neutralization. Nat Biotechnol. 2010;28:1203–7. doi: 10.1038/nbt.1691. [DOI] [PubMed] [Google Scholar]
- 60.Jansen S, Collins A, Yang C, Rebowski G, Svitkina T, Dominguez R. Mechanism of actin filament bundling by fascin. J Biol Chem. 2011;286:30087–96. doi: 10.1074/jbc.M111.251439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ji CG, Zhang JZ. Understanding the molecular mechanism of enzyme dynamics of ribonuclease A through protonation/deprotonation of HIS48. J Am Chem Soc. 2011;133:17727–37. doi: 10.1021/ja206212a. [DOI] [PubMed] [Google Scholar]
- 62.Jiang P, Wei L, Pervushin K, Mu Y. pH-Dependent interactions of human islet amyloid polypeptide segments with insulin studied by replica exchange molecular dynamics simulations. The journal of physical chemistry. B. 2010;114:10176–83. doi: 10.1021/jp101811u. [DOI] [PubMed] [Google Scholar]
- 63.Karve S, Bandekar A, Ali MR, Sofou S. The pH-dependent association with cancer cells of tunable functionalized lipid vesicles with encapsulated doxorubicin for high cell-kill selectivity. Biomaterials. 2010;31:4409–16. doi: 10.1016/j.biomaterials.2010.01.064. [DOI] [PubMed] [Google Scholar]
- 64.Karydis A, Jimenez-Vidal M, Denker SP, Barber DL. Mislocalized scaffolding by the Na-H exchanger NHE1 dominantly inhibits fibronectin production and TGF-beta activation. Molecular biology of the cell. 2009;20:2327–36. doi: 10.1091/mbc.E08-08-0842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khaled AR, Kim K, Hofmeister R, Muegge K, Durum SK. Withdrawal of IL-7 induces Bax translocation from cytosol to mitochondria through a rise in intracellular pH. Proc Natl Acad Sci U S A. 1999;96:14476–81. doi: 10.1073/pnas.96.25.14476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Knaus KJ, Morillas M, Swietnicki W, Malone M, Surewicz WK, Yee VC. Crystal structure of the human prion protein reveals a mechanism for oligomerization. Nature structural biology. 2001;8:770–4. doi: 10.1038/nsb0901-770. [DOI] [PubMed] [Google Scholar]
- 67.Knight JD, Williamson JA, Miranker AD. Interaction of membrane-bound islet amyloid polypeptide with soluble and crystalline insulin. Protein Sci. 2008;17:1850–6. doi: 10.1110/ps.036350.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kooijman EE, King KE, Gangoda M, Gericke A. Ionization properties of phosphatidylinositol polyphosphates in mixed model membranes. Biochemistry. 2009;48:9360–71. doi: 10.1021/bi9008616. [DOI] [PubMed] [Google Scholar]
- 69.Kundrotas PJ, Alexov E. Electrostatic properties of protein-protein complexes. Biophys J. 2006;91:1724–36. doi: 10.1529/biophysj.106.086025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Landry CR, Levy ED, Michnick SW. Weak functional constraints on phosphoproteomes. Trends Genet. 2009;25:193–7. doi: 10.1016/j.tig.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 71.Lee HM, Chmielewski J. Liposomal cargo unloading induced by pH-sensitive peptides. J Pept Res. 2005;65:355–63. doi: 10.1111/j.1399-3011.2005.00229.x. [DOI] [PubMed] [Google Scholar]
- 72.Lee SA, Eyeson R, Cheever ML, Geng J, Verkhusha VV, et al. Targeting of the FYVE domain to endosomal membranes is regulated by a histidine switch. Proc Natl Acad Sci U S A. 2005;102:13052–7. doi: 10.1073/pnas.0503900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leithe E, Rivedal E. Ubiquitination of gap junction proteins. J Membr Biol. 2007;217:43–51. doi: 10.1007/s00232-007-9050-z. [DOI] [PubMed] [Google Scholar]
- 74.Leyman S, Sidani M, Ritsma L, Waterschoot D, Eddy R, et al. Unbalancing the phosphatidylinositol-4,5-bisphosphate-cofilin interaction impairs cell steering. Molecular biology of the cell. 2009;20:4509–23. doi: 10.1091/mbc.E09-02-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li S, Sato S, Yang X, Preisig PA, Alpern RJ. Pyk2 activation is integral to acid stimulation of sodium/hydrogen exchanger 3. J Clin Invest. 2004;114:1782–9. doi: 10.1172/JCI18046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li W, Nicol F, Szoka FC., Jr. GALA: a designed synthetic pH-responsive amphipathic peptide with applications in drug and gene delivery. Adv Drug Deliv Rev. 2004;56:967–85. doi: 10.1016/j.addr.2003.10.041. [DOI] [PubMed] [Google Scholar]
- 77.Lietzke S, Bose S, Cronin T, Klarlund J, Chawla A, et al. Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains. Mol Cell. 2000;6:385–94. doi: 10.1016/s1097-2765(00)00038-1. [DOI] [PubMed] [Google Scholar]
- 78.Liu B, Westhead DR, Boyett MR, Warwicker J. Modelling the pH-dependent properties of Kv1 potassium channels. J Mol Biol. 2007;368:328–35. doi: 10.1016/j.jmb.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 79.Lundback T, van Den Berg S, Hard T. Sequence-specific DNA binding by the glucocorticoid receptor DNA-binding domain is linked to a salt-dependent histidine protonation. Biochemistry. 2000;39:8909–16. doi: 10.1021/bi000231i. [DOI] [PubMed] [Google Scholar]
- 80.Maciver SK, Pope BJ, Whytock S, Weeds AG. The effect of two actin depolymerizing factors (ADF/cofilins) on actin filament turnover: pH sensitivity of F-actin binding by human ADF, but not of Acanthamoeba actophorin. European journal of biochemistry/FEBS. 1998;256:388–97. doi: 10.1046/j.1432-1327.1998.2560388.x. [DOI] [PubMed] [Google Scholar]
- 81.Magalhaes MA, Larson DR, Mader CC, Bravo-Cordero JJ, Gil-Henn H, et al. Cortactin phosphorylation regulates cell invasion through a pH-dependent pathway. J Cell Biol. 2011;195:903–20. doi: 10.1083/jcb.201103045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mandell DJ, Chorny I, Groban ES, Wong SE, Levine E, et al. Strengths of hydrogen bonds involving phosphorylated amino acid side chains. J Am Chem Soc. 2007;129:820–7. doi: 10.1021/ja063019w. [DOI] [PubMed] [Google Scholar]
- 83.Marttila AT, Hytonen VP, Laitinen OH, Bayer EA, Wilchek M, Kulomaa MS. Mutation of the important Tyr-33 residue of chicken avidin: functional and structural consequences. Biochem J. 2003;369:249–54. doi: 10.1042/BJ20020886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mason AC, Jensen JH. Protein-protein binding is often associated with changes in protonation state. Proteins. 2008;71:81–91. doi: 10.1002/prot.21657. [DOI] [PubMed] [Google Scholar]
- 85.Matsuyama S, Llopis J, Deveraux QL, Tsien RY, Reed JC. Changes in intramitochondrial and cytosolic pH: early events that modulate caspase activation during apoptosis. Nat Cell Biol. 2000;2:318–25. doi: 10.1038/35014006. [DOI] [PubMed] [Google Scholar]
- 86.McLachlan GD, Cahill SM, Girvin ME, Almo SC. Acid-induced equilibrium folding intermediate of human platelet profilin. Biochemistry. 2007;46:6931–43. doi: 10.1021/bi0602359. [DOI] [PubMed] [Google Scholar]
- 87.Meyer RK, McKinley MP, Bowman KA, Braunfeld MB, Barry RA, Prusiner SB. Separation and properties of cellular and scrapie prion proteins. Proc Natl Acad Sci U S A. 1986;83:2310–4. doi: 10.1073/pnas.83.8.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mishra R, Geyer M, Winter R. NMR spectroscopic investigation of early events in IAPP amyloid fibril formation. Chembiochem. 2009;10:1769–72. doi: 10.1002/cbic.200900237. [DOI] [PubMed] [Google Scholar]
- 89.Mitra RC, Zhang Z, Alexov E. In silico modeling of pH-optimum of protein-protein binding. Proteins. 2011;79:925–36. doi: 10.1002/prot.22931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mongan J, Case DA. Biomolecular simulations at constant pH. Current opinion in structural biology. 2005;15:157–63. doi: 10.1016/j.sbi.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 91.Mongan J, Case DA, McCammon JA. Constant pH molecular dynamics in generalized Born implicit solvent. J Comput Chem. 2004;25:2038–48. doi: 10.1002/jcc.20139. [DOI] [PubMed] [Google Scholar]
- 92.Morag E, Bayer EA, Wilchek M. Reversibility of biotin-binding by selective modification of tyrosine in avidin. Biochem J. 1996;316(Pt 1):193–9. doi: 10.1042/bj3160193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moseley JB, Okada K, Balcer HI, Kovar DR, Pollard TD, Goode BL. Twinfilin is an actin-filament-severing protein and promotes rapid turnover of actin structures in vivo. Journal of cell science. 2006;119:1547–57. doi: 10.1242/jcs.02860. [DOI] [PubMed] [Google Scholar]
- 94.Murphy RM. Peptide aggregation in neurodegenerative disease. Annual review of biomedical engineering. 2002;4:155–74. doi: 10.1146/annurev.bioeng.4.092801.094202. [DOI] [PubMed] [Google Scholar]
- 95.Murtaugh ML, Fanning SW, Sharma TM, Terry AM, Horn JR. A combinatorial histidine scanning library approach to engineer highly pH-dependent protein switches. Protein Sci. 2011;20:1619–31. doi: 10.1002/pro.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Musgaard M, Thogersen L, Schiott B. Protonation states of important acidic residues in the central Ca(2)(+) ion binding sites of the Ca(2)(+)-ATPase: a molecular modeling study. Biochemistry. 2011;50:11109–20. doi: 10.1021/bi201164b. [DOI] [PubMed] [Google Scholar]
- 97.Nanga RP, Brender JR, Vivekanandan S, Ramamoorthy A. Structure and membrane orientation of IAPP in its natively amidated form at physiological pH in a membrane environment. Biochimica et biophysica acta. 2011;1808:2337–42. doi: 10.1016/j.bbamem.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nanga RP, Brender JR, Xu J, Veglia G, Ramamoorthy A. Structures of rat and human islet amyloid polypeptide IAPP(1-19) in micelles by NMR spectroscopy. Biochemistry. 2008;47:12689–97. doi: 10.1021/bi8014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Narayanan A, Jacobson MP. Computational studies of protein regulation by post-translational phosphorylation. Current opinion in structural biology. 2009;19:156–63. doi: 10.1016/j.sbi.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 100.Nichols JW, Deamer DW. Net proton-hydroxyl permeability of large unilamellar liposomes measured by an acid-base titration technique. Proc Natl Acad Sci U S A. 1980;77:2038–42. doi: 10.1073/pnas.77.4.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Olkhova E, Hunte C, Screpanti E, Padan E, Michel H. Multiconformation continuum electrostatics analysis of the NhaA Na+/H+ antiporter of Escherichia coli with functional implications. Proc Natl Acad Sci U S A. 2006;103:2629–34. doi: 10.1073/pnas.0510914103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, et al. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci U S A. 1993;90:10962–6. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Patil SM, Xu S, Sheftic SR, Alexandrescu AT. Dynamic alpha-helix structure of micelle-bound human amylin. The Journal of biological chemistry. 2009;284:11982–91. doi: 10.1074/jbc.M809085200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Peitzsch RM, McLaughlin S. Binding of acylated peptides and fatty acids to phospholipid vesicles: pertinence to myristoylated proteins. Biochemistry. 1993;32:10436–43. doi: 10.1021/bi00090a020. [DOI] [PubMed] [Google Scholar]
- 105.Pepys MB. Amyloidosis. Annual review of medicine. 2006;57:223–41. doi: 10.1146/annurev.med.57.121304.131243. [DOI] [PubMed] [Google Scholar]
- 106.Perier A, Chassaing A, Raffestin S, Pichard S, Masella M, et al. Concerted protonation of key histidines triggers membrane interaction of the diphtheria toxin T domain. J Biol Chem. 2007;282:24239–45. doi: 10.1074/jbc.M703392200. [DOI] [PubMed] [Google Scholar]
- 107.Pey AL, Rodriguez-Larrea D, Gavira JA, Garcia-Moreno B, Sanchez-Ruiz JM. Modulation of buried ionizable groups in proteins with engineered surface charge. J Am Chem Soc. 2010;132:1218–9. doi: 10.1021/ja909298v. [DOI] [PubMed] [Google Scholar]
- 108.Pintsch T, Zischka H, Schuster SC. Hisactophilin is involved in osmoprotection in Dictyostelium. BMC Biochem. 2002;3:10. doi: 10.1186/1471-2091-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pope BJ, Zierler-Gould KM, Kuhne R, Weeds AG, Ball LJ. Solution structure of human cofilin: actin binding, pH sensitivity, and relationship to actin-depolymerizing factor. J Biol Chem. 2004;279:4840–8. doi: 10.1074/jbc.M310148200. [DOI] [PubMed] [Google Scholar]
- 110.Preisig PA. The acid-activated signaling pathway: starting with Pyk2 and ending with increased NHE3 activity. Kidney Int. 2007;72:1324–9. doi: 10.1038/sj.ki.5002543. [DOI] [PubMed] [Google Scholar]
- 111.Putney LK, Barber DL. Na-H exchange-dependent increase in intracellular pH times G2/M entry and transition. J Biol Chem. 2003;278:44645–9. doi: 10.1074/jbc.M308099200. [DOI] [PubMed] [Google Scholar]
- 112.Rapedius M, Haider S, Browne KF, Shang L, Sansom MS, et al. Structural and functional analysis of the putative pH sensor in the Kir1.1 (ROMK) potassium channel. EMBO Rep. 2006;7:611–6. doi: 10.1038/sj.embor.7400678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wuthrich K. NMR structure of the mouse prion protein domain PrP(121-231) Nature. 1996;382:180–2. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 114.Rodnin MV, Kyrychenko A, Kienker P, Sharma O, Posokhov YO, et al. Conformational switching of the diphtheria toxin T domain. J Mol Biol. 2010;402:1–7. doi: 10.1016/j.jmb.2010.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rossman K, Der C, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–80. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 116.Russo C, Gao Y, Mancini P, Vanni C, Porotto M, et al. Modulation of oncogenic DBL activity by phosphoinositol phosphate binding to pleckstrin homology domain. J Biol Chem. 2001;276:19524–31. doi: 10.1074/jbc.M009742200. [DOI] [PubMed] [Google Scholar]
- 117.Sackin H, Nanazashvili M, Palmer LG, Krambis M, Walters DE. Structural locus of the pH gate in the Kir1.1 inward rectifier channel. Biophys J. 2005;88:2597–606. doi: 10.1529/biophysj.104.051474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sarkar CA, Lowenhaupt K, Horan T, Boone TC, Tidor B, Lauffenburger DA. Rational cytokine design for increased lifetime and enhanced potency using pH-activated “histidine switching”. Nat Biotechnol. 2002;20:908–13. doi: 10.1038/nbt725. [DOI] [PubMed] [Google Scholar]
- 119.Scheel J, Ziegelbauer K, Kupke T, Humbel BM, Noegel AA, et al. Hisactophilin, a histidine-rich actin-binding protein from Dictyostelium discoideum. J Biol Chem. 1989;264:2832–9. [PubMed] [Google Scholar]
- 120.Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–5. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schreiber R. Ca2+ signaling, intracellular pH and cell volume in cell proliferation. J Membr Biol. 2005;205:129–37. doi: 10.1007/s00232-005-0778-z. [DOI] [PubMed] [Google Scholar]
- 122.Seo K, Chung SW, Byun Y, Kim D. Paclitaxel loaded nano-aggregates based on pH sensitive polyaspartamide amphiphilic graft copolymers. Int J Pharm. 2012;424:26–32. doi: 10.1016/j.ijpharm.2011.12.047. [DOI] [PubMed] [Google Scholar]
- 123.Sheng H, Niu B, Sun H. Metabolic targeting of cancers: from molecular mechanisms to therapeutic strategies. Curr Med Chem. 2009;16:1561–87. doi: 10.2174/092986709788186255. [DOI] [PubMed] [Google Scholar]
- 124.Simpson LL. Identification of the major steps in botulinum toxin action. Annu Rev Pharmacol Toxicol. 2004;44:167–93. doi: 10.1146/annurev.pharmtox.44.101802.121554. [DOI] [PubMed] [Google Scholar]
- 125.Smith MT, Meissner J, Esmonde S, Wong HJ, Meiering EM. Energetics and mechanisms of folding and flipping the myristoyl switch in the {beta}-trefoil protein, hisactophilin. Proc Natl Acad Sci U S A. 2010;107:20952–7. doi: 10.1073/pnas.1008026107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Soppa J. Protein acetylation in archaea, bacteria, and eukaryotes. Archaea 2010. 2010 doi: 10.1155/2010/820681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Srivastava J, Barber DL, Jacobson MP. Intracellular pH sensors: design principles and functional significance. Physiology (Bethesda) 2007;22:30–9. doi: 10.1152/physiol.00035.2006. [DOI] [PubMed] [Google Scholar]
- 128.Srivastava J, Barreiro G, Groscurth S, Gingras AR, Goult BT, et al. Structural model and functional significance of pH-dependent talin-actin binding for focal adhesion remodeling. Proc Natl Acad Sci U S A. 2008;105:14436–41. doi: 10.1073/pnas.0805163105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sriwilaijaroen N, Suzuki Y. Molecular basis of the structure and function of H1 hemagglutinin of influenza virus. Proc Jpn Acad Ser B Phys Biol Sci. 2012;88:226–49. doi: 10.2183/pjab.88.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Stock C, Schwab A. Protons make tumor cells move like clockwork. Pflugers Arch. 2009;458:981–92. doi: 10.1007/s00424-009-0677-8. [DOI] [PubMed] [Google Scholar]
- 131.Stouffer AL, Acharya R, Salom D, Levine AS, Di Costanzo L, et al. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451:596–9. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Swietach P, Leem CH, Spitzer KW, Vaughan-Jones RD. Experimental generation and computational modeling of intracellular pH gradients in cardiac myocytes. Biophys J. 2005;88:3018–37. doi: 10.1529/biophysj.104.051391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Syntichaki P, Samara C, Tavernarakis N. The vacuolar H+ -ATPase mediates intracellular acidification required for neurodegeneration in C. elegans. Curr Biol. 2005;15:1249–54. doi: 10.1016/j.cub.2005.05.057. [DOI] [PubMed] [Google Scholar]
- 134.Takeuchi K, Takahashi H, Kawano S, Shimada I. Identification and characterization of the slowly exchanging pH-dependent conformational rearrangement in KcsA. J Biol Chem. 2007;282:15179–86. doi: 10.1074/jbc.M608264200. [DOI] [PubMed] [Google Scholar]
- 135.Tawfik DS, Chap R, Eshhar Z, Green BS. pH on-off switching of antibody-hapten binding by site-specific chemical modification of tyrosine. Protein Eng. 1994;7:431–4. doi: 10.1093/protein/7.3.431. [DOI] [PubMed] [Google Scholar]
- 136.Thompson AN, Posson DJ, Parsa PV, Nimigean CM. Molecular mechanism of pH sensing in KcsA potassium channels. Proc Natl Acad Sci U S A. 2008;105:6900–5. doi: 10.1073/pnas.0800873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tominaga T, Barber DL. Na-H exchange acts downstream of RhoA to regulate integrin-induced cell adhesion and spreading. Molecular biology of the cell. 1998;9:2287–303. doi: 10.1091/mbc.9.8.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tracz SM, Abedini A, Driscoll M, Raleigh DP. Role of aromatic interactions in amyloid formation by peptides derived from human Amylin. Biochemistry. 2004;43:15901–8. doi: 10.1021/bi048812l. [DOI] [PubMed] [Google Scholar]
- 139.Tresguerres M, Buck J, Levin LR. Physiological carbon dioxide, bicarbonate, and pH sensing. Pflugers Archiv: European journal of physiology. 2010;460:953–64. doi: 10.1007/s00424-010-0865-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.van der Kamp MW, Daggett V. Influence of pH on the human prion protein: insights into the early steps of misfolding. Biophysical journal. 2010;99:2289–98. doi: 10.1016/j.bpj.2010.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.van Rheenen J, Song X, van Roosmalen W, Cammer M, Chen X, et al. EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J Cell Biol. 2007;179:1247–59. doi: 10.1083/jcb.200706206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Varkouhi AK, Scholte M, Storm G, Haisma HJ. Endosomal escape pathways for delivery of biologicals. J Control Release. 2011;151:220–8. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 143.Vartiainen MK, Mustonen T, Mattila PK, Ojala PJ, Thesleff I, et al. The three mouse actin-depolymerizing factor/cofilins evolved to fulfill cell-type-specific requirements for actin dynamics. Molecular biology of the cell. 2002;13:183–94. doi: 10.1091/mbc.01-07-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Veith NM, Plattner H, Stuermer CA, Schulz-Schaeffer WJ, Burkle A. Immunolocalisation of PrPSc in scrapie-infected N2a mouse neuroblastoma cells by light and electron microscopy. European journal of cell biology. 2009;88:45–63. doi: 10.1016/j.ejcb.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 145.Wallace JA, Shen JK. Predicting pKa values with continuous constant pH molecular dynamics. Methods Enzymol. 2009;466:455–75. doi: 10.1016/S0076-6879(09)66019-5. [DOI] [PubMed] [Google Scholar]
- 146.Wang F, Sampogna RV, Ware BR. pH dependence of actin self-assembly. Biophysical journal. 1989;55:293–8. doi: 10.1016/S0006-3495(89)82804-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Webb BA, Chimenti M, Jacobson MP, Barber DL. Dysregulated pH: a perfect storm for cancer progression. Nature Reviews Cancer. 2011 doi: 10.1038/nrc3110. submitted. [DOI] [PubMed] [Google Scholar]
- 148.Wei L, Jiang P, Yau YH, Summer H, Shochat SG, et al. Residual structure in islet amyloid polypeptide mediates its interactions with soluble insulin. Biochemistry. 2009;48:2368–76. doi: 10.1021/bi802097b. [DOI] [PubMed] [Google Scholar]
- 149.Weis WI, Brunger AT, Skehel JJ, Wiley DC. Refinement of the influenza virus hemagglutinin by simulated annealing. J Mol Biol. 1990;212:737–61. doi: 10.1016/0022-2836(90)90234-D. [DOI] [PubMed] [Google Scholar]
- 150.Westermark P, Andersson A, Westermark GT. Islet amyloid polypeptide, islet amyloid, and diabetes mellitus. Physiological reviews. 2011;91:795–826. doi: 10.1152/physrev.00042.2009. [DOI] [PubMed] [Google Scholar]
- 151.Westheimer FH. Why nature chose phosphates. Science. 1987;235:1173–8. doi: 10.1126/science.2434996. [DOI] [PubMed] [Google Scholar]
- 152.Wijesinghe D, Engelman DM, Andreev OA, Reshetnyak YK. Tuning a polar molecule for selective cytoplasmic delivery by a pH (Low) insertion peptide. Biochemistry. 2011;50:10215–22. doi: 10.1021/bi2009773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wiltzius JJ, Landau M, Nelson R, Sawaya MR, Apostol MI, et al. Molecular mechanisms for protein-encoded inheritance. Nature structural & molecular biology. 2009;16:973–8. doi: 10.1038/nsmb.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Xiao F, Weng J, Fan K, Wang W. Mechanism of Ser88 phosphorylation-induced dimer dissociation in dynein light chain LC8. J Phys Chem B. 2010;114:15663–72. doi: 10.1021/jp1048869. [DOI] [PubMed] [Google Scholar]
- 155.Xu P, Van Kirk EA, Murdoch WJ, Zhan Y, Isaak DD, et al. Anticancer efficacies of cisplatin-releasing pH-responsive nanoparticles. Biomacromolecules. 2006;7:829–35. doi: 10.1021/bm050902y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yokoyama T, Mizuguchi M, Nabeshima Y, Kusaka K, Yamada T, et al. Hydrogen-bond network and pH sensitivity in transthyretin: Neutron crystal structure of human transthyretin. Journal of Structural Biology. 2012;177:283–90. doi: 10.1016/j.jsb.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 157.Young JA, Collier RJ. Anthrax toxin: receptor binding, internalization, pore formation, and translocation. Annual review of biochemistry. 2007;76:243–65. doi: 10.1146/annurev.biochem.75.103004.142728. [DOI] [PubMed] [Google Scholar]
- 158.Yuan X, Yu X, Lee TH, Essex M. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. Journal of virology. 1993;67:6387–94. doi: 10.1128/jvi.67.11.6387-6394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zahn R, Liu A, Luhrs T, Riek R, von Schroetter C, et al. NMR solution structure of the human prion protein. Proc Natl Acad Sci U S A. 2000;97:145–50. doi: 10.1073/pnas.97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhao H, Hakala M, Lappalainen P. ADF/cofilin binds phosphoinositides in a multivalent manner to act as a PIP(2)-density sensor. Biophysical journal. 2010;98:2327–36. doi: 10.1016/j.bpj.2010.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zimmer J, Doyle DA, Grossmann JG. Structural characterization and pH-induced conformational transition of full-length KcsA. Biophys J. 2006;90:1752–66. doi: 10.1529/biophysj.105.071175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zimmerle CT, Frieden C. Effect of pH on the mechanism of actin polymerization. Biochemistry. 1988;27:7766–72. doi: 10.1021/bi00420a027. [DOI] [PubMed] [Google Scholar]