Abstract
Tethering proteins play a key role in vesicular transport, ensuring that cargo arrives at a specific destination. The bacterial effector protein SidC and its paralog SdcA have been described as tethering factors encoded by the intracellular pathogen Legionella pneumophila. Here, we demonstrate that SidC proteins are important for early events unique to maturation of vacuoles containing Legionella and discover mono-ubiquitination of Rab1 as a new SidC-dependent activity. The crystal structure of the SidC N-terminus revealed a novel fold that is important for function and could be involved in Legionella adaptations to evolutionarily divergent host cells it encounters in natural environments.
Vesicle transport is the fundamental process in eukaryotic cells used for the delivery of material between membrane-bound organelles. At the donor organelle, cargo is packaged into vesicles, which travel to another compartment. There the vesicle is recognized and fuses to deliver its cargo. The last stages of vesicle transport, after the vesicle arrives at the acceptor compartment, are orchestrated by a number of proteins, including not only the SNARE proteins, which are the core membrane fusion machinery, but also small GTPases and their regulators, SM proteins, and tethering complexes. Tethering complexes themselves are multi-component assemblies with as many as eight subunits (1). They are thought to physically link the vesicle and the target membranes and to function in vesicle recognition, so that vesicles are recruited only to the correct compartment, as well as subsequently in the regulation of SNARE mediated vesicle fusion (1, 2). The large number of proteins involved has complicated a mechanistic analysis of the later stages in membrane transport, and the function of the so-called “tethering” complexes is not well defined at the molecular level.
Microbial pathogens subvert host cell processes to survive and replicate intracellularly, and studies of how they do so have often led to insights regarding the host processes themselves. The bacterium Legionella pneumophila, in particular, has proven an excellent model system to study modulation of membrane transport by bacterial pathogens. This gram negative intracellular bacterium is the causative agent of a severe form of pneumonia called Legionnaires’ disease (3, 4). During infection, L. pneumophila is phagocytosed into a plasma membrane-derived organelle, and the pathogen directs the remodeling of this compartment into a specialized compartment called the Legionella-containing vacuole (LCV) (5, 6). Legionella uses a type IV secretion apparatus to secrete ~300 effectors into the host cell (7–9) and, in a complex process involving recruitment and fusion of ER-derived vesicles with the vacuole, the coordinated activity of these effectors lead to the formation of the LCV (10, 11). Among these Legionella effectors, the protein SidC and its paralog SdcA (71.7 % identity) were recently identified as tethering factors that function in ER-to-LCV trafficking (12). To better understand tethering events at the molecular level, and intrigued by the apparent simplicity of SidC and SdcA as compared to the large multimeric cellular tethering complexes, and to better understand their role during Legionella infection, we undertook a further characterization of these proteins.
We find that SidC proteins play a role in early LCV maturation and in the recruitment of host cell proteins to the LCV. Specifically, we found that SidC/SdcA are required for the early recruitment of LCV markers such as Arf1 and ubiquitin, and that Rab1 at the LCV, while not dependent on SidC/SdcA for localization, is ubiquitinated in a SidC/SdcA-dependent manner. Arf1 and Rab1 are two host small GTPases that play key roles in LCV maturation. Legionella is known to modify Rab1 to regulate its association with the LCV, and although mono-ubiquitination of Rab1 has not been previously reported, we speculate that it may serve a similar purpose.
In parallel, we determined the crystal structure of the N-terminal portion of SidC (SidCNT, residues 1-608) at 2.8 Å resolution. The PI4P-binding domain at the C-terminus of SidC (12) was not part of the crystallization construct. We find that SidCNT comprises three domains (A, B, C) which bear no resemblance to any known tethering factor or other protein characterized to date. In an extended conformation, the three domains of SidCNT are ~180 Å in length, similar to known tethering complexes or individual subcomplexes thereof (1, 2). We used our finding that SidC/SdcA mediate Rab1 ubiquitination to probe function, discovering that domain C plays a critical role. For SidC or SdcA to function as tethers, we therefore propose that domain C interacts with a still unidentified factor on ER-derived vesicles, while the C-terminal PI4P-binding domain interacts with the LCV, as reported (12).
Results and Discussion
SidC and SdcA function in LCV establishment and are required for optimal growth
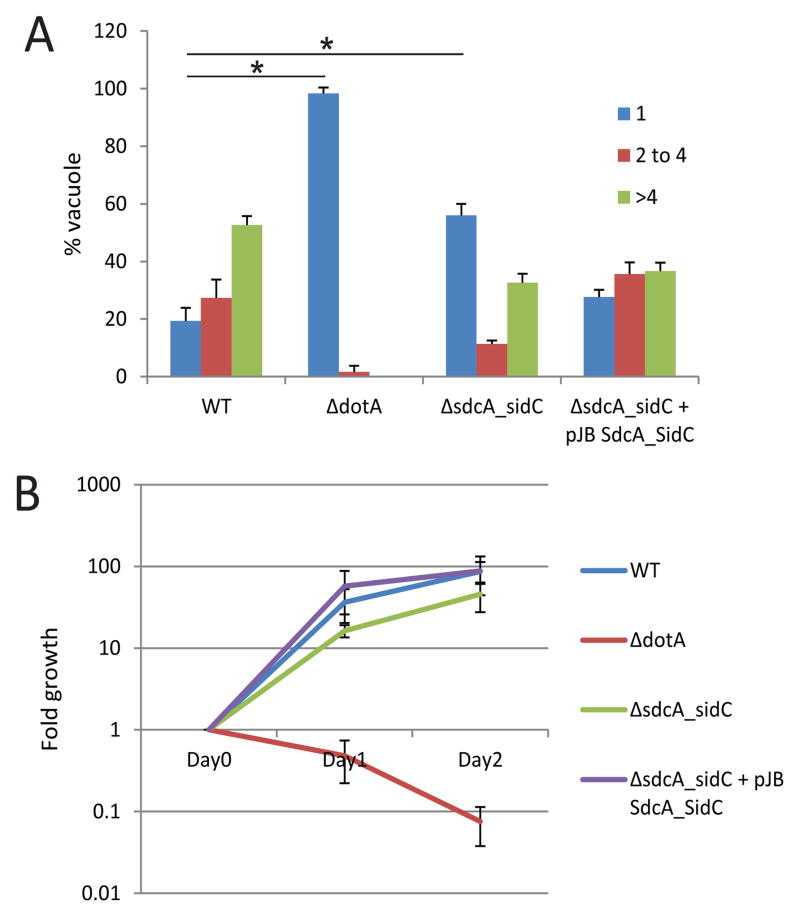
To assess the role of SidC/SdcA in LCV formation, we determined the efficiency by which Legionella mutants lacking these proteins are able to create vacuoles that support bacterial replication. We infected bone marrow-derived macrophages from NOD like receptor family CARD domain-containing protein 4 (NLRC4) −/− mice with L. pneumophila wild-type, ΔdotA (an isogenic strain of Legionella lacking the functional Dot/Icm secretion system), ΔsdcA-sidC (an isogenic strain having a chromosomal deletion that eliminates both sidC and the neighboring sdcA gene) and ΔsdcA-sidC complemented with a plasmid-encoded sdcA-sidC operon. At 8 h post-infection vacuoles containing replicating Legionella were more frequent in macrophages containing WT Legionella compared to macrophages infected with the ΔsdcA-sidC mutant. This defect in formation of vacuoles that support replication was complemented upon introduction of a plasmid expressing the sdcA-sidC region. Cells infected with the ΔsdcA-sidC mutant showed a kinetic delay in establishing vacuoles that support replication (Fig. 1A and S1). WT infection resulted in 19% vacuoles with a single bacterium at 8 h, while 31% had 2–4 bacteria and 50% cells had more than 4 bacteria. In contrast, 96% of cells infected with a ΔdotA strain showed only one bacterium per cell. Infection with the ΔsdcA-sidC strain resulted in 52% cells with a single bacterium, 12% with 2–4 bacteria and 36% with more than 4 bacteria. However, a growth curve assay revealed only a modest growth defect in the ΔsdcA-sidC (Fig. 1B). This result is consistent with the idea that absence of SidC/SdcA results in a delay in establishment of the replicative vacuole. Once the vacuole is formed, however, there is no defect in replication. One difficulty in studying Legionella is the redundancy in its replication strategies, and other pathways likely compensate for SidC/SdcA deletion.
Figure 1. SdcA/SidC are required for optimal Legionella growth.
A and B) NLRC4 −/− bone marrow derived macrophages were infected with L. pneumophila wild-type, ΔdotA, ΔsdcA-sidC and ΔsdcA-sidC complemented with a plasmid-encoded sdcA-sidC operon. A) Cells were fixed 8 h after infection. Intracellular replication of L. pneumophila was assessed by counting the number of bacteria residing in a single vacuole in infected cells. The data are presented as vacuoles containing 1 (blue), 2–4 (red) or >4 bacteria (green). Average ± standard deviations were obtained from 3 independent experiments in which 300 vacuoles were scored for each strain. Statistical significance was obtained by comparing the number of cells showing a single bacteria between WT and ΔdotA or WT and ΔsdcA-sidC (* P<0.01). B) Intracellular replication of L. pneumophila strains in NLRC4−/− cells were determined by counting the number of colony forming units (cfus) on charcoal yeast extract agar plates. Values are the mean ± SEM of four independent experiments.
SidC, SdcA and host protein recruitment to the LCV
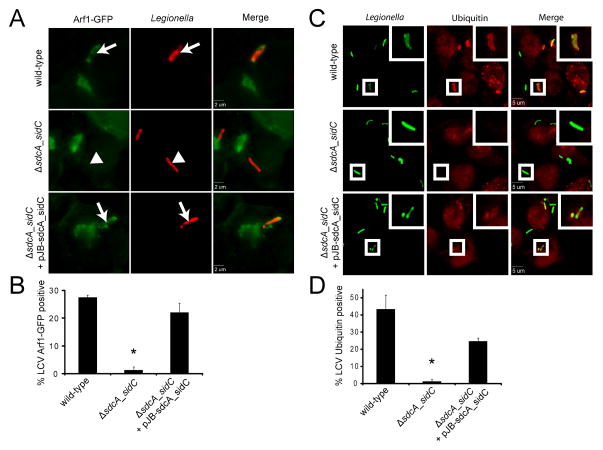
To understand why the absence of SidC/SdcA results in delayed formation of the mature LCV, we investigated the effect of these proteins in the recruitment of host proteins. It was previously shown that some host proteins recruited uniquely to the mature LCV, such as the ER proteins calnexin and calreticulin (12), are lacking at the LCV in a ΔsdcA-sidC mutant, although the level of host proteins found on the early LCV, such as the small GTPase Rab1, were unchanged (12). We extended this study to other host proteins that are recruited to the LCV early following infection. In particular, ubiquitin (13) and Arf1 (14) are both recruited to the LCV by WT L. pneumophila but not by a ΔdotA mutant lacking a functional type IV secretion system. We found that both proteins were largely absent at the LCV 1 h post-infection following infection with a Legionella ΔsdcA-sidC mutant. Whereas after infection by WT, 27% and 43% of LCVs were positive for Arf1 and ubiquitin, respectively, with the ΔsdcA-sidC mutant only 1% and 1.3% of the LCVs were positive for these two markers (Fig. 2). This defect was complemented by expression of the sdcA-sidC operon from a plasmid. The mutation of either sdcA or sidC alone did not significantly modify Arf1 recruitment at the LCV (Fig. S2), consistent with SidC and SdcA having redundant functions during infection of macrophages. Thus, SidC/SdcA play a role in a process important for recruiting Arf1 and ubiquitin to the LCV.
Figure 2. SdcA/SidC are required for early recruitment of specific LCV markers.
A) and B) HEK293 cells stably expressing GFP-tagged Arf1 were infected with L. pneumophila wild-type, ΔsdcA-sidC and ΔsdcA-sidC complemented with a plasmid-encoded sdcA-sidC operon. A) Cells were fixed 1 h post-infection. Intracellular bacteria are shown in red. Arrows show Arf1-positive vacuoles, arrowheads show Arf1-negative vacuoles. B) Quantification of the proportion of Arf1 positive vacuoles. Average ± standard deviations were obtained from 3 independent experiments (* P<0.01). C) and D) HEK293 cells were infected with L. pneumophila wild-type, ΔsdcA-sidC and ΔsdcA-sidC complemented with a plasmid-encoded sdcA-sidC operon. C) Cells were fixed 1 h post-infection, and stained for ubiquitin with FK2 antibodies. Intracellular bacteria are shown in green. D) Quantification of the proportion of ubiquitin positive vacuoles. Average ± standard deviations were obtained from 3 independent experiments (* P<0.01).
SidC and SdcA mediate a novel Rab1 modification: ubiquitination
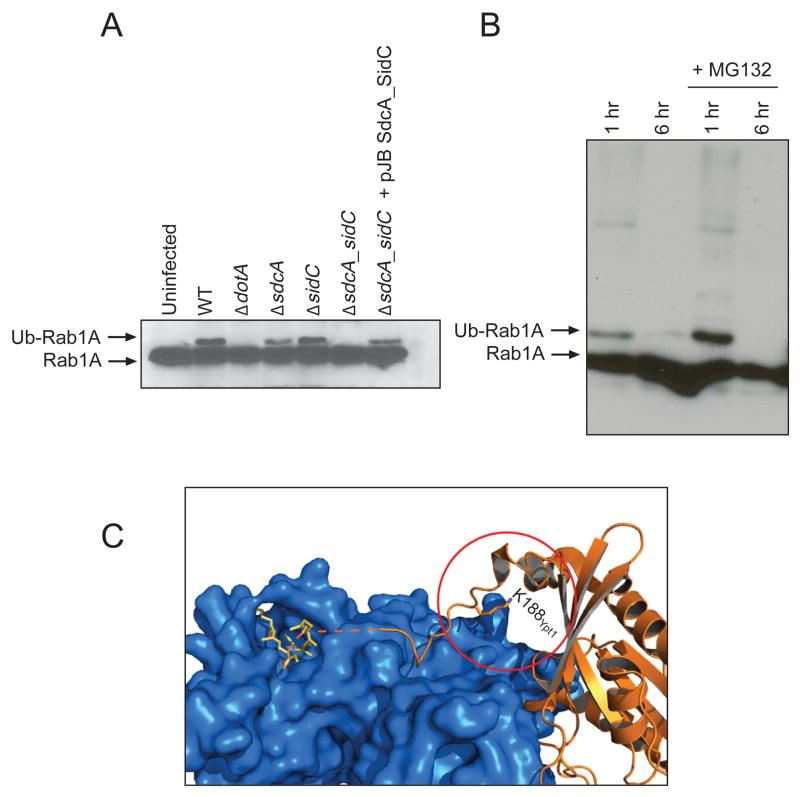
Even though the absence of SidC/SdcA does not cause a defect in Rab1A recruitment to the LCV, Rab1A western blots showed a single additional band above the Rab1A band with a mass shift (~ 8 kDa) consistent with the addition of a single ubiquitin moiety in cells infected with the WT Legionella strain; this mass shift was not present in uninfected cells or cells infected with the dotA mutant or the ΔsdcA-sidC double mutant, suggesting that this modification required SidC/SdcA (Fig. 3A). We confirmed that the modification was indeed ubiquitin by immunoblot analysis using an ubiquitin-specific antibody (Fig. S3). The single mutants ΔsdcA or ΔsidC did not show any defect in Rab1 ubiquitination (Fig. 3A), confirming the functional redundancy of these two proteins. To identify the Lys residue ubiquitinated during infection, Rab1A was immunoprecipitated from cells infected with WT and ΔdotA Legionella strains. Rab1A was digested completely with trypsin by following established protocols. The digested products were analyzed by LCMS analysis. A peptide showing the ubiquitin signature at Lys 187 (bold) (R.MGPGATAGGAEK.S + GlyGly (K)) was detected in the WT but not in the ΔdotA strain (data not shown).
Figure 3. Rab1 is ubiquitinated during infection.<.
br>A) HEK293 cells stably expressing Flag-tagged Rab1 were either uninfected or infected with L. pneumophila wild-type, ΔdotA, ΔsdcA, ΔsidC, ΔsdcA-sidC and ΔsdcA-sidC complemented with a plasmid-encoded sdcA-sidC operon. Cell lysates were prepared 1 h post-infection, and Rab1A was probed with anti-Rab1A antibody. B) HEK293 cells stably expressing Flag-tagged Rab1 were infected with L. pneumophila wild-type for 1 h or 6 h with or without the proteasome inhibitor MG132 and probed with anti-Rab1A antibody. C) Structure of the doubly prenylated (yellow) yeast Rab1 homologue Ypt1 (orange) in complex with GDI (blue) (PDB: 2BCG) (18). K188Ypt1 is the Ypt1 residue homologous to the Rab1A ubiquitination site K187. Addition of a ubiquitin moiety at this position would interfere with the Rab1:GDI interaction. The radius of the red circle around K188Ypt1 is 10 Å.
We next tested whether SidC or SdcA act as ubiquitin ligases. Ectopic expression of SidC or SdcA alone in HEK293 cells did not result in Rab1 ubiquitination, which suggests that neither SdcA nor SidC are E3 ligases. This finding is consistent with the idea that these proteins are tethers, and therefore may bring ubiquitin ligases to Rab1A to mediate this modification.
Interestingly, ubiquitination of Rab1A is temporally regulated; it is evident at 1 h but is not present at 6 h post-infection (Fig. 3B). To analyze if ubiquitinated Rab1A was targeted for proteasomal degradation, MG132 was added during infection to inhibit the proteasome. Addition of MG132 was not able to accumulate ubiquitinated Rab1A at 6 h, suggesting that ubiquitinated Rab1A was not degraded by the proteasome.
What is the purpose of Rab1A ubiquitination? Although mono-ubiquitination of Rab1 is novel, other forms of Rab1 modification have been described previously (15). In particular, the Legionella effectors DrrA and AnkX respectively AMPylate and phosphocholinate residues in the switch II region of Rab1. Rab regulator and effector proteins like Rab1 GEF as well as GDP dissociation inhibitor (GDI) interact with the switch regions. These modifications presumably alter Rab1 function by blocking access of host cell proteins to Rab1 (16, 17). In the case of SidC/SdcA-mediated ubiquitination, however, the Rab1 C-terminus is modified. Interestingly, the host protein GDI which is the Rab regulatory protein that interacts with membrane anchored Rabs to extract them from the membrane and to solubilize them, interacts not only with the switch regions but also the Rab C-terminus (Fig. 3C) (18). Analysis of the crystal structure of Rab1 complexed to GDI (18) suggests that C-terminal Rab1 ubiquitination would interfere with the Rab1:GDI interaction (Fig. 3C). It is therefore possible that ubiquitination constitutes a mechanism for regulating the time that Rab1 spends at the LCV before it is extracted by obstructing the Rab1:GDI interaction; but the role of Rab1 ubiquitination in Legionella infection remains to be further explored.
Arf1 recruitment and SidC/SdcA
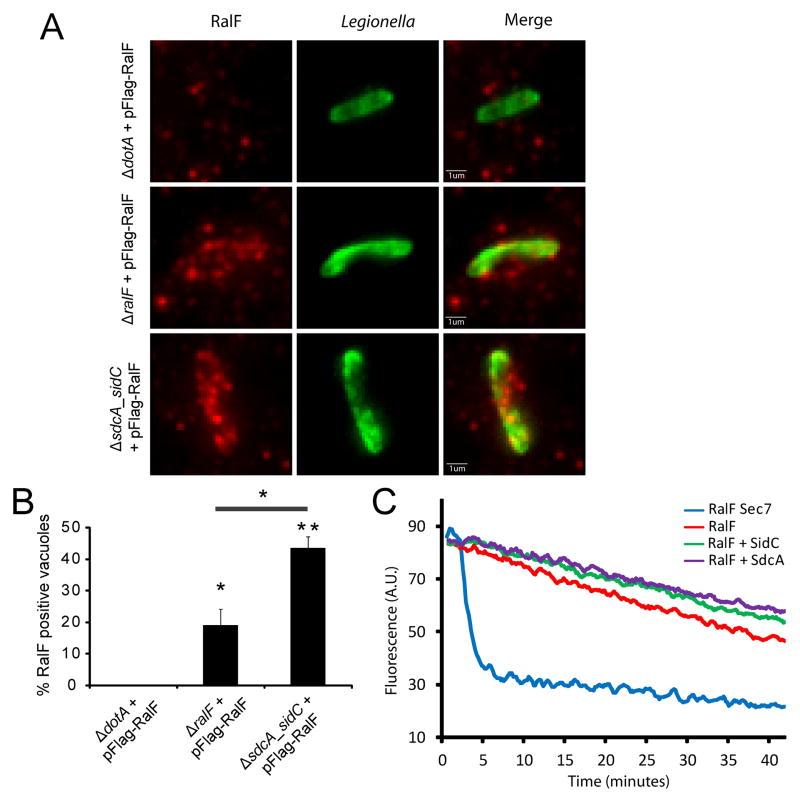
Guanine nucleotide exchange factors (GEFs) are involved in small GTPase recruitment to specific subcellular compartments. The L. pneumophila effector RalF (14) acts as a GEF for Arf1 and RalF is essential for recruitment of Arf1 to the LCV (14, 19), and mislocalization of RalF results in a defect in Arf1 recruitment at the LCV (20). The absence of Arf1 recruitment in a ΔsdcA-sidC mutant could therefore be explained by a mislocalization of RalF. RalF localization was examined in cells infected with ΔralF and ΔsdcA-sidC Legionella, both expressing Flag-tagged RalF from a plasmid (Fig. 4A). As expected, Flag-RalF was found to localize at the LCV in the ΔralF mutant. The staining was specific, as confirmed with the absence of signal using the ΔdotA mutant, which is unable to translocate RalF into host cells. We found, however, that RalF clearly localized at the LCV in cells infected with the ΔsdcA-sidC mutant, indicating that absence of SidC/SdcA does not perturb RalF localization. In these conditions, the presence of the ArfGEF at the LCV is therefore not sufficient to mediate the recruitment of its substrate Arf1. Interestingly, the number of LCVs positive for RalF was higher in the ΔsdcA-sidC mutant (45% vs 20% for the ΔralF strain; Fig. 4B). This suggests that the ArfGEF RalF accumulates on the LCV when its substrate is not present. Because RalF is autoinhibited in solution (19), we also explored the possibility that SidC/SdcA might act by relieving RalF autoinhibition. As we have shown previously (19) using a mant-GDP release assay, the RalF Sec7 domain exhibits an ArfGEF activity, whereas full length RalF is inactive in vitro (Fig. 4C). The addition of purified GST-SidC or GST-SdcA did not lead to an increased ArfGEF activity of full length RalF (Fig. 4C), suggesting SidC/SdcA do not act by relieving RalF autoinhibition.
Figure 4. SidC does not alter RalF function.
A) and B) RalF is recruited at the surface of the L pneumophila ΔsdcA-sidC LCV. HEK293 cells were infected with L. pneumophila ΔdotA, ΔralF and ΔsdcA-sidC expressing 3*Flag-tagged RalF from a plasmid. A) Cells were fixed 2 h post-infection, and RalF was probed with anti-Flag antibodies (red). Intracellular bacteria are shown in green. B) Quantification of the proportion of RalF positive vacuoles. Average ± standard deviations were obtained from one experiment done in triplicate and are representative of 3 independent experiments (* P=0.01; ** P=0.001). C) SidC or SdcA do not relieve full length RalF ArfGEF activity autoinhibition. His-ΔN17Arf1 nucleotide exchange was measured by mant-GDP release fluorescence assay in the presence of indicated MBP-RalF and GST-SdcA or SidC proteins. The MBP-RalF Sec7 domain was used as a positive control for ArfGEF activity.
These results suggest that the role of SidC/SdcA in Arf1 and ubiquitin recruitment is indirect. This is consistent with their proposed roles as tethering proteins. SidC/SdcA likely have a global effect on the LCV, changing its composition and thereby altering its maturation. That no dramatic growth defect was observed in a strain lacking sidC and sdcA suggests that L. pneumophila uses alternative mechanisms to circumvent the absence of SidC/SdcA. Consistently, ubiquitin is found at the vacuole at 8 h post-infection in a ΔsdcA-sidC mutant, indicating the absence of SidC/SdcA only results in a delay in LCV maturation (Fig. S1). Other Legionella type IV secretion system effectors, such as DrrA (21), were proposed to act as tethers, and could function redundantly with SidC/SdcA. Indeed, the extensive functional redundancy of Legionella effectors greatly complicates genetic approaches aimed at evaluating function. In these conditions, in vitro approaches are useful for characterizing effector function.
Structure of the SidC N-Terminal Fragment
For insights as to how SidC/SdcA carry out their tethering function, we crystallized an N-terminal fragment of the L. pneumophila SidC protein (SidCNT), comprising residues 1-608. This fragment is sufficient for binding to ER-derived vesicles (12), whereas a C-terminal fragment (residues 609-917) contains a PI4P binding domain that targets SidC/SdCA to the LCV (12).
SidCNT was expressed in E. coli, purified, crystallized, and its structure was solved by single anomalous wavelength dispersion method using data from a selenomethionine-substituted crystal. The final model contains two molecules of SidC in the asymmetric unit and includes all residues from 7 to 608, except for a turn close to the C-terminus (residues 595-598) and a flexible linker region (residues 239-248) in one of two SidC molecules. The final model additionally contains 166 water molecules and eight barium ions. The structure was refined to 2.8 Å resolution with Rwork/Rfree = 19.62/23.91 and good stereochemistry (Table 1).
Table 1.
Data collection and refinement statistics
| Data collection | |
| Space group | P 21 21 21 |
| Unit cell dimension | a = 105.46 Å, b = 107.58 Å. c = 177.54 Å, α = β = γ = 90° |
| Wavelength (Å) | 0.9792 |
| Resolution (Å) | 50–2.77 (3.0–2.77) |
| Rmerge | 0.171 (1.057) |
| I/σ | 17.8 (2.6) |
| Completeness (%) | 99.8 (98.9) |
| Redundancy | 13 (12.2) |
| Refinement | |
| No. of unique reflections | 52251 |
| Rwork/Rfree (%) | 19.62/23.91 |
| No of protein atoms (without H) | 18699 (9442) |
| No of water molecules | 166 |
| Average B (Å2) | 32.61 |
| Rmsd | |
| Bond length (Å) | 0.0058 |
| Bond angle (°) | 0.879 |
| Ramachandran plot | |
| Allowed (% residues) | 98.3 |
| Generously allowed (% residues) | 1.7 |
| Dissallowed (% residues) | 0 |
Numbers in parentheses indicate values in highest resolution bin.
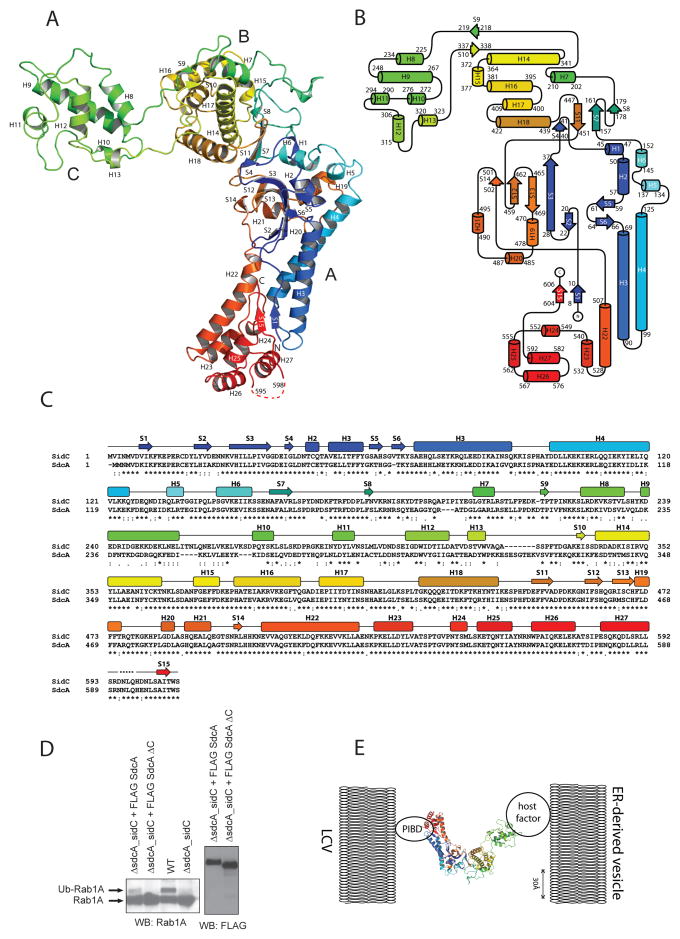
SidCNT comprises three domains arranged as a hook shape in the crystal (Fig. 5A). It contains 27 α-helices and 15 β-strands which form 5 β-sheets (Fig. 5B). The domains are formed in a non-sequential order, so that domain A at the base of the hook contains the most N- and C-terminal portions (residues 7-180, 447-608), the middle domain B includes residues 200-218 and 341-439, and domain C at the tip of the hook contains residues 225-323. Domain A is a mixed α-helical/β-sheet fold with helices H2 and H3 forming a central coiled coil. The final six helices H22-H27 in the C-terminal portion of domain A form a compact bundle at the very base of the hook, in which the N-terminal end of H22 assembling with a portion of the H2–H3 coiled-coil. Domain B consists of a bundle of α-helices (H7 and H14–H18) sandwiched between β-sheets (S4, S7, S8, S11 and S9, S10). Domain C is all α-helical, with helices H8–H13.
Figure 5. Structure of SidCNT (residue 1-608).
A) SidCNT, colored from blue at the N-terminus to red at the C-terminus. The domains are labeled A–C. B) Secondary structure topology of SidCNT. C) Sequence alignment of SidC and SdcA. D) HEK293 cells stably expressing 3XFlag-tagged Rab1 were transfected with the cDNAs encoding 3XFLAG-tagged SdcA full-length (3XFLAG-SdcAFL) or SdcA lacking residues 221-329 (3XFLAG-SdcAΔC). 14 hours after transfection, cells were infected with a L. pneumophila wild-type strain or ΔsdcA-sidC strain for 1 h, following which they were lysed. Rab1A was immunoprecipitated using FLAG-antibody coated beads. After incubation, beads were boiled and subjected to SDS-PAGE analysis and subsequently probed with either a Rab1A specific antibody or a FLAG antibody. E) Model of SdcA/SidC-mediated vesicle tethering at the LCV.
Within the crystal, two SidCNT molecules dimerize, with domain C from one SidCNT interacting with a portion of domain A in the second, and vice-versa (Fig. S4). Nevertheless, analytical size exclusion chromatography experiments with full-length SidC (SidCFL), SidCNT and SidCΔC, a deletion construct lacking domain C (Δ225-323), which should be unable to dimerize, indicate that SidC is a monomer in solution (Fig. S5).
The structure shows that the domains are linked by long stretches of residues in random coiled conformation that allow for flexibility between the domains. SidCNT, when fully extended, would span ~180 Å: ~100 Å from the base to the turn of the hook and additionally 80 Å from the turn to the tip. SidCFL is presumably somewhat longer as the PI4P binding domain at its C-terminus is not present in the crystal structure. SidCFL would then be ~220 Å long and similar in dimension to building blocks of characterized host cell tethering complexes (1, 2). Conceivably, then, SidC could bridge apposing membranes much as has been postulated for host cell tethers.
Although the length of SidC is consistent with dimensions observed for host cell tethers, there is no further structural similarity between SidC and known tethering proteins, suggesting that there is no evolutionary relationship between SidC and these tethers. We searched for structural homologues in the PDB and CATH 1.73 databases using the DALI-server and pdbeFOLD (22, 23) with the whole structure of SidCNT as well as the individual domains as queries. No structural homologues could be identified, and to our knowledge SidCNT therefore represents a novel fold.
Tethering by SidC
We were unable to identify direct interaction partners for SidC in protein-protein interaction experiments. We tested the interaction of SidC/SdcA with cellular factors that are important for ER-derived vesicle transport (Sec23/Sec24 of the COPII coat) or implicated in LCV formation (Rab1, Arf1, syntaxin2, syntaxin3, Sec22b) or else other Legionella effectors that are reported to play a role in early LCV formation (RalF, AnkX, DrrA, LepB). Binding studies were performed by either co-immunoprecipitation of in vivo co-expressed proteins or in vitro pull down experiments with recombinantly expressed and isolated proteins. However, SidC/SdcA did not show specific binding to any of the proteins tested.
We therefore used SidC/SdcA-mediated ubiquitination as a functional assay to investigate portions of SidC/SdcA critical for tethering. The sequence alignment (Fig. 5C) between SidC and SdcA shows that the two proteins are conserved overall (82% homology), but conservation in domain C (residues 225-323) is significantly lower (53% homology). This may indicate that SidC and SdcA target different isoforms of a protein or homologue proteins with their C-domain. Moreover the structure reveals that domain C is most distant from the C-terminal membrane-binding domain and attached to the rest of the protein by long flexible linkers. This would position domain C ideally for tethering. We therefore designed SidC/SdcA constructs in which domain C was excised. We observed that SdcA mediated Rab1A ubiquitination more efficiently than SidC, and for this reason we used the SdcAΔC construct (Δ222-315) rather than a SidC construct to test ubiquitination of Rab1A during infection. We transfected HEK293 cells stably expressing 3XFLAG Rab1A with plasmids encoding either 3XFLAG-SdcAFL or 3XFLAG-SdcAΔc (Fig. 5D). Following infection with Legionella strains lacking sdcA-sidc for 1 h, Rab1A was immunoprecipitated. Ubiquitination of Rab1A was only observed in cells expressing 3XFLAG-SdcAFL but not 3XFLAG-SdcAΔc (Fig. 5D). The relative expressions of both the proteins were similar, suggesting that this was not due to misfolding or degradation of 3XFLAG-SdcAΔc. The results clearly demonstrate that domain C is important for SidC/SdcA function.
It had previously been demonstrated that the C-terminal portion of Sidc/SdcA interacts with the PI4P-enriched LCV while the N-terminal portion recruits ER-derived vesicles (12). Our structural studies have shown that SidC/SdcA domain C would be furthest away from the PI4P binding domain that interacts with the LCV membrane while at the same time being flexibly attached to the rest of the protein. Domain C is therefore ideally positioned for vesicle capture. Further, we showed that SidC/SdcA domain C is required for SidC/SdcA function. We therefore propose a model in which SidC/SdcA domain C mediates the interaction with ER-derived vesicles (Fig. 5E). Although SidC differs in fold from all host cell tethering factors characterized to date, it is similar in its overall length and could therefore bridge apposing membranes in a similar fashion to that proposed for host cell tethers. Future work will determine with which proteins or complexes on these vesicles SidC and SdcA interact.
Materials and Methods
Protein Expression and Isolation
Recombinant proteins were expressed in E. coli BL21(DE3) cells as GST-fusion proteins with a TEV protease cleavage site after the GST-tag. Cells were grown at 37°C to an OD600 of 0.6 when protein expression was induced with 0.3 mM IPTG and the cultures were shifted to 25°C. Cells were harvested 20 h after induction. Selenomethionine substituted SidC (residues 1-608) for structure determination was expressed similarly according to (24). Cells were collected by centrifugation and resuspended in lysis buffer (20 mM Hepes pH 7.8, 300 mM NaCl, 1 mM PMSF, 0.5 mM TCEP) supplemented with protease inhibitors (complete EDTA-free, Roche) and lysed using a cell disruptor (Avestin).
Cells lysates were cleared by centrifugation and the GST-fusion proteins isolated by affinity chromatography using glutathione-sepharose 4B resin (GE Life Science). The proteins were eluted by on column cleavage from the GST-tag with His-TEV protease at 4°C overnight. The TEV protease was removed by subsequent Ni-NTA chromatography. SidC constructs for crystallization were further purified by size exclusion chromatography (Superdex S200 16/60) in 20 mM Hepes pH 7.8, 300 mM NaCl, 0.5 mM TCEP and concentrated to ~24 mg/ml.
MBP-RalF fusion proteins and His-tagged ΔN17Arf1 protein were purified as described in (19).
Protein Crystallization and Structure Determination
Crystals were grown by hanging drop vapor diffusion by mixing protein with reservoir solution 1:1 at 4°C as follows: SidCNT (10 mg/ml) with 0.1 M Hepes pH 7.4, 8% PEG 8000, 0.2 M BaCl2. Crystals were quick soaked in mother liquor supplemented with 20% ethylene glycol, loop mounted and flash frozen in liquid nitrogen.
Diffraction data was collected at beam line NE-CAT 24C at APS Argonne National Laboratory and processed using XDS (25). Phases were obtained by SAD at the selenium edge using seleno-methinone substituted crystals. Using the phenix AutoSol pipeline (26) we identified the selenium positions and eight additional barium positions, calculated experimental phases and density modified maps and build an initial model. After an initial simulated annealing (torsion angle) in phenix.refine (26) the model was refined by multiple cycles of manual model rebuilding in coot (27) followed by positional and TLS refinement with phenix.refine (26). Figures were made using PyMOL (28).
Bacterial strains and infections
E. coli DH5α was cultivated in LB media with ampicillin (100 μg/ml) when necessary. Legionella pneumophila serogroup 1, strain Lp01 (29), and the isogenic ΔdotA mutant strain (30) were used for infection experiments. The ΔsdcA, ΔsidC clean deletion mutant was generated for this study as described in (30). Legionella strains were grown on charcoal yeast extract (CYE) plates (1% yeast extract, 1% N-(2-acetamido)-2-aminoethanesulfonic acid (ACES; pH 6.9), 3.3 mM l-cysteine, 0.33 mM Fe(NO3)3, 1.5% bacto-agar, 0.2% activated charcoal), supplemented with 10 μg/mL chloramphenicol when required (31). Legionella were harvested from 2-day heavy patch, and used to infect HEK293 cells. Cells were stably expressing the receptor FcγRII, allowing L. pneumophila opsonized with anti-Legionella antibodies to be internalized efficiently by non-phagocytic cells. Bacteria were first opsonized with rabbit anti-Legionella antibody diluted 1/1000 for 30 min at 37°C. Bacteria were then added to the cells at an MOI of 1. The cells were centrifuged 5 min at 1000 rpm and incubated for the desired time at 37°C. Cells were then fixed with PFA for 20 min at room temperature before staining.
Preparation of Cell Extracts and Western Blotting
Cells were lysed in medium containing 20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% TX-100, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and a mixture of protease inhibitors (Roche). Each sample was loaded on an appropriate polyacrylamide gel, followed by transfer to Immobilon-P membranes (Millipore Corp). Membranes were blocked in 5% nonfat milk in phosphate-buffered saline containing 0.1% Tween 20 (PBST) for 1 h. Subsequently, they were probed with appropriate antibodies in PBST, washed extensively, and the immunoreactive bands were visualized by Enhanced Chemiluminescence (Amersham Biosciences).
Cell culture, transfection and immunoprecipitation
Human Embryonic Kidney cells (HEK293) stably expressing the receptor Fcγ, stably expressing both the receptor Fcγ and Arf1-GFP (19) or stably expressing both the receptor Fcγ st 3XFLAG-Rab1A (16) were used for Legionella infections. Cells were maintained in minimal Dulbecco’s Modified Eagle’s Medium, supplemented with 10% heat inactivated fetal bovine serum, 100 μg/mL penicillin and 10 μg/mL streptomycin at 37°C with 5% CO2. For immunoprecipitation studies, lysates were incubated with anti-FLAG M2 (Sigma) beads for 1 h at 4°C. After incubation, beads were washed extensively and 3x-FLAG fusion proteins were eluted using a 3x-FLAG peptide. Mouse monoclonal antibodies to FLAG were obtained from Sigma Aldrich. Rabbit polyclonal antibodies to Rab1A were purchased from Santa Cruz. Rabbit polyclonal antibody to Legionella was described previously (16).
Immunofluorescence Microscopy
Cells were grown on coverslips and treated as indicated, after which they were fixed in 4% paraformaldehyde and processed for immunofluorescence microscopy as described previously (16). Extracellular bacteria were stained with Alexa Fluor 350 anti-rabbit (Invitrogen). Permeabilization was performed by a methanol treatment 1 min at RT. Ubiquitin was probed with anti-ubiquitinylated proteins antibody, clone FK2 (Millipore), followed by secondary staining with Alexa Fluor 594 anti-mouse. For detection of RalF on the LCV, the indicated L. pneumophila strains expressing 3*Flag-RalF from a plasmid (20) were used to infect HEK293-FcγR cells. After fixation and permeabilization, cells were stained with monoclonal anti-Flag antibodies (Sigma) followed by secondary staining with Alexa Fluor 594 anti-mouse. Total bacteria were stained with Alexa fluor conjugated anti-rabbit antibodies. Cells were washed in PBS and mounted on plain microscope slides. Cells were subsequently visualized by fluorescence microscopy using a Nikon Eclipse TE2000-S microscope and a 100×/1.40 oil objective (Nikon Plan Apo). Z-stacks were acquired using a Hamamatsu ORCA-ER camera and 3D max was generated. Images were processed with the software Slidebook for the creation of figures.
ArfGEF assay
Purified 6XHis-tagged ΔN17Arf1 protein was incubated in loading buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 5 mM EDTA, and 1 mM DTT) containing a 10 fold molar excess of N-methylanthraniloyl (mant)-GDP (Invitrogen) for 30 min at 37°C. To terminate the loading reaction, MgCl2 was added to a final concentration of 10 mM, and free mant-GDP was removed using a polyacrylamide Desalting Column (Piercenet), previously equilibrated with column buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 2 mM MgCl2). Exchange kinetics were monitored using the decrease in emission intensity accompanying release of mant-GDP as described in (32). Exchange reactions were initiated by mixing mant-GDP loaded ΔN17Arf1 at a final concentration of 2 μM with 1 μM of the MBP-RalF and 1 μM of GST-SdcA or GST-SidC when indicated in the presence of 200 μM GTP. MBP-RalF Sec7 domain was used as a positive control for ArfGEF activity under identical conditions. Data were collected over 45 min with an infinite M1000 microplate reader (Tecan) using excitation and emission wavelengths of 360 nm and 440 nm.
Replicative vacuole assay and growth curve
NLRC4 −/− primary bone marrow-derived mouse macrophages were prepared and cultured as described previously (33). For replicative vacuole formation, cells were infected with Legionella strains at an MOI of 25 for 1 h, after which cells were washed three times with sterile PBS. RPMI media + 10% FBS was added to the cells and infection was allowed to proceed for an additional 7 hr. Cells were fixed with 4% PFA and processed for immunofluorescence as indicated above. Growth curve of Legionella strains in NLRC4 −/− primary bone marrow-derived mouse macrophages were done as described previously (33).
Statistics
Differences between control and experimental groups were analyzed by Student’s T-test. Treshold P-value below the statistical significance value of 0.01 are presented in figures.
Databank Deposition
Coordinates and structure factors have been deposited in the PDB (ID 4OM6).
Supplementary Material
Supplemental Figure S1: Replicative vacuole assay in NLRC4 −/− cells. NLRC4 −/− bone marrow derived macrophages were infected with L. pneumophila wild-type, ΔdotA and ΔsdcA-sidC strains for 1 h (A) or 8 h (B). Cells were fixed and stained with antibodies against ubiquitin (red) or Legionella (green). Nuclei were stained with Hoechst. (C) Quantification of ubiquitin positive vacuoles after 1 h and 8 h. Average ± standard deviations were obtained from 3 independent experimentsin which 300 vacuoles were scored for each strain. Statistical significance was obtained by comparing the number of cells showing ubiquitin positive bacteria at 1 h and 8 h between WT and ΔsdcA-sidC mutant. (* P<0.01).
Supplemental Figure S2: Single deletion mutants ΔsidC and ΔsdcA are not impaired for Arf1 recruitment. HEK293 cells stably expressing GFP-tagged Arf1 were infected with L. pneumophila wild-type, ΔsdcA, ΔsidC and ΔsidC_sdcA. A) Cells were fixed 1 h post-infection. Intracellular bacteria are shown in red. B) Quantification of the proportion of Arf1 positive vacuoles. Average ± standard deviations were obtained from 3 independent experiments. (* P<0.01).
Supplemental Figure S3. Rab1 is mono-ubiquitinated. HEK293 cells stably expressing Flag-tagged Rab1 were either uninfected or infected with L. pneumophila wild-type or ΔdotA for 1 h, following which they were lysed. FLAG-tagged Rab1 was immunoprecipitated using FLAG-antibody coated beads. Immunoprecipitated proteins were boiled and subjected to SDS-PAGE analysis and subsequently probed with either a Rab1A antibody or an ubiquitin specific antibody FK2. The monoubiquitinted Rab1 band has been marked with an arrow in both blots.
Supplemental Figure S4: Dimeric SidCNT assembly found in the crystal. One SidCNT is shown in surface representation the second SidCNT in ribbon representation.
Supplemental Figure S5: Analytical Size-exclusion chromatography of SidC shows that SidC is a monomer in solution. Analytical size exclusion chromatography (Superdex S75 10/30) of SidCFL, SidCΔC and SidCNT shows that SidC is a monomer in solution.
Synopsis.
The bacterial effector protein SidC and its paralog SdcA have been described as tethering factors encoded by the intracellular pathogen Legionella pneumophila. Here we demonstrate that SidC proteins are important for early events unique to maturation of vacuoles containing Legionella and discover mono-ubiquitination of Rab1 as a new SidC-dependent activity. The crystal structure of the SidC N-terminus revealed a novel fold comprising three domains (A–C), where domain C plays a critical role in SidC/SdcA mediated Rab1 ubiquitination.
Acknowledgments
We are grateful to the staff at beamline ID24-C at Argonne National Laboratory for their help in data collection and thank J. and E. Goldberg for providing mammalian COPII proteins for use in protein-protein interaction experiments. K.M.R. and C.R.R. were supported by the NIH (GM080616, AI041699). F.H. was recipient of a Brown-Coxe Fellowship. S.M and E.A were supported by Anna Fuller and EMBO Fellowships respectively.
Footnotes
Conflict of Interest: The authors declare NO conflict of interest.
References
- 1.Yu IM, Hughson FM. Tethering factors as organizers of intracellular vesicular traffic. Annu Rev Cell Dev Biol. 2010;26:137–156. doi: 10.1146/annurev.cellbio.042308.113327. [DOI] [PubMed] [Google Scholar]
- 2.Brocker C, Engelbrecht-Vandre S, Ungermann C. Multisubunit tethering complexes and their role in membrane fusion. Current biology: CB. 2010;20(21):R943–952. doi: 10.1016/j.cub.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 3.Rowbotham TJ. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. Journal of clinical pathology. 1980;33(12):1179–1183. doi: 10.1136/jcp.33.12.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horwitz MA, Silverstein SC. Legionnaires’ disease bacterium (Legionella pneumophila) multiples intracellularly in human monocytes. The Journal of clinical investigation. 1980;66(3):441–450. doi: 10.1172/JCI109874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horwitz MA. Formation of a novel phagosome by the Legionnaires’ disease bacterium (Legionella pneumophila) in human monocytes. The Journal of experimental medicine. 1983;158(4):1319–1331. doi: 10.1084/jem.158.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kagan JC, Roy CR. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nature cell biology. 2002;4(12):945–954. doi: 10.1038/ncb883. [DOI] [PubMed] [Google Scholar]
- 7.Segal G, Feldman M, Zusman T. The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii. FEMS Microbiol Rev. 2005;29(1):65–81. doi: 10.1016/j.femsre.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Nagai H, Cambronne ED, Kagan JC, Amor JC, Kahn RA, Roy CR. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc Natl Acad Sci U S A. 2005;102(3):826–831. doi: 10.1073/pnas.0406239101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci U S A. 2004;101(3):841–846. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruggemann H, Cazalet C, Buchrieser C. Adaptation of Legionella pneumophila to the host environment: role of protein secretion, effectors and eukaryotic-like proteins. Current opinion in microbiology. 2006;9(1):86–94. doi: 10.1016/j.mib.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Nagai H, Roy CR. Show me the substrates: modulation of host cell function by type IV secretion systems. Cellular Microbiology. 2003;5(6):373–383. doi: 10.1046/j.1462-5822.2003.00285.x. [DOI] [PubMed] [Google Scholar]
- 12.Ragaz C, Pietsch H, Urwyler S, Tiaden A, Weber SS, Hilbi H. The Legionella pneumophila phosphatidylinositol-4 phosphate-binding type IV substrate SidC recruits endoplasmic reticulum vesicles to a replication-permissive vacuole. Cell Microbiol. 2008;10(12):2416–2433. doi: 10.1111/j.1462-5822.2008.01219.x. [DOI] [PubMed] [Google Scholar]
- 13.Ivanov SS, Roy CR. Modulation of ubiquitin dynamics and suppression of DALIS formation by the Legionella pneumophila Dot/Icm system. Cell Microbiol. 2009;11(2):261–278. doi: 10.1111/j.1462-5822.2008.01251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science. 2002;295(5555):679–682. doi: 10.1126/science.1067025. [DOI] [PubMed] [Google Scholar]
- 15.Goody RS, Itzen A. Modulation of small GTPases by legionella. Current topics in microbiology and immunology. 2013;376:117–133. doi: 10.1007/82_2013_340. [DOI] [PubMed] [Google Scholar]
- 16.Mukherjee S, Liu X, Arasaki K, McDonough J, Galan JE, Roy CR. Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature. 2011;477(7362):103–106. doi: 10.1038/nature10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller MP, Peters H, Blumer J, Blankenfeldt W, Goody RS, Itzen A. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science. 2010;329(5994):946–949. doi: 10.1126/science.1192276. [DOI] [PubMed] [Google Scholar]
- 18.Pylypenko O, Rak A, Durek T, Kushnir S, Dursina BE, Thomae NH, Constantinescu AT, Brunsveld L, Watzke A, Waldmann H, Goody RS, Alexandrov K. Structure of doubly prenylated Ypt1:GDI complex and the mechanism of GDI-mediated Rab recycling. The EMBO journal. 2006;25(1):13–23. doi: 10.1038/sj.emboj.7600921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alix E, Chesnel L, Bowzard BJ, Tucker AM, Delprato A, Cherfils J, Wood DO, Kahn RA, Roy CR. The capping domain in RalF regulates effector functions. PLoS pathogens. 2012;8(11):e1003012. doi: 10.1371/journal.ppat.1003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Folly-Klan M, Alix E, Stalder D, Ray P, Duarte LV, Delprato A, Zeghouf M, Antonny B, Campanacci V, Roy CR, Cherfils J. A Novel Membrane Sensor Controls the Localization and ArfGEF Activity of Bacterial RalF. PLoS pathogens. 2013;9(11):e1003747. doi: 10.1371/journal.ppat.1003747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arasaki K, Toomre DK, Roy CR. The Legionella pneumophila effector DrrA is sufficient to stimulate SNARE-dependent membrane fusion. Cell host & microbe. 2012;11(1):46–57. doi: 10.1016/j.chom.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta crystallographica Section D, Biological crystallography. 2004;60(Pt 12 Pt 1):2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 23.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic acids research. 2010;38(Web Server issue):W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Budisa N, Steipe B, Demange P, Eckerskorn C, Kellermann J, Huber R. High-level biosynthetic substitution of methionine in proteins by its analogs 2-aminohexanoic acid, selenomethionine, telluromethionine and ethionine in Escherichia coli. European journal of biochemistry / FEBS. 1995;230(2):788–796. doi: 10.1111/j.1432-1033.1995.tb20622.x. [DOI] [PubMed] [Google Scholar]
- 25.Kabsch W. Automatic Processing of Rotation Diffraction Data from Crystals of Initially Unknown Symmetry and Cell Constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 26.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.The PyMOL Molecular Graphics System VS, LLC. The PyMOL Molecular Graphics System, Version 1.5.0.4. Schrödinger, LLC; [Google Scholar]
- 29.Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Molecular microbiology. 1993;7(1):7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 30.Zuckman DM, Hung JB, Roy CR. Pore-forming activity is not sufficient for Legionella pneumophila phagosome trafficking and intracellular growth. Mol Microbiol. 1999;32(5):990–1001. doi: 10.1046/j.1365-2958.1999.01410.x. [DOI] [PubMed] [Google Scholar]
- 31.Feeley JC, Gibson RJ, Gorman GW, Langford NC, Rasheed JK, Mackel DC, Baine WB. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. Journal of clinical microbiology. 1979;10(4):437–441. doi: 10.1128/jcm.10.4.437-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DiNitto JP, Delprato A, Gabe Lee MT, Cronin TC, Huang S, Guilherme A, Czech MP, Lambright DG. Structural basis and mechanism of autoregulation in 3-phosphoinositide-dependent Grp1 family Arf GTPase exchange factors. Molecular cell. 2007;28(4):569–583. doi: 10.1016/j.molcel.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choy A, Dancourt J, Mugo B, O’Connor TJ, Isberg RR, Melia TJ, Roy CR. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science. 2012;338(6110):1072–1076. doi: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1: Replicative vacuole assay in NLRC4 −/− cells. NLRC4 −/− bone marrow derived macrophages were infected with L. pneumophila wild-type, ΔdotA and ΔsdcA-sidC strains for 1 h (A) or 8 h (B). Cells were fixed and stained with antibodies against ubiquitin (red) or Legionella (green). Nuclei were stained with Hoechst. (C) Quantification of ubiquitin positive vacuoles after 1 h and 8 h. Average ± standard deviations were obtained from 3 independent experimentsin which 300 vacuoles were scored for each strain. Statistical significance was obtained by comparing the number of cells showing ubiquitin positive bacteria at 1 h and 8 h between WT and ΔsdcA-sidC mutant. (* P<0.01).
Supplemental Figure S2: Single deletion mutants ΔsidC and ΔsdcA are not impaired for Arf1 recruitment. HEK293 cells stably expressing GFP-tagged Arf1 were infected with L. pneumophila wild-type, ΔsdcA, ΔsidC and ΔsidC_sdcA. A) Cells were fixed 1 h post-infection. Intracellular bacteria are shown in red. B) Quantification of the proportion of Arf1 positive vacuoles. Average ± standard deviations were obtained from 3 independent experiments. (* P<0.01).
Supplemental Figure S3. Rab1 is mono-ubiquitinated. HEK293 cells stably expressing Flag-tagged Rab1 were either uninfected or infected with L. pneumophila wild-type or ΔdotA for 1 h, following which they were lysed. FLAG-tagged Rab1 was immunoprecipitated using FLAG-antibody coated beads. Immunoprecipitated proteins were boiled and subjected to SDS-PAGE analysis and subsequently probed with either a Rab1A antibody or an ubiquitin specific antibody FK2. The monoubiquitinted Rab1 band has been marked with an arrow in both blots.
Supplemental Figure S4: Dimeric SidCNT assembly found in the crystal. One SidCNT is shown in surface representation the second SidCNT in ribbon representation.
Supplemental Figure S5: Analytical Size-exclusion chromatography of SidC shows that SidC is a monomer in solution. Analytical size exclusion chromatography (Superdex S75 10/30) of SidCFL, SidCΔC and SidCNT shows that SidC is a monomer in solution.