Abstract
A subset of cutaneous and superficial soft tissue myoepithelial (ME) tumors displays a distinct ductal component and closely resembles mixed tumors/pleomorphic adenomas of salivary gland. As PLAG1 and HMGA2 rearrangements are the most common genetic events in pleomorphic adenomas, we sought to investigate if these abnormalities are also present in the skin/soft tissue ME lesions. In contrast, half of the deep-seated soft tissue ME tumors lacking ductal differentiation are known to be genetically unrelated, showing EWSR1 rearrangements. FISH analysis to detect PLAG1 and HMGA2 abnormalities was performed in 35 ME tumors, nine skin and 26 soft tissue, lacking EWSR1 and FUS rearrangements. For the PLAG1-rearranged tumors, FISH and RACE were performed to identify potential fusion partners, including CTNNB1 (beta-catenin) on 3p21 and LIFR (leukemia inhibitory factor receptor) on 5p13. Recurrent PLAG1 rearrangement by FISH was detected in 13 (37%) lesions, including three (33%) in the skin and 10 (38%) in the soft tissue. All were classified as benign and all except one showed abundant tubulo-ductal differentiation (comprising 12/24 [50%] of all tumors with ductal structures). A LIFR-PLAG1 fusion was detected by RACE and then confirmed by FISH in one soft tissue ME tumor with tubular formation. No CTNNB1 or LIFR abnormalities were detected in any of the remaining PLAG1-rearranged tumors. No structural HMGA2 abnormalities were detected in any of the 22 ME lesions tested. A subset of cutaneous and soft tissue ME tumors appears genetically linked to their salivary gland counterparts, displaying frequent PLAG1 gene rearrangements and occasionally LIFR-PLAG1 fusion.
INTRODUCTION
Pleomorphic adenoma is the prototypical benign myoepithelial (ME) tumor arising in the salivary gland, with predilection for the parotid. The hallmark morphologic appearance of pleomorphic adenoma is a mixture of myoepithelial cells arranged in solid or reticular pattern, as well as well-formed tubulo-ductal structures. Rearrangements of PLeomorphic Adenoma Gene 1 (PLAG1), a developmentally regulated zinc-finger proto-oncogene, located on chromosomal band 8q12, are common abnormalities in these tumors, leading to aberrant expression of its protein and thus implicated in its pathogenesis (Kas et al., 1997; Voz et al., 2000; Martins et al., 2005). Additionally, HMGA2 gene rearrangements with or without amplifications have been described in both benign pleomorphic adenoma, as well as in carcinoma ex-pleomorphic adenoma (Persson et al., 2009).
In contrast, soft tissue ME lesions have been only more recently recognized and their classification remains somewhat controversial due to their rarity at extra-salivary sites and diverse histologic spectrum (Hornick and Fletcher, 2003). Similar to the salivary counterpart, the soft tissue ME tumors are defined based on their co-expression of EMA/cytokeratin and S100 protein and/or GFAP, in keeping with a myoepithelial phenotype. At the molecular level, however, most soft tissue ME lesions appear unrelated to the pleomorphic adenoma common PLAG1/HMGA2 genetic alterations, instead, being associated with EWSR1 gene rearrangements in half of the cases (Antonescu et al., 2010). In our previous study, a higher proportion of negative-EWSR1 gene rearranged tumors occurred in the skin or superficial soft tissue and showed evidence of tubulo-ductal and/or chondroid matrix differentiation (Antonescu et al., 2010). In fact this latter subset recapitulates the closest the histology of salivary gland pleomorphic adenoma. Thus, we sought to examine the hypothesis of a potential genetic link between these histologically similar tumors at different sites, and selected a group of EWSR1- negative cutaneous and soft tissue ME tumors for analysis of PLAG1 and HMGA2 gene rearrangements.
MATERIALS AND METHODS
Tumor Characteristics
The files of the corresponding authors (CRA, CDMF) were searched for the diagnosis of cutaneous and soft tissue myoepithelial tumors. As previously published, the inclusion criteria included, in addition to typical morphologic appearance, the presence of immunohistochemical evidence of myoepithelial differentiation, i.e. positivity for EMA and/or cytokeratin AE1:AE3 and for S100 protein and/or GFAP. The tumors were assessed for their anatomic location (skin versus soft tissue), nuclear pleomorphism, mitotic activity, presence of tubulo-ductal differentiation, and chondroid matrix formation.
Fluorescence In Situ Hybridization (FISH)
FISH on interphase nuclei from paraffin embedded 4-micron sections was performed applying custom probes using bacterial artificial chromosomes (BAC), covering and flanking EWSR1 in 22q12, FUS in 16p11, PLAG1 in 8q12, HMGA2 in 12q14, CTNNB1 in 3p21, LIFR in 5p13 and FGFR1 in 8p11 (Supplementary Table 1; Antonescu et al., 2010). BAC clones were chosen according to USCS genome browser (http://genome.uscs.edu) and obtained from BACPAC sources of Children's Hospital of Oakland Research Institute (CHORI) (Oakland, CA) (http://bacpac.chori.org). DNA from individual BACs was isolated according to the manufacturer's instructions, labeled with different fluorochromes in a nick translation reaction, denatured, and hybridized to pretreated slides. Slides were then incubated, washed, and mounted with DAPI in an antifade solution, as previously described (Antonescu et al., 2010). The genomic location of each BAC set was verified by hybridizing them to normal metaphase chromosomes. Two hundred successive nuclei were examined using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (Metasystems; Watertown, MA, USA). A positive score was interpreted when at least 20% of the nuclei showed a break-apart signal. Nuclei with incomplete set of signals were omitted from the score.
Rapid Amplification of cDNA Ends (RACE) and Reverse Transcription Polymerase Chain Reaction (RT-PCR)
Adequate RNA extracted from frozen tissue (Trizol Reagent; Invitrogen; Grand Island, NY, USA) was available in one case with PLAG1 gene rearrangement to investigate possible fusion transcripts. RNA quality was determined by Eukaryote Total RNA Nano Assay and cDNA quality was tested for PGK housekeeping gene (247 bp amplified product).
Three microgram of total RNA was prepared for 5’ RACE cDNA by using the SMARTer™ RACE cDNA Amplification Kit (Clontech; Mountain View, CA, USA). Reverse transcription reaction was initiated at poly(A) tail by 3’ RACE CDS Primer A according to manufacture's protocol. Primary PCR was performed by Advantage 2 PCR kit (Clontech) with PLAG1 reverse primer exon 2.2 (5’- GAGGCTTTTTCCATCAGTAATCAG -3’) and SMART™ RACE Universal Primer A Mix. Nested PCR was then carried on with PLAG1 reverse primer exon 2.1 (5’-AAGTCTGAGGCTTTTTCCATC -3’) and SMART™ RACE Nested Universal Primer A Mix. Amplified PCR products were cloned by TOPOVR TA Cloning® Kit for Sequencing with One Shot® TOP10 Chemically Competent E. coli (Invitrogen). The constructed plasmid DNA was sequenced using Sanger's method.
Three microgram of total RNA was prepared for confirmatory RT-PCR, with oligo(dT) primer under SuperScript® III system (Invitrogen) used for first-strand cDNA synthesis, followed by Advantage® 2 PCR Kit (Clontech) with forward LIFR ex1.1 (5’-AAGGGAGCCTCTGCGACTCATTCATC-3’) and reverse PLAG1 ex3.2 (5’-AAACTTTCCTTGCCAACTGTGTGAC-3’).
RESULTS
Clinical and Pathologic Characteristics
Thirty-five cases lacking EWSR1 and FUS gene rearrangement (19 cases were included in our prior study; Antonescu et al., 2010) with available material for further molecular analysis were included in the study. None of the cases was included in the prior study of Bahrami et al., (2012), in which one of the corresponding authors has been involved as well. Among them nine arose in the skin, while the remaining 26 occurred in the soft tissue. On microscopic examination, 28 were considered histologically benign, with bland cytologic features and no detectable mitotic figures, and 24 had evidence of tubulo-ductal differentiation. The remaining seven tumors were deemed as histologically malignant, based on the presence of moderate degree of nuclear pleomorphism, hyperchromasia and increased mitotic activity (range 3-5MF/10HPFs).
The review of the immunohistochemical studies showed that all except two cases were positive for both cytokeratin and S100 protein. Two cases (Table 1) were negative for S100 protein, but instead expressed GFAP. In addition 25/31 cases tested showed EMA reactivity.
TABLE 1.
Clinicopathologic Characteristics of PLAG1-Rearranged Myoepithelial Tumors
| ME | Age/sex | Location | Skin/ST | Ductal structures | Benign/malignant | CK/S100 |
|---|---|---|---|---|---|---|
| 1 | 48/F | foot | s-c | yes | Benign | +/+ |
| 2 | 66/F | lower leg | s-c | yes | Benign | +/+ |
| 3 | 58/M | buttock | s-c | no | Benign | +/+ |
| 4a | 64/F | hand | s-c | yes | Benign | +/+ |
| 5 | 53/F | 5th toe | s-c | yes | Benign | +/+ |
| 6 | 52/F | foot | s-c | yes | Benign | +/+ |
| 7 | 52/M | post neck | s-c | yes | Benign | +/+ |
| 8 | 45/F | foot | s-c | yes | Benign | +/+ |
| 9 | 37/F | hand | skin | yes | Benign | +/+ |
| 10 | 65/F | thigh | s-c | yes | Benign | +/+ |
| 11 | 18/F | scalp | skin | yes | Benign | +/-b |
| 12 | 32/F | foot | skin | yes | Benign | +/+ |
| 13 | 29/F | foot | s-c | yes | Benign | +/+ |
ST, soft tissue; F, female; M, male; s-c, subcutaneous tissue; post, posterior;
LIFR-PLAG1 fusion positivity confirmed by RACE, RT-PCR and FISH;
positive for EMA, P63 and GFAP
In addition a control group of 12 salivary gland myoepithelial tumors, including nine salivary myoepithelial carcinomas ex-pleomorphic adenoma and three de novo myoepithelial carcinoma were included for study and comparison. All except one case of myoepithelial carcinoma ex-pleomorphic adenoma arose in the parotid, with one case originating from the floor of mouth. The myoepithelial carcinoma arising in the absence of pleomorphic adenoma varied widely in anatomic distribution, with one case each arising from the parotid, submandibular, and tongue, respectively. Their diagnosis of malignancy was established based on the infiltrative growth outside the capsule as well as cytologic atypia.
FISH Results
Thirteen (37%) out of 35 tumors showed the presence of PLAG1 gene rearrangement by FISH, without accompanying gene amplification. The PLAG1-positive group included three (33%) of the nine cutaneous and 10 (38%) of the 26 soft tissue myoepithelial tumors. All tumors in this group were deemed as benign morphologically, and all except one showed evidence of tubulo-ductal differentiation (Fig. 1).
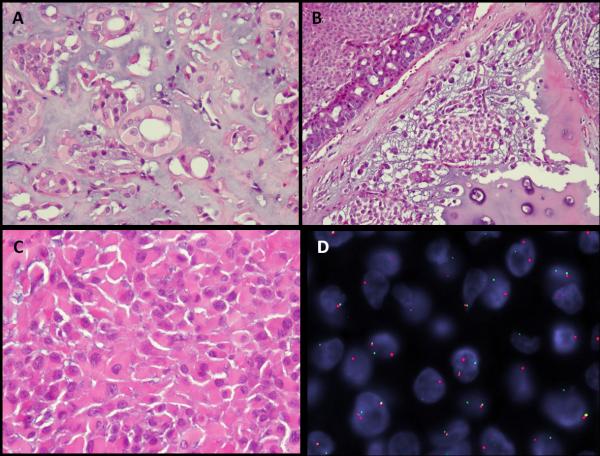
Figure 1.
Morphologic spectrum of PLAG1-rearranged skin and soft tissue myoepithelial tumors is quite variable, but resembling the salivary gland pleomorphic adenoma phenotype. (A) Ductal structure lined by bland cuboidal epithelial cells with moderate amount of pale, eosinophilic cytoplasm embedded in a myxoid background (ME5, subcutaneous soft tissue, left 5th toe, 53 year old female). (B) Cribriform arrangement of tubulo-ductal structures adjacent to areas of more solid squamoid appearance as well as chondroid matrix differentiation (ME6, subcutaneous soft tissue, right foot, 52 year old female). C. Diffuse solid sheets of myoepithelial cells with a distinctive plasmacytoid appearance (ME9, skin, left hand, 37 year old female). (D) PLAG1 gene rearrangement by FISH showing break-apart signals (telomeric probe, green; centromeric probe, orange; ME9).
The PLAG1-positive tumors were further investigated by FISH for potential rearrangements with frequent PLAG1-fusion partners, previously described in pleomorphic adenoma, including CTNNB1 and LIFR. All except one case did not show abnormalities of these two genes. One case showed break-apart signals for LIFR, which was initially detected by RACE (see below) as a potential fusion partner with PLAG1 (Fig. 2; Table 1). No structural abnormalities were detected of the HMGA2 or FGFR1 gene in any of the cases tested.
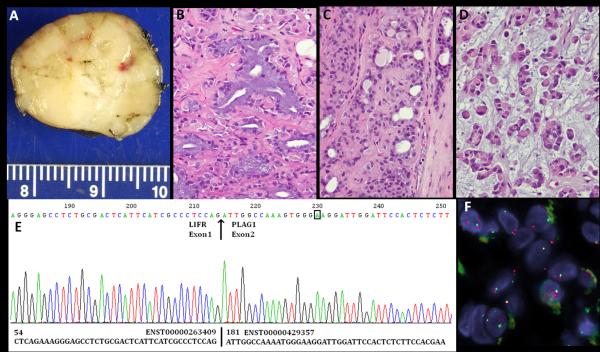
Figure 2.
LIFR-PLAG1 gene fusion in a soft tissue myoepithelial tumor (ME4). (A) Gross appearance of this well-circumscribed, yellow-tan, firm, hand subcutaneous lesion, arising in a 64 year-old female. The histologic spectrum of this tumor is quite broad, including: (B) ducts lined by columnar cells with abundant mucin-containing cytoplasm and basally-located nuclei; C. tubular structures lined by bland cuboidal epithelial cells; (D) Sheets of myoepithelial cells, with a distinctive rhabdoid or plasma-cell phenotype, showing densely eosinophilic cytoplasm and eccentric round nuclei. E.ABI sequence of 5’RACE product showed the presence of a fusion between LIFR exon 1 to PLAG1 exon 2; F. FISH showing a LIFR gene break-apart signal (telomeric probe , green; centromeric probe, orange).
In the PLAG1-negative group, we examined the copy number of both PLAG1 and HMGA2 (Table 2). Only one case each showed the presence of three copies of PLAG1 (ME21) and four copies of HMGA2 (ME27), respectively, in the absence of increased copy number signals for other probes tested, i.e. EWSR1 and FUS.
TABLE 2.
Clinicopathologic Characteristics of Tumors Lacking PLAG1-Rearrangements
| ME | Age/sex | Location | Skin/ST | Ductal structures | Benign/malignant | CK/S100 | PLAG1 CNA FISH |
|---|---|---|---|---|---|---|---|
| 14 | 46/F | chest/breast | skin | no | Benign | +/+ | normal |
| 15 | 57/F | elbow | s-c | no | Benign | +/+ | polysomyd |
| 16 | 45/M | chest wall | deep | no | Malignant | +/+ | normal |
| 17 | 3mo/M | Knee | deep | no | Malignant | +/+ | normal |
| 18 | 3/M | mediastinum | deep | no | Malignant | +/+ | normal |
| 19 | 8/F | thigh | deep | no | Malignant | +/+ | normal |
| 20 | 6mo/M | mediastinum | deep | no | Malignant | +/+ | normal |
| 21 | 45/M | chest wall | deep | no | Malignant | +/+ | trisomyb |
| 22 | 58/M | groin | s-c | yes | Benign | +/+ | normal |
| 23 | 67/M | axilla | s-c | yes | Benign | +/+ | normal |
| 24 | 52/F | Foot | s-c | yes | Benign | +/+ | normal |
| 25 | 53/F | abdominal wall | s-c | no | Benign | +/+ | normal |
| 26 | 84/M | forehead | skin | yes | Benign | +/-a | normal |
| 27 | 6mo/M | scalp | skin | yes | Benign | +/+ | normalc |
| 28 | 74/M | ankle | skin | yes | Benign | +/+ | normal |
| 29 | 29/M | Shin | skin | yes | Benign | +/+ | normal |
| 30 | 48/F | Arm | s-c | yes | Benign | +/+ | normal |
| 31 | 70/M | shoulder | s-c | yes | Malignant | +/+ | polysomyd |
| 32 | 42/F | axilla | s-c | yes | Benign | +/+ | normal |
| 33 | 10/F | Heel | s-c | yes | Benign | +/+ | normal |
| 34 | 66/F | scalp | skin | yes | Benign | +/+ | normal |
| 35 | 38/F | Foot | s-c | no | Benign | +/+ | normal |
ST, soft tissue; F, female; M, male; mo, months; s-c, subcutaneous tissue; CNA, copy number alterations;
positive for GFAP;
three copies of PLAG1 were detected by FISH, but two copies of HMGA2, EWSR1 and FUS;
two copies of PLAG1 were detected by FISH but 4 copies of HMGA2;
trisomy/polysomy of PLAG1, HMGA2, and EWSR1
In the control group, all except one myoepithelial carcinoma ex-pleomorphic adenoma arising in salivary gland showed abnormalities of either PLAG1 or HMGA2 by FISH (Table 3). Four tumors showed PLAG1 gene rearrangements and amplification, which were consistently associated with FGFR1 gene amplifications (Fig. 3). Two tumors showed evidence of gene rearrangement without PLAG1 copy number alterations, while two other lesions had concomitant HMGA2 gene rearrangement and amplification. Interestingly, none of the de novo myoepithelial carcinoma showed abnormalities in EWSR1, FUS, PLAG1 or HMGA2 genes.
TABLE 3.
Pathologic and FISH Results on the Salivary Gland Myoepithelial Tumors Included in the Control Group.
| ME | Diagnosis | Location | PLAG1 | HMGA2 | FGFR1 |
|---|---|---|---|---|---|
| 36 | ME carcinoma ex-PA | parotid | rearrangeda | ND | normal |
| 37 | ME carcinoma ex-PA | parotid | rearrangeda | ND | normal |
| 38 | ME carcinoma ex-PA | parotid | rearranged & C'amplified | ND | rearranged & C'amplified |
| 39 | ME carcinoma ex-PA | parotid | rearranged & amplified | ND | rearranged & amplified |
| 40 | ME carcinoma ex-PA | parotid | rearranged & C'amplified | Normal | rearranged & C'amplified |
| 41 | ME carcinoma ex-PA | parotid | rearranged & C'amplified | ND | rearranged & C'amplified |
| 42 | ME carcinoma ex-PA | parotid | normal | rearranged & C'amplifiedb | ND |
| 43 | ME carcinoma ex-PA | parotid | normal | rearranged & C'amplified | ND |
| 44 | ME carcinoma ex-PA | floor of mouth | normal | Normal | ND |
| 45 | ME carcinoma | parotid | normal | Normal | ND |
| 46 | ME carcinoma | submand | normal | Normal | ND |
| 47 | ME carcinoma | tongue | normal | Normal | ND |
ME, myoepithelial, PA, pleomorphic adenoma; submand, submandibular; ND, not done;
rearrangement present in both PA and ME carcinoma components (also negative for LIFR and CTNNB1 gene rearrangements); C'amplified, centromeric portion is amplified, while telomeric part is not;
HMGA2 rearrangement and C'amplification noted only in the ME-carcinoma, but not in the PA component.
Figure 3.
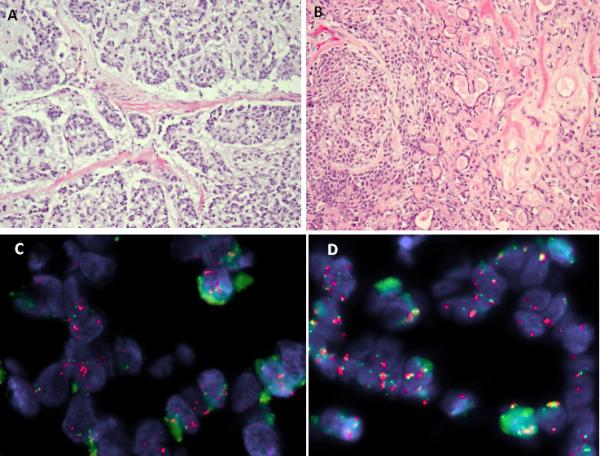
Myoepithelial carcinoma ex-pleomorphic adenoma. A. Morphologic appearance showing cords and nests of epithelial cells with an infiltrative pattern within a myxoid stroma (ME41). B. Small component of benign pleomorphic adenoma seen adjacent to the malignant myoepithelial component (ME41). C. FISH showing break-apart PLAG1 signals, associated with amplification of the centromeric portion (red); D. similar finding for FGFR1 which show rearrangement and centromeric portion amplification (orange), while the telomeric part is not (green).
5’RACE and RT-PCR Results
In the single case of PLAG1-rearanged ME tumor with available frozen tissue, 5’RACE showed the presence of a fusion between LIFR exon 1 to PLAG1 exon 2 (Fig. 2E). This result was further validated by RT-PCR as well as by FISH showing a break-apart signal for LIFR (Fig. 2F).
DISCUSSION
In this study we present a group of benign cutaneous and soft tissue myoepithelial tumors showing PLAG1 gene rearrangements by FISH. This genetic abnormality was noted predominantly in tumors characterized by distinct ductal structures, suggesting that myoepithelial neoplasms with tubulo-glandular differentiation, so-called mixed tumors of skin and soft tissue, are genetically linked to their salivary gland counterpart.
Soft tissue and cutaneous myoepithelial tumors demonstrate heterogeneous morphologic and immunophenotypic features. EWSR1 gene rearrangement is a common event in ME tumors arising outside salivary gland, irrespective of anatomic location (Antonescu et al., 2010). Their histologic spectrum is quite wide, ranging from either undifferentiated small blue cells, to rhabdoid, plasmacytoid or epithelioid cells with moderate amount of clear or eosinophilic cytoplasm (Antonescu et al., 2010). Slightly more than half of the EWSR1-positive ME tumors show morphologic features of malignancy, based on increased mitotic activity and/or prominent nucleoli or nuclear pleomorphism (Antonescu et al., 2010). In 8% of cases the ME tumors harbor a EWSR1-POU5F1 fusion and are characterized by a distinctive phenotype, with epithelioid cells with abundant clear cytoplasm, arranged in a nested pattern, presenting in the deep soft tissues of the extremities in children or young adults (Antonescu et al., 2010). In a similar proportion of cases, soft tissue ME tumors display an alternative EWSR1-PBX1 fusion and are composed of a deceptively bland mixture of epithelioid and spindle cells, embedded in a fibrotic stroma (Brandal et al., 2008; Antonescu et al., 2010). In contrast, EWSR1-negative tumors are more often benign, superficially located, and show ductal differentiation, suggesting an alternative pathogenesis (Antonescu et al., 2010).
In a study by Hallor et al., (2008) whole genome DNA copy number analysis was used to investigate imbalances and the genomic status of the PLAG1 gene in 5 benign soft tissue myoepitheliomas. Their results showed a heterogeneous genetic profile and only one significant recurrent aberration in these tumors, the CDKN2A tumor suppressor gene deletion. A PLAG1 gene rearrangement was identified only in a single soft tissue tumor; however, given the previous history of salivary pleomorphic adenoma in the patient, it remained unclear whether the soft tissue tumor was a primary or a morphologically benign metastasis from the patient's prior salivary gland lesion.
Previous genetic studies on pleomorphic adenoma have implicated a highly specific pattern of chromosomal translocations, affecting the DNA-binding transcription factor genes PLAG1 or HMGA2, resulting in gene fusions in which the 3’-part of PLAG1 or the 5’-part of HMGA2 are linked to various fusion partner genes (Geurts et al., 1997; Geurts et al., 1998; Persson et al., 2009). PLAG1 activation through translocation typically occurs due to promoter swapping from various fusion partner genes that are ubiquitously expressed, i.e. CTNNB1 and LIFR (Kas et al., 1997; Voz et al., 1998). Fusions occur in the 5’-non-coding regions of both partner genes, exchanging regulatory control elements, while preserving the coding sequences (Kas et al., 1997). Despite their common involvement in pleomorphic adenoma, only one of the soft tissue PLAG1-rearranged ME tumors showed the presence of a LIFR-PLAG1 fusion, detected by 5’RACE, and subsequently confirmed by FISH and RT-PCR. The lesion occurred in a 64 year-old female in the subcutis of the hand and showed abundant glandular differentiation.
Additionally, a subgroup of pleomorphic adenoma with ring chromosomes contains amplification of a peri-centromeric segment of chromosome 8, with consistent breakpoints in PLAG1 in 8q12.1 and in FGFR1 in 8p12 and with formation of a novel FGFR1-PLAG1 gene fusion (Persson et al., 2008). In support of these findings all except one myoepithelial carcinoma ex-pleomorphic adenoma included in our control group showed abnormalities of either PLAG1 or HMGA2 by FISH. Among them, four tumors showed PLAG1 gene rearrangements and amplification, which were consistently associated with FGFR1 gene amplifications, in keeping with amplification of the fusion gene. In contrast, none of the PLAG1-rearranged cutaneous and soft tissue ME tumors showed evidence of gene amplification. Furthermore, among the PLAG1-non-rearranged ME tumors, only one case each showed the presence of three copies of PLAG1 and HMGA2, respectively, in the absence of a polysomic phenotype as tested by other chromosomal sites. No structural HMGA2 gene abnormalities were detected in any of the ME lesions tested. Interestingly, none of the de novo salivary gland myoepithelial carcinoma included in our control group showed abnormalities in either PLAG1 or HMGA2.
In this larger group of cases, our results are in full agreement with the recent study by Bahrami et al., (2012), which describes a high incidence of PLAG1 gene rearrangement by FISH in eight of the 11 cases of skin and soft tissue ME tumors. Similar to our findings, all except one PLAG1-rearranged tumors showed histologic evidence of tubulo-ductal differentiation. PLAG1 overexpression as determined by immunohistochemistry was seen in nearly all PLAG1-rearranged tumors, but was however, weakly expressed in one EWSR1-rearranged ME tumor. In addition to the findings by Bahrami et al., (2012), pointing to shared PLAG1-gene rearrangements between a subset of soft tissue ME tumors and pleomorphic adenoma, our in-depth molecular analysis further identifies important differences among these two groups, such as the lack of PLAG1 or HMGA2 gene amplifications, and the infrequent PLAG1 fusions with CTNNB1 or LIFR in the soft tissue ME tumors. Of further interest are the distinct genetic alterations identified in the control group, between de novo salivary gland myoepithelial carcinoma versus the secondary subset, occurring in the setting of a pre-existent pleomorphic adenoma. Despite their common morphology and immunohistochemical profile, the first subgroup lacks PLAG1 or HMGA2 structural or numerical abnormalities, suggesting a different pathogenesis compared to pleomorphic adenoma-related malignancies.
In summary, we report the presence of PLAG1 gene rearrangements in a significant proportion of skin and soft tissue ME tumors associated with tubulo-ductal differentiation morphologically. This finding together with their shared histologic appearance establishes a pathogenetic link with the salivary gland counterpart, i.e. pleomorphic adenoma.
Supplementary Material
Acknowledgments
Supported in part by: P01CA47179 (CRA), P50 CA 140146-01 (CRA).
REFERENCES
- Antonescu CR, Zhang L, Chang NE, Pawel BR, Travis W, Katabi N, Edelman M, Rosenberg AE, Nielsen GP, Dal Cin P, Fletcher CD. EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes Cancer. 2010;49:1114–1124. doi: 10.1002/gcc.20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrami A, Dalton JD, Krane JF, Fletcher CD. A subset of cutaneous and soft tissue mixed tumors are genetically linked to their salivary gland counterpart. Genes Chromosomes Cancer. 2012;51:140–148. doi: 10.1002/gcc.20938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandal P, Panagopoulos I, Bjerkehagen B, Gorunova L, Skjeldal S, Micci F, Heim S. Detection of a t(1;22)(q23;q12) translocation leading to an EWSR1-PBX1 fusion gene in a myoepithelioma. Genes Chromosomes Cancer. 2008;47:558–564. doi: 10.1002/gcc.20559. [DOI] [PubMed] [Google Scholar]
- Geurts JM, Schoenmakers EF, Roijer E, Astrom AK, Stenman G, van de Ven WJ. Identification of NFIB as recurrent translocation partner gene of HMGIC in pleomorphic adenomas. Oncogene. 1998;16:865–872. doi: 10.1038/sj.onc.1201609. [DOI] [PubMed] [Google Scholar]
- Geurts JM, Schoenmakers EF, Roijer E, Stenman G, Van de Ven WJ. Expression of reciprocal hybrid transcripts of HMGIC and FHIT in a pleomorphic adenoma of the parotid gland. Cancer Res. 1997;57:13–17. [PubMed] [Google Scholar]
- Hallor KH, Teixeira MR, Fletcher CD, Bizarro S, Staaf J, Domanski HA, von Steyern FV, Panagopoulos I, Mandahl N, Mertens F. Heterogeneous genetic profiles in soft tissue myoepitheliomas. Mod Pathol. 2008;21:1311–1319. doi: 10.1038/modpathol.2008.124. [DOI] [PubMed] [Google Scholar]
- Hornick JL, Fletcher CD. Myoepithelial tumors of soft tissue: a clinicopathologic and immunohistochemical study of 101 cases with evaluation of prognostic parameters. Am J Surg Pathol. 2003;27:1183–1196. doi: 10.1097/00000478-200309000-00001. [DOI] [PubMed] [Google Scholar]
- Kas K, Voz ML, Roijer E, Astrom AK, Meyen E, Stenman G, Van de Ven WJ. Promoter swapping between the genes for a novel zinc finger protein and beta-catenin in pleiomorphic adenomas with t(3;8)(p21;q12) translocations. Nat Genet. 1997;15:170–174. doi: 10.1038/ng0297-170. [DOI] [PubMed] [Google Scholar]
- Martins C, Fonseca I, Roque L, Pereira T, Ribeiro C, Bullerdiek J, Soares J. PLAG1 gene alterations in salivary gland pleomorphic adenoma and carcinoma ex-pleomorphic adenoma: a combined study using chromosome banding, in situ hybridization and immunocytochemistry. Mod Pathol. 2005;18:1048–1055. doi: 10.1038/modpathol.3800386. [DOI] [PubMed] [Google Scholar]
- Persson F, Andren Y, Winnes M, Wedell B, Nordkvist A, Gudnadottir G, Dahlenfors R, Sjogren H, Mark J, Stenman G. High-resolution genomic profiling of adenomas and carcinomas of the salivary glands reveals amplification, rearrangement, and fusion of HMGA2. Genes Chromosomes Cancer. 2009;48:69–82. doi: 10.1002/gcc.20619. [DOI] [PubMed] [Google Scholar]
- Persson F, Winnes M, Andren Y, Wedell B, Dahlenfors R, Asp J, Mark J, Enlund F, Stenman G. High-resolution array CGH analysis of salivary gland tumors reveals fusion and amplification of the FGFR1 and PLAG1 genes in ring chromosomes. Oncogene. 2008;27:3072–3080. doi: 10.1038/sj.onc.1210961. [DOI] [PubMed] [Google Scholar]
- Voz ML, Agten NS, Van de Ven WJ, Kas K. PLAG1, the main translocation target in pleomorphic adenoma of the salivary glands, is a positive regulator of IGF-II. Cancer Res. 2000;60:106–113. [PubMed] [Google Scholar]
- Voz ML, Astrom AK, Kas K, Mark J, Stenman G, Van de Ven WJ. The recurrent translocation t(5;8)(p13;q12) in pleomorphic adenomas results in upregulation of PLAG1 gene expression under control of the LIFR promoter. Oncogene. 1998;16:1409–1416. doi: 10.1038/sj.onc.1201660. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.