Abstract
Distinct differences between how model proteins interact in-cell and in vitro suggest that cytosol may have a profound effect in modulating protein-protein and/or protein-ligand interactions that are not observed in vitro. Analyses of in-cell NMR spectra of target proteins interacting with physiological partners are further complicated by low signal to noise ratios, and the long over-expression times used in protein-protein interaction studies may lead to changes in the in-cell spectra over the course of the experiment. To unambiguously resolve the principal binding mode between two interacting species against the dynamic cellular background, we analyzed in-cell spectral data of a target protein over the time course of over-expression of its interacting partner by using Single Value Decomposition, SVD. SVD differentiates between concentration-dependent and concentration-independent events and identifies the principal binding mode between the two species. The analysis implicates a set of amino acids involved in the specific interaction that differs from previous NMR analyses but are in good agreement with crystallographic data.
Keywords: in-cell NMR spectroscopy, protein-protein interactions, statistical analysis
In-cell NMR spectra are inherently noisier than spectra acquired in vitro due to the myriad of interactions between the target protein and components of the cytosol.[1] The in-cell spectra of interacting proteins are further complicated by peak broadening and changes in chemical shifts, which often confound straightforward identification of the amino acids on the target protein that are involved in binding to its physiological interactor, i.e. the principal binding mode. Time dependent degradation of the target protein inside the cell or differences in sample preparations can also lead to changes in the resulting NMR spectra. A rigorous objective analysis of spectral changes is needed to unambiguously differentiate between signals that result from interactor-concentration-dependent and –independent processes.
Specific high affinity interactions between a target and interactor protein are studied by using STINT-NMR,[2] which elucidates STructural INTeractions between proteins within their native environment by using in-cell NMR.[2b, 3] After over-expressing the target protein on [U-15N] medium, the binding partner protein is sequentially over-expressed inside the cells grown on unlabeled medium for up to 18 hours. Samples are collected at various times and the 1H{15N}-HSQC, NMR spectra of the [U-15N] target protein is acquired. Changes in the in-cell spectrum of the free target correspond to increasing concentrations of interactor and afford a structural titration of the binding process.
Conventional analyses of interacting proteins tend to incorrectly estimate the number of residues involved in the interaction because of the widespread signal broadening associated with the formation of a stoichiometric complex. The process of distinguishing which spectral changes are due to specific binding generally considers only the spectrum of free target and the final target spectrum following full over-expression of the interactor to assess the change in intensity of a given peak resonance[4]. This difference is used to infer whether or not the corresponding amino acid contributes to the principal binding mode of the target. Because individual signals change at different rates as the concentration of the interactor protein increases (Figure 1A), this analysis ignores changes in the in-cell spectrum that arise over the time course of the experiment and from non-interactor and concentration-independent binding processes. This problem can be overcome by analyzing in-cell NMR data by using Single Value Decomposition, SVD.
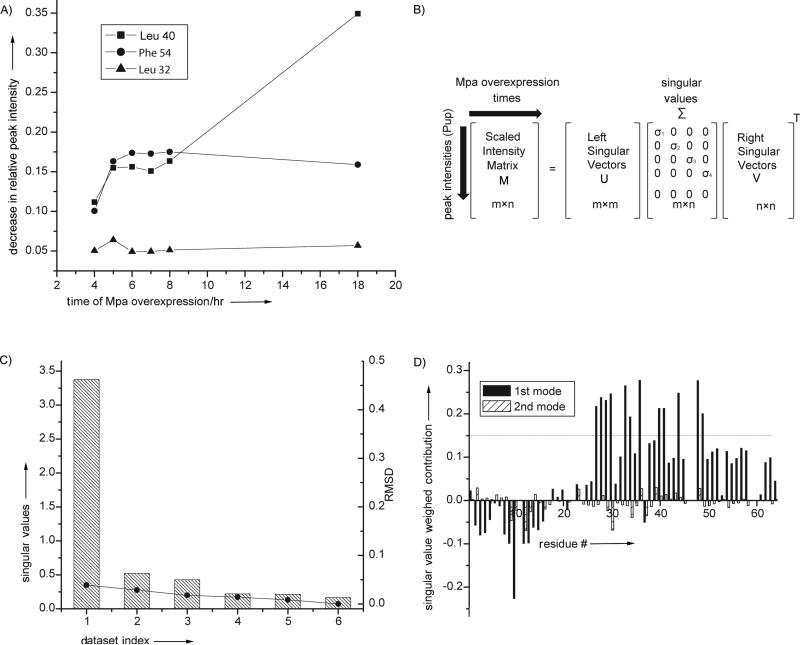
Figure 1. SVD analysis of in-cell protein-protein interactions.
A). The intensity of individual cross peaks of Pup change at different rates during interactor protein over-expression, Mpa. SVD analysis evaluates the magnitude of the contribution of an intensity change to the NMR data over the experimental time course to identify MPA concentration-dependent interactions. B). SVD of the experimental data matrix M of size m x n yields the matrices U, Σ, and VT,[5a] with sizes m × m, m × n, and n × n, respectively, where m is the number of target protein amino acid residues used in the NMR analysis, n is the number of time course NMR datasets, and VT is the transpose of matrix V. C). The Scree plot shows the distribution of singular values for each dataset index (binding mode) from 1 to 6. The root mean square deviation, RMSD, values between respective components and the complete dataset are indicated by solid circles. D). The weighted contribution of each Pup amino acid residue to the Mpa principal binding modes, calculated as a product of a corresponding singular value and left singular vector (see Experimental Section), is shown for the 1st (black) and 2nd (hatched) binding modes. The threshold of 0.14 is chosen to highlight the 12 amino acids that exhibit the largest singular value weighted. Negative values are due to spectral overlap between the target protein and cellular metabolites.
SVD is a mathematical technique used to identify the principal components of an arbitrary matrix that contribute maximally to the variance of its elements.[5] Over the course of a STINT-NMR titration, a series of in-cell NMR spectra are collected, and a matrix, M, is created that contains the changes in target protein peak intensities versus the expression time of the unlabeled binding partner (Figure 1B). SVD analysis of matrix M discriminates between changes in the in-cell NMR spectrum of a target protein due to specific and non-specific binding interactions and changes due to the presence of the complex cellular environment over the time course of interactor over-expression. The analysis identifies the amino acid residues involved in the principal binding mode of a target protein with its interactor.
SVD was previously used to process in vitro NMR spectra,[6] to identify an allosteric interaction network within a protein-ligand complex,[7] and to determine the binding modes of proteins based on chemical shift perturbations.[8] Here, we used Singular Value Decomposition (SVD) to analyze STINT-NMR data previously collected[9] to investigate the in-cell interaction of two proteins involved in Mycobacterium tuberculosis (Mtb) proteasome degradation, the prokaryotic ubiquitin-like protein, Pup, and mycobacterial proteasome ATPase Mpa.[10] The crystal structure of Pup-Mpa is known and the system has been extensively studied in E. coli by using both functional cellular assays[11], and biochemical[12] and structural biology approaches,[13] making this system ideal for evaluating the results of SVD analysis.
The target protein, Pup, is a 64 amino acid protein that modifies and tags Mtb proteins for degradation, and is functionally similar to eukaryotic Ubiquitin. Pup and Ubiquitin have little structural or sequence homology and the mechanism of pupylation is markedly different from that of ubiquitination.[10b] A critical step in degrading pupylated proteins requires recognition of Pup by Mpa. Free Pup is an intrinsically disordered protein, upon binding to Mpa it acquires a helical structure.[13b] The crystal structure of Mpa-bound Pup reveals that the central region spanning amino acids 21-51 becomes helical upon complex formation and is involved in the interaction with Mpa,[13b] the N- and C-termini remain unstructured.
For our purposes, [U-15N] Pup was over-expressed in E. coli BL21(DE3) for two hours followed by sequential over-expression of unlabeled Mpa[2a, 9] for up to sixteen hours; subsequent spectra were collected at time intervals corresponding to increasing Mpa concentrations (Figure S1, S2, and S3 see also Supplementary Information). After collecting the sample the cells were subjected to in-cell NMR analysis without prior freezing. The 1H{15N}-HSQC NMR spectrum of free Pup was used as a control. The acquisition time for an in-cell NMR experiment was less than two hours. To verify that the protein signals originated from the cellular protein, after each in-cell NMR experiment, the cells were recovered from the tube, the in-cell NMR sample was centrifuged and the NMR spectrum of the supernatant was recorded; no protein signals were detected.
To minimize the signal to noise ratio, the amplitudes of crosspeak intensities in the in-cell NMR spectra of free [U-15N] Pup and [U-15N] Pup-Mpa complexes were determined and scaled to define matrix M (Table S1). The data indicate differential broadening of Pup resonances (positive matrix values) over the course of the titration due to interactions with Mpa. The data also show peaks that increase in intensity (negative matrix values) because of spectral overlap between cellular metabolites and Pup.
To determine how Pup binds to Mpa we applied an SVD analysis to matrix M. The analysis indicates the existence of only one principal-binding mode (Table S2). The Scree plot[5b, 14] of singular values (Figure 1C) shows a clear drop after the first singular value, which contributes to 97% of the Frobenius norm of Σ. The root mean square deviation, RMSD, of the contribution of the principal-binding mode to M does not improve by including additional binding modes (Figure 1C and Figure 1D).
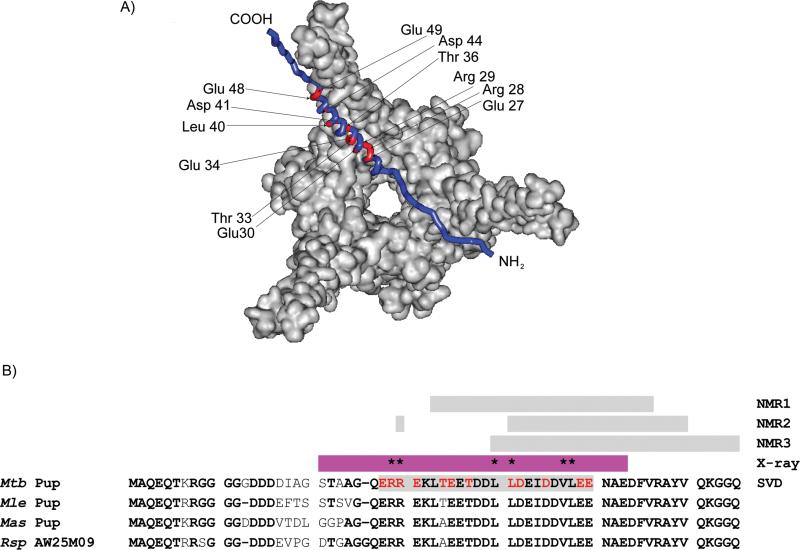
Physically, the first principal binding mode corresponds to Mpa-concentration dependent specific and non-specific interactions between Pup and Mpa. All concentration-independent interactions between Pup-Mpa appear as noise reflected in the five remaining diminishing singular values (Figure 1C, Table S2). Figure 1D shows the contribution of Pup amino acids to the first and second principal-binding modes. The 12 residues exhibiting the greatest singular value weighted contributions are confined to the structured portion of Pup, the α-helix. These residues, E27, R28, R29, E30, T33, E34, T36, L40, D41, D44, E48, and E49, are mapped onto the Mpa-Pup crystal structure (Figure 2A).
Figure 2. Only a conserved segment of the Pup helix is involved in in-cell interactions with Mpa.
A). Residues comprising the principle-binding mode (red) between Pup and Mpa are mapped onto a Pup-Mpa complex (PDB code 3M9D).[13b] B). Sequence alignments of Pup show conserved residues in the mycobacterial genome (bold). Pup residues involved in the first principal-binding mode are in red. The gray bars show the regions of Pup that were implicated in the Pup-Mpa interaction by using in vitro NMR (NMR1[13c] and NMR2),[13a] by in-cell NMR (NMR3),[9] and by SVD analysis of the in-cell data; individual residues involved are omitted for ease of viewing. The α-helical region identified in the crystallographic study (X-ray)[13b] is shown in magenta. Residues that abolish Mtb proteasome function when mutated are marked with an asterisk.[13b]
A second class of 17 amino acids with smaller singular value weighted contributions are located in the C-terminal half of the α-helix through the disordered C-terminus. The reduced signal broadening associated with these amino acids suggests that these residues bind non-specifically to Mpa. The two classes of binding identified in the SVD analysis likely correspond to two separate sets of determinants, ordered and disordered first proposed in.[13a]
The original in-cell analysis of the NMR data[9] and previous in vitro NMR analyses [13c, 15] implicated the C-terminal half of the α-helix and the C-terminus of Pup in the specific interaction with Mpa (Figure 2B). SVD analysis reveals that Pup residues involved in specfic interactions are confined to the conserved region of the helix, in strikingly good agreement with the crystal structure (Figure 2B). The line broadening observed for residues 26-30 was not as dramatic as for residues 31-49 but was strongly dependent on the Mpa concentration; this may explain why the contribution of those residues was not observed in previous NMR analyses. Mutating residues R28 and R29 abolishes degradation of known proteasome substrates in mycobacteria,[13b] confirming the importance of these residues in the physiological interaction. Previous structural studies show that the N-terminus of the Pup helix is not affected by the in-cell interaction with Mpa and remains unstructured in the presence of Mpa.[13b]
Conventional analysis of the original in-cell NMR data[9] overestimated the number of amino acid residues in the C-terminal half of the α-helix and in the C-terminal region of the protein and underestimated the number of residues in the N-terminal half of the α-helix that contribute to the specific interaction between Pup and Mpa. Inspection of the V matrix (Table S2), which determines the weight that each dataset contributes to the principal binding mode, shows the deficiency of the conventional approach; datasets from long Mpa over-expression times dominate the first principal binding mode. Conventional analyses employing only two datasets (e.g. early and late protein over-expression) cannot compensate for variations in the level of protein over-expression and changes in the metabolic state of the cell, which occur over the time course of the experiment, to unambiguously identify the amino acids that are involved in the principal binding between the two species (Figure 1A).
The strength of SVD is its ability to identify consistent spectral changes that track with the increase in concentration of the interactor, resulting in a mathematically rigorous, objective analysis that define the amino acid residues involved in the principal interaction. The differences between the amino acid residues of Pup that interact specifically with Mpa, identified in previous in vitro studies and the current work, emphasize the importance of cytosol in modulating the interactions that give rise to protein complexes in general and the need to be able to resolve these signals from the excessively noisy in-cell background, thus underscoring the importance of using SVD analysis to interpret in-cell protein-protein and also protein-ligand interactions.
Experimental Section
NMR spectroscopy
Cells containing free [U-15N] Pup-GGQ or [U-15N] Pup-GGQ/Mpa were re-suspended in NMR buffer (0.5 mL), potassium phosphate (10 mM, pH 6.5), 90%/10% H2O/D2O, and transferred to an NMR tube. All NMR experiments were performed at 293K using a Bruker Avance 700 MHz NMR spectrometer equipped with a cryoprobe. We used a Watergate version of an 1H{15N}-edited HSQC.[16] Data were recorded with 32 transients as 512 x 64 complex points in the proton and nitrogen dimensions, respectively, apodized with a squared cosine-bell window function and zero-filled to 1k{128}[11a] points prior to Fourier transformation. The corresponding sweep widths were 12 and 35 ppm in the 1H and 15N dimensions, respectively. Chemical shifts of [U-15N] Pup-GGQ inside the cell are slightly different from purified Pup.[13a] The backbone chemical shifts of Pup-GGQ were reassigned using a clarified lysate of [U-13C, 15N]-Pup-GGQ and a standard suite of triple resonance experiments.[9] To reassign the [U-15N] Pup-GGQ peaks that changed their positions due to complex formation, we assumed minimum chemical shift changes,[17] calculated as Δmin = (δmin2 + (δN/4)2)1/2, where δH(N) represents the change in hydrogen and nitrogen chemical shifts. After each NMR experiment, the cells were pelleted and the 1H{15N}-HSQC spectrum of the supernatant was collected. No NMR signal was observed above the noise level implying that no leakage or cell lysis was occurring during the experimental acquisition time.[9] Cell viability after in-cell NMR experiments was tested by plating bacteria at dilutions of 1:10,000, 1:100,000, and 1:1000,000 on plates containing the appropriate antibiotics before and after in-cell NMR experiments. After counting the colonies, it was established that cell viability was 92 ± 7%.
Data analysis
The amplitudes of the intensities of the cross peaks in the in-cell 1H{15N}-HSQC spectra of free target protein and target protein with binding partner over-expressed for different times were determined. Experimental intensities were scaled for the entire data and changes in intensity were calculated by using ΔI = (I/Iref)bound – (I/Iref)free, where (I/Iref)free is the scaled intensity of an individual peak in the in-cell spectrum of free Pup-GGQ, (I/Iref)bound is the scaled intensity of individual peaks in the in-cell spectrum of the Pup Msm Mpa complex, and Iref is a glutamine peak at 7.45 ppm and 112.5 ppm in proton and nitrogen dimensions, respectively, that does not shift during titration. Positive changes in relative intensities denote peak broadening due to binding interactions. Negative changes in intensities are due to overlapping peaks in the bound state. Peak overlap was resolved as described in.[9]
The data are represented by an m × n matrix, M, in which the column index n represents different time points in the over-expression of a binding partner, corresponding to increasing concentration, and the row index m represents the scaled intensities of amino acid peaks on the target protein (Figure 1A). M is transformed into three matrices:[5] two unitary matrices, U = (U1, U2, ..., Um) and V = (V1, V2, ..., Vn) of sizes m × m and n × n, respectively, and a singular values matrix, Σ, of size m × n. In these matrices Ui are eigenvectors of MMT, or so-called left singular vectors, Vi are eigenvectors of MTM, or so-called right singular vectors, and MT is the transpose of M. Furthermore, the Σ matrix has a form Σ = diag(σ1, σ2,...., σn), where σi are eigenvalues of both MTM and MMT arranged in order of decreasing magnitude. SVD is unique when the singular values, σi, are not degenerate.[5a]
In this construction, the left singular vector, Ui represents the ith principal component or principal binding mode of M, and Vi determines the contribution of an individual dataset (i = 1...n) to each principal binding mode, MVi = σiUi. The strength of each principal binding mode is indicated by the respective singular value, σi, so that the most dominant principal binding mode has the highest singular value. Since the data matrix M can be reconstituted in terms of principal binding modes as M = σ1U1V1 T + σ2U2V2 T + ...+ σnUnVn T, the normalized difference between M and the sum of k principal binding modes describes the goodness of fit, or RMSD, of M by using only k principal binding modes: RMSD = ||M − σ1U1V1 T − σ2U2V2 T − ...− σkUkVk T ||F/(k × m)1/2, where k < n and ||...||F denotes a standard Frobenius norm.[5b]
Matrix M was assembled in Excel (Microsoft, Inc), exported as an ASCII text file, and read into MATLAB 6.5 (Mathworks, Inc). Singular value decomposition of M was accomplished by using the [U,Σ,V]=svd[M] command.[5a] The generated output matrices U, Σ, V are left singular vectors, singular value matrix and right singular vectors, respectively. The transpose of V is calculated by using the command T=V’.
Supplementary Material
Acknowledgements
This work was supported by NIH grant GM085006 to A.S.
References
- 1.Maldonado AY, Burz DS, Shekhtman A. Prog Nucl Magn Reson Spectrosc. 2011;59:197–212. doi: 10.1016/j.pnmrs.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a Burz DS, Dutta K, Cowburn D, Shekhtman A. Nat Methods. 2006;3:91–93. doi: 10.1038/nmeth851. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Selenko P, Serber Z, Gadea B, Ruderman J, Wagner G. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:11904–11909. doi: 10.1073/pnas.0604667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a Serber Z, Dotsch V. Biochemistry. 2001;40:14317–14323. doi: 10.1021/bi011751w. [DOI] [PubMed] [Google Scholar]; b Inomata K, Ohno A, Tochio H, Isogai S, Tenno T, Nakase I, Takeuchi T, Futaki S, Ito Y, Hiroaki H, Shirakawa M. Nature. 2009;458:106–109. doi: 10.1038/nature07839. [DOI] [PubMed] [Google Scholar]; c Bertrand K, Reverdatto S, Burz DS, Zitomer R, Shekhtman A. Journal of the American Chemical Society. 2012;134:12798–12806. doi: 10.1021/ja304809s. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Banci L, Barbieri L, Bertini I, Luchinat E, Secci E, Zhao Y, Aricescu AR. Nature chemical biology. 2013 doi: 10.1038/nchembio.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Hamatsu J, O'Donovan D, Tanaka T, Shirai T, Hourai Y, Mikawa T, Ikeya T, Mishima M, Boucher W, Smith BO, Laue ED, Shirakawa M, Ito Y. Journal of the American Chemical Society. 2013;135:1688–1691. doi: 10.1021/ja310928u. [DOI] [PubMed] [Google Scholar]
- 4.Burz DS, Dutta K, Cowburn D, Shekhtman A. Nat Protoc. 2006;1:146–152. doi: 10.1038/nprot.2006.23. [DOI] [PubMed] [Google Scholar]
- 5.a Golub GH, Van Loan CF. Matrix Computations. 4 ed. The Johns Hopkins University Press; Baltimore, USA: 2012. [Google Scholar]; b Demmel JW. Applied Numerical Linear Algebra. Society for Industrial and Applied Mathematics; Philadelphia, USA: 1997. [Google Scholar]
- 6.a Trbovic N, Smirnov S, Zhang F, Bruschweiler R. Journal of magnetic resonance. Vol. 171. Calif; San Diego: 2004. pp. 277–283. 1997. [DOI] [PubMed] [Google Scholar]; b Losonczi JA, Andrec M, Fischer MW, Prestegard JH. Journal of magnetic resonance. Vol. 138. Calif; San Diego: 1999. pp. 334–342. 1997. [DOI] [PubMed] [Google Scholar]
- 7.Selvaratnam R, Chowdhury S, VanSchouwen B, Melacini G. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6133–6138. doi: 10.1073/pnas.1017311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a Arai M, Ferreon JC, Wright PE. Journal of the American Chemical Society. 2012;134:3792–3803. doi: 10.1021/ja209936u. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Caillet-Saguy C, Piccioli M, Turano P, Izadi-Pruneyre N, Delepierre M, Bertini I, Lecroisey A. Journal of the American Chemical Society. 2009;131:1736–1744. doi: 10.1021/ja804783x. [DOI] [PubMed] [Google Scholar]
- 9.Maldonado AY, Burz DS, Reverdatto S, Shekhtman A. PLoS One. 2013;8:e74576. doi: 10.1371/journal.pone.0074576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a Striebel F, Imkamp F, Ozcelik D, Weber-Ban E. Biochimica et biophysica acta. 2013 doi: 10.1016/j.bbamcr.2013.03.022. [DOI] [PubMed] [Google Scholar]; b Samanovic MI, Li H, Darwin KH. Sub-cellular biochemistry. 2013;66:267–295. doi: 10.1007/978-94-007-5940-4_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Science. 2008;322:1104–1107. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Burns KE, Liu WT, Boshoff HI, Dorrestein PC, Barry CE., 3rd The Journal of biological chemistry. 2009;284:3069–3075. doi: 10.1074/jbc.M808032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Striebel F, Imkamp F, Sutter M, Steiner M, Mamedov A, Weber-Ban E. Nature structural & molecular biology. 2009;16:647–651. doi: 10.1038/nsmb.1597. [DOI] [PubMed] [Google Scholar]
- 13.a Chen X, Solomon WC, Kang Y, Cerda-Maira F, Darwin KH, Walters KJ. J Mol Biol. 2009;392:208–217. doi: 10.1016/j.jmb.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wang T, Darwin KH, Li H. Nature structural & molecular biology. 2010;17:1352–1357. doi: 10.1038/nsmb.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Liao S, Shang Q, Zhang X, Zhang J, Xu C, Tu X. Biochem J. 2009;422:207–215. doi: 10.1042/BJ20090738. [DOI] [PubMed] [Google Scholar]
- 14.Cattell RB. Multivariate Behav. Res. 1966;1:245–276. doi: 10.1207/s15327906mbr0102_10. [DOI] [PubMed] [Google Scholar]
- 15.Sutter M, Striebel F, Damberger FF, Allain FH, Weber-Ban E. FEBS Lett. 2009;583:3151–3157. doi: 10.1016/j.febslet.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 16.Piotto M, Saudek V, Sklenar V. J Biomol NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 17.Farmer BT, 2nd, Constantine KL, Goldfarb V, Friedrichs MS, Wittekind M, Yanchunas J, Jr., Robertson JG, Mueller L. Nature structural biology. 1996;3:995–997. doi: 10.1038/nsb1296-995. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.