Abstract
We analyzed highly vancomycin-resistant Gram-positive bacteria isolated from the saliva of migratory songbirds captured, sampled, and released from a birdbanding station in western Kansas. Individual bacterial isolates were identified by partial 16S rRNA sequencing. Most of the bacteria in this study were shown to be Staphylococcus succinus with the majority being isolated from the American Robin. Some of these bacteria were shown to carry vanA, vanB, and vanC vancomycin-resistance genes and have the ability to form biofilms. One of the van gene-carrying isolates is also coagulase positive, which is normally considered a virulence factor. Other organisms isolated included Staphylococcus saprophyticus as well as Enterococcus gallinarum. Given the wide range of the American Robin and ease of horizontal gene transfer between Gram-positive cocci, we postulate that these organisms could serve as a reservoir of vancomycin-resistance genes capable of transferring to human pathogens.
Introduction
Six resistance patterns for vancomycin resistance have been reported in the genus Enterococcus designated VanA through VanE and VanG [17]. Vancomycin resistance can arise in bacteria through the presence of two different types of operons. The majority of resistance is conferred by operons which encode enzymes which alter the cellular vancomycin target, thereby reducing the affinity of vancomycin for the target. A second known type of operon encodes enzymes that rid cells of precursors of high-affinity vancomycin targets [10]. The first type of vancomycin resistance described was the VanA type, which confers high levels of resistance to both vancomycin and teicoplanin. The VanA type is conferred by genes contained within the transposon Tn1546. The VanB-type mediates variable resistance to both vancomycin and teicoplanin, and has been shown to be associated with either transposon Tn1547 or Tn1549. Since both VanA and VanB resistance types are associated with mobile genetic elements, they can be found in either the bacterial chromosome or within plasmids, and maybe passed to other bacteria via conjugation. Where VanA resistance is inducible by both vancomycin and teicoplanin, the VanB type is only inducible by vancomycin [10].
The VanC type has been shown to express intrinsic resistance capability encoded by genes located only within the bacterial chromosome. Unlike VanA and VanB, the resistance conferred by VanC is only to low levels of vancomycin and does not confer resistance to teicoplanin. VanC was first reported in clinical isolates of Enterococcus gallinarum, and was later shown to also exist in Enterococcus casseliflavus and Enterococcus flavescens [10]. The VanD resistance type confers moderate levels of intrinsic resistance to vancomycin and teicoplanin. Like the VanC operon, VanD has only been reported in bacterial chromosomes, and not associated with mobile elements such as transposons or plasmids [10]. The vancomycin-resistance operons contain genes of common function, but the actual genes in each operon differ. For instance, the VanB operon contains the vanW gene, of unknown function, which the VanA operon does not carry [11].
Birds have been shown to carry infectious diseases such as West Nile virus, avian influenza, and pathogenic bacteria, such as Salmonella. Studies indicate songbirds can carry various genera of bacteria such as Escherichia, Pseudomonas, Staphylococcus, Streptococcus, and Yersinia, all of which can be pathogenic in birds [7]. One of the songbird species captured in our study, Swainson's Thrush, has been shown to carry West Nile virus, Borrelia burgdorferi, and the blood parasites Haemoproteus and Plasmodium [3]. Prukner-Radovcic et al. [23] isolated Escherichia coli, Staphylococcus, and Streptococcus from birds in both urban and rural locations of Croatia; these bacteria were isolated from pharyngeal swabs of migratory species. However, little is known about the normal flora of birds, and most studies of bacteria in birds have focused on fecal samples of passerines [7, 21].
The songbirds that migrate through western Kansas use many different habitats as stopover points, and have a diverse range of dietary requirements that include grains, fruits, and insects [25]. Migrants will forage throughout trees, in agricultural fields, grasslands, along streams, and on the ground. Since birds make contact with much of the environment, this opens the opportunity to spread bacteria they are carrying into their surroundings. Conversely, the opportunity also exists for bacteria in the environment to be transferred to the birds. If resistance genes are already in the environment, the birds then could potentially ingest bacteria carrying resistance genes into their mouths, or expel bacteria carrying resistance genes.
The purpose of this study was to determine whether migratory songbirds harbored Gram-positive, vancomycin-resistant cocci, and whether such bacteria harbored known vancomycin-resistance genes, as well as possess the ability to produce biofilms.
Materials and Methods
Study Site
The Fort Hays State University Wildlife Sanctuary (38°87′3′’N, 99°34′8′’W), located along Big Creek in Ellis County, Kansas, served as the study site. This 10-ha site is composed of riparian habitat with a mixture of mature hardwoods, forbs, and shrubs. An alfalfa field lies on the north and west side and Big Creek runs on the east of the study site. The banding site contains 12 individual mist nets, each 12 m in length by 2.6 m in height, set up in standard locations throughout the study area.
Collection of Saliva Samples
The mist nets in the study site were open from approximately 0630 CST to 1100 CST six days a week, from 20 August to 31 October 2006, to passively capture individual birds. The nets were checked every 10–15 min to minimize stress and possible mortality. Once an individual was captured, the bird was transported to a central location for processing, where mass, body fat index, molt, age, and sex of each individual were determined. In addition, a saliva sample was taken by inserting a sterile swab into the mouth of the individual; the swab was then placed into a sterile bag where it was later taken to the lab for culturing. After the collection of the saliva sample, the individual bird was released.
Culture and Isolation
The saliva sample from each bird was streaked on Brain Heart Infusion (BHI) agar (Difco Laboratories, Detroit, Michigan) and incubated for 24 h at 37 °C to allow for colony growth and formation. The bacterial colonies were examined for the isolated differences in morphology, specifically color, growth types, and other visual variables. Each type of bacterial colony growth was then streaked for isolation on a BHI agar plate to obtain pure colonies. Isolated colonies were restreaked until pure cultures were obtained. The isolates were subsequently Gram stained according to the kit manufacturers' protocols (BBL Becton– Dickinson Microbiology Systems, Cockeysville, Maryland) to determine cell morphology and grouping, as well as Gram reactions. Gram-positive cocci isolates were assigned a unique number and frozen at −80 °C in sterile phosphate buffered saline (PBS) and glycerol media at a ratio of 60 % PBS to 40 % glycerol.
Coagulase Production
Tellurite glycine agar (Difco Laboratories, Detroit, Michigan) was used to screen for coagulase production by the vancomycin-resistant Gram-positive cocci.
Antibiotic Resistance Screening
The Gram-positive isolates were streaked on Mueller– Hinton agar (Difco Laboratory, Detroit, Michigan) containing 100 μg/ml of vancomycin hydrochloride (Acros Organics, Geel, Belgium). Bacterial isolates that showed colony formation were again Gram stained to confirm the Gram reactions as well as cell morphology and grouping.
Following manufacturer's protocols, minimum inhibitory concentrations (MIC) were determined by vancomycin Etests (AB Biodisk, Solna Sweden) for each antibiotic-resistant organism. Staphylococcus aureus ATCC 25923 and Escherichia coli ATCC 25922 were used as control organisms.
Bacterial Identification
The resulting vancomycin-resistant Gram-positive cocci isolates were submitted to MIDI Labs (Newark, Delaware) for the identification based on sequencing analysis of the first 500 base pairs of the 16S rRNA gene.
Genomic DNA Isolation
Pure cultures of the bacteria were inoculated in 4.0 mL of tryptic soy broth (Difco, Detroit, Michigan, USA) and incubated for 12 h at 37 °C. Bacterial genomic DNA was isolated from the overnight cultures by previously described methods [5]. The presence of genomic DNA was confirmed by 1 % agarose gel electrophoresis.
van Gene Detection
The presence of van genes was detected by the polymerase chain reaction (PCR) using the extracted genomic DNA as the template, and specific primers for the vanA, vanB, vanC, and vanD genes as designed by Depardieu et al. [11]. The PCR was performed using the following cycle conditions: 94 °C for three min, 40 cycles of 94 °C for one min, 54 °C for one min, and 72 °C for one min, the final elongation step was 72 °C for seven min. PCR products were verified via 1 % agarose gel electrophoresis. Bacteria cultures known to carry vanA, vanB, vanC, and vanD genes were used as positive controls for PCR detection. These were Enterococcus faecalis SFV1, Enterococcus faecalis V583, Enterococcus gallinarum ATCC 49579, and Enterococcus faecium N97-0330, respectively.
Biofilm Detection by Crystal Violet Assay
Biofilm presence was detected via the Crystal Violet Assay (CVA) as described by Toledo-Arana et al. [27]. Pure cultures of each organism were inoculated in 4.0 mL of tryptic soy broth (TSB) (Difco, Detroit, Michigan, USA) and grown for 12 h at 37 °C. Staphylococcus epidermidis ATCC 35984 and Staphylococcus epidermidis ATCC 12228 were used as positive and negative controls, respectively.
Biofilm Detection by Scanning Electron Microscopy
Specimens were prepared for scanning electron microscopy as described by Thurlow and Gillock [26] and examined for the presence of biofilm. Staphylococcus epidermidis ATCC 35984 and Staphylococcus epidermidis ATCC12228 were used as positive and negative biofilm controls, respectively.
Results and Discussion
Throughout the 2006 banding season, a total of 22 saliva samples were collected from 38 netted American Robin, and 8 of these carried vancomycin-resistant Gram-positive cocci. Of the 80 Orange-crowned Warbler netted, three of twenty-two individuals sampled also carried these microorganisms. Two of the ten sampled Swainson's Thrush sampled harbored vancomycin-resistant Gram-positive cocci, as did one Carolina Wren, and one Least Flycatcher of the two individuals of each species sampled. One saliva sample was collected from a Mourning Warbler with that one captured individual also harboring vancomycin-resistant Gram-positive cocci (Table 1).
Table 1. Summary of results indicating bacterial species, isolate number, coagulase activity, MIC (μg/mL), bird host (AR, American Robin; CW, Carolina Wren; LF, Least Flycatcher; MW, Mourning Warbler; OCW, Orange-crowned Warbler; ST, Swainson's Thrush), the presence of van genes, and biofilm-forming ability.
| Bacterial ID | Isolate | Coagulase | MIC | Bird host | van | Biofilm |
|---|---|---|---|---|---|---|
| S. succinus | V45 | + | ≥256 | AR | A | – |
| S. succinus | V46 | – | ≥256 | AR | A, B, C | – |
| S. succinus | V47 | – | ≥256 | AR | – | – |
| S. succinus | V54 | – | ≥256 | AR | A | – |
| S. succinus | V56 | – | ≥256 | AR | – | + |
| S. succinus | V67 | – | ≥256 | AR | – | – |
| S. succinus | V185 | – | 1.5 | CW | A | – |
| S. succinus | V163 | – | ≥256 | LF | – | + |
| S. succinus | V154 | + | 1.7 | MW | – | + |
| S. succinus | V187 | – | ≥256 | OCW | A | – |
| S. succinus | V166 | – | ≥256 | OCW | – | – |
| S. succinus | V170 | – | ≥256 | OCW | – | – |
| S. succinus | V186 | + | 2.0 | ST | – | – |
| S. succinus | V183 | – | ≥256 | ST | – | – |
| S. saprophyticus | V22 | – | ≥256 | AR | A | – |
| E. gallinarum | V49 | – | ≥256 | AR | C | – |
The collected saliva samples produced 260 bacterial isolates cultured and isolated for further testing. Sixteen vancomycin-resistant Gram-positive cocci isolates were identified from the partial sequencing of the 16S rRNA gene. Fourteen of the isolates were determined to be Staphylococcus succinus, one was shown to be Staphylococcus saprophyticus and the other was identified as Enterococcus gallinarum (Table 1).
Staphylococcus succinus was carried by six American Robin, three Orange-crowned Warbler, two Swainson's Thrush, one Mourning Warbler, one Least Flycatcher, and one Carolina Wren. Staphylococcus saprophyticus and Enterococcus gallinarum were each carried by two different individual American Robins. Of the organisms isolated, only three were shown to be positive for coagulase production. It is interesting to note that all three coagulase-positive isolates were identified as Staphylococcus succinus and all were sampled from different individual birds (Table 1).
The Etest revealed that many bacterial isolates had extremely high MICs relative to vancomycin with thirteen of the sixteen exhibiting MICs of ≥256 μg/mL. The majority of the organisms with high vancomycin MICs (eleven out of thirteen) were Staphylococcus succinus; however, the single Staphylococcus saprophyticus and Enterococcus gallinarum isolates also showed MICs of ≥256 μg/mL (Table 1).
According to the PCR assay for van genes, four of the fourteen Staphylococcus succinus isolates, as well as the single Staphylococcus saprophyticus sample, carried the vanA gene alone. One of the Staphylococcus succinus isolates was demonstrated to carry the vanA, vanB, and vanC genes. In addition, the single Enterococcus gallinarum was shown to carry vanC alone. The vanD gene was not detected in any of the isolates (Table 1).
Three of the fourteen Staphylococcus succinus isolates were shown to have the ability to form biofilms according to the results of both the CVA as well as scanning electron microscopy. Biofilm production was not observed in the negative control strain, Staphylococcus epidermidis ATCC12228, using either of these methods. Conversely, biofilm production was readily seen in the positive control strain, Staphylococcus epidermidis ATCC35984, by both methods; the electron microscopy method produced the most striking results (Fig. 1). The three Staphylococcus succinus cultures that exhibited biofilm production were isolated from three different birds: an American Robin, Mourning Warbler, and Least Flycatcher (Table 1).
Fig. 1.
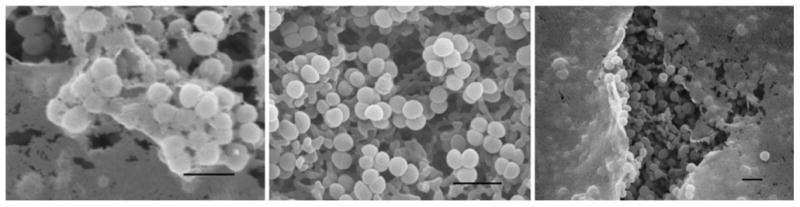
Scanning electron microscopy method for viewing biofilm production. Left, positive control (Staphylococcus epidermidis ATCC 35984); Center, negative control (Staphylococcus epidermidis ATCC 12228); Right, isolate V56 (Staphylococcus succinus). Scale bar 20 microns in all panels
Most birds migrating through western Kansas and much of North America move seasonally from the northern United States and Canada south to Mexico or Central America for the winter. During this migration, several billion individual birds move across the landscape and may stop multiple times to acquire resources and rest [14]. During the several-hour to several-day intervals when they are relatively stationary, migratory birds can acquire or spread novel bacteria. We did not determine where the vancomycin-resistant genes the birds were carrying originated or how long they had been carrying them.
The bird species detected to carry the antibiotic-resistant bacteria normally occur in large trees, in the understory, in litter, or downed wood on the ground. Owing to their feeding ecology, they often come into contact with soil and various plant surfaces. Any of these sites are possible direct or indirect sources where they could acquire the antibiotic-resistant bacteria. Alternatively, all species of birds identified to carry the antibiotic-resistant bacteria consume invertebrates; insects could be the mechanism of transport [20].
American Robin was the most common bird species to carry antibiotic-resistant bacteria, and it was one of the most abundant species on the site. Most of the antibiotic-resistant bacteria were sampled during only a few days of high capture, where one of more individual robin was captured from a large flock. Because this species is more likely to inhabit urban and suburban areas, it is possible that it would come into contact with environmental sources contaminated by humans. This could include environmental sources harboring bacteria resistant to antibiotics that have been used only in humans. American Robins in this study carried several bacterial species which have been isolated from a variety of sources [16, 19, 22].
Few published studies identify bacteria from saliva samples of birds, in contrast to surveys of normal gut flora and fecal sampling. Both Gram-positive and Gram-negative species of bacteria have been isolated from cloacal samples of birds [7]. Staphylococcus and Streptococcus species have been identified from pharyngeal swabs of birds in Croatia [23]. Thus, it is not known if the identified antibiotic-resistant isolates are normal flora, opportunistic pathogens, or pathogenic strains.
The vancomycin-resistant Gram-positive cocci were isolated from birds throughout the banding season. Most bacteria were isolated during the month of September, the month when the majority of the migrating birds were passing through the banding site. Capture dates of individuals carrying the antibiotic-resistant bacteria ranged from 20 August, the first date of bird banding to 12 October 2006, 1 week before we ceased netting efforts.
The most prevalent of the vancomycin-resistant Gram-positive cocci identified in this study were Staphylococcus succinus. This microorganism is normally associated with foods, or plays a major role in the food processing industry, and is not usually considered a pathogen [24]. However, opportunistic coagulase-negative staphylococci (CNS) can cause severe infections since several are relatively multi-drug resistant. Resch et al. [24] chose 21 antibiotics for the assessment of resistance in the CNS organisms Staphylococcus succinus and Staphylococcus xylosus. They reported that 90 % of the assayed strains of these organisms exhibited resistance to the chosen antibiotics. Furthermore, some of the Staphylococcus succinus strains were resistant to up to four different antibiotics. It is interesting to note, however, none of their Staphylococcus succinus strains were resistant to vancomycin [24]. The MIC against vancomycin of most of the Staphylococcus succinus strains from this study was ≥256 μg/mL, indicative of extreme vancomycin resistance since vancomycin-resistant microorganisms are characterized as those in which the MIC is ≥32 μg/mL [30].
One vancomycin-resistant organism from this study was Staphylococcus saprophyticus, another CNS, which is usually considered normal microbial flora, but can also cause infections in humans. It causes acute urinary tract infections [15] and can be an agent of nosocomial and bloodstream infections [31]. Staphylococcus saprophyticus accounts for up to 42 % of all urinary tract infections in young women [29]. Several studies have reported that some Staphylococcus saprophyticus strains are vancomycin resistant [1]. To our knowledge, however, this is the first report of a Staphylococcus saprophyticus strain which carries the vanA gene and has a vancomycin MIC of ≥256 μg/mL.
The third vancomycin-resistant Gram-positive coccus isolated in this study was Enterococcus gallinarum, which is also not typically considered a major human pathogen. However, there is a report of vancomycin-resistant Enterococcus gallinarum causing a nosocomial outbreak in 2004 in a Colombian-teaching hospital. In this case, the main clinical syndromes were bloodstream infections and surgical site infections. In these organisms, the virulence genes normally associated with either Enterococcus faecalis or Enterococcus faecium were not detected [9]. Another study suggested that Enterococcus gallinarum might be the original source of the vanC gene [13]. The Enterococcus gallinarum isolate from our study may harbor other means of resisting vancomycin besides the vanC gene, given the MIC is ≥256 μg/mL, a much higher value than the MIC of 2–32 μg/mL normally conferred by this gene [8]. To the best of our knowledge, this is the first report of any van genes being found in a staphylococcal species other than Staphylococcus aureus, as well as the first report of the vanB gene in a bacterial genus other than Enterococcus.
Three of the isolated vancomycin-resistant Gram-positive cocci exhibited the ability to form biofilms. Biofilm formation is usually considered a virulence factor in pathogens especially in infections related to the implanted medical devices (IMDs) [18]. Biofilms adhere to abiotic objects such as IMDs and allow bacteria to flourish, while avoiding the host immune system and antibiotic therapy. In some instances, biofilm-encased bacteria are dislodged into the circulatory system, leading to bacteremia. In the case of Staphylococcus epidermidis, antibiotics may kill planktonic bacteria shed from the biofilm surface, but they fail to eradicate bacteria embedded within the biofilm, which can then act subsequently as a reservoir for recurrent infection. Following antibiotic treatment, a minority of drug-resistant bacteria survive that repopulate the biofilm with the survivors becoming much more antibiotic resistant [18].
A major concern raised from this study is the isolation of extremely antibiotic-resistant organisms from highly mobile wild songbirds. The majority of birds which carried the highly vancomycin-resistant bacteria from this study were the American Robins, which are migratory and cover the entire United States, Mexico, and Canada [2]. Since many American Robins appear to carry Staphylococcus succinus, it is conceivable that the bacterium is a normal flora microorganism of American Robin. If this is the case, the American Robin could act as a vector animal to spread vancomycin-resistance genes over a wide range. Birds acting as vector animals for pathogens are not unusual, as there are numerous examples of organisms such as bacteria, viruses, fungi, and parasites being spread over wide distances by birds [28]. Vancomycin-resistant Enterococcus species have been isolated from the newly hatched Northern Bobwhite chicks raised in captivity and fed commercial game bird feed that contained no antibiotics [22]. In addition to the concern that birds could spread antibiotic-resistant bacteria to humans, a clear link has been demonstrated in which humans have spread antibiotic-resistant bacteria into bird populations [6]. Other reports indicate that antibiotic-resistant bacteria can be isolated from birds in remote locations such as Easter Island [4] and the Beringia region of Alaska, which have low to non-existent human populations [12].
Since bacteria freely engage in horizontal gene transfer, it is possible that the relatively innocuous highly vancomycin-resistant isolates from this study could transfer resistance genes to other more virulent microorganisms such as MRSA. According to Resch et al. [24], CNS strains could have a high incidence of transfer of antibiotic-resistance genes to other staphylococci because of phylogenetic relationship.
Additional research is needed to verify whether freeliving wild song birds are really vector animals for vancomycin-resistance genes, or whether vancomycin-resistant Staphylococcus succinus should be considered normal flora in the American Robin. Further research is required to identify and understand the mechanisms of vancomycin resistance used by bacteria in the absence of van genes, biofilm formation, or cell wall thickening. It would also be of interest to determine whether the VanA and VanB resistance types found in our isolates could be transferred to Staphylococcus aureus, or other potential pathogens, under laboratory conditions.
Acknowledgments
This study was supported by the NIH Grant Number P20 RR016475 from the INBRE Program of the National Center for Research Resources. The authors thank Dr. Susan Flannagan for Enterococcus faecalis SFV1, Dr. Ludek Zurek for Enterococcus faecalis V583, and Dr. Mike Mulvey for Enterococcus faecium N97-0330. Field assistance was provided by many volunteers, especially Terry Mannell and Matt Sexson.
References
- 1.Alamo L, Cereda RF, Tosin I, Miranda EA, Sader HS. Antimicrobial susceptibility of coagulase negative staphylococci and characterization of isolates with reduced susceptibility to glycopeptides. Diagn Microbiol Infect Dis. 1999;34:185–191. doi: 10.1016/s0732-8893(99)00034-6. [DOI] [PubMed] [Google Scholar]
- 2.Aldrich JW, James FC. Ecogeographic variation in the American Robin (Turdus migratorius) Vol. 108. University of California Press; California: 1991. pp. 230–249. [Google Scholar]
- 3.Altizer S, Bartel R, Han BA. Animal migration and infectious disease risk. Science. 2011;331:296–302. doi: 10.1126/science.1194694. [DOI] [PubMed] [Google Scholar]
- 4.Ardiles-Villegas K, Gonzáles-Acuña D, Waldenström J, Olsen B, Hernández J. Antibiotic resistance patterns in fecal bacteria isolated from Christmas Shearwater (Puffinus nativitatis) and Masked Booby (Sula dactylatra) at remote Easter Island. Avian Dis. 2011;55:486–489. doi: 10.1637/9619-122010-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Short protocols in molecular biology. Wiley; New York, NY: 1997. [Google Scholar]
- 6.Bonnedahl J, Drobni M, Gauthier-Clerc M, Hernandez J, Granholm S, Kayser Y, Melhus Å, Kahlmeter G, Waldenström J, Johansson A, Olsen B. Dissemination of Escherichia coli with CTX-M type ESBL between humans and Yellow-legged gulls in the south of France. PLoS ONE. 2009;4:1–6. doi: 10.1371/journal.pone.0005958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brittingham MC, Temple SA, Duncan RM. A survey of the prevalence of selected bacteria in wild birds. J Wild Dis. 1988;23:299–307. doi: 10.7589/0090-3558-24.2.299. [DOI] [PubMed] [Google Scholar]
- 8.Cetinkaya Y, Falk P, Mayhall G. Vancomycin-resistant enterococci. Clin Microbiol Rev. 2000;13:686–707. doi: 10.1128/cmr.13.4.686-707.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Contreras GA, DiazGranados CA, Cortes L, Reyes J, Vanegas S, Panesso D, Rincon S, Diaz L, Prada G, Murray BE, Arias CA. Nosocomial outbreak of Enterococcus gallinarum: untaming of rare species of enterococci. J Hosp Inf. 2008;70:346–352. doi: 10.1016/j.jhin.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 10.Courvalin P. Vancomycin resistance in Gram-positive cocci. Clin Infect Dis. 2006;42:S24–S25. doi: 10.1086/491711. [DOI] [PubMed] [Google Scholar]
- 11.Depardieu F, Perichon B, Courvalin P. Detection of the van alphabet and identification of enterococci and staphylococci at the species level by multiplex PCR. J Clin Microbiol. 2004;42:5857–5860. doi: 10.1128/JCM.42.12.5857-5860.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drobni M, Bonnendahl J, Hernandez J, Haemig P, Olsen B. Vancomycin-resistant enterococci, Point Barrow, Alaska, USA. Emerging Infect Dis. 2009;15:838–839. doi: 10.3201/eid1505.081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gholizadeh Y, Courvalin P. Acquired and intrinsic glycopeptide resistance in enterococci. Int J Antimicrob Agents. 2000;16:S11–S17. doi: 10.1016/s0924-8579(00)00300-9. [DOI] [PubMed] [Google Scholar]
- 14.Gill FB. Ornithology. 3rd. W.H. Freeman and Co.; New York: 2007. [Google Scholar]
- 15.Irlinger F. Safety assessment of dairy microorganisms: coagulase-negative staphylococci. Int J Food Microbiol. 2008;126:302–310. doi: 10.1016/j.ijfoodmicro.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 16.Jennes W, Dicks LMT, Verwoerd DJ. Enterocin 012, a bacteriocin produced by Enterococcus gallinarum isolated from the intestinal tract of ostrich. J Appl Microbiol. 2000;88:349–357. doi: 10.1046/j.1365-2672.2000.00979.x. [DOI] [PubMed] [Google Scholar]
- 17.Levine DP. Vancomycin: a history. Clin Infect Dis. 2006;42:s5–s12. doi: 10.1086/491709. [DOI] [PubMed] [Google Scholar]
- 18.McCann MT, Gilmore BF, Gorman SP. Staphylococcus epidermidis device-related infections: pathogenesis and clinical management. J Pharm Pharmacol. 2008;60:1551–1571. doi: 10.1211/jpp/60.12.0001. [DOI] [PubMed] [Google Scholar]
- 19.Morot-Bizot SC, Leroy S, Talon R. Staphylococcal community of a small unit manufacturing traditional dry fermented sausages. Int J Food Microbiol. 2006;108:210–217. doi: 10.1016/j.ijfoodmicro.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Nandi S, Maurer JJ, Holfacre C, Summers AO. Gram-positive bacteria are a major reservoir of class 1 antibiotic resistance integrons in poultry litter. Proc Natl Acad Sci U S A. 2004;101:7118–7122. doi: 10.1073/pnas.0306466101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nascimento AMA, Cursino L, Goncalves-Dornelas H, Reis A, Charone-Souza E, Marini MA. Antibiotic-resistant gramnegative bacteria in birds from the Brazilian Atlantic forest. The Condor. 2003;105:358–361. [Google Scholar]
- 22.Nováková D, Sedláček I, Pantůček R, Štětina V, Švek P, Petráš P. Staphylococcus equorum and Staphylococcus succinus isolated from human clinical specimens. J Med Microbiol. 2006;55:523–528. doi: 10.1099/jmm.0.46246-0. [DOI] [PubMed] [Google Scholar]
- 23.Prukner-Radovcic E, Horvatek D, Jeren M. Bacteria and fungi of the digestive system in the wild birds. Slovenian-Croatian congress on exotic pets and wild animals. 2005:112. [Google Scholar]
- 24.Resch M, Nagel V, Hertel C. Antibiotic resistance of coagulase-negative staphylococci associated with food and used in starter cultures. Int J Food Microbiol. 2008;127:99–104. doi: 10.1016/j.ijfoodmicro.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 25.Thompson MC, Ely CA, Gress B, Otte C, Patti ST, Seibel D, Young EA. Birds of Kansas. University Press of Kansas; Lawrence: 2011. [Google Scholar]
- 26.Thurlow LR, Gillock ET. Characterization of two environmental bacterial isolates by 16S rRNA sequence analysis, fatty acid methyl ester analysis, and scanning electron microscopy. Trans KS Acad Sci. 2005;108:22–31. [Google Scholar]
- 27.Toledo-Arana A, Valle J, Solano C, Arrizubieta MJ, Cucarella C, Lamata M, Amorena B, Leiva J, Penades JR, Lasa I. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl Environ Microbiol. 2001;67:4538–4545. doi: 10.1128/AEM.67.10.4538-4545.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsiodras S, Kelesidis T, Kelesidis I, Bauchinger U, Falagas ME. Human infections associated with Wild Birds. J Infect. 2008;56:83–98. doi: 10.1016/j.jinf.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Eiff C, Peters G, Heilmann C. Pathogenesis of infections due to coagulase negative staphylococci. The Lancet Inf Dis. 2002;2:677–685. doi: 10.1016/s1473-3099(02)00438-3. [DOI] [PubMed] [Google Scholar]
- 30.Walsh TR, Howe RA. The prevalence and mechanisms of vancomycin resistance in Staphylococcus aureus. Annu Rev Microbiol. 2002;56:657–675. doi: 10.1146/annurev.micro.56.012302.160806. [DOI] [PubMed] [Google Scholar]
- 31.Zell C, Resch M, Rosenstein R, Albrecht T, Hertel C, Gotz F. Characterization of toxin production of coagulase-negative staphylococci isolated from food and starter cultures. Int J Food Microbiol. 2008;127:246–251. doi: 10.1016/j.ijfoodmicro.2008.07.016. [DOI] [PubMed] [Google Scholar]


