Abstract
We introduce the Ontology of Craniofacial Development and Malformation (OCDM) as a mechanism for representing knowledge about craniofacial development and malformation, and for using that knowledge to facilitate integrating craniofacial data obtained via multiple techniques from multiple labs and at multiple levels of granularity. The OCDM is a project of the NIDCR-sponsored FaceBase Consortium, whose goal is to promote and enable research into the genetic and epigenetic causes of specific craniofacial abnormalities through the provision of publicly accessible, integrated craniofacial data. However, the OCDM should be usable for integrating any web-accessible craniofacial data, not just those data available through FaceBase. The OCDM is based on the Foundational Model of Anatomy (FMA), our comprehensive ontology of canonical human adult anatomy, and includes modules to represent adult and developmental craniofacial anatomy in both human and mouse, mappings between homologous structures in human and mouse, and associated malformations. We describe these modules, as well as prototype uses of the OCDM for integrating craniofacial data. By using the terms from the OCDM to annotate data, and by combining queries over the ontology with those over annotated data, it becomes possible to create “intelligent” queries that can, for example, find gene expression data obtained from mouse structures that are precursors to homologous human structures involved in malformations such as cleft lip. We suggest that the OCDM can be useful not only for integrating craniofacial data, but also for expressing new knowledge gained from analyzing the integrated data
Keywords: Ontologies, data integration, craniofacial development and malformation, knowledge representation
Introduction
Craniofacial abnormalities comprise over half of all congenital malformations. Even for the most common group of craniofacial malformations, orofacial clefts, the etiology is unknown for most affected individuals, the phenotypic variability is great, and the treatment outcomes vary considerably. These gaps in knowledge impact our ability to counsel patients regarding recurrence risks, and to optimize management practices and long-term treatment outcomes. Even for well-established syndromes associated with clefting with known genetic mutations, the phenotypic variability can significantly impact clinical management.
For example, the well-recognized phenotype associated with Apert syndrome [Online Mendelian Inheritance in Man, 2010; Robin et al., 1993] is characterized by craniosynostosis, midface hypoplasia, syndactyly and cleft palate. It is almost exclusively caused by one of two point mutations in the fibroblast growth receptor 2 (FGFR2) gene [Wilkie et al., 1995]; yet phenotypic variation can significantly affect clinical outcome among patients with the same condition. For example, some affected individuals have relatively mild midface hypoplasia with minimal impact on respiratory status, while other individuals have such severe midface hypoplasia they require significant surgical intervention. Identification of the genetic and epigenetic factors that determine sub-phenotypes in this or other craniofacial conditions would lead to a better understanding of pathogenesis, allow clinicians to tailor treatment to optimize outcomes, enable further research into possible prevention and therapeutic strategies, and facilitate more effective counseling for families.
To understand the genetic and epigenetic contributions to the clinical variability of craniofacial conditions requires precise and detailed phenotypic and genotypic data from multiple sources. This may include temporally restricted gene expression data from microarray or RNAseq experiments, and 3D shape information from stereophotogrammetry or Computed Tomography, acquired by multiple labs around the world. However, until recently, there was no systematic method for integrating all these data in order to more completely and precisely characterize craniofacial phenotypes.
Recognizing this problem, the National Institute of Dental and Craniofacial Research (NIDCR) sponsored the creation of FaceBase in 2009. FaceBase is a consortium of biology and technology projects, the purpose of which is to systematically acquire and integrate multiple forms of data from human and model organisms in order to facilitate research aimed at a systems level understanding of the causes and possible treatments for craniofacial abnormalities. The FaceBase Hub coordinates consortium activities and provides data to the research community through a public web repository [Hochheiser et al., 2011].
The benefits of an ontology for data integration
In order to maximize the utility of existing and future data in repositories such as FaceBase, it is essential to link the data through standardized terminology [Olson et al., 2008]. Once data are annotated (or associated) with common terminology, they may be integrated by searching for common terms, in a manner similar to a Google search. However, such keyword-based searches still require considerable user knowledge, since only the investigators know the associations between terms. To achieve integration at the level of meaning (semantics) rather than simple term matching requires that the terms be organized in an ontology, which among other things defines specific relations among terms [Sowa, 1999].
The importance of ontologies to accomplish this type of semantically-based integration is highlighted by an active research community in the development of both ontological content and computational techniques for using ontologies, often in conjunction with genetic and genomic data, to derive novel insights. The National Center for Biomedical Ontology provides access to several hundred such ontologies through the Bioportal archive [Musen et al., 2012; Whetzel et al., 2011]. Two of the many examples of the benefits of ontologies include using the Gene Ontology [Ashburner et al., 2000] to identify classes of genes that may be over-represented in expression data sets [Shah et al., 2012] and using cross-species phenotype descriptions, potentially in combination with genomic data, to infer relationships between human diseases and animal models [Chen et al., 2012; Washington et al., 2009; Doelken et al., 2012; Bauer et al., 2012].
Why a dedicated ontology for the craniofacial community?
Many existing ontologies include terms that are relevant for craniofacial research, such as the Foundational Model of Anatomy Ontology (FMA) [Rosse and Mejino, 2003, 2007] for canonical adult human anatomy; and the Human Developmental Anatomy [Bard, 2012] for embryological development; SNOMED-CT [Spackman and Campbell, 1998], the Human Phenotype Ontology [Robinson and Mundlos, 2010] Disease Ontology [Du et al., 2009] and Elements of Morphology [Allanson et al., 2009] for phenotypic abnormalities; and the Phenotype Trait Ontology [Mungall et al., 2010] for description of phenotypic qualities.
In addition, translating between human anatomy, development, and malformation and both analogous and homologous counterparts in other species is an important goal for the research community. Ontologies for adult mouse anatomy [Hayamizu et al., 2005]; developmental mouse anatomy [Armit et al., 2012; Baldock et al., 1999], and mammalian phenotype [Smith and Eppig, 2012] parallel the human ontologies. A number of efforts have developed cross-species ontological links for both anatomic [Travillian et al., 2011b; Dahdul et al., 2012; Mungall et al., 2012; Hayamizu et al., 2012; Niknejad et al., 2012] and phenotypic [Kohler et al., 2013] descriptors.
Despite the breadth of these efforts, these ontologies and mappings currently do not provide sufficient anatomical, developmental, and phenotypic detail to meet all of the needs of the craniofacial research community for describing human development and anatomy, and for relating these descriptions to relevant counterparts in model organisms such as mice. For example, whereas craniofacial research requires a comprehensive representation of head anatomical entities and their relations, an ontology such as the mouse anatomy ontology [Hayamizu et al., 2005] represents only two types of relation (part_of and is_a), with limited depth for craniofacial structures. Similarly, whereas the mapping ontology UBERON [Mungall et al., 2012] was deliberately designed to be homology-neutral, craniofacial researchers often need to link orthologous genes (i.e. inherited from a common ancestor, and distinguished from one another through speciation events) across species. It is for these reasons that the NIDCR sponsored the creation of the OCDM.
The purpose of this article is to introduce the OCDM to the craniofacial research community, to describe its current status and future plans, and to provide examples of how the OCDM will be of use not only to FaceBase but also to geneticists, dysmorphologists, and craniofacial investigators. We begin by describing our overall approach, and then provide specific details on our driving use cases, the organizational framework and content entered to-date, and prototype examples of the potential use of the OCDM.
Overall approach
When complete, the OCDM will include normal and abnormal craniofacial structure and function, developmental anatomy and processes, syndromes, and other conditions relevant to craniofacial research and clinical practice. It will include links to corresponding model organism knowledge sources, as well as relevant genomic knowledge sources such as the gene ontology [Ashburner et al., 2000].
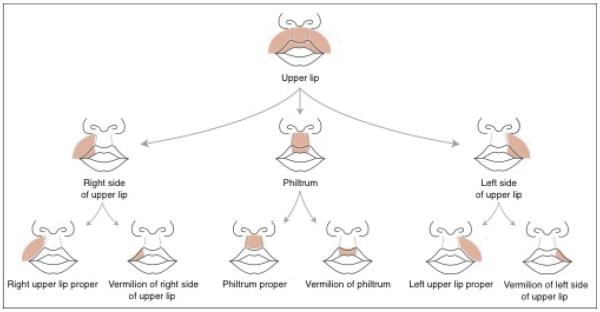
To provide a framework for this large and diverse amount of information we organize the OCDM primarily around human adult and developmental anatomy, as provided by our Foundational Model of Anatomy (FMA) [Rosse and Mejino, 2003]. The FMA is a symbolic representation of the phenotypic structure of the body ranging from macroscopic anatomy to molecules, which is by design limited to canonical structure. The FMA includes a level of detail that is not found in anatomy textbooks like Gray's Anatomy [Gray, 1918]. For example, as shown in Figure 1, in order to establish ontological relationships between the adult lip, a cleft lip, embryologic development of the lip, and gene expression in the lip region, the FMA includes definitions for many subcomponents and bounds of the lip that are unnamed in standard texts.
Figure 1.
Schematic diagram of the regional parts of the upper lip
The FMA currently comprises over 93,000 classes (e.g. “Head”, “Mouth”) represented by about 170,000 terms including preferred names, synonyms, eponyms and non-English equivalents, as well as over 2.3 million relations between classes (is_a, part_of, etc). The FMA is currently the largest and most comprehensive human anatomy ontology, and is becoming widely accepted as the canonical anatomy reference ontology, with most of the major ontologies and controlled terminologies, such as SNOMED-CT [Spackman and Campbell, 1998], RadLex [Kundu et al., 2009], and Terminologia Anatomica [Whitmore, 1999] aligning to or incorporating the FMA as the anatomy axis of their own ontologies.
Given this anatomical framework we are building the OCDM by first enhancing the FMA to provide detail on both adult and developmental craniofacial anatomy as needed by a series of use cases (described in the next section) that we have elicited from craniofacial researchers. We then extract a view (or “slim” in GO terms) [Gene Ontology Consortium, 2013] of just the craniofacial components of the FMA needed to create the canonical craniofacial human sub-ontology (CHO), and utilize this view as a template for developing other OCDM components (or sub-ontologies) dealing with human malformations, normal and abnormal mouse development and malformations, and mappings between homologous anatomical structures in human and mouse. Our current focus is on human and mouse, and a similar approach of mapping to canonical human anatomy as the fundamental organizing principle could be used with other relevant model organisms.
Content creation for the OCDM is done in the Protégé ontology authoring tool [Gennari et al., 2003], and is made available as a semantic web service [McIlraith et al., 2001], which allows the ontology to be queried by end-user applications. Whereas the OCDM content and the web services are “under-the-hood”, of interest mainly to ontology authors, it is the end-user applications that will provide the functionality and graphical interfaces that allow craniofacial researchers to utilize the OCDM in their own work.
In the following sections we provide more detail on the use cases, the current OCDM content, example queries that can currently be answered by the OCDM, and a prototype application that accesses the OCDM via the web service.
Use Cases
The development of the OCDM is informed by a series of use cases, where for our purposes a “use case” is a task or set of tasks that a craniofacial researcher would carry out in his or her work, and that would use the OCDM. Based both on our experience in building the FaceBase Hub and on input from craniofacial researchers in the consortium, these use cases describe tasks and scenarios that might use OCDM-based descriptions to support data navigation, exploration, and interpretation.
Each use case is characterized by a description, a list of potential users (e.g. clinician, researcher), a list of tasks that need to be accomplished to implement the use case, a list of information needs that should be met by the ontology, the data or both; and a list of user interface issues that need to be addressed in order to make the implemented use case accessible to craniofacial researchers. Current use cases are organized in five categories – visualization, annotation, searching/browsing, gene expression display, and analytics (Table 1).
Table 1.
Categories of use cases
| Data annotation | Associating OCDM terms with images or other data that are uploaded to the FaceBase Hub. The annotations can be associated with sets of related data files, with individual files or even subsets, such as selected regions in images. |
| Searching/Browsing | Using terms and relations from the OCDM to search for data, and to navigate between data points based on relations. |
| Visualization | Graphical and navigable displays of ontological concepts and relations. Possibilities includes graph-based browsing and reference atlas displays that depict anatomic or phenotypic characteristics directly on 2D or 3D renderings of human or animal images. |
| Gene Expression Display | Organize gene expression results based on anatomic or phenotypic annotations from the OCDM, along with other metadata such as developmental stage and species. Results might be summarized automatically in a manner similar to the Gene Wiki [Huss et al., 2010]) or contrasted across closely-related samples (e.g., adjacent anatomic regions at a common developmental time point]. |
| Analytics | Support enrichment analyses or other data mining algorithms that use ontological structure to infer relationships. Examples include finding associations between genes and specific precursor structures associated with a malformation, and cross-species phenotype analyses [Chen et al., 2012]2] [Washington et al., 2009]] [Doelken et al., 2012]] [Bauer et al., 2012]. |
As one specific example Table 2 describes the use case, Finding structures associated with syndromes, which is classified under “Searching/Browsing”. In this scenario, a user is interested in finding relevant clinical or basic research data that might be relevant to Apert syndrome. Rather than having to tediously search through the literature or online databases the user in this scenario would simply type “Apert Syndrome” into a user interface. The system would then consult the OCDM to find the reported malformations associated with Apert. The user might then select those malformations relevant to a specific case of interest. The system would then find the associated anatomic structures, the developmental precursors to those structures, and the homologous structures in the mouse or other model organisms. For each of these structures it would then consult online databases like FaceBase, and return a list of publically available data (such as gene expression data) that have been acquired about each of these structures. At the same time the system might also search Pubmed to find articles indexed by these structures, pruned by other factors such as species and developmental stage.
Table 2.
Sample use case
| Finding structures associated with syndromes |
|---|
| A clinician or researcher is interested in learning more about anatomical structures in Apert syndrome (or some other craniofacial syndrome]. Starting from the name of the syndrome of interest, the clinician identifies associated anatomic structures. For each associated structure: |
| • Find the homologous structure in mouse or other organisms |
| • Find developmental progenitors - early structures that led to one or more structures invovled in the syndrome of interest. |
| • Find associated gene expression data that's been uploaded to Facebase. |
| • Organize results by structure and developmental stage |
| The inverse - finding syndromes associated with structures - is also possibly relevant. |
| Roles |
| • Clinician |
| • Translational Researcher |
| Tasks |
| • Mapping syndrome descriptions to affected structures |
| • Examining corresponding anatomic structures in developing organisms |
| Information Needs |
| • Association of malformation syndromes with associated anatomic structures |
| • Links between human and animal anatomic structures |
| • Anatomic precursors and successors |
| • Links between structures and gene expression data |
| • Annotation of structures with developmental stages |
| Interface Implications |
| • Graphical anatomic region selection |
| • Developmental stage timeline |
| • Set-based relations |
| • Dynamic data tables |
This and other OCDM use cases can be viewed on the FaceBase Hub at https://www.facebase.org/ocdm/. Registered FaceBase users are invited to use the Hub's comment facility to discuss use cases and suggest additions and revisions.
OCDM content
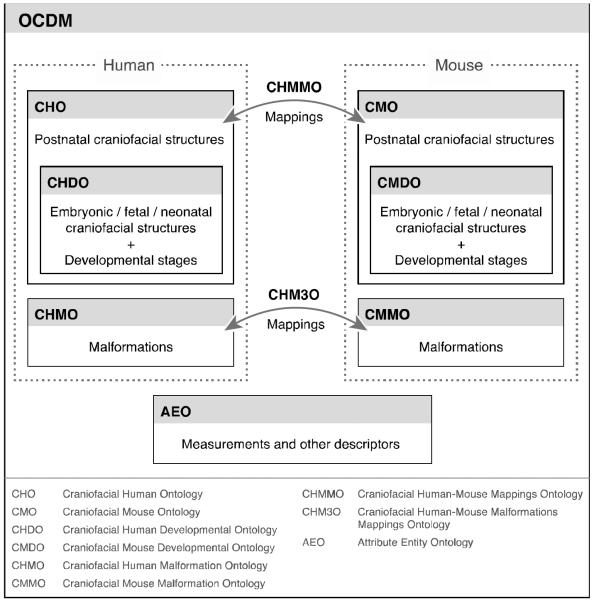
Figure 2 shows that the OCDM is organized as a series of components (also called sub-ontologies) that are included in the overall ontology. Each is developed somewhat independently, although coordination is required. The current sub-ontologies are:
Craniofacial Human Ontology (CHO): canonical human adult craniofacial anatomy, derived from the FMA.
Craniofacial Human Development Ontology (CHDO): canonical human developmental craniofacial anatomy, derived from the FMA as a subset of the CHO.
Craniofacial Mouse Ontology (CMO): canonical mouse adult craniofacial anatomy, based on the CHO, and augmented with mouse-specific terms.
Craniofacial Mouse Development Ontology (CMDO): canonical mouse developmental craniofacial anatomy, based on the CHO, and augmented with mouse-specific terms.
Craniofacial Human-Mouse Mappings Ontology (CHMMO): structure-based mappings for homologous anatomical entities.
Craniofacial Human Malformation Ontology (CHMO): phenotypic human craniofacial malformations.
Craniofacial Mouse Malformation Ontology (CMMO): phenotypic mouse craniofacial malformations.
Craniofacial Human-Mouse Malformation Mappings Ontology (CHM3O): mappings between human and mouse malformations
Attribute Entity Ontology (AEO): quantitative and qualitative physical descriptors of the attributes and properties expressed in other components of the OCDM.
Figure 2.
OCDM component ontologies
The initial focus for FaceBase was on the midface and cleft lip/palate (CL/P), so most of the current content of the OCDM to-date is related to these areas. Expansion to other malformations or syndromes should be faster in the future now that the organizational framework is in place. The CHO is nearing completion for CL/P, and the CMO, CHMMO and AEO are in active development. We thus describe these in more detail in the next sections. The mouse malformations ontology (CMMO) and mappings from human to mouse malformations (CHM3O) are currently only in rudimentary form so we defer their description to future reports. Note that the modular organization of the OCDM will permit additional components to be developed in the future (such as gene to phenotype mappings, other model organisms, or physiological manifestations keyed to the associated anatomical entities).
Craniofacial Human Ontology (CHO, CHDO)
The CHO and CHDO are the subset of the FMA that deals with the canonical representation of both adult (CHO) and developmental (CHDO) craniofacial structures. In the following sections, classes (as for example, anatomical structures) are represented in Courier New font and relations between classes are in italic.
Canonical adult anatomy (CHO)
The craniofacial content of the FMA was extended to include all the spatio-structural relationships necessary to comprehensively represent the structural components involved in the genesis of orofacial clefts. CL/P is the most common type of orofacial cleft, so the content enhancement largely focused on all anatomical entities and structural relationships pertaining to the lips, nose, oral vestibule, gingiva, alveolar ridge, palate, and orbital structures. We augmented the ontological description of canonical adult anatomy to granularity levels that closely correspond to developmental information that specifies the structures affected during dysmorphogenesis. For example, a lateral cleft upper lip is characterized by a gap between the philtrum and a part of the upper lip that is unnamed in textbooks. We identified the unnamed parts by extending the partition of the upper lip into three major regions: Philtrum in the middle, flanked by Right side of upper lip and Left side of upper lip (Figure 1). Figure 3, which is generated by our OCDM ontology viewer, illustrates how this partition is captured in the ontology. In this case, both regional parts (partition by topography, e.g. lateral region or zone of organ) and constitutional parts (partition by composition, e.g. tissues) are represented. Each regional part derives from different embryological structures. A cleft results from either lack of, or partial fusion between the precursor of the philtrum (intermaxillary process) and that of either the right or the left side of the upper lip (right or left maxillary prominence). Another partition of clinical importance is subdividing the upper lip into Vermilion of upper lip and Upper lip proper to distinguish between two constitutionally distinct parts, which can serve as landmarks for incomplete clefts. Hence, we have formally associated the developmental structures with their adult versions by enhancing the granular partition of the upper lip at different stages of transformation.
Figure 3.
Regional (R) and constitutional (C) parts of the upper lip as shown in our OCDM ontology viewer. Clicking a horizontal arrow opens up more detail.
Developmental anatomy (CHDO)
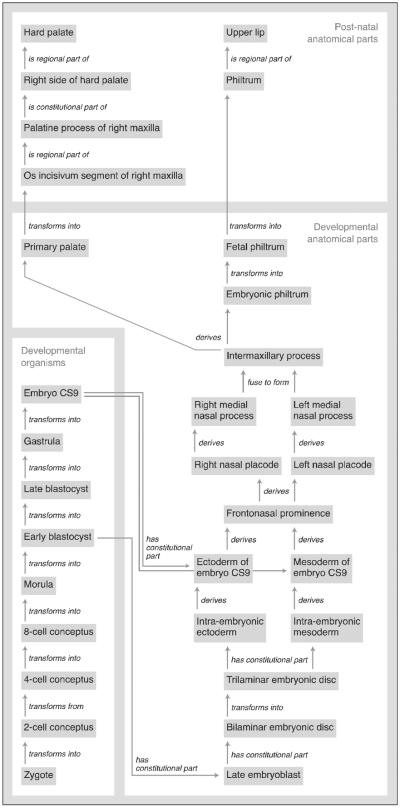
The Craniofacial Human Development Ontology is a subset of the CHO. It is an implementation of the Anatomical Transformation Abstraction component (ATA) of the FMA [Rosse and Mejino, 2007], which was created to provide the human development framework for the FMA. The ATA accounts for normal human development from the zygote to the organism's post-gestational stage. The high-level class structure of this human developmental ontology parallels the adult counterpart, where developmental anatomical entities are represented and defined in terms of their physical properties, which explicitly specify their structural types. Embryological structures are classified according to the FMA definitions of the different classes of anatomical structures (Figure 4) such as Organ (e.g. Neural tube is_a Embryonic organ, which in turn is_a Developmental organ), or Organ part (e.g. Rhombomere is_a Region of neural tube, which in turn is_a Embryonic organ part).
Figure 4.
Class inheritance (is_a) hierarchy of developmental structures
The ontology framework we have built leverages existing developmental ontologies, such as the Edinburgh Human Developmental Anatomy (EHDA) [Bard, 2012]. However, unlike the EHDA, we extended the ontological representation to other relationships between the developing structures at different stages of development. For example, we instantiated spatial-structural relationships such as parthood (e.g. Palatopterygoquadrate bar part_of Maxillary prominence), adjacency (e.g. Cytotrophoblast surrounds Extra–embryonic mesoblast), connectivity (e.g. Bilaminar embryonic disc continuous_with Amnion). In addition, we introduced temporal properties (Carnegie stage, gestational age, days post-ovulation) and developmental properties (transforms, derives, fuses, merges), as for example, Frontonasal prominence derives Right nasal placode and Left nasal placode. Figure 5 shows this relation as well as many others that allow us to represent in the OCDM the developmental lineage of structures involved in CL/P. This level of detail, which is not present in other developmental ontologies, will allow us to identify properties (e.g. gene expression) that are involved in each stage of development, as well as determine what structures at what stage are affected when malformation occurs. The CHO and CHDO together include 11,451 classes, 23389 terms and 196 relations (properties), many of which were already present in the FMA.
Figure 5.
Schematic diagram of the development of the hard palate and upper lip as represented in the OCDM. Grey boxes are classes, labeled lines are relations.
Craniofacial Mouse Ontology (CMO, CMDO)
The Craniofacial Mouse Ontology (CMO) and Craniofacial Mouse Development Ontology (CMDO) are representations of canonical mouse craniofacial anatomy, both adult and developmental. We used the Craniofacial Human Ontology (CHO) as a template, pruning and editing as needed to represent anatomically homologous mouse structures, and then integrated MA and EMAP structures. We used inference of the existence of missing structures from the vertebrate Bauplan and FMA, and verified these structures, as well as finding omitted ones, from domain experts and the translational research literature. As a result the taxonomy, property, and annotation property structures in the CMO are almost the same as in the CHO. Additionally, the derivation of mouse structures from existing FMA/CHO structures necessitated some additional annotation property slots to provide a sound representation. To date there are 11063 classes, 23383 terms and 198 relations in the CMO/CMDO. Of these, 482 classes were acquired from MA or EMAP.
Craniofacial Human-Mouse Mappings Ontology (CHMMO)
The Craniofacial Human-Mouse Mappings Ontology (CHMMO) contains structure-based mappings for homologous anatomical entities. CHMMO mappings are explicit hypotheses of anatomical homology between two species-specific structures. This was an explicit design decision, made for the purpose of enabling linkages to be drawn among anatomic and phenotypic structures derived from orthologous genes.
The CHMMO was built from the ground up, although it operates on the same mapping principles as the Comparative Anatomy Information System (CAIS) [Travillian et al., 2011b] and the Vertebrate Bridging Ontology [Travillian et al., 2011a]. Initially, structures with similar or the same names between species are provisionally assumed to be homologous. Homologies between structures with dissimilar names across species are added as needed.
These provisional mappings are validated by domain experts. To-date we have used a single expert, Dr. Cox (one of the co-authors); in the future we will establish a mechanism for others to comment on the mappings as well as other aspects of the OCDM. To validate the provisional mappings the domain expert and content developers consult the appropriate research literature, as, for example, Incus (Homo sapiens) <--> Incus (Mus musculus), Malleus (Homo sapiens) <--> Malleus (Mus musculus), Stapes (Homo sapiens) <--> Stapes (Mus musculus) [Kanzaki et al., 2011]. In general validation is based on explicit molecular evidence when available [Brugmann et al., 2006; Laue et al., 2011]; in the absence of direct molecular evidence, mappings are inferred on the basis of the vertebrate Bauplan [Depew and Compagnucci, 2008; Fish et al., 2011].
The result of these validations is a confidence level assigned to the mapping, together with the provenance of the mapping, the expert's confidence in the mapping, and the number of returned results in the literature.
The resulting mappings and confidence levels are represented as instances of classes in the CHMMO, where each represents a type of mapping. For example, Alveolar bone of maxilla maps 1-1 with Alveolar bone of maxilla (Mus musculus) [355 such mappings]. Vibrissa (Mus musculus) is a null mapping, because there is no such corresponding structure in humans [38 such mappings], and Snout (Mus musculus) maps to a corresponding region of the face that is yet to be named (nasomaxillary region) in humans [2 such mappings]. An advantage of these specific types of mappings is that even though there are null mappings such as Vibrissa, the OCDM will be able to retrieve the structures from which it is derived, which will have a 1-1 mapping, thus highlighting points of developmental divergence.
Attribute Entity Ontology (AEO)
The AEO provides quantitative and qualitative physical descriptors of the attributes expressed by classes in the OCDM. These are the physical object properties of anatomical structures and pathological entities, such as size, shape, distance and mass. We developed the AEO based on the Phenotypic Quality Ontology (PATO), which is designed to represent “phenotypic qualities” of “quality-bearing entities” [Mungall et al., 2010]. An example of such an entity is the “vermilion of upper lip” which could be the bearer of the quality “thin”. The combination therefore describes the “thin vermilion” phenotype. This approach allows us to extend the phenotypic descriptions to a level of detail that is necessary to sort out sub-phenotypes and to accommodate quantitative measurements, without requiring an apriori distinction between normal and abnormal. Thus, the AEO provides an ideal framework within which we incorporated the standards used to describe human morphology that were created and published by the Elements of Morphology Working Group (http://elementsofmorphology.nih.gov/). We are also populating this ontology with other attributes for the anatomical entities related to CL/P. There are currently 106 classes, 119 terms and 7 relations in AEO, of which 43 high level classes were derived from PATO.
Craniofacial Human Malformation Ontology (CHMO)
Many cleft classification systems exist [Marazita and Mooney, 2004]. In developing the CHMO for CL/P, we first identified and compared over a dozen published classification systems for CL/P phenotypes. Based on this comparison, a full description of which is in preparation, we concluded that there is as yet no universal classification system that captures all variant presentations of CL/P. Given the existence of so many approaches, each of which has merit in its own right, our goal is not to invent yet another classification scheme but rather to create a unifying framework that will accommodate and reconcile existing classification schemes. To create such a framework requires that we explicitly represent the assumptions underlying each classification scheme in terms of the basic anatomy, pathology and developmental processes.
To create this framework we first consulted several existing ontologies or controlled vocabularies relating to malformations, namely: SNOMED-CT [Spackman and Campbell, 1998] Human Disease Ontology (HDO) [Du et al., 2009], Human Phenotype Ontology (HPO) [Robinson and Mundlos, 2010], ICD-10CM [NCHS, 2013], and Elements of Morphology [Allanson et al., 2009]. In general, most sources do not include representation of the underlying basic science in the phenotypic descriptions. For example, HPO classifies “cleft upper lip” as an abnormality of the upper lip but does not clearly specify whether the term pertains to the pathological structure itself or its phenotypic abnormality. It is important to distinguish between the two because the kinds of basic information that are associated with each are significantly different from one another. That is, pathological structures may be described by morphometric measurements (e.g. upper lip height, philtrum width) whereas phenotypic abnormalities may be described by processual properties (e.g. transforms, derives, fusion, or partial fusion).
The framework we have created therefore consists of building separate sub-ontologies for pathological structures and phenotypic abnormalities, using canonical anatomy as the basis for classifying both types of entities, and ontology best practices to ensure consistency with high level classification systems [Rosse et al., 2005; Grenon et al., 2004].
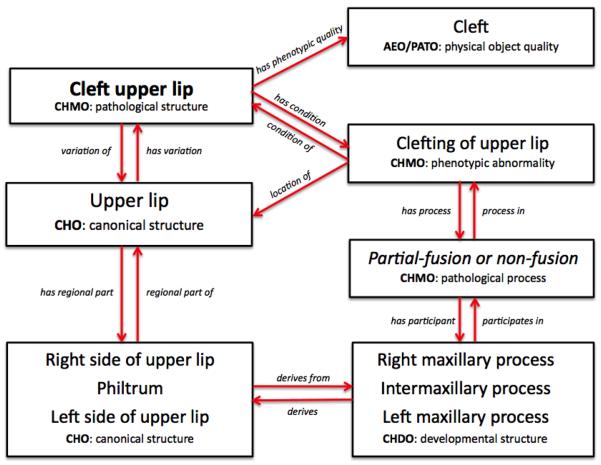
Figure 6 illustrates this dual representation for cleft upper lip. Because we regard cleft upper lip as a pathological structure (as opposed to a phenotypic abnormality) we say that Cleft upper lip is a Pathological structure. We say that Cleft upper lip has_condition Clefting of upper lip, which is a Phenotypic abnormality.
Figure 6.
Classes and relations involved in the genesis of cleft upper lip, as represented in the OCDM.
We then associate these entities with their corresponding canonical structures in the CHO. Thus, Cleft upper lip is a variation_of Upper lip, and Clefting of upper lip is located_in the Upper lip, wherein a gap exists between certain regions of the upper lip. We also associate morphometric measurements and descriptors with canonical structures. These measurements, which come from the AEO, include such properties as “cleft” and length (“philtrum length”), that are the basis for classifying an Upper lip as a Pathological structure. Similarly, we plan to associate phenotypic abnormalities such as Clefting of upper lip, with pathological processes such as Partial-fusion or Non-fusion of precursor developmental structures.
By associating these types of entities with each other and with canonical developmental structures via specific relations whose basic biological meaning is well-defined, we can represent qualitative knowledge of developmental biology in enough detail that we can trace the morphogenesis of clinical malformations, and hence search the appropriate databases for experimental evidence of genetic or epigenetic factors that could be associated with their pathogenesis. Although these detailed relations may seem unnecessary to a clinician or researcher, they are needed in order to make the knowledge “computable” so that it can not only be used for reconciling different classification schemes, but also for “intelligent” searches that can traverse these links to find data relevant to malf rmations. We illustrate such a search in the next section. Currently there are 280 classes and 21 relations in the CHMO, with 16 classes derived from HPO.
Prototype uses
To be of greatest utility the ontology needs to be “under the hood”, embedded in applications that allow end users to perform the kind of tasks required by the use cases described earlier. The ontology could be distributed with various applications if it is small enough, or could be made available as a web service that can accept queries from both human users and software applications. Advantages of this approach are that the ontology remains up-to-date, and the answers to queries may more easily be combined with other web-accessible ontology and data sources.
We are taking the latter approach by periodically making the OCDM available as a queryable semantic web [Berners-Lee et al., 2001] service. Queries over this service are created and saved in our Query Integrator application (QI) [Brinkley and Detwiler, 2012]. The QI allows different queries to be integrated, as for example, a query over the ontology with a query over data, as we describe in the next section. In addition, saved queries can be accessed by end-user applications that hide the details of the underlying ontology and query engine.
In the next two sections we describe examples of saved queries over the OCDM, as well as an example application that accesses saved queries, but presents the results in a graphical form that is more attuned to end users.
Queries
There are currently about 30 saved queries over the OCDM in the Query Integrator database. Links to executable versions of several of these queries can be found on the OCDM page available at the FaceBase Hub https://www.facebase.org/content/ocdm. For example, a query on the “human nose” finds the parts of the human nose, and then for each of these parts, finds the mapping (if any) to the corresponding homologous structures in the mouse. Similarly, a query on the “human right nasal bone” retrieves the facial landmarks associated with that bone. These and similar landmarks will be useful for retrieving specific measured distances from morphometric data, as for example, the normative data for facial measures that are currently available through the FaceBase website.
The above queries, however, are only over the ontology. A third query integrates two queries: one over the ontology and one over a data source, in order to perform an “intelligent” query. In this case the data source is the FaceBase Hub, which currently houses over 200 datasets contributed by FaceBase consortium members.
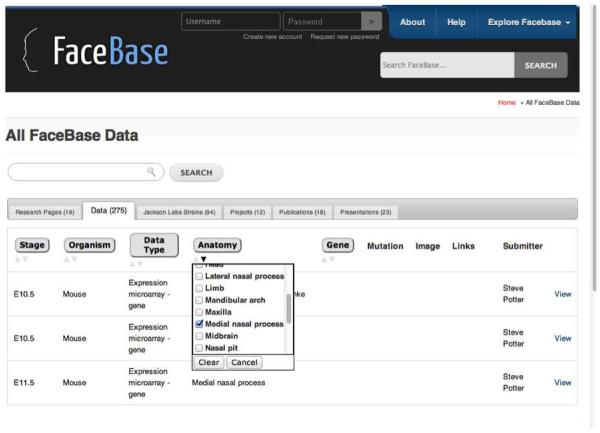
When datasets are uploaded to the Hub they are annotated with terms from a set of controlled vocabularies that are accessible via pull-down menus. These terms can then be u ed to search for specific datasets by clicking checkboxes in the search interface shown in Figure 7. One of the controlled vocabularies is Anatomy, which has been clicked to open up the list of Anatomy terms from the OCDM. The user has then checked Medial nasal process, which causes the search engine to retrieve all those datasets that have been annotated with Medial nasal process, in this case gene expression data from the developing mouse. A user looking for data relevant to cleft lip might know that the left and right medial nasal processes are developmental precursors to the philtrum (Figure 5), which is in turn part of the upper lip and not fused in some kinds of cleft lip. But if the user didn't have this knowledge, they wouldn't know to click on medial nasal process in the Hub search. However, this knowledge is present in the ontology, so we have used the Query Integrator to integrate a query over the ontology with one over the Hub annotated data, allowing the user to start just with cleft lip and return all those datasets annotated with a developmental precursor of cleft lip.
Figure 7.
FaceBase search interface. Clicking on one of the categories in the top row (e.g. Anatomy) brings up a set of terms from the OCDM. The user has checked one of these terms (Medial nasal process), which retrieves the three mouse datasets annotated with this term.
The ontology query first examines relations like those shown in Figure 6 to determine that Cleft lip has subclass Cleft upper lip, which is a variation_of Upper lip. The query then examines relations like those shown in Figure 5 to find the developmental lineage of Upper lip, returning all structures in this lineage, including the right and left medial nasal process. This list of precursor structures is then used in a second query that searches the Hub database for those datasets annotated with an Anatomy term that is one of these precursor structures. The result, which is only available in raw form (and hence not shown here) since it is not yet integrated with the Hub search interfaces, is the set of data annotated with any precursor structures to a structure involved in cleft lip, without requiring the user to know what those structures are.
Although the knowledge embodied in this example is straightforward and likely already known by most craniofacial researchers, this query nevertheless demonstrates the potential of combining ontological knowledge with queries over annotated data. As the knowledge in the OCDM becomes more detailed, and as relevant data become more precisely annotated, this potential should become increasingly useful for finding data relevant to specific phenotypes, not only from the Hub, but also from other web accessible craniofacial data sources.
OCDM Viewer
The queries presented in the last section are not ideal for end users since the queries and their results are only available in raw form. However, these queries can be accessed by applications that present a clickable user interface, thereby hiding the details of the query from the user. As one example of such an application, a simple web-based viewer (available at http://purl.org/sig/ocdm/viewer) displays content from the OCDM. The viewer has been developed for two purposes: (1) it exposes the structure and content of the OCDM to members of the craniofacial research community as the OCDM is being developed, because feedback will be necessary to ensure that the OCDM accurately and completely represents concepts in craniofacial research and in clinical settings; and (2) it serves as a demonstration of how a user interface can retrieve content of the OCDM using saved queries in the Query Integrator. In fact Figure 3 and Figure 4 are screenshots of this viewer in action. Each time a user clicks a structure a new query is sent to the QI, with that structure as the argument. The viewer currently displays class hierarchies and part hierarchies for several components of the OCDM, as well as mappings of the CHMMO.
Although the current OCDM viewer is designed for ontology authors rather than for end-user tasks like annotation or search, it nevertheless demonstrates the potential to build usable applications over the underlying ontology. In our current work we are examining additional user interface requirements in order to add such features as selection from a developmental timeline, graphs depicting developmental relations, and many others. Our next steps will be to design the user interfaces for these features, enter any new required knowledge in the OCDM, and write additional queries over the OCDM and/or data available on the web.
Discussion
In this report we have introduced the Ontology of Craniofacial Development and Malformation, which includes both human and mouse adult and developmental anatomy, as well as mappings between them. The use of model organisms to enhance understanding of genetic determinants of human developmental processes is the essence of translational medicine. Although genetic experiments in developing mice provide detailed insight into the regulatory processes that guide craniofacial development, these models may not always present with the anticipated phenotype or it may be difficult to appreciate or predict how the homologous structures will be affected because of subtle differences in development between species. Understanding these differences not only aids in recognition of more appropriate animal models but also helps to better decipher the translational impact of genetic and epigenetic factors on facial development and susceptibility to malformation. Detailed descriptions of correspondences between anatomic regions throughout development are needed to localize genetic activity with enough precision to create meaningful correspondences between species. The need for ontological precision similarly extends to descriptions of malformations, as terms that are imprecise regarding the location, nature, and timing of a malformation might lead to incorrect inferences regarding the fitness of various animal models for specific human diseases. The goal of the OCDM is therefore to provide fine-grained ontological models of craniofacial anatomy, development, and malformation, in support of high-precision translational craniofacial research.
The modular design of the OCDM follows the tradition of various “slim” ontology approaches [Gene Ontology Consortium, 2013; Courtot et al., 2011], providing applications or other ontologies with the option of using some or all of the OCDM components as necessary. This approach will support the evolution of the OCDM, including planned components for modeling mouse malformations (CMMO – the Craniofacial Mouse Malformation Ontology) and mappings between human and mouse malformations (CHM3O Craniofacial Human-Mouse Malformation Mappings Ontology). It will also support interoperability of the OCDM with related ontologies like EMAP [Baldock et al., 1999], especially as more ontologies move toward the common Web Ontology Language (OWL) [World Wide Web Consortium, 2010], which is designed to allow different ontologies to interoperate in a worldwide web of knowledge (the semantic web). In addition, the availability of the OCDM via a web service makes it readily accessible to any application, such as the Query Integrator and the ontology browser described earlier, or to other groups who would like to use the OCDM to annotate their own data.
In our future work we will take advantage of these capabilities to implement applications for craniofacial researchers, who will not need to understand the underlying ontology. Example applications include more intuitive ontology viewers, tools for annotating data, and “intelligent” search interfaces for finding data that are currently only related by knowledge in the user's head. As one example, the use case illustrated in Table 2, finding relevant mouse gene expression data for Apert or other syndromes, could be implemented as a generalized search interface that allows a user to select any syndrome, not just Apert, and have the system return relevant mouse gene expression data for that syndrome. Or the user could select a set of syndromes and have the system return expression data that are relevant for all members of the set.
Such intelligent search capabilities, initially over the FaceBase Hub, but later over other web accessible craniofacial data sources, should make it much easier than it is now for both clinicians and researchers to find candidate genes and other factors that might be involved in a particular malformation. This type of retrieved information could in turn suggest further experiments, the results of which could then be made available through the FaceBase Hub or through other web accessible repositories, for later intelligent searches. If the results of these experiments demonstrate new relations between genes and phenotypes, and if these results are verified by others, then this new information could be entered into the OCDM, likely with probabilities to indicate the degree of confidence in the relations as is currently done with gene annotation. The OCDM and related ontologies could therefore gradually become repositories for the knowledge gained by endeavors such as FaceBase.
With our growing recognition of the complexities involved in normal and abnormal development and malformations of the human organism it is becoming increasingly clear to many that no single human mind is capable of grasping all this information at once. Thus, “computable knowledge”, of the type represented in the OCDM and other ontologies, will become increasingly essential, not only for integrating data, but also for representing the scientific theories that result from analyzing these integrated data [Karp, 2001].
Acknowledgements
This work was funded by NIH FaceBase Consortium grant U01DE020050.
References
- Allanson JE, Biesecker LG, Carey JC, Hennekam RCM. Elements of morphology: introduction. Am. J. Med. Genet. A. 2009;149A:2–5. doi: 10.1002/ajmg.a.32601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armit C, Venkataraman S, Richardson L, Stevenson P, Moss J, Graham L, Ross A, Yang Y, Burton N, Rao J, Hill B, Rannie D, Wicks M, Davidson D, Baldock R. eMouseAtlas, EMAGE, and the spatial dimension of the transcriptome. Mamm. Genome. 2012;23:514–524. doi: 10.1007/s00335-012-9407-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldock RA, Dubreuil C, Hill B, Davidson D. Bioinformatics Databases and Systems. Kluwer Academic Press; 1999. The Edinburgh Mouse Atlas: Basic Structure and Informatics; pp. 102–115. [Google Scholar]
- Bard J. A new ontology (structured hierarchy) of human developmental anatomy for the first 7 weeks (Carnegie stages 1–20) J. Anatomy. 2012;221:406–416. doi: 10.1111/j.1469-7580.2012.01566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer S, Kohler S, Schulz MH, Robinson PN. Bayesian ontology querying for accurate and noise-tolerant semantic searches. Bioinformatics. 2012;28:2502–2508. doi: 10.1093/bioinformatics/bts471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berners-Lee T, Hendler J, Lassila O. The semantic web. Scientific American. 2001;284:34–43. [Google Scholar]
- Brinkley JF, Detwiler LT. A query integrator and manager for the Query Web. Journal of Biomedical Informatics. 2012;45:975–991. doi: 10.1016/j.jbi.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugmann SA, Tapadia MD, Helms JA. The molecular origins of species-specific facial pattern. Curr. Top. Dev. Biol. 2006;73:1–42. doi: 10.1016/S0070-2153(05)73001-5. [DOI] [PubMed] [Google Scholar]
- Chen C, Mungall CJ, Gkoutos GV, Doelken SC, Köhler S, Ruef BJ, Smith C, Westerfield M, Robinson PN, Lewis SE, Schofield PN, Smedley D. MouseFinder: Candidate disease genes from mouse phenotype data. Human Mutation. 2012;33:858–866. doi: 10.1002/humu.22051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtot M, Gibson F, Lister A, Malone J, Schober D, Brinkman R, Ruttenberg A. MIREOT: The minimum information to reference an external ontology term. Applied Ontology. 2011;6:23–33. [Google Scholar]
- Dahdul WM, Balhoff JP, Blackburn DC, Diehl AD, Haendel MA, Hall BK, Lapp H, Lundberg JG, Mungall CJ, Ringwald M, Segerdell E, Van Slyke CE, Vickaryous MK, Westerfield M, Mabee PM. A unified anatomy ontology of the vertebrate skeletal system. PLoS ONE. 2012;7:e51070. doi: 10.1371/journal.pone.0051070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew MJ, Compagnucci C. Tweaking the hinge and caps: testing a model of the organization of jaws. J. Exp. Zool. B Mol. Dev. Evol. 2008;310:315–335. doi: 10.1002/jez.b.21205. [DOI] [PubMed] [Google Scholar]
- Doelken SC, Kohler S, Mungall CJ, Gkoutos GV, Ruef BJ, Smith C, Smedley D, Bauer S, Klopocki E, Schofield PN, Westerfield M, Robinson PN, Lewis SE. Phenotypic overlap in the contribution of individual genes to CNV pathogenicity revealed by cross-species computational analysis of single-gene mutations in humans, mice and zebrafish. Disease Models & Mechanisms. 2012;6:358–372. doi: 10.1242/dmm.010322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Feng G, Flatow J, Song J, Holko M, Kibbe WA, Lin SM. From disease ontology to disease-ontology lite: statistical methods to adapt a general-purpose ontology for the test of gene-ontology associations. Bioinformatics. 2009;25:i63–68. doi: 10.1093/bioinformatics/btp193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish JL, Villmoare B, Köbernick K, Compagnucci C, Britanova O, Tarabykin V, Depew MJ. Satb2, modularity, and the evolvability of the vertebrate jaw. Evol. Dev. 2011;13:549–564. doi: 10.1111/j.1525-142X.2011.00511.x. [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium . GO Slim and Subset Guide. 2013. [Google Scholar]
- Gennari JH, Musen MA, Fergerson RW, Grosso WE, Crubezy M, Eriksson H, Noy NF, Tu S. The evolution of Protege: an environment for knowledge-based systems development. Int. J. Human-Computer Studies. 2003;58:89–123. [Google Scholar]
- Gray H. Anatomy of the Human Body. Lea & Febiger; Philadelphia: 1918. [Google Scholar]
- Grenon P, Smith B, Goldberg L. Biodynamic ontology: applying BFO in the biomedical domain. Stud Health Technol Inform. 2004;102:20–38. [PubMed] [Google Scholar]
- Hayamizu TF, de Coronado S, Fragoso G, Sioutos N, Kadin JA, Ringwald M. The mouse-human anatomy ontology mapping project. Database (Oxford) 2012;2012:bar066. doi: 10.1093/database/bar066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayamizu TF, Mangan M, Corradi JP, Kadin JA, Ringwald M. The Adult Mouse Anatomical Dictionary: a tool for annotating and integrating data. Genome Biol. 2005;6:R29. doi: 10.1186/gb-2005-6-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheiser H, Aronow BJ, Artinger K, Beaty TH, Brinkley JF, Chai Y, Clouthier D, Cunningham ML, Dixon M, Donahue LR, Fraser SE, Iwata J, Marazita ML, Murray JC, Murray S, Postlethwait J, Potter S, Shapiro LG, Spritz R, Visel A, Weinberg SM, Trainor PA. The FaceBase Consortium: A comprehensive program to facilitate craniofacial research. Developmental Biology. 2011;355:175–82. doi: 10.1016/j.ydbio.2011.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huss JW, 3rd, Lindenbaum P, Martone M, Roberts D, Pizarro A, Valafar F, Hogenesch JB, Su AI. The Gene Wiki: community intelligence applied to human gene annotation. Nucleic Acids Res. 2010;38:D633–639. doi: 10.1093/nar/gkp760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzaki S, Takada Y, Niida S, Takeda Y, Udagawa N, Ogawa K, Nango N, Momose A, Matsuo K. Impaired vibration of auditory ossicles in osteopetrotic mice. Am. J. Pathol. 2011;178:1270–1278. doi: 10.1016/j.ajpath.2010.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp P. Pathway databases: a case study in computational symbolic theories. Science. 2001;293 doi: 10.1126/science.1064621. [DOI] [PubMed] [Google Scholar]
- Kohler S, Doelken SC, Ruef BJ, Bauer S, Washington N, Westerfield M, Gkoutos G, Schofield P, Smedley D, Lewis SE, Robinson PN, Mungall CJ. Construction and accessibility of a cross-species phenotype ontology along with gene annotations for biomedical research. F1000Research 2. 2013 doi: 10.12688/f1000research.2-30.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu S, Itkin M, Gervais DA, Krishnamurthy VN, Wallace MJ, Cardella JF, Rubin DL, Langlotz CP. The IR Radlex Project: an interventional radiology lexicon--a collaborative project of the Radiological Society of North America and the Society of Interventional Radiology. J Vasc Interv Radiol. 2009;20:S275–277. doi: 10.1016/j.jvir.2009.04.031. [DOI] [PubMed] [Google Scholar]
- Laue K, Pogoda H-M, Daniel PB, van Haeringen A, Alanay Y, von Ameln S, Rachwalski M, Morgan T, Gray MJ, Breuning MH, Sawyer GM, Sutherland-Smith AJ, Nikkels PG, Kubisch C, Bloch W, Wollnik B, Hammerschmidt M, Robertson SP. Craniosynostosis and multiple skeletal anomalies in humans and zebrafish result from a defect in the localized degradation of retinoic acid. Am. J. Hum. Genet. 2011;89:595–606. doi: 10.1016/j.ajhg.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazita ML, Mooney MP. Current concepts in the embryology and genetics of cleft lip and cleft palate. Clin Plast Surg. 2004;31:125–140. doi: 10.1016/S0094-1298(03)00138-X. [DOI] [PubMed] [Google Scholar]
- McIlraith SA, Son TC, Zeng Honglei. Semantic Web services. IEEE Intelligent Systems. 2001;16:46–53. [Google Scholar]
- Mungall CJ, Gkoutos GV, Smith CL, Haendel MA, Lewis SE, Ashburner M. Integrating phenotype ontologies across multiple species. Genome Biol. 2010;11:R2. doi: 10.1186/gb-2010-11-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungall CJ, Torniai C, Gkoutos GV, Lewis SE, Haendel MA. Uberon, an integrative multi-species anatomy ontology. Genome Biol. 2012;13:R5. doi: 10.1186/gb-2012-13-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musen MA, Noy NF, Shah NH, Whetzel PL, Chute CG, Story M-A, Smith B. The National Center for Biomedical Ontology. J Am Med Inform Assoc. 2012;19:190–195. doi: 10.1136/amiajnl-2011-000523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCHS . Clinical Modification. Tenth Revision 2013. ICD - ICD-10-CM - International Classification of Diseases. [Google Scholar]
- Niknejad A, Comte A, Parmentier G, Roux J, Bastian FB, Robinson-Rechavi M. vHOG, a multispecies vertebrate ontology of homologous organs groups. Bioinformatics. 2012;28:1017–1020. doi: 10.1093/bioinformatics/bts048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson J, Hofer E, Box N, Zimmerman A, Olson G, Cooney D. Scientific Collaboration on the Internet. MIT Press; Cambridge, MA: 2008. A theory of remote scientific collaboration. [Google Scholar]
- Online Mendelian Inheritance in Man . Apert Syndrome. 2010. MIM ID #101200. [Google Scholar]
- Robin NH, Falk MJ, Haldeman-Englert CR. FGFR-Related Craniosynostosis Syndromes. In: Pagon RA, Bird TD, Dolan CR, Stephens K, Adam MP, editors. GeneReviews™. University of Washington, Seattle; Seattle (WA): 1993. [Google Scholar]
- Robinson PN, Mundlos S. The human phenotype ontology. Clin. Genet. 2010;77:525–534. doi: 10.1111/j.1399-0004.2010.01436.x. [DOI] [PubMed] [Google Scholar]
- Rosse C, Kumar A, Mejino JLV, Cook DL, Detwiler LT, Smith B. A strategy for improving and integrating biomedical ontologies. Proceedings, AMIA Fall Symposium; Washington, D.C. 2005. pp. 639–643. [PMC free article] [PubMed] [Google Scholar]
- Rosse C, Mejino JLV. A reference ontology for bioinformatics: the Foundational Model of Anatomy. Journal of Biomedical Informatics. 2003;36:478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Rosse C, Mejino JLV. The Foundational Model of Anatomy Ontology. In: Burger A, Davidson D, Baldock R, editors. Anatomy Ontologies for Bioinformatics: Principles and Practice. Springer; 2007. pp. 59–117. [Google Scholar]
- Shah NH, Cole T, Musen MA. Chapter 9: Analyses using disease ontologies. PLoS Comput Biol. 2012;8:e1002827. doi: 10.1371/journal.pcbi.1002827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Eppig JT. The Mammalian Phenotype Ontology as a unifying standard for experimental and high-throughput phenotyping data. Mamm. Genome. 2012;23:653–668. doi: 10.1007/s00335-012-9421-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa JF. Knowledge Representation: Logical, Philosophical, and Computational Foundations. Brooks Cole; Pacific Grove, Ca: 1999. [Google Scholar]
- Spackman KA, Campbell KE. Compositional concept representation using SNOMED: towards further convergence of clinical terminologies. Proc AMIA Symp. 1998:740–744. [PMC free article] [PubMed] [Google Scholar]
- Travillian R, Malone J, Pang C, Hancock J, Holand P, Schofield P, Parkinson H. The Vertebrate Bridging Ontology (VBO) 2011a. [Google Scholar]
- Travillian RS, Diatchka K, Judge TK, Wilamowska K, Shapiro LG. An ontology-based comparative anatomy information system. Artif Intell Med. 2011b;51:1–15. doi: 10.1016/j.artmed.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington NL, Haendel MA, Mungall CJ, Ashburner M, Westerfield M, Lewis SE. Linking human diseases to animal models using ontology-based phenotype annotation. PLoS Biol. 2009;7:e1000247. doi: 10.1371/journal.pbio.1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetzel PL, Noy NF, Shah NH, Alexander PR, Nyulas C, Tudorache T, Musen MA. BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acids Res. 2011;39:W541–545. doi: 10.1093/nar/gkr469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore I. Terminologia anatomica: new terminology for the new anatomist. Anat. Rec. 1999;257:50–53. doi: 10.1002/(SICI)1097-0185(19990415)257:2<50::AID-AR4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Wilkie AO, Slaney SF, Oldridge M, Poole MD, Ashworth GJ, Hockley AD, Hayward RD, David DJ, Pulleyn LJ, Rutland P, et al. Apert syndrome results from localized mutations of FGFR2 and is allelic with Crouzon syndrome. Nat Genet. 1995;9:165–72. doi: 10.1038/ng0295-165. [DOI] [PubMed] [Google Scholar]
- World Wide Web Consortium . The OWL Web Ontology Language. 2010. [Google Scholar]