Abstract
Although the number of new cases of Basal Cell Carcinoma (BCC) has increased rapidly in the last few decades, the molecular basis of its pathogenesis is not completely understood. Activation of Hedgehog (Hh) signaling pathway has been shown to be a key factor driving the development of BCC. The Wnt/β-catenin signaling pathway was also shown to be activated in BCCs and to perhaps modulate the activity of Hh pathway. We have previously identified a novel mechanism by which Wnt signaling regulates the transcriptional outcome of Hh signaling pathway. We demonstrated that CRD-BP, a direct target of the Wnt/β-catenin signaling, binds to GLI1 mRNA, stabilizes it, and consequently upregulates its levels (mRNA and protein) and activities. We hypothesized that Wnt-induced and CRD-BP-dependent regulation of GLI1 expression and activities is important to the development of BCC. In this study, we show that CRD-BP is over-expressed in BCC and that its expression positively correlates with the activation of both Wnt and Hh signaling pathways. We also describe the generation and characterization of a human BCC cell line. This cell line was utilized to demonstrate the importance of CRD-BP-dependent regulation of GLI1 expression and activities in the development of BCC.
Introduction
Basal cell carcinoma (BCC) is the most common form of skin cancer, affecting approximately one million Americans each year. This cancer arises in the basal cells lining the deepest layer of the epidermis. The vast majority of BCCs occur sporadically, but immuno-compromised patients and patients with the rare inherited disorder, basal cell nevus syndrome (BCNS), are more susceptible to develop BCC (Epstein, 2008). BCC is more common in people over age 40. The majority of BCC occurs on those areas of the skin that are regularly exposed to sunlight or other ultraviolet radiation. This includes the face, ears, neck, scalp, shoulders, and back, although virtually every part of the skin has been reported to be involved (Rippey, 1998). Anyone with a history of sun exposure can develop BCC, however, people with fair skin are at highest risk. Although the death rate from BCC is low, as BCC rarely metastasizes (Rippey, 1998), this malignancy is locally aggressive and if untreated can destroy tissues, penetrating deeply to bone and causing ulceration, loss of function, and disfiguration. BCC therefore causes considerable morbidity and places a huge burden on healthcare service worldwide. The cost of care for BCC is the fifth highest for all cancers in the Medicare population in the United States (Epstein, 2008). Furthermore, people who have BCC are at high risk of developing further BCC lesions and other malignancies (Wong et al., 2003). Although the number of new cases of BCC has increased rapidly each year in the last few decades, the molecular basis of its pathogenesis is not well understood. Aberrant regulation of the Hh pathway contributes to the development of BCC. Activating mutations of Smo or suppressing mutations of Ptch have been shown to constitutively activate the Hh signaling pathway (Wicking et al., 1999), and these mutations have been observed in BCC. Patients with the autosomal dominant nevoid basal cell carcinoma syndrome carry disabling germline mutations in one allele of the Ptch1 gene, and BCC from these patients lack the remaining, normal Ptch1 gene (Oro et al., 1997). Activation of Hh signaling pathway is also a hallmark of sporadic BCC (Epstein, 2008).
Transgenic mouse models have provided further evidence that activation of Hh signaling pathway is a key step in the initiation of the tumorigenic program leading to BCC (Grachtchouk et al., 2000; Nilsson et al., 2000a; Nilsson et al., 2000b; Tang et al., 2004; Xie et al., 1998). In addition to Hh signaling, Wnt/β-catenin signaling pathway has been shown to be activated in BCC (Doglioni et al., 2003; El-Bahrawy et al., 2003). Canonical Wnt/β-catenin signaling is involved in the BCC tumorigenesis perhaps by modulating the Hh pathway activity (Yang et al., 2008). Wnt and Hh are two major pathways that are critical in embryonic development, stem cell maintenance, and tumorigenesis. These two pathways have been postulated to interact or cross-regulate at multiple levels, yet the mechanisms of these interactions are not clear. In our previous studies, we identified a novel mechanism by which Wnt signaling regulates the transcriptional outcome of Hh signaling pathway. We demonstrated that Wnt/β-catenin signaling induces expression of the Hh transcriptional activator GLI1. We showed that CRD-BP, a direct target of the Wnt/β-catenin signaling (Noubissi et al., 2006), binds to the segment of the coding region of GLI1 mRNA and stabilizes it. We also showed that Wnt/β-catenin signaling induces the expression and transcriptional activity of GLI1 in a CRD-BP dependent manner (Noubissi et al., 2009). In this study, we address the question of whether CRD-BP-dependent regulation of GLI1 expression and activities is important to the development of BCC. We show that CRD-BP is over-expressed in BCC and that its expression positively correlates with the activation of both Wnt and Hh signaling pathways. One of the hurdles in elucidating the biology of BCC is the lack of human BCC cell lines. We developed and characterized a novel human BCC cell line in our lab. Using this cell line, we demonstrated that CRD-BP-dependent regulation of GLI1 expression and activities is important to the development of BCC.
Results and Discussion
CRD-BP is over-expressed in BCC
In order to assess CRD-BP levels in BCC samples, RNA isolated from 13 superficial BCCs was analyzed by RT-qPCR. We observed an over-expression of CRD-BP mRNA in 12 out of the 13 BCC samples compared to matched controls (Fig 1a, b, S1a, S1b). In the same 12 samples characterized by over-expression of CRD-BP, we also observed activation of both Wnt and Hh signaling pathways indicated by the expression levels of β-TrCP1, c-myc, AXIN, and TCF1 for the activation of Wnt signaling, and the expression levels of GLI1, PTCH1 and PTCH2 for the activation of the Hh signaling (Fig 1a, b, S1a). Our results showed a strong positive correlation between CRD-BP expression and the expression of the genes that it regulates (β-TrCP1, c-myc and GLI1) in the BCC samples (Fig 1a). This observation was further validated in another set of BCC samples where the range of expression levels of CRD-BP was also higher in the ten BCC samples when compared to the range of its expression in the unmatched normal skin controls (Fig S1a). The expression levels of CRD-BP also correlated with the activation of the Wnt and Hh signaling pathways in these samples (Fig S1a).
Figure 1. Analysis of the expression of CRD-BP and the activity of Wnt and Hh signaling pathways in BCC.
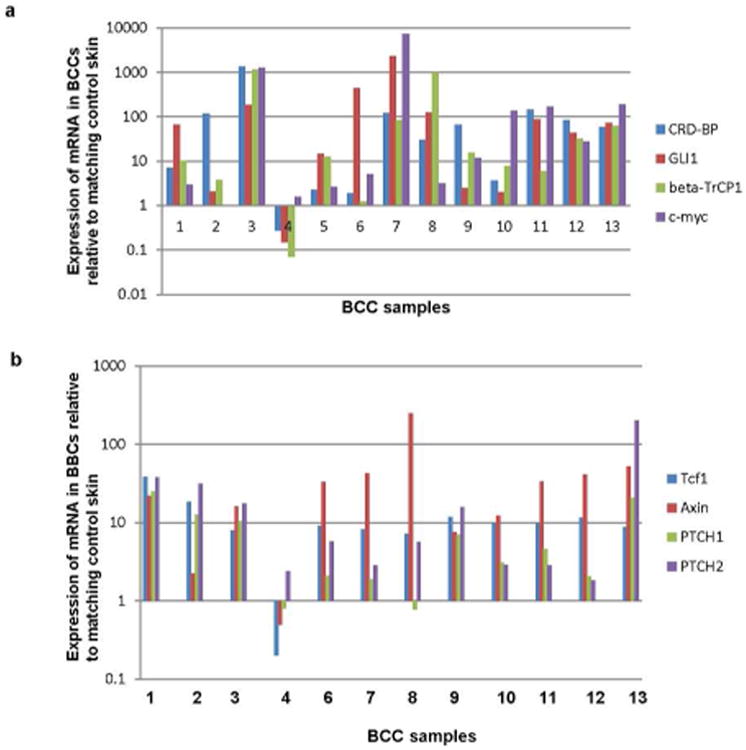
a. Relative expression of CRD-BP, GLI1, β-TrCP1, and c-myc in mRNA isolated from 13 superficial BCCs, determined by quantitative RT-PCR. Each value represents the ratio between the expression of each mRNA in BCC and its expression in matching normal skin.
b. Relative expression of Tcf1, Axin, PTCH1, and PTCH2 in mRNA isolated from 12 superficial BCCs, determined by quantitative RT-PCR. Each value represents the ratio between the expression of each mRNA in BCC and its expression in matching normal skin.
Role of CRD-BP in the regulation of GLI-driven transcription, growth and apoptosis in hTERT immortalized normal keratinocytes
We previously showed that Wnt/β-catenin signaling stimulates the transcriptional outcome of Hh signaling pathway (Noubissi et al., 2009). As expected, activation of the Wnt/β-catenin signaling increases GLI promoter-dependent luciferase activity in a CRD-BP-dependent manner in hTERT immortalized normal keratinocytes (Fig 2a). Our data also show that over-expression of the PTCH mutant, which constitutively activates the Hh signaling pathway (Barnes et al., 2005), induces GLI promoter-driven luciferase activity in the keratinocytes. Interestingly, knockdown of CRD-BP by specific shRNA also inhibits PTCH mutant dependent increase in the GLI promoter-driven luciferase activity (Fig 2a). These data suggest that stabilization of GLI1 mRNA by CRD-BP is important for the regulation of GLI1 expression by both Wnt and Hh signaling pathways. Conversely, over-expression of CRD-BP leads to an increase in the GLI promoter-driven luciferase activity in the same cells (Fig 2b). We also observed accelerated proliferation (Fig 2c-e) and inhibition of apoptosis (Fig 2f) of the hTERT immortalized normal keratinocytes when CRD-BP was over-expressed.
Figure 2. CRD-BP over-expression stimulates growth and inhibits apoptosis of hTERT-immortalized normal keratinocytes.
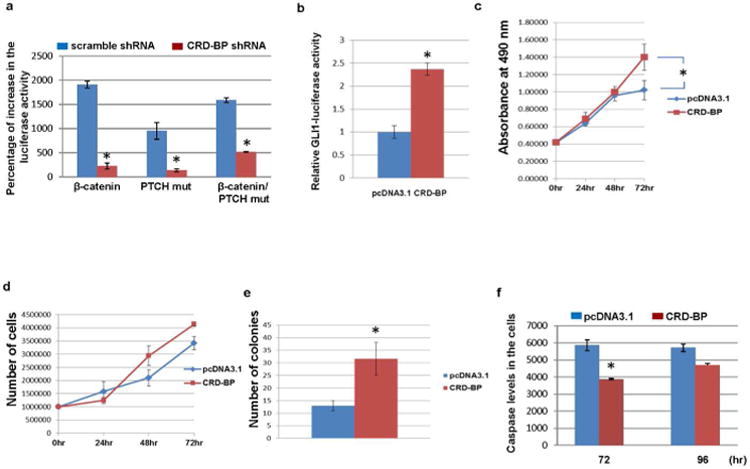
(a) Normal keratinocytes were grown in six-well plates and co-transfected with 8×3’GLI BS-LucII reporter plasmid, pSV-40-galactosidase and pcDNA3.1, β-catenin, PTCH mut, or β-catenin and PTCH-mut plasmids in the presence of scramble small hairpin RNA (shRNA) or CRD-BP shRNA as indicated. In total, 48 hours after transfection, the luciferase activity was estimated using luciferase reporter assay reagent (Promega). β-Galactosidase was used for normalization and estimated using β-galactosidase assay reagent. (b) The immortalized normal keratinocytes were grown in six-well plates and co-transfected with 8×3’GLI BS-LucII reporter plasmid, pSV-40-galactosidase and pcDNA3.1 or CRD-BP plasmid. Forty-eight hours after transfection, the luciferase activity was estimated using luciferase reporter assay reagent (Promega). β-Galactosidase was used for normalization and estimated using β-galactosidase assay reagent. (c) Immortalized normal keratinocytes grown in 100-mm plates were transfected with pcDNA3.1 (10 μg) or CRD-BP (10 μg) as indicated. Twenty-four hours post transfection, 2,000 cells from each plate were seeded in 96-well plates. Growth assay was measured at 24, 48, and 72 hours post seeding. (d) A total of 1 million immortalized normal keratinocytes transfected as in c were seeded in 100-mm plates and the cell growth was measured at 24, 48, and 72 hours using the TC10 automated cell counter from BioRad. (e) hTERT-immortalized normal keratinocytes grown in 100-mm plates were co-transfected with pTK-puro plasmid (0.5 μg) and pcDNA3.1 (9.5 μg) or pTK-puro plasmid (0.5 μg) and CRD-BP (9.5 μg) as indicated. Forty-eight hours after transfection, equal number of cells from each plate was seeded in five 100-mm plates and treated with puromycin (5 μg/ ml) for 10 days. The colonies were counted under light microscope. (f) The immortalized normal keratinocytes were transfected as in c. Twenty four hours after transfection 2,000 cells from each plate was seeded in 96 well plates and Caspase 3/7 was measured at 48 and 72hours post seeding. Each experiment was done at least twice and in triplicates. Each value represents mean±SD. *P≤0.05
Characterization of an established long-term human cell line (UW-BCC1)
The absence of characterized long-term human BCC cell line has hindered the research progress in the study of BCC biology. We, in our lab, have successfully established a long-term human BCC cell line (UW-BCC1) (Fig 3a) (Materials and Methods). The cell line is cultured in low calcium media used to establish normal epidermal keratinocytes. At passage six, the karyotype of the cells was abnormal near triploid with complex structural and numerical aberrations. This karyotype remained unchanged at passage 39 (Fig S2a). The assessment of the activity of the Hh pathway in UW-BCC1 showed high levels of expression of the Hh transcriptional activator GLI1 (Fig 3b, d), a remarkable GLI promoter-driven luciferase activity (Fig 3e), and a significant expression of endogenous GLI1 targets PTCH1 and PTCH2 (Fig 3d). In addition, UW-BCC1 responded well to the treatment with vismodegib, a small molecule inhibitor of smoothened recently approved by the FDA for the treatment of BCC (Fig 3f). These data indicate that the Hh pathway is activated in this cell line. UW-BCC1 shares some features of normal epidermal keratinocytes including the polygonal shape of the cells (Fig 3a) and the significant expression of high molecular weight keratins (K5 and K14) (Fig 3c). Injection of UW-BCC1 in nude mice induced tumor growth in 100% animals with even as little as 100,000 cells per injection and addition of matrigel enhanced in vivo growth (Table 1, Fig S2b). Histologically, the xenografts grew in sheets and large nests with sharp borders and central areas of necrosis. The tumors fulfill cytologic criteria for basal cell carcinoma including: 1) high intercellular cohesion in tissue, 2) morphologically uniform tumor cells, 3) high nuclear-cytoplasm ratio of tumor cells with the cytoplasm forming a very narrow rim around the nucleus, 4) oval or fusiform and sometimes round nuclei with a smooth outline and evenly dispersed, finely dotted chromatin, and 5) inconspicuous nucleoli (Fig S2c). The Hh pathway is activated in the tumors as shown by the expression levels of GLI1, PTCH1 and PTCH2 (Fig S2d). The Wnt pathway is also activated in these tumors as indicated by the expression levels of CRD-BP, β-TrCP1 and c-myc which are Wnt targets (Fig S2d-g). Keratins 5 and 14 are significantly expressed in these tumors as well (Fig S2e-f). Our data altogether show that we have established a bona fide human BCC cell line.
Figure 3. Characterization of a human BCC cell line (UW-BCC1).
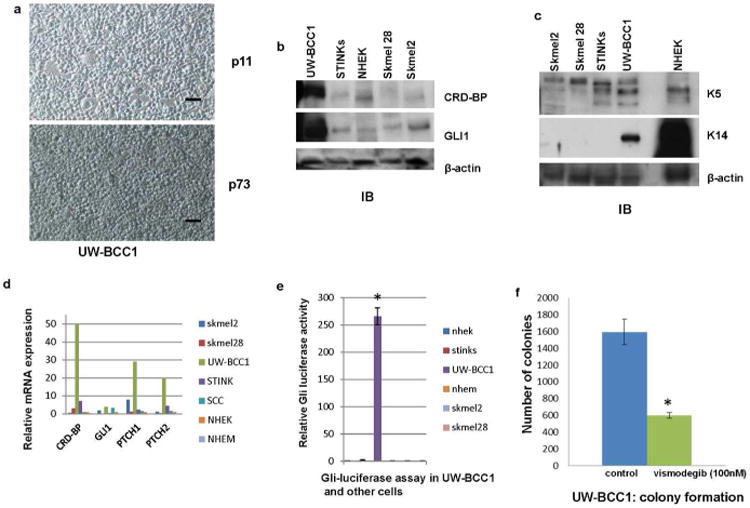
a. UW-BCC1 in culture at passage 11 and at passage 73. A BCC sample was sequentially trypsinized for 6, 12 and 18 hrs in a humidified cell culture incubator with 5% CO2 and grown in low calcium media as described in the supplemental information. Scale bar = 30 μm.
b. Protein expression of CRD-BP and GLI1 in UW-BCC1 determined by immunoblot analyses. β-actin was used as internal control. This is a representative of three independent experiments.
c. Protein expression of keratin 5 and keratin 14 in UW-BCC1 determined by immunoblot analyses. β-actin was used as internal control. This is a representative of three independent experiments.
d. Relative expression of CRD-BP, GLI1, PTCH1, and PTCH2 in mRNA isolated from UW-BCC1 and other cell lines determined by quantitative RT-PCR.
e. UW-BCC1 and other cells as indicated were grown in 6 well plates and co-transfected with 8×3′GLI BS-LucII reporter plasmid and pSV-40 -galactosidase plasmid. 48 hrs after transfection, the luciferase activity was estimated using luciferase reporter assay reagent (Promega). β-galactosidase was used for normalization and estimated using β-galactosidase assay reagent (Pierce). Each experiment was done at least twice and in triplicates. Each value represents Mean+/-SD. *P≤0.05
f. UW-BCC1 was grown in two 100 mm plates and transfected with pTK-puro plasmid. Equal number of cells from each plate was seeded in five 100 mm plates. One set of plates was treated with Vismodegib (100 nM) for 72 hrs and puromycin (5 μg) for 10 days. The other set of plates was treated with puromycin (5 μg) alone for 10 days. Colonies were counted under the microscope. Each experiment was done at least twice and in triplicates. Each value represents Mean+/-SD.
*P≤0.05.
Table 1. average tumor size in mm3 induced by UW-BCC1 injection in nude mice.
| Week3 | Week4 | Week5 | Week6 | Week7 | |
|---|---|---|---|---|---|
| 1 million cells (UW-BCC1) | 249+/-18 | 943+/-188 | - | - | - |
| 500,000 cells | 121+/-11 | 247+/-58 | 911+/-111 | - | - |
| 100,000 cells | Very tiny | small | 39+/-4 | 229+/-24 | 874+/-140 |
Dependence of BCC cells on CRD-BP
We hypothesized that CRD-BP-dependent regulation of GLI1 expression and activities is important to the development of BCC. CRD-BP is highly expressed in BCCs and in UW-BCC1 (Fig 1a, 3b, 3d, S1a, S1b, S1c, and S3). Knock down of CRD-BP with specific shRNA (Noubissi et al., 2006) significantly inhibits the ability of UW-BCC1 to form colonies (Fig 4a), and considerably slows down its growth (Fig 4b). Knock down of CRD-BP also increases apoptosis in UW-BCC1 (Fig 4c), and inhibits its ability to migrate through the membrane in the invasion assay (Fig 4d). The effects of CRD-BP inhibition are more pronounced than the effects of GLI1 inhibition. This is expected as in addition to GLI1, CRD-BP regulates other genes involved in proliferation, invasion, and inhibition of apoptosis including β-TrCP1, c-myc, and other oncogenes that might contribute to the development of BCC. Future studies will delineate the role of CRD-BP in the pathogenesis of BCC development in vivo.
Figure 4. Dependence of BCC cells on CRD-BP.
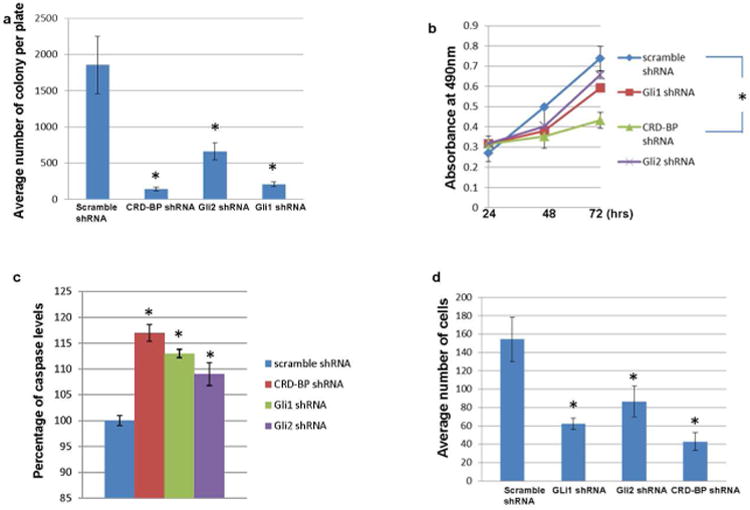
a. UW-BCC1 cells grown in 100 mm plates were co-transfected with pTK-puro plasmid (0.5 μg) and irrelevant shRNA (9.5 μg), pTK-puro plasmid and CRD-BP shRNA (9.5 μg), pTK-puro plasmid and GLI2 shRNA (9.5 μg), or pTK-puro plasmid and GLI1 shRNA (9.5 μg) as indicated. 48 hrs after transfection, equal number of cells from each plate was seeded in five 100 mm plates and treated with puromycin (5μg/ml) for 10 days. The colonies were counted under the light microscope.
b. UW-BCC1 cells grown in 100 mm plates were transfected with irrelevant shRNA (10 μg), CRD-BP shRNA (10 μg), GLI2 shRNA (10 μg) or GLI1 shRNA (10 μg) as indicated. 24hrs post transfection 2000 cells from each plate was seeded in 96 well plates. Growth assay was measured at 24, 48 and 72hrs post seeding.
c. UW-BCC1 cells were transfected as in b. 24hrs after transfection 2000 cells from each plate was seeded in 96 well plates. Caspase 3/7 was measured at 48hrs post seeding.
d. UW-BCC1 cells were transfected as in b. 2.5×104 cells from each plate was used for the invasion assay. 24hrs after transfection cells suspended in low serum media (1% FBS) were added to the insert of the invasion plate and incubated for 48hrs in a humidified incubator. The cells were fixed with 10% formalin for 10 min at room temperature and lightly stained with 0.5% crystal violet for 30 min. The non-invading cells/collagen layer from the interior of the inserts was removed and the cells bound to the membrane of the insert were air dried and counted under the microscope.
Each experiment was done at least twice and in triplicates. Each value represents Mean+/-SD.
*P≤0.05
In summary our data suggest that CRD-BP may play a critical role in BCC development: it is over-expressed in human BCCs, it regulates the expression and activities of GLI1 and other targets important for BCC tumorigenesis, and its down-regulation suppresses growth and tumorigenic properties of BCC cells. Notably, the cell line generated in this study is an asset for studying the biology of BCC and may be critical for developing novel therapies.
Although it is absent or scarce in adult tissues, CRD-BP is de novo activated and/or over-expressed in various neoplastic and pre-neoplastic tumors (Doyle et al., 1998; Ioannidis et al., 2004; Ioannidis et al., 2003a, b; Ioannidis et al., 2001; Leeds et al., 1997; Ross et al., 2001) and its expression has been shown to be associated with the most aggressive form of some cancers (Gu et al., 2004; Ioannidis et al., 2004; Ioannidis et al., 2003a, b). Our data indicate that CRD-BP is important in BCC development, making CRD-BP an attractive target for developing novel therapeutic approaches in the treatment of BCC. Developing effective drugs for BCC treatment would alleviate the cost of care for BCCs, and be a better alternative to repeated surgical procedures, especially in the case of BCNS and immuno-compromised patients who are more prone to develop hundreds of BCCs in their lifetime.
Material and Methods
BCC samples collection
BCC samples were collected from the UW Dermatology clinics. They are biopsy-proven collected from BCC patients during MOHS surgery. The matching normal skin tissues resulted from clear margin skin obtained after closure of the wound. The samples were snap-frozen in liquid nitrogen for RNA isolation and kept in culture media for cell isolation. The protocol was reviewed and fully approved by the University of Wisconsin, Health Science Institutional Review Board (IRB).
RNA isolation and RT-qPCR
Total RNA from tissues or cells was isolated using TRI-Reagent (Molecular Research Center, Inc., Cincinnati, OH).
Real-time PCR for quantitative measurement of CRD-BP, GLI1, β-TrCP1, c-myc, TCF1, and AXIN was performed using Advantage RT-for-PCR kit (Clontech, Mountain View, CA) and SYBR Green Core PCR reagents (Applied Biosystems, Foster city, CA). Primer sequences are listed in Supplementary Table S1. GAPDH was used as a reference gene. Quantitative measurement of PTCH1 and PTCH2 was done using pre-designed TaqMan gene expression assays (Applied Biosystems, Foster city, CA).
Establishment of UW-BCC1
UW-BCC1 was isolated from a superficial BCC that was removed from the leg of a patient. The BCC sample was sequentially trypsinized for 6, 12 and 18 hrs in a humidified cell culture incubator with 5% CO2 using 0.25% Trypsin/2.21mM EDTA (Mediatech, Inc., Herndon, VA). The trypsinized cells were collected, pooled together and grown on an uncoated plate in Eagle's minimum essential medium (EMEM) with nonessential amino acids and L-glutamine, but without calcium (Lonza, Walkersville, MD). The media was further supplemented with epidermal growth factor (EGF; 5 ng/mL), bovine pancreas insulin (5 μg/mL), transferrin (10 ng/mL), ethanolamine (10 μM), phospho-ethanolamine (10 μM), 0.02 mM calcium chloride, 7% fetal bovine serum (Sigma, St Louis, MO) and 1% penicillin-streptomycin-amphotericin B (Mediatech, Inc., Herndon, VA).
We started with the serum media and we alternated between serum media and serum free media (EpiLife Cat# MEPI500CA supplemented with human keratinocyte growth supplement from Life Technologies EDGS Cat# S-012-5). The media was changed every other day. When the cells reached 70% confluency (∼1.67 ×106 cells), they were plated at a density of ∼5.57 × 105/100mm plate (1/3 of the cells). After the third passage, the cells were plated at the density of ∼1.67 × 105/100mm plate (1/10 of the cells) for maintenance in the serum media. Using this method our success rate in establishing a BCC cell line was one out of a total of two attempts.
Cell lines and transfection
UW-BCC1, the melanoma cell lines sk-mel2, sk-mel28 (ATCC, Manassas, VA), and the hTERT immortalized keratinocytes (Bhatia et al., 2008) were transfected using Lipofectamine 2000 (Invitrogen Corporation, Carlsbad, CA). Normal Human Epidermal Keratinocytes (NHEK) (Invitrogen Corporation, Carlsbad, CA) and Normal Human Epidermal Melanocytes (NHEM) (gift from Dr. Setaluri) were electroporated using Lonza nucleofector kit (Lonza, Walkersville, MD) according to the manufacturer recommendations.
Immunoblotting
The following antibodies were used for western blot analysis: anti- GLI1, -K5, -K14, -β-actin (Santa Cruz Biotechnology, Santa Cruz, CA), anti-CRD-BP mouse monoclonal antibody (generated in our lab); secondary antibody conjugated with horseradish peroxidase (Chemicon, Billerica, MA).
In order to obtain whole cell lysates for western blot analysis, the cells were lysed using a denaturing RIPA buffer containing PBS (pH 7.4), 0.5% sodium deoxycholate, 0.1% SDS, 1% (v/v) IGEPAL, 100 mM sodium orthovanadate, and proteinase inhibitor cocktail (Sigma, St. Louis, MO). Frozen tissues were ground in pestle and mortar to fine powder and RIPA lysis buffer was added to it for homogenization and the lysates were used for western blot. Immunoblotting procedures were performed as described previously (Spiegelman et al., 2000).
Luciferase assay
Cells were co-transfected with 8×3′GLI BS-LucII reporter plasmid and pSV-40 β-galactosidase plasmid. GLI promoter-driven luciferase activity was estimated 48 hrs after transfection using luciferase reporter assay reagent (Promega, Madison WI). β-galactosidase used for normalization was estimated by the β-galactosidase assay reagent (Pierce, Rockford, IL).
Immunostaining
Tissue sections cut from formalin-fixed paraffin-embedded tumors were de-paraffinized in xylene (Fisher, NJ) and rehydrated in grades of alcohol. The sections were subjected to antigen retrieval by incubation in 10 mM citrate buffer in a microwave oven for 20 min. The sections were blocked with normal serum, probed with specific primary antibodies to CRD-BP, β-catenin, or GLI1, followed by the appropriate fluorescent labeled secondary antibody (Invitrogen Corporation, Carlsbad, CA). Tissue sections were mounted with Prolong Gold Antifade with Dapi mounting media (Invitrogen Corporation, Carlsbad, CA).
Cytostaining
Cells were cultured on chamber slides and fixed with 10% formalin. Nonspecific binding was averted by blocking in 10% BSA. Cells were incubated at 4°C overnight with CRD-BP, β-catenin (Millipore, Billerica, MA) or GLI1 (Santa Cruz Biotechnology, Santa Cruz, CA) antibodies. Slides were incubated with the appropriate secondary antibody (Invitrogen Corporation, Carlsbad, CA)) and mounted with Prolong Gold Antifade with Dapi mounting media (Invitrogen Corporation, Carlsbad, CA).
Cell proliferation assay
Cells were counted using the TC10™ automated cell counter from BioRad (BioRad, Hercules, CA) and equal number of viable cells was plated in 96 well plates. To measure cell proliferation, the MTS-based cell proliferation assay kit was used (Promega, Madison, WI). The assay was performed every 24 hrs and incubation time was kept constant. The intensity of the chromogenic substrate was measured at 490 nm.
Clonogenic assay
UW-BCC1 and the immortalized keratinocytes grown in 100-mm plates were co-transfected with pTK-puro plasmid and any other appropriate plasmid for the experiment. Forty-eight hours after transfection, equal number of cells from each plate were seeded in five 100-mm plates and treated with puromycin (5μg/mL) (Sigma, St Louis, MO) for 14 days. Colonies were counted under the light microscope.
Detection of apoptosis
Apoptosis in UW-BCC1 and in the immortalized keratinocytes was measured using the Caspase-Glo 3/7 assay kit (Promega, Madison, WI) according to the manufacturer's recommendations.
Invasion assay
The invasion assay was performed using the BD BioCoat™ Growth Factor Reduced BD Matrigel™Invasion Chamber, 8.0 μm PET Membrane 24-well Cell Culture Inserts, packaged ready-to-use in BD Falcon™ Companion Plates (BD Bioscience, San Jose, CA). The chemo-attractant used was 10% FBS in the culture media. 2.5×104 cells suspended in low serum media (1% FBS) were added to the insert and incubated in a humidified incubator for 48hrs. The cells were fixed with 10% formalin at room temperature for 10 min and lightly stained with 0.5% crystal violet for 30 min. The non-invading cells/collagen layer from the interior of the inserts was removed and the cells bound to the membrane of the insert were air dried and counted under the microscope.
Generation of tumors in nude mice
5 million UW-BCC1 cells at passage 6 were injected sc with or without matrigel on the back of athymic nude mice (Hsd:Athymic Nude-Foxn1nu) (Harlan, Madison, WI). Each group was composed of five 4 week old male mice. Tumor growth was monitor weekly and tumor width, length and depth was measured using a caliper. The volume was calculated using the formula described in (Siddiqui et al., 2006).
Statistical Analyses
The T-test was used for statistical analyses. P≤0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank the Department of Dermatology at UW Madison for providing logistics during the collection of the BCC samples, Dr. I. Siddiqui for helping the generation of tumors in nude mice, and Dr. K. Spiegelman for editing the manuscript. This work was supported by Dermatology Foundation Career Development Award (to F.K.N.), NIH grants CA153102 (to F.K.N.), CA121851 and AR063361 (to V.S.S.).
Abbreviations
- CRD-BP
- BCC
- Hh
- GLI
- Smo
- PTCH
- BCNS
Footnotes
Conflict of Interest: The authors state no conflict of interest.
References
- Barnes EA, Heidtman KJ, Donoghue DJ. Constitutive activation of the shh-ptc1 pathway by a patched1 mutation identified in BCC. Oncogene. 2005;24:902–15. doi: 10.1038/sj.onc.1208240. [DOI] [PubMed] [Google Scholar]
- Bhatia N, Demmer TA, Spiegelman VS. Inhibition of beta-TrCP function potentiates UVB-induced apoptosis in hTERT-immortalized normal human keratinocytes. Photochem Photobiol. 2008;84:376–81. doi: 10.1111/j.1751-1097.2007.00272.x. [DOI] [PubMed] [Google Scholar]
- Doglioni C, Piccinin S, Demontis S, et al. Alterations of beta-catenin pathway in non-melanoma skin tumors: loss of alpha-ABC nuclear reactivity correlates with the presence of beta-catenin gene mutation. Am J Pathol. 2003;163:2277–87. doi: 10.1016/s0002-9440(10)63585-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle GA, Betz NA, Leeds PF, et al. The c-myc coding region determinant-binding protein: a member of a family of KH domain RNA-binding proteins. Nucleic Acids Res. 1998;26:5036–44. doi: 10.1093/nar/26.22.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Bahrawy M, El-Masry N, Alison M, et al. Expression of beta-catenin in basal cell carcinoma. Br J Dermatol. 2003;148:964–70. doi: 10.1046/j.1365-2133.2003.05240.x. [DOI] [PubMed] [Google Scholar]
- Epstein EH. Basal cell carcinomas: attack of the hedgehog. Nat Rev Cancer. 2008;8:743–54. doi: 10.1038/nrc2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grachtchouk M, Mo R, Yu S, et al. Basal cell carcinomas in mice overexpressing Gli2 in skin. Nat Genet. 2000;24:216–7. doi: 10.1038/73417. [DOI] [PubMed] [Google Scholar]
- Gu L, Shigemasa K, Ohama K. Increased expression of IGF II mRNA-binding protein 1 mRNA is associated with an advanced clinical stage and poor prognosis in patients with ovarian cancer. International journal of oncology. 2004;24:671–8. [PubMed] [Google Scholar]
- Ioannidis P, Kottaridi C, Dimitriadis E, et al. Expression of the RNA-binding protein CRD-BP in brain and non-small cell lung tumors. Cancer Lett. 2004;209:245–50. doi: 10.1016/j.canlet.2003.12.015. [DOI] [PubMed] [Google Scholar]
- Ioannidis P, Mahaira L, Papadopoulou A, et al. 8q24 Copy number gains and expression of the c-myc mRNA stabilizing protein CRD-BP in primary breast carcinomas. Int J Cancer. 2003a;104:54–9. doi: 10.1002/ijc.10794. [DOI] [PubMed] [Google Scholar]
- Ioannidis P, Mahaira L, Papadopoulou A, et al. CRD-BP: a c-Myc mRNA stabilizing protein with an oncofetal pattern of expression. Anticancer Res. 2003b;23:2179–83. [PubMed] [Google Scholar]
- Ioannidis P, Trangas T, Dimitriadis E, et al. C-MYC and IGF-II mRNA-binding protein (CRD-BP/IMP-1) in benign and malignant mesenchymal tumors. Int J Cancer. 2001;94:480–4. doi: 10.1002/ijc.1512. [DOI] [PubMed] [Google Scholar]
- Leeds P, Kren BT, Boylan JM, et al. Developmental regulation of CRD-BP, an RNA-binding protein that stabilizes c-myc mRNA in vitro. Oncogene. 1997;14:1279–86. doi: 10.1038/sj.onc.1201093. [DOI] [PubMed] [Google Scholar]
- Nilsson LO, Gustafsson A, Mannervik B. Redesign of substrate-selectivity determining modules of glutathione transferase A1-1 installs high catalytic efficiency with toxic alkenal products of lipid peroxidation. Proc Natl Acad Sci U S A. 2000a;97:9408–12. doi: 10.1073/pnas.150084897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson M, Unden AB, Krause D, et al. Induction of basal cell carcinomas and trichoepitheliomas in mice overexpressing GLI-1. Proc Natl Acad Sci U S A. 2000b;97:3438–43. doi: 10.1073/pnas.050467397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noubissi FK, Elcheva I, Bhatia N, et al. CRD-BP mediates stabilization of betaTrCP1 and c-myc mRNA in response to beta-catenin signalling. Nature. 2006;441:898–901. doi: 10.1038/nature04839. [DOI] [PubMed] [Google Scholar]
- Noubissi FK, Goswami S, Sanek NA, et al. Wnt signaling stimulates transcriptional outcome of the Hedgehog pathway by stabilizing GLI1 mRNA. Cancer research. 2009;69:8572–8. doi: 10.1158/0008-5472.CAN-09-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oro AE, Higgins KM, Hu Z, et al. Basal cell carcinomas in mice overexpressing sonic hedgehog. Science. 1997;276:817–21. doi: 10.1126/science.276.5313.817. [DOI] [PubMed] [Google Scholar]
- Rippey JJ. Why classify basal cell carcinomas? Histopathology. 1998;32:393–8. doi: 10.1046/j.1365-2559.1998.00431.x. [DOI] [PubMed] [Google Scholar]
- Ross J, Lemm I, Berberet B. Overexpression of an mRNA-binding protein in human colorectal cancer. Oncogene. 2001;20:6544–50. doi: 10.1038/sj.onc.1204838. [DOI] [PubMed] [Google Scholar]
- Siddiqui IA, Zaman N, Aziz MH, et al. Inhibition of CWR22Rnu1 tumor growth and PSA secretion in athymic nude mice by green and black teas. Carcinogenesis. 2006;27:833–9. doi: 10.1093/carcin/bgi323. [DOI] [PubMed] [Google Scholar]
- Spiegelman VS, Slaga TJ, Pagano M, et al. Wnt/beta-catenin signaling induces the expression and activity of betaTrCP ubiquitin ligase receptor. Mol Cell. 2000;5:877–82. doi: 10.1016/s1097-2765(00)80327-5. [DOI] [PubMed] [Google Scholar]
- Tang X, Kim AL, Feith DJ, et al. Ornithine decarboxylase is a target for chemoprevention of basal and squamous cell carcinomas in Ptch1+/- mice. J Clin Invest. 2004;113:867–75. doi: 10.1172/JCI20732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicking C, Smyth I, Bale A. The hedgehog signalling pathway in tumorigenesis and development. Oncogene. 1999;18:7844–51. doi: 10.1038/sj.onc.1203282. [DOI] [PubMed] [Google Scholar]
- Wong CS, Strange RC, Lear JT. Basal cell carcinoma. Bmj. 2003;327:794–8. doi: 10.1136/bmj.327.7418.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Murone M, Luoh SM, et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391:90–2. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- Yang SH, Andl T, Grachtchouk V, et al. Pathological responses to oncogenic Hedgehog signaling in skin are dependent on canonical Wnt/beta3-catenin signaling. Nat Genet. 2008;40:1130–5. doi: 10.1038/ng.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


