Abstract
Background
Recent studies show that various inflammatory diseases are regulated at the level of RNA translation by small non-coding RNAs, termed microRNAs (miRNAs). We sought to determine whether sarcoidosis tissues harbor a distinct pattern of miRNA expression and then considered their potential molecular targets.
Methods and Results
Genome-wide microarray analysis of miRNA expression in lung tissue and peripheral blood mononuclear cells (PBMCs) was performed and differentially expressed (DE)-miRNAs were then validated by real-time PCR. A distinct pattern of DE-miRNA expression was identified in both lung tissue and PBMCs of sarcoidosis patients. A subgroup of DE-miRNAs common to lung and lymph node tissues were predicted to target transforming growth factor (TGFβ)-regulated pathways. Likewise, the DE-miRNAs identified in PBMCs of sarcoidosis patients were predicted to target the TGFβ-regulated “wingless and integrase-1” (WNT) pathway.
Conclusions
This study is the first to profile miRNAs in sarcoidosis tissues and to consider their possible roles in disease pathogenesis. Our results suggest that miRNA regulate TGFβ and related WNT pathways in sarcoidosis tissues, pathways previously incriminated in the pathogenesis of sarcoidosis.
Keywords: microRNA, granuloma, gene, TGFβ, peripheral blood mononuclear cells, lung
1 Introduction
Sarcoidosis is a granulomatous disease, frequently involving the lungs and presumed to be promoted by an abnormal host immune response to common environmental antigens. In support of this premise, host genetic variability, particularly in the form of DNA polymorphisms, has been linked to specific disease phenotypes [1–3]. However, phenotypic variability cannot be fully explained by DNA polymorphisms. In this regard, there is a growing interest in the role of specific small non-coding RNAs, referred to as microRNAs (miRNAs) as natural suppressors of gene translation. Abnormal patterns of tissue miRNA expression have been strongly linked to the pathogenesis of various cancers and recent data suggest that these molecules could promote fibrotic and obstructive lung disease through the regulation of fundamental biological processes [4–6]. The role, if any, of altered tissue miRNA expression in the pathogenesis of sarcoidosis remains unknown.
Microarray-based assays allow for the quantitative detection of thousands of transcripts from relatively small samples for the purpose of hypothesis generation. Previous studies have applied microarray-based mRNA expression analysis of diseased tissues, including lung, lymph nodes, and blood cells, to discover novel mechanisms for sarcoidosis pathogenesis [7–9]. In addition to identifying novel biomarkers, such as MMP12, unsupervised bioinformatic analyses of gene expression data can identify highly regulated molecular pathways that are likely to promote abnormal granulomatous inflammation [7]. In this regard, the additional effects of miRNA could explain why gene expression often fails to correlate with protein expression [10].
In order to identify miRNAs that are likely to contribute to the pathogenesis of sarcoidosis, we conducted microarray analyses in both lung tissue and peripheral blood mononuclear cells (PBMCs) derived from patients with sarcoidosis and controls. The array data were then validated by quantitative real-time (qRT)-PCR in an extended group of patients and controls. We demonstrated a common profile of miRNA expression in diseased tissue. This group of miRNAs was predicted to target molecular pathways regulated by TGFβ and WNT, signaling pathways known to cooperatively regulate common pathological conditions [11]. To our knowledge, this study is the first attempt to characterize tissue miRNA expression in sarcoidosis and to link dysregulated miRNAs identified in this study to a mechanistically plausible biological pathway.
2 Material and Methods
2.1 Human subject diagnosis and characteristics
Gene expression analysis was performed on lung and lymph node tissues, and peripheral blood cells from patients meeting the operational diagnosis of sarcoidosis. Specifically, sarcoidosis diagnosis was based upon the identification of well-formed, non-necrotizing, epithelioid granulomas in diseased tissue in the absence of identifiable infection or foreign objects, in agreement with the diagnostic criteria described in the American Thoracic Society’s Joint Statement on Sarcoidosis [12]. Samples exhibiting signs of infection or atypical pathological features, such as necrosis or fibrosis, were excluded from analysis. Disease-free lung and lymph node tissues were obtained during surgical lung resections or in the immediate post-mortem period from patients who had submitted for organ donation for the purposes of medical research. Each sample was verified to have normal lung histology by a certified pathologist. Informed consent was provided for the blood samples while all other samples were de-identified and provided by an “honest broker”. All recruitment for this study was performed in compliance with the universities’ institutional review boards. The demographic features of the sarcoidosis patients and controls are summarized in Table S1.
2.2 RNA isolation from fresh, fixed and frozen samples
Briefly, total RNA was extracted from frozen tissue by the Trizol method (Invitrogen, Carlsbad, California). RNA from FFPE samples was extracted from a 50 micron tissue slice using RecoverAll Total Nucleic Acid Isolation kit (Ambion, Austin Texas), following kit protocol. A single slice of FFPE mediastinal lymph node tissue yields 0.42–1.8 µg of RNA. Quantification of RNA integrity was performed utilizing the Bioanalyzer 2100 capillary electrophoresis system (Agilent Technologies; Santa Clara, CA). Subsequent quantitative analysis of electropherograms for degradation products was performed with proprietary Degradometer software [13]. RNA of insufficient quality was excluded from microarray analysis based upon RIN scores < 7 or 28s:18s ratio <1.0.
2.3 MiRNA Array Analysis
A customized miRNA microarray chip, generated by Carlo Croce, M.D. at The Ohio State University [14], was used for all miRNA expression analyses. Signal intensities were quantified by Genepix Pro software. Local background intensities were subtracted from spot intensities. A filtering method based on percentage of arrays above noise cutoff was applied to filter out low expression miRNAs. Quantile normalization method was used for normalization across arrays [15]. Linear models were performed to detect differentially expressed (DE)-miRNAs across groups. In order to improve the estimates of variability and statistical tests for differential expressions, variance shrinkage methods were employed for this study [16, 17]. The significance level was adjusted by controlling the mean number of false positives (i.e., 5 false discoveries per analysis) [18].
Principle component analysis (PCA) was used to discriminate samples, sarcoidosis vs controls, based upon miRNA expression profiling [19]. All of the miR data was projected onto multiple dimensions (i.e., PC1, PC2, and PC3) for each sample, where PC1, PC2, and PC3 are the first three dimensions with the largest variation the expression data and they are the linear combinations of all miRNAs’ expression. Heat-maps were drawn for statistically significant DE-miRNAs with hierarchical clustering performed on both samples and miRNAs.
2.4 Quantitative real-time (qRT)-PCR validation of DE-miRNA
Subsets of DE-miRNA transcripts identified by tissue microarray were validated by qRT-PCR using established techniques [6]. Production of cDNA from 20 ng total RNA was accomplished using TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems) following the manufacturer’s protocol. qRT-PCR was performed on the Applied Biosystems 7900HT Fast Real-Time PCR System, using specific primers for each miR (see Table S2). Results are presented as 2^-(dCt), and statistical p-values were generated using Student’s t-test on the calculated dCt.
2.5 DIANA-mirPath analysis
The DIANA-mirPath is a web-based computational tool designed to identify molecular pathways potentially altered by the expression of single or multiple miRNAs (http://diana.cslab.ece.ntua.gr/pathways/). The software performs enrichment analysis of multiple miRNA target genes comparing each set of miRNA to all available pathways provided by the Kyoto Encyclopedia of Genes and Genomes (KEGG) and assigns an overall priority score (enrichment P-value) based upon the predicted strength of the miRNA interactions with components of the target pathway. MiRNA target genes found to be implicated in a given pathway are graphically annotated as an overlay of the pathway wiring diagram provided by the KEGG database [20]. The pathways identified were not influenced by investigator bias (unsupervised).
3 Results
3.1 DE-miRNA Expression Patterns in Sarcoidosis Tissues
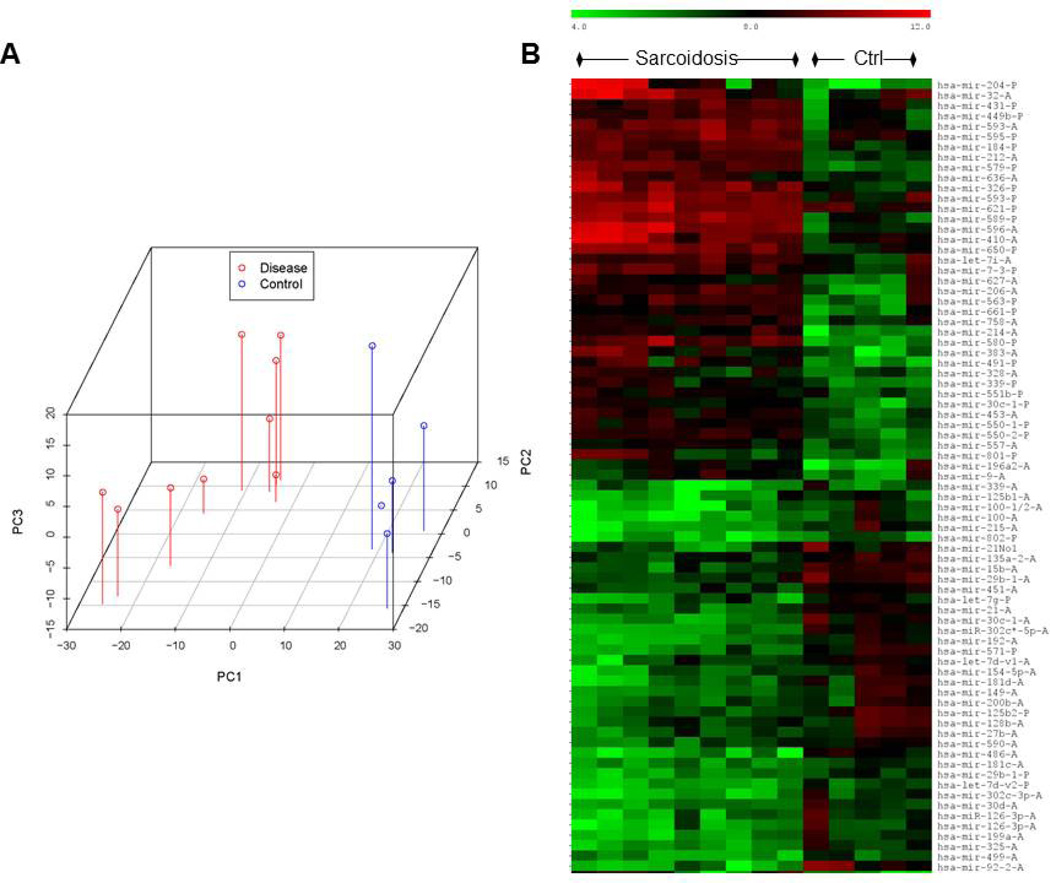
MiRNA array analysis of a test set of lung tissues derived from patients with active pulmonary sarcoidosis and controls identified 60 DE-miRNAs. With the exception of one control outlier, the DE-miRNA expression pattern of sarcoidosis lung tissues was readily distinguished from controls, as reflected by PCA and heat map analyses (Figures 1A and B). The DE-miRNAs associated with sarcoidosis with the greatest change in expression relative to controls are provided in Table S3. The tissue expression of a select group of miRNAs identified by array analysis, for which the fold change was highly significant, were further validated by qPCR in lung and lymph node tissues. In general, the PCR results for miRNA expression in lung tissue (Figure 1C) were consistent with the miRNA array analysis. Although DE-miRNAs identified in lung tissues were also differentially expressed in lymph node tissues relative to matching controls (Figure S1), comparison of DE-miRNA expression in lung and lymph node tissues were not identical. For instance, expression levels of miR-20a and miR-302c were reduced in lung and elevated in lymph node tissues relative to matching controls. In contrast, expression of miR-92b and miR-206 were elevated in both lung and lymph node tissues of sarcoidosis patients. Of note, fixed and frozen tissues yielded similar miRNA expression results.
Figure 1. Differentially expressed (DE)-miRNAs in sarcoidosis lung tissues.
MiRNA arrays were performed on lung tissue samples derived from patients with untreated active pulmonary sarcoidosis and controls (n=4 for each group). Panel A: Principal component analysis demonstrates a distinct pattern of DE-miRNAs in sarcoidosis tissues (red) compared to controls (blue), with the exception of one control outlier. Panel B: Supervised heat map analysis of the array data indicates that 3 out of 4 sarcoidosis samples were readily distinguished from controls based upon the relative expression values of a subset of DE-miRNAs.
3.2 Predicted DE-miRNA Molecular Targets of DE-miRNA Identified in Sarcoidosis Tissues
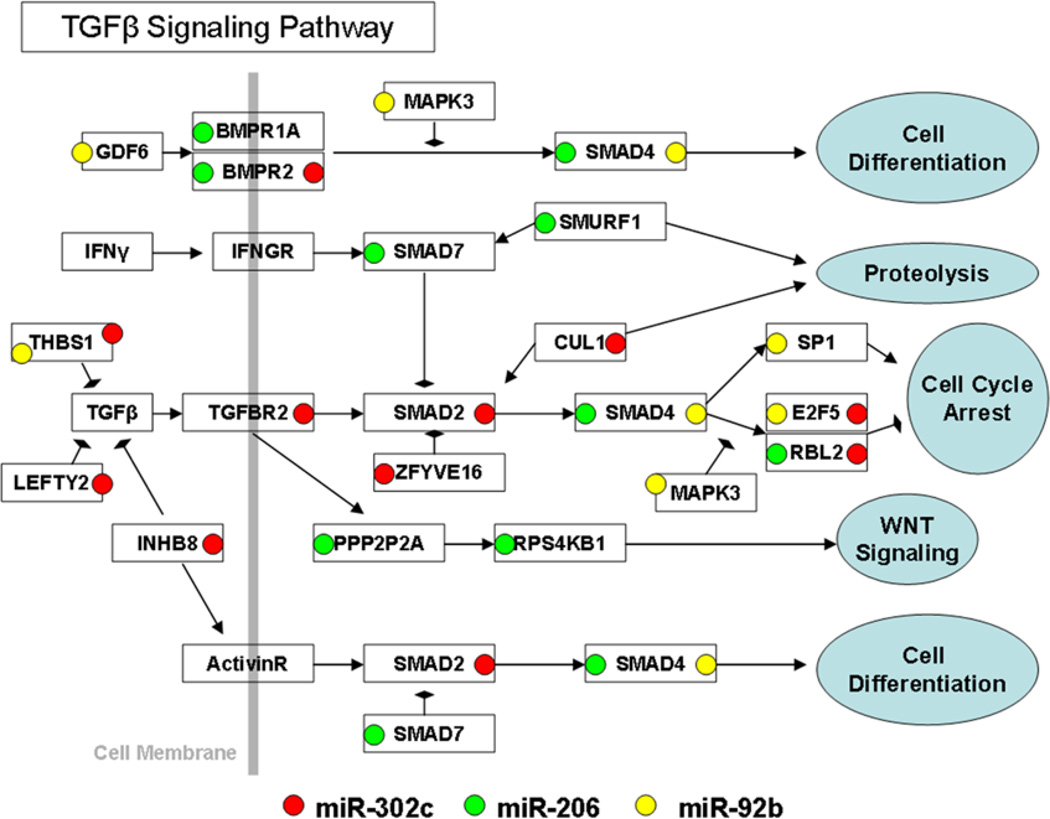
The DIANA-mirPath program was employed to determine the molecular pathways that are most highly predicted to be regulated by the PCR-validated DE-miRNAs identified by tissue array analysis. A number of molecules within a molecular pathway regulated by TGFβ were predicted to be targeted by one or more of the DE-miRNA identified in diseased tissues, as graphically represented in Figure 2.
Figure 2. DE-miRNAs identified in lung sarcoidosis tissues are predicted to target the TGFβ molecular pathway.
DIANA-mirPath was used to predict the most likely molecular pathways targeted by PCR-validated DE-miRNAs in sarcoidosis lung tissue and identified the TGFβ pathway as the most likely candidate. The arrows represent activation, whereas the hammerheads represent inhibition. The molecular components of the TGFβ signaling pathway and their putative interactions were derived from the KEGG pathway database.
3.3 DE-miRNA Expression Patterns in Peripheral Blood Mononuclear Cells Derived from Patients with Pulmonary Sarcoidosis
We next sought to determine if the miRNA profile of PBMCs, some of which are presumed to traffic to and from diseased tissues [21], was consistent with the tissue array results. MiRNA array analysis identified 214 DE-miRNAs in PBMCs from sarcoidosis patients. MiRNAs exhibiting the greatest fold change in sarcoidosis PBMCs are shown in Table S4. As for lung tissue, the profile of PBMC DE-miRNAs was readily distinguished from controls based upon PCA and heat map analyses (Figures 3A and 3B). No significant overlap between lung tissue and PBMC DE-miRNA was observed. Specifically, the selected DE-miRNAs validated in tissues, particularly miR-92b and miR-206 (Figure S2), were unchanged in PBMCs from sarcoidosis patients compared to controls (data not shown). While the DE-miRNA profile of PBMC was generally distinct from that of the tissues in sarcoidosis patients, DIANA-mirPath analysis of PCR-validated DE-miRNAs identified in sarcoidosis PBMCs (Figure S2) predicted targeting of the WNT-pathway (Figure 4), which is highly interactive with the TGFβ pathway [11, 22]. Thus, the predicted targets of DE-miRNA in all sarcoidosis samples tested were commonly linked to TGFβ-regulated molecular pathways.
Figure 3. Differential (DE)-miRNA expression in PBMCs in sarcoidosis.
MiRNA arrays were performed on PBMCs derived from patients with sarcoidosis (n=10) and controls (n=5). Panel A: Principal component analysis demonstrates distinct clustering of DE-miRNA expression in sarcoidosis (red) and controls (blue). Panel B: Supervised heat map analysis of the PBMC data identified a distinct pattern of DE-miRNA in sarcoidosis compared to controls.
Figure 4. DE-miRNAs identified in PBMCs from sarcoidosis patients are predicted to target the TGFβ/WNT molecular pathway.
DIANA-mirPath was used to predict the most likely molecular pathways targeted by PCR-validated DE-miRNAs in sarcoidosis PBMCs and identified the TGFβ/WNT pathway as the most likely candidate. The arrows represent activation, whereas the hammerheads represent inhibition. The molecular components of the WNT signaling pathway and their putative interactions were derived from the KEGG pathway database.
4 Discussion
To our knowledge this is the first published report of miRNA expression patterns in diseased tissues in the context of pulmonary sarcoidosis. Whereas common DE-miRNA expression patterns were detected in lung and lymph node tissues, in which granulomas were present, a distinct profile of DE-miRNA was identified in PBMCs derived from sarcoidosis patients. Despite distinct miRNA expression profiles in diseased tissue and PBMCs, an unsupervised bioinformatic analysis of DE-miRNA targets predicts that miRNAs in diseased tissue and PBMCs commonly target TGFβ/WNT–regulated molecular pathways. This finding has important implications for modulation of immune cell proliferation and activity.
The biological implications of altered miRNA expression are often difficult to predict. MiRNAs have multiple potential targets and their effects can be additive in that many miRNAs can act on a single transcript. Thus, each miRNA has the capacity to regulate tens to hundreds of genes in many different pathways, and in most cases the expression of any given mRNA transcript is influenced by multiple miRNA. In this regard, co-regulated polycistronic miRNA clusters are shown to modify complex biological systems and are important regulators of immune cell functions [23–25]. The DE-miRNAs identified in this analysis of sarcoidosis tissues and PBMCs were derived from distinct chromosome locations and did not conform to a cluster. Nonetheless, the predicted binding affinities of DE-miRNAs with mRNA targets, based upon sequence characteristics, and the associated probability of suppressing mRNA translation, as determined by the DIANA-mirPath web-based computational tool [20], strongly indicated a common functional consequence of DE-miRNAs in patients with sarcoidosis.
It is interesting that the primary predicted target of DE-miRNAs identified in sarcoidosis tissues is a pathway primarily regulated by TGFβ. TGFβ has many functions, including suppression of fibrosis [26] and T-cell-mediated immunity [27]. TGFβ is expressed at high levels within granulomas derived from patients with active pulmonary tuberculosis [28] or sarcoidosis [29]. In the context of sarcoidosis, an inverse relationship between TGFβ release by alveolar cells and their IL-2 release has been observed, and higher TGFβ expression corresponds with spontaneous regression of sarcoidosis [30]. This would explain why various polymorphisms of TGFβ strongly correlate with sarcoidosis disease severity [3, 31], particularly in patients progressing to lung fibrosis [32]. With respect to the latter, a recent study links changes in miR-21 expression to the pro-fibrogenic actions of TGFβ in the context of idiopathic pulmonary fibrosis [5]. In ddition to fibroproliferation, TGFβ is critical for the development and survival of T-cells [33], which are considered to be critical for granuloma formation in the context of sarcoidosis [34]. To the extent that the DE-miRNAs identified in the current analysis alter TGFβ-related functions, it is mechanistically plausible that the DE-miRNAs influence disease outcomes in the context of pulmonary sarcoidosis.
We observed no overlap in miRNA expression in PBMCs relative to diseased lung tissue. Recent studies indicate that plasma miRNA profiles often reflect diseased tissue miRNA expression patterns, presumably relating to liberation of miRNA from the latter [35]. While components represented in PBMC samples (e.g., T cells) may transit to and from sarcoidosis tissues [21], their contribution to the miRNA signal in tissue may be altered by the tissue microenvironment or obscured by other parenchymal cells. Nonetheless, the biological significance of the DE-miRNAs identified in PBMCs and lung tissues are likely to be related. In this regard, array analysis of PBMCs identified a distinct pattern of DE-miRNA expression predicted to target the WNT signaling pathway, which is strongly influenced by TGFβ [11, 22]. Thus, TGFβ -pathways emerge as a common miRNA target in both tissues.
Recent studies indicate that the WNT signaling pathway is intimately engaged in inflammatory responses in the context of granulomatous diseases. The WNT pathway is strongly linked to antigen-mediated inflammation where it has been shown to regulate effector T-cell development, regulatory T-cell activation and dendritic cell maturation [36]. WNT genes and molecules expressed within granulomatous tissues are induced by infectious antigens, thereby providing a bridge between innate and adaptive immunity [37]. The WNT pathway further reprograms macrophages to induce anergy [38], a well-documented consequence of granulomatous diseases such as tuberculosis [39] and sarcoidosis [40, 41]. In support of the evidence provided herein linking DE-miRNA expression in sarcoidosis PBMCs to predicted WNT signaling targets, Levänen et al recently demonstrated increased expression of WNT receptors and activation of specific WNT signaling pathway components, including β-catenin in sarcoidosis tissue [42]. Further evidence supporting miRNA trgeting of the TGFβ and WNT pathways is provided by recent studies linking specific DE-miRNAs identified in sarcoidosis PBMCs, particularly members of let-7, miR-21, miR-29, miR-30, and miR-92, with TGFβ-induced lung fibrosis [5, 43, 44]. Further investigation is necessary to determine whether targeting of TGFβ and WNT pathways by miRNA fundamentally influences inflammatory responses or disease phenotype in patients with sarcoidosis. For instance, it is interesting to speculate that the fibrotic pulmonary sarcoidosis phenotype is associated with a specific profile of miRNAs targeting the TGFβ/WNT molecular pathway.
There are several study limitations that could influence our results. The likelihood of false discovery increases when small sample sizes are used to identify disease-specific DE-miRNA. This limitation was addressed by utilization of variance shrinkage methods, as described in the Methods, and by PCR confirmation of DE-miRNAs identified by lung tissue array analysis in a larger sample (i.e., fixed and frozen lymph nodes). Failure to validate all DE-miRNAs identified by microarray is not unexpected and could relate to false discovery, albeit <5% chance based upon study design, or intrinsic challenges relating to miRNA profiling, as detailed recently by Benes and Castoldi [45]. In general, the results of PCR are considered to be most definitive in terms of confirming DE-miRNAs. Furthermore, functional validation of predicted miRNA and mRNA interactions are ultimately required to support the predicted link between miRNAs and TGFβ-regulated pathways in sarcoidosis. Despite these limitations, and recognizing that the cell profiles of blood and diseased tissues are very different, the functional relatedness of the validated miRNAs identified in sarcoidosis tissues and PBMCs strongly suggests that TGFβ and related WNT pathways are highly regulated by miRNA in this disease.
4.1 Conclusions
To our knowledge, this is the first study to profile miRNA expression in diseased tissue in the context of active pulmonary sarcoidosis. An unbiased bioinformatic analysis of DE-miRNA function identified TGFβ/WNT-regulated molecular pathways as probable targets. These findings are in keeping with recent investigations linking altered TGFβ and WNT pathways to sarcoidosis disease activity [3, 30–32]. Further investigation is required to determine whether miRNA regulate immune cell activation and disease phenotype in a larger sarcoidosis cohort.
Supplementary Material
Highlights.
This is the first study to profile microRNA expression in sarcoidosis tissues
MicroRNA in diseased sarcoidosis tissue shares a common microRNA profile
Peripheral blood cells of sarcoidosis patients exhibit a unique microRNA expression profile
MicroRNA expressed in sarcoidosis tissue and blood are predicted to target the WNT/TGFβ pathway
Acknowledgements
We would like to thank David Cosmar for his assistance with tissue and clinical data procurement.
Supported in part by a joint grant from the American Thoracic Society and the Foundation for Sarcoidosis Research (ECD) and HL095425 (SPN)
Abbreviations
- DE-miRNA
differentially expressed microRNA
- FFPE
formalin-fixed, paraffin-embedded
- miRNA
microRNA
- PBMCs
peripheral blood mononuclear cells
- TGFβ
transforming growth factor beta
- WNT
wingless and integrase-1
References
- 1.Wijnen PA, Voorter CE, Nelemans PJ, Vershakelen JA, Bekers O, Drent M. Butyrophilin-like 2 in pulmonary sarcoidosis: a factor for susceptibility and progression? Hum. Immunol. 2011;72:342. doi: 10.1016/j.humimm.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 2.Grunewald J. Review: role of genetics in susceptibility and outcome of sarcoidosis. Semin. Respir. Crit. Care Med. 2010;31:380. doi: 10.1055/s-0030-1262206. [DOI] [PubMed] [Google Scholar]
- 3.Jonth AC, Silveira L, Fingerlin TE, et al. TGF-beta 1 variants in chronic beryllium disease and sarcoidosis. J. Immunol. 2007;179:4255. doi: 10.4049/jimmunol.179.6.4255. [DOI] [PubMed] [Google Scholar]
- 4.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009;10:704. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu G, Friggeri A, Yang Y, et al. MiR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J. Exp. Med. 2010;207:1589. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ezzie ME, Crawford M, Cho J, et al. Gene expression networks in COPD: microRNA and mRNA regulation. Thorax. 2011 doi: 10.1136/thoraxjnl-2011-200089. (PMID: 21940491) [DOI] [PubMed] [Google Scholar]
- 7.Crouser ED, Culver DA, Knox KS, et al. Gene expression profiling identifies MMP-12 and ADAMDEC1 as potential pathogenic mediators of pulmonary sarcoidosis. Am. J. Respir. Crit. Care Med. 2009;179:929. doi: 10.1164/rccm.200803-490OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenbaum JT, Pasadhika S, Crouser EC, et al. Hypothesis: sarcoidosis is a STAT1-mediated disease. Clin. Immunol. 2009;132:174. doi: 10.1016/j.clim.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koth LL, Soldberg OD, Peng JC, Bhakta NR, Nguyen CP, Woodruff PG. Sarcoidosis blood transcriptome reflects lung inflammation and overlaps with tuberculosis. Am. J. Respir. Crit. Care Med. 2011 doi: 10.1164/rccm.201106-1143OC. (PMID: 21852540) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Labbé E, Lock L, Letamendia A, et al. Transcriptional cooperation between the transforming growth factor-beta and Wnt pathways in mammary and intestinal tumorigenesis. Cancer Res. 2007;67:75. doi: 10.1158/0008-5472.CAN-06-2559. [DOI] [PubMed] [Google Scholar]
- 12.American Thoracic Society Statement on Sarcoidosis. Am. J. Respir. Crit. Care Med. 1999;160:736. doi: 10.1164/ajrccm.160.2.ats4-99. [DOI] [PubMed] [Google Scholar]
- 13.Auer H, Liyanarachchi S, Newsom D, Klisovic MI, Marcucci G, Kornacker K. Chipping away at the chip bias: RNA degradation in microarray analysis. Nat. Genet. 2003;35:292. doi: 10.1038/ng1203-292. [DOI] [PubMed] [Google Scholar]
- 14.Liu CG, Calin GA, Volinia S, Croce CM. MicroRNA expression profiling using microarrays. Nat. Protoc. 2008;3:563. doi: 10.1038/nprot.2008.14. [DOI] [PubMed] [Google Scholar]
- 15.Irizarry R, Hobbs B, Collin F, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 16.Smyth GK. Linear models and empirical Bayes methods for assessing differentially expression in microarray experiments. Stat. Appl. Genet. Molec. Biol. 2004;3:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 17.Sartor AM, Tomlinson RC, Wesselkamper CS, Sivaganesan S, Leikauf DG, Medvedovic M. Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC Bioinformatics. 2006;7:358. doi: 10.1186/1471-2105-7-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordon A, Glazko G, Qiu X, Yakovlev A. Control of the mean number of false discoveries, Bonferroni and stability of multiple testing. Ann. Appl. Stat. 2007;1:179. [Google Scholar]
- 19.Johnson RA, Wichern DW. Applied multivariate statistical analysis. fourth ed. New Jersey: Prentice Hall; 2000. [Google Scholar]
- 20.Papadopoulos GL, Alexious P, Maraqkakis M, Reczko M, Hatzigeorgiou AG. DIANA-mirPath: Integrating human and mouse microRNAs in pathways. Bioinformatics. 2009;25:1991. doi: 10.1093/bioinformatics/btp299. [DOI] [PubMed] [Google Scholar]
- 21.Hunninghake GW, Gadek JE, Young RC, Kawanami O, Ferrans VJ, Crystal RG. Maintenance of granuloma formation in pulmonary sarcoidosis by T lymphocytes within the lung. N. Engl. J. Med. 1980;302:594. doi: 10.1056/NEJM198003133021102. [DOI] [PubMed] [Google Scholar]
- 22.Pera MF, Tam PP. Extrinsic regulation of pluripotent stem cells. Nature. 2010;465:713. doi: 10.1038/nature09228. [DOI] [PubMed] [Google Scholar]
- 23.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao C, Rajewsky R. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 25.Hassan MQ, Gordon JA, Beloti MM, et al. A network connecting Runx2, SATB2 and the miR-23a~27a~24-2 cluster regulates the osteoblastic differentiation program. Proc. Natl. Acad. Sci. USA. 2010;107:19879. doi: 10.1073/pnas.1007698107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wynn TA. Cellular and molecular mechanisms of fibrosis. J. Pathol. 2008;214:199. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flavell RA, Sanjabi S, Wrzesinski SH, Licona-Limon P. The polarization of immune cells in the tumour environment by TGFbeta. Nat. Rev. Immunol. 2010;10:554. doi: 10.1038/nri2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toossi Z, Gogate P, Shiratsuchi H, Young T, Ellner JJ. Enhanced production of TGF-beta by blood monocytes from patients with active tuberculosis and presence of TGF-beta in tuberculous granulomatous lung lesions. J. Immunol. 1995;154:465. [PubMed] [Google Scholar]
- 29.Limper AH, Colby TV, Sanders MS, Asakura S, Roche PC, DeRemee RA. Immunohistochemical localization of transforming growth factor-beta 1 in the nonnecrotizing granulomas of pulmonary sarcoidosis. Am. J. Respir. Crit. Care Med. 1994;149:197. doi: 10.1164/ajrccm.149.1.8111583. [DOI] [PubMed] [Google Scholar]
- 30.Zissel G, Homolka J, Schlaak J, Schlaak M, Müller-Quernheim J. Anti-inflammatory cytokine release by alveolar macrophages in pulmonary sarcoidosis. Am. J. Respir. Crit. Care Med. 1996;154:713. doi: 10.1164/ajrccm.154.3.8810610. [DOI] [PubMed] [Google Scholar]
- 31.Pabst S, Fränken T, Schönau J, et al. Transforming growth factor-{beta} gene polymorphisms in different phenotypes of sarcoidosis. Eur. Respir. J. 2011;38:169. doi: 10.1183/09031936.00120410. [DOI] [PubMed] [Google Scholar]
- 32.Kruit A, Grutters JC, Ruven HJ, et al. Transforming growth factor-beta gene polymorphisms in sarcoidosis patients with and without fibrosis. Chest. 2006;129:1584. doi: 10.1378/chest.129.6.1584. [DOI] [PubMed] [Google Scholar]
- 33.Licona-Limón P, Soldevila G G. The role of TGF-β superfamily during T cell development: new insights. Immunol. Letters. 2007;109:1. doi: 10.1016/j.imlet.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 34.Facco M, Cabrelle A, Teramo A, et al. Sarcoidosis is a Th1/Th17 multisystem disorder. Thorax. 2011;66:144. doi: 10.1136/thx.2010.140319. [DOI] [PubMed] [Google Scholar]
- 35.Wang K, Zhang S, Marzolf B, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc. Natl. Acad. Sci. USA. 2009;106:4402. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staal FJ, Luis TC, Tiemessen MM. WNT signaling in the immune system: WNT is spreading its wings. Nat. Rev. Immunol. 2008;8:581. doi: 10.1038/nri2360. [DOI] [PubMed] [Google Scholar]
- 37.Blumenthal A, Ehlers S, Lauber J, et al. The Wingless homolog WNT5A and its receptor Frizzled-5 regulate inflammatory responses of human mononuclear cells induced by microbial stimulation. Blood. 2006;108:965. doi: 10.1182/blood-2005-12-5046. [DOI] [PubMed] [Google Scholar]
- 38.Neumann J, Schaale K, Farhat K, et al. Frizzled1 is a marker of inflammatory macrophages, and its ligand Wnt3a is involved in reprogramming Mycobacterium tuberculosis-infected macrophages. FASEB J. 2010;24:4599. doi: 10.1096/fj.10-160994. [DOI] [PubMed] [Google Scholar]
- 39.Kleinhenz ME, Ellner JJ, Spagnuolo PJ, Daniel TM. Suppression of lymphocyte responses by tuberculous plasma and mycobacterial arabinogalactan. Monocyte dependence and indomethacin reversibility. J. Clin. Invest. 1981;68:153. doi: 10.1172/JCI110231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyara M, Amoura Z, Parizot C, et al. The immune paradox of sarcoidosis and regulatory T cells. J. Exp. Med. 2006;203:359. doi: 10.1084/jem.20050648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hudspith BN, Flint KC, Geraint-James D, Brostoff J, Johnson NM. Lack of immune deficiency in sarcoidosis: compartmentalisation of the immune response. Thorax. 1987;42:250. doi: 10.1136/thx.42.4.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levanen B, Wheelock AM, Eklund A, Grunewald J, Nord M. Increased pulmonary Wnt (wingless/integrated)-signaling in patients with sarcoidosis. Respir. Med. 2011;105:282. doi: 10.1016/j.rmed.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 43.Pandit KV, Milosevic J, Kaminski N N. MicroRNAs in idiopathic pulmonary fibrosis. Transl. Res. 2011;157:191. doi: 10.1016/j.trsl.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 44.Pandit KV, Corcoran D, Yousef H, et al. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care. Med. 2010;182:220. doi: 10.1164/rccm.200911-1698OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benes V, Castoldi M. Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available. Methods. 2010;50:244. doi: 10.1016/j.ymeth.2010.01.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.