Abstract
Ribonucleotide reductase (RNR) supplies cellular deoxyribonucleotide triphosphates (dNTP) pools by converting ribonucleotides to the corresponding deoxy forms using radical-based chemistry. Eukaryotic RNR comprises α and β subunits: α contains the catalytic and allosteric sites; β houses a diferric-tyrosyl radical cofactor (FeIII2-Y•) that is required to initiates nucleotide reduction in α. Cells have evolved multi-layered mechanisms to regulate RNR level and activity in order to maintain the adequate sizes and ratios of their dNTP pools to ensure high-fidelity DNA replication and repair. The central role of RNR in nucleotide metabolism also makes it a proven target of chemotherapeutics. In this review, we discuss recent progress in understanding the function and regulation of eukaryotic RNRs, with a focus on studies revealing the cellular machineries involved in RNR metallocofactor biosynthesis and its implication in RNR-targeting therapeutics.
Keywords: ribonucleotide reductase (RNR), diferric-tyrosyl radical (FeIII2-Y•), iron homeostasis
Introduction: RNR enzymes
Ribonucleotide reductases (RNRs) catalyze the reduction of ribonucleoside 5′-di- or triphosphates (NDPs or NTPs) to deoxyribonucleoside 5′-di- or triphosphates (dNDPs or dNTPs), the only pathway for de novo synthesis of the monomeric building blocks for both DNA replication and DNA repair (Reichard, 1993; Stubbe et al., 2001). All cellular organisms that synthesize their own DNA require RNR. Some viruses even carry their own copy of RNR in their genomes, likely to facilitate viral replication in the infected host cells (Boehmer and Lehman, 1997; Kurita and Nakajima, 2012). All RNRs share a common catalytic mechanism in which a metal co factor is involved in oxidation of a conserved cysteine within the active site to a thiyl radical to initiate nucleotide reduction (Stubbe and van Der Donk, 1998; Hofer et al., 2012).
RNRs are grouped into three main classes based on their different metallocofactor requirements. Class I RNRs are oxygen-dependent enzymes that occur in both eukaryotes and eubacteria and contain a dinuclear metal cluster (Fe or Mn) (Bollinger et al., 2008; Cotruvo and Stubbe, 2011). Class II RNRs, found in both aerobic and anaerobic microbes, are oxygen-independent and use a cobalt-containing cobalamin (vitamin B12) cofactor (Eklund et al., 2001; Kolberg et al., 2004). Class III RNRs, found in anaerobic microorganisms, are oxygen-sensitive anduse a [4Fe-4S]1 +/2+ cluster coupled to S-adenosylmethionine (SAM) for catalytic activity (Fontecave et al., 2002; Logan et al., 2003).
Class I RNRs are further grouped into three subclasses according to their metal cluster identities. All class I RNRs share the same structural fold consisting of two subunits, α and β. α binds the substrates NDPs/NTPs, while β houses the dinuclear metal cluster required for catalysis (Reichard, 1993; Mowa et al., 2009; Cotruvo and Stubbe, 2011). In class Ia RNRs it is a diferric-tyrosyl radical (FeIII2-Y•) (Sjöberg and Reichard, 1977), in class Ib a dimanganese-tyrosylradical (MnIII2-Y•) (Willing et al., 1988; Cotruvo and Stubbe, 2010), and in class Ic a proposed MnIVFeIII cofactor (Jiang et al., 2007). While all three subclasses of class I RNRs occur in prokaryotes, most eukaryotic genomes from yeast to human only encode class Ia RNRs (Nordlund and Reichard, 2006; Cotruvo and Stubbe, 2011). This review will discuss recent studies in understanding the mechanisms underlying regulation of the eukaryotic class Ia RNRs, especially the biosynthesis of the metallocofactor in β.
The active form of class Ia RNR holoenzymes has been proposed to have a quaternary structure of (α2)n(β2)m (n = 2, 4, 6, m = 1 or 3, depending on the organisms) (Kashlan and Cooperman, 2003; Rofougaran et al., 2006; Wang et al., 2007; Aye et al., 2012a; Minnihan et al., 2013a). In addition to the active site, α also contains allosteric sites for controlling overall activity and substrate specificity (Jordan and Reichard, 1998; Eklund et al., 2001). During each turnover cycle, a transient thiyl radical is generated within the active site of α by oxidation of a conserved cysteine residue. This oxidation occurs by the Y• in β over a long distance (~35Å), via a specific proton-coupled election transfer pathway involving conserved aromatic residues in both α and β subunits (Reece et al., 2006; Minnihan et al., 2013b).
Although the prototype class Ia RNR comprises homo-dimeric α2 and β2 subunits, some eukaryotic genomes encode multiple α and β isoforms, adding another layer of complexity for configurations of the RNR holoenzyme. The human genome encodes one a (RRM1) and two βs (RRM2 and RRM2B or p53R2) (Tanaka et al., 2000). The budding yeast Saccharomyces cerevisiae has two α isoforms (encoded by RNR1 and RNR3) and two β isoforms (encoded by RNR2 and RNR4) (Elledge et al., 1993; Huang and Elledge, 1997).
RNR regulation, dNTP pools, and genomic integrity
The central role of RNR in controlling cellular dNTP pool highlights its importance in the maintenance of genome stability. Both the sizes and the relative ratios of cellular dNTP pools are critical for optimal DNA replication and repair. Abnormally elevated or imbalanced dNTP pools can lead to increased mutagenesis by enhancing mis-incorporation and inhibiting proofreading of DNA polymerases (Kunkel et al., 1982; Loeb and Kunkel, 1982; Reichard, 1988). Imbalance in dNTP pools can cause mutations, DNA breaks, and cell death (Ke et al., 2005; Song et al., 2005; Mathews, 2006; Kumar et al., 2010; Saldivar et al., 2012).
The regulation of RNRs is complicated and multifaceted, involving allosteric control, RNR subunit levels as well as subcellular localization. Allosteric regulation allows RNR to maintain an adequate and balanced supply of all four dNTPs and to adapt quickly to perturbation in cellular dNTP pools. The α subunit has two separate allosteric sites, S site and A site, which bind specific NTPs and dNTPs as effectors. The S site regulates specificity for each of the four substrates through binding of ATP, dATP, dTTP or dGTP. The A site controls overall enzyme activity through binding of ATP and dATP, with ATP binding activates, whereas dATP inhibits activity (Kashlan and Cooperman, 2003; Nordlund and Reichard, 2006).
The RNR subunit protein levels are tightly regulated both by the cell cycle and the DNA damage checkpoint In proliferating cells, the dNTP pools increase 5- to 10-fold as cells transit from G0/G1 to S phase, which is mainly achieved by increased RNR levels at the G1-to-S transition point (Mathews, 2006; Nordlund and Reichard, 2006). DNA replication occurs during the S phase and requires the continued synthesis of all four dNTPs. The RRM1 and RRM2 genes in mammals and plants are both maximally transcribed during S phase through the activity of the E2F family transcription factors (Björklund et al., 1990; Chabes et al., 2003b; Chabouté et al., 2000, 2002). In addition, RRM2 is also subjected to ubiquitin mediated pioteasomal degradation once cells completed DNA replication and/or repair, through two E3 ubiquitin ligase complexes, the Skp1/Cullin/F-box (SCF) and the anaphase-promoting complex (APC) (Chabes et al., 2003b; D’Angiolella et al., 2012). In budding yeast, transcription of the RNR genes peaks at the beginning of the S phase through regulation of the transcription factor pairs Mbp1/Swi6 and Swi4/Swi6 (Elledge et al., 1993; Sanvisens et al., 2013). In response to DNA damage, mammalian cells increase transcription of p53R2 (RRM2B) to facilitate DNA repair (Nakano et al., 2000; Tanaka et al., 2000) via activation of the ATM/ATR-CHK checkpoint pathway (Harper and Elledge, 2007). Similarly, DNA damage and replication blockage in budding yeast increase transcription of the RNR genes through activation of the Mec1-Rad53-Dun1 damage checkpoint kinase cascade. The activated checkpoint also removes two negative regulators of RNR in yeast cells, the Rnr1 inhibitor Sml1 and the Rnr2 nuclear transport facilitator Dif1 (Zhao et al., 2001; Lee et al., 2008; Wu and Huang, 2008), leading to a 6- to 8-fold increase of dNTP pool sizes (Chabes et al., 2003a; Chabes and Thelander, 2003).
Another mode of RNR regulation involves subcellular compartmentalization of the α and β subunits. In both the budding yeast and the fission yeast Schizosaccharomyces pombe, β is sequestered in the nucleus during most part of the cell cycle except for the S phase or in response to genotoxic stress, during which β relocalizes to the cytoplasm where a resides to allow holoenzyme formation (Liu et al., 2003; Yao et al., 2003). The nucleus-to-cytoplasm redistribution of β is mediated by the DNA damage checkpoint, which degrades Dif1 and Spd1, two small size proteins involved in nuclear import of β in S. cerevisiae and S. pombe, respectively. Regulation of RNR subunit subcellular localization has also been reported in mammalian and plant cells (Xue et al., 2003; Halimi et al., 2011). Interestingly, RRM1 and RRM2 have recently been shown to accumulate at sites of DNA damage in mammalian cells, presumably to facilitate optimal localized dNTP supplies for repair (Niida et al., 2010; Hu et al., 2012).
The RNR metallocofactor: biosynthesis and maintenance
An important and yet much less understood layer of RNR regulation, given the fact that level of the FeIII2-Y• is directly correlated with nucleotide reduction activity of the holoenzyme (Ortigosa et al., 2006; Zhang et al., 2011), involves the assembly and maintenance of its metallocofactor. Early studies from the 1970s demonstrated that the FeIII2-Y• cluster in class Ia RNR can be self-assembled in vitro with apo-β2, FeII, O2, and reductant (Eq. (1)) (Atkin et al., 1973). The mechanism of self-assembly has since been elucidated by various rapid kinetics studies with wild-type and specifically engineered mutant β2 proteins (Bollinger et al., 1991; Ormö et al., 1992; Salowe et al., 1993; Bollinger et al., 1995). A simplified model of RNR cofactor assembly is shown in Fig. 1 based on studies of the self-assembly process in vitro. The FeIII2-Y• cofactor assembly process involves delivery of FeII into β, reaction of the resulting FeII2-β2 with O2 to form a diferric peroxide, and a series of intermediate steps that eventually require a one-electron reductant leading to oxidization of a conserved tyrosine, located near the di-iron center within β, to generate a tyrosyl radical Y• that can be measured by electron paramagnetic resonance (EPR) (Sjöberg and Reichard, 1977; Stubbe, 2003; Stubbe and Cotruvo, 2011). In vitro FeII serves both as the metal ligand and the reductant that provide the source of reducing equivalent for RNR cluster assembly. However, FeII is unlikely the election donor in vivo because it is highly reactive and has to be chaperoned inside the cell. The steps of FeII loading and election delivery are also poorly understood because of the inability to control these processes in vitro. The lack of control and general inefficiency of in vitro self-assembly argue for the importance of a cellular biosynthesis pathway for controlled cofactor formation.
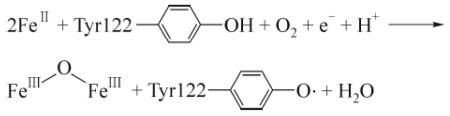 |
(1) |
Figure 1.
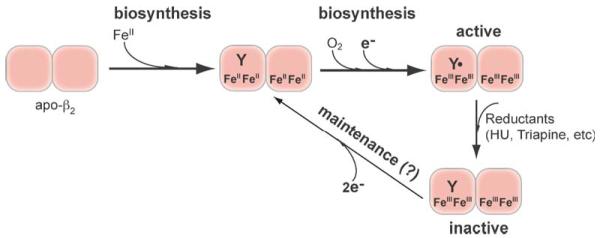
The proposed pathways for biosynthesis and maintenance of the FeIII2-Y• cofactor of class Ia RNR. The biosynthetic pathway requires delivery of two FeII and a reducing equivalent (electron) per β subunit to carry out the four-electron reduction of O2 to H2O (Eq. (1)). The other three electrons come from the two FeII and Tyr residue to form the FeIII2-Y•. The maintenance pathway may use the same source of reducing equivalents to convert the inactive FeIII2-Y cluster to FeII2-Y, which subsequently forms FeIII2-Y• in the presence of O2 via the biosynthetic pathway.
The FeIII2-Y• cofactor of eukaryotic RNR is inherently unstable, which raises the issue of cofactor repair, or namely, maintenance pathway. The half-life of the E. coli β2 Y• is four days, whereas that in S. cerevisiae β2 is several hours, the human β2 25 min and the mouse β2 10 min (Atkin et al., 1973; Thelander et al., 1983; Ortigosa et al., 2006; Hristova et al., 2008). The half-life of the RNR Y• is much shorter than the duration of the S phase of the cell cycle in mammalian cells. In addition, the RNR Y• is also susceptible to one electron reduction by endogenous and exogenous reductants such as hydroxyurea, resulting an inactive β2 (met-β2 with diferrictyrosine, i.e. FeIII2-Y). As such, there is a continual need for regeneration of the RNR Y• during S phase to meet the demand of dNTP production for genome replication during S phase (Barlow et al., 1983; Thelander et al., 1983). To reactivate β2 to produce deoxynucleotides, the FeIII2-Y• cofactor would need to be assembled either de novo (biosynthesis pathway in Fig. 1) from apo-β2 or from half-way from met-β2 (maintenance pathway in Fig. 1). It is conceivable that a cofactor maintenance/repair process may play an important role for in vivo RNR regulation by modulating the levels of Y• /β2 and consequently the nucleotide reduction activity.
The mechanism of RNR cofactor maintenance is under active investigation. The current model for the E. coli class Ia RNR maintenance pathway is that FeIII2-Y in met-β2 can be reactivated by reduction of diferric (FeIII2) to diferious (FeII2) form, which can subsequently form the active-β2 through the biosynthetic pathway (Wu et al., 2007; Hristova et al., 2008). Early studies have demonstrated that the reduction of FeIII2 to FeII2 could be achieved by a feriedoxin reductase Fie, NADPH, flavin (FAD or FMN), DTT, and a “Fraction B,” the components of which remain to be defined (Coves et al., 1997; Fontecave et al., 1987, 1989). Recently, the [2Fe-2S]-ferredoxin protein YfaE has been shown to be involved in FeIII2-Y• cofactor maintenance in the E. coli class Ia RNR (Wu et al., 2007). It is unclear whether eukaryotic class Ia RNR employs similar cellular machinery for cofactor biosynthesis and maintenance. A problem with this notion is that the eukaryotic ferredoxin and ferredoxin reductase are primarily localized in the mitochondria (Lill et al., 2012).
RNR metallocofactor assembly: sources of iron and electron
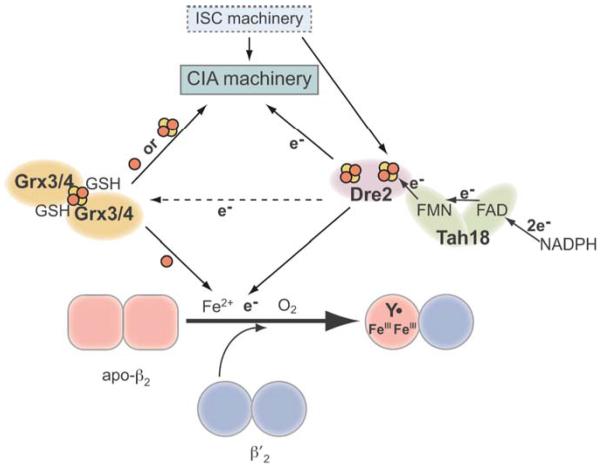
The two key issues of in vivo RNR cluster assembly are the sources of iron and electron. Iron is an essential element for all eukaryotic organisms because iron cofactors, such as heme, iron-sulfur cluster, and di-iron clusters, are required for the activity of many cellular enzymes involved in a wide range of central cellular pathways including oxidative phosphorylation, ribosomal biogenesis, and DNA replication and repair (Rouault and Klausner, 1997; Kaplan et al., 2006; Ye and Rouault, 2010; White and Dillingham, 2012;). The facile ability of iron to gain and lose electrons allows it to participate in various oxidation-reduction reactions but on the other hand can lead to iron overload toxicity at higher levels (Rouault and Klausner, 1997). As such, cells have evolved sophisticated mechanisms for tightly regulated iron uptake, intracellular iron trafficking, and delivery of iron to specific proteins. The nature of bioavailable iron, namely the “labile iron pool,” has been under debate since the 1960s (Pollycove and Maqsood, 1962). Recent studies have shown that two evolutionary conserved cytosolic monothiol glutaredoxins, Grx3/Grx4, are required to provide bioavailable iron to assembly of many iron-containing cofactors such as heme, Fe-S, and the di-iron cluster in RNR (Mühlenhoff et al., 2010; Zhang et al., 2011; Haunhorst et al., 2013). The monothiol glutaredoxins form a [2Fe-2S] bridged and GSH-liganded dimer, which mediates intracellular iron trafficking and utilization (Mühlenhoff et al., 2010). Cells depleted of Grx3/4 exhibit deficiencies in most iron-requiring reactions including the RNR nucleotide reduction activity, although it is unclear how Grx3/4 directly or indirectly participate in iron delivery into diverse iron-requiring proteins.
As for the source of the obligatory electron for RNR cofactor assembly, recent studies of the S. cerevisiae RNR suggest that the Fe-S cluster-containing protein Dre2 may act as the electron donor (Zhang et al., 2011). Dre2 and its binding partner, the diflavin NADPH oxidoreductase Tah18, (Netz et al., 2010) are conserved from yeast to human; their mammalian counterparts are CIAPIN1 (or anamorsin) and NDOR1, respectively (Shibayama et al., 2004; Hao et al., 2005; Pujol-Carrion et al., 2006; Zhang et al., 2008). The two proteins constitute an electron transfer chain that transfers electrons from NADPH via its FAD and FMN to die Fe-S clusters of Dre2 (Netz et al., 2010). The cytosolic iron-sulfur cluster assembly (CIA) pathway, which is unique to eukaryotes and is for maturation of all cytosolic and nuclear Fe-S proteins, also require the Dre2-Tah18 complex (Netz et al., 2010), although the electron-requiring step requiring Dre2-Tah18 is yet to be identified. It would thus appear that the Dre2-Tah18 complex functions as the cytosolic equivalent of the mitochondrial ferredoxin-ferredoxin reductase pair to deliver electrons to multiple and divergent biosynthesis pathways of iron-containing cofactors. The findings that Grx3/4 and Dre2-Tah18 are required for RNR and the CIA pathway raise the intriguing perspective that biosynthesis of two apparently quite different iron cofactors, the diferric-Y• in RNR and the Fe-S cluster in nuclear and cytoplasmic proteins, share the same sources of iron and electrons (Fig. 2). It is conceivable that additional cytosolic and nuclear iron-requiring proteins also utilize these conserved protein complexes for cofactor maturation. In addition, physical and genetic interactions have been observed between Grx3 and Dre2 in yeast and mammalian cells consistent with a model that Dre2-Tah18 may also be involved in Grx3/4 Fe-S cluster maturation (Saito et al., 2011; Zhang et al., 2011).A mechanistic understanding of the contribution of Grx3/4 and Dre2-Tah18 to RNR cluster assembly would entail combination of genetic dissection and biochemical reconstitution studies.
Figure 2.
A model depicting the role of the Dre2-Tah18 complex as the electron donor for cluster assembly in both RNR and in cytosolic and nuclear Fe-S cluster containing proteins. Grx3/4 is the source of iron for all cellular iron-requiring pathways including Fe-S cluster assembly in mitochondria (ISC) and cytosol (CIA), and FeIII2-Y• assembly in RNR (Mühlenhoff et al., 2010). Electrons from NADPH are transferred via FAD and FMN, the two flavin cofactors in Tah18, to the Fe-S cluster(s) in Dre2, which subsequently deliver the electrons to proteins in the CIA pathway and β in RNR. Dre2-Tah18 may also provide reducing equivalents to facilitate iron release from the [2Fe2S]-GSH2 cluster in the Grx3/Grx4 dimer (Zhang et al., 2011).
Targeting the RNR metallocofactor in cancer treatment
The hallmark of cancer cells is uncontrolled proliferation (Hanahan and Weinberg, 2011). Cancer cells have heightened metabolic load and demand for energy and dNTP pools to replicate DNA (Cairns et al., 2011). RNR is a critical target in chemotherapy and increased RNR level and activity is associated with tumor growth, metastasis, and resistance to RNR-targeting drugs (Yen et al., 1994; Liu et al., 2007; Souglakos et al., 2008; Shao et al., 2013).
Although the human RRM1 (α) and RRM2 (β) are both required for the RNR holoenzyme activity, the two subunits exhibit different effect on cancer cells. In general, increased RRM2 expression is associated with enhanced invasive and metastatic potential and drug resistance of tumor cells, whereas RRM2 knockdown reverses drug resistance and suppresses proliferation (Fan et al., 1998; Souglakos et al., 2008; Zuckerman et al., 2011). In contrast, the role of RRM1 in cancer is more complex. Increased RRM1 expression is observed in many types of cancer. However, RRM1 overexpressionis often associated with reduced transformation and suppression of tumorigenicity and metastasis (Shao et al., 2013). The differences may reflect the potentially opposing effects of RNR-dependent dNTP biosynthesis on cancer cells at different stages of tumorigenesis. Improperly increased dNTP pools can reduce DNA repair fidelity and cause genomic instability. On the other hand, augmented dNTP pools may also enhance DNA repair capacity and survival of cancer cells, which are faced with increased replicational stress (Hanahan and Weinberg, 2011; Burrell et al., 2013).
RNR has been an important target in cancer treatment since the 1970s because of its central role in nucleotide metabolism (Brockman et al., 1970). Both small-molecule inhibitors and antisense RNA therapy approaches have been successfully employed in RNR inhibition, which can enhance sensitivity to other chemotherapeutics and radiation therapy. Chemical inhibitors targeting the α or β subunit are the primary clinical strategy for RNR inhibition, α inhibitors are nucleoside analogs that inactivate enzyme activity through interaction with the substrate binding site or allosteric sites, whereas β inhibitors inactivate the metallocofactor (Cerqueira et al., 2007; Shao et al., 2013). In addition, the α–β interaction can be also targeted by using oligopeptides with similar sequences to the β C terminus, which interacts with a (Yang et al., 1990; Climent et al., 1991). Here, we will focus on several β metallocofactor targeting RNR inhibitors that are in clinical application or trials. These β inhibitors act by quenching the tyiosyl free radical (Hydroxyurea), depleting intracellular iron (Desferoxamine), or disrupting the di-iron center (Triapine).
Hydroxyurea is a simple small-molecule radical scavenger that has been used in cancer therapy for over five decades (Fishbein and Carbone, 1963). It inactivates RNR by directly reducing the tyrosyl radical via one-electron transfer from hydroxyurea to Y• and causing structural perturbation of the di-iron center of β (Lassmann et al., 1992; Offenbacher et al., 2009). Hydroxyurea treatment depletes cellular dNTP pools leading to stalled replication forks and activation of the DNA damage checkpoint (Bianchi et al., 1986; Allen et al., 1994; Davies et al., 2009). Hydroxyurea has been widely used for treatment of acute and chronic myeloid leukemias (AML and CML), head and neck cancers, and brain tumors (Donehower, 1992; Kennedy, 1992; Madaan et al., 2012). Furthermore, the synergism of hydroxyurea with nucleoside analogs didanosine (DDI) and 3′-azido-3′-deoxythymidine (AZT) has shown effectiveness for treatment of HIV-1 infectionin clinical trials, likely due to their effect on alteration of the cellular dNTP pools (Johns and Gao, 1998; Mayhew et al., 2005). Although β is a major target of hydroxyurea, other metalloenzymes can also be inhibited by hydroxyurea (Scozzafava and Supuran, 2003; Campestre et al., 2006). In addition to its promiscuity, the effectiveness of hydroxyurea is limited by its low affinity for RNR short half-life and the development of resistance due to β overexpression (Yen et al., 1994; Gwilt and Tracewell, 1998; Ren et al., 2002).
Desferoxamine is a bacterial siderophore that is used clinically as an iron chelator in treatment of iron over-load disorders and acute iron poisoning. It acts by binding free iron in the bloodstream and facilitating iron removal (Miyajima et al., 1997). Desferoxamine has antiproliferative effect both in vitro and in vivo, which is mediated by its depleting effect on intracellular iron pools (Dayani et al., 2004). Since iron is an essential component of the RNR FeIII2-Y• cofactor, depletion of intracellular iron pool would impair RNR cofactor biosynthesis. In mammalian cells, iron deprivation results in decreased RNR activity, leading to cell growth arrest (Nyholm et al., 1993). It has been shown that desferoxamine inhibits RNR activity through chelating intracellular iron pools and preventing formation of the RNR FeIII2-Y• cofactor, which can be reversed by addition of iron (Chaston et al., 2003). Since iron is a cofactor of many enzymes in addition to RNR, the effect of desferoxamine treatment is pleiotropic. The clinical efficacy of desferoxamine is also limited by its poor membrane permeability and a very short half-life in the bloodstream (Chaston and Richardson, 2003; Lovejoy and Richardson, 2003). This limitation has encouraged the development of more effective iron chelators with novel properties, one of which is Triapine.
Triapine (3-AP) is a potent β inhibitor that is currently in phase II clinical trials for treating CML and various solid tumors (Finch et al., 1999; Kunos et al., 2012; Zeidner et al., 2013). It belongs to the heterocyclic carboxaldehyde thiosemicabazone family of iron chelators, which exhibit wide-spectmm antitumor activity and cytotoxicity (Buss et al., 2003; Yu et al., 2006). Studies to date suggest that the cytotoxicity of 3-AP involved multiple mechanisms. 3-AP chelates iron by forming complexes with both FeII and FeIII (Enyedy et al., 2011). The FeII-(3-AP) can react with O2 to produce reactive oxygen species (ROS) (Richardson et al., 2009). Based on these properties of 3-AP, several models have been proposed for the mechanism of β inhibition by 3-AP. One model suggests that 3-AP chelate FeIII directly from the β FeIII2-Y• cofactor leading to RNR inactivation. Another model proposes that 3-AP chelates iron from the intracellular iron pool (s), thus interfering with the β cofactor biosynthesis. A third model posits that FeIII-(3-AP) resulted from FeIII chelation is reduced to FeII-(3-AP) by endogenous reductants, which subsequently leads to ROS in the presence of O2 causing β inactivation an DNA degradation. Recent biochemical studies have revealed that the loss of the Y• signal, measured by EPR, occurs independently of O2 and ROS and always precedes the loss of iron when β2 is incubated with 3-AP (Aye et al., 2012b). These results indicate that the observed rapid inhibition of β activity by 3-AP results from reduction of Y• by FeII-(3-AP), although the source of the FeII-(3-AP) remains to be determined (Aye et al., 2012b). 3-AP can inhibit both RRM2 and p53R2 of the mammalian RNR, whereas hydroxyurea is ineffective at inhibiting p53R2 (Shao et al., 2004; Yu et al., 2006). The FeII-(3-AP) complex has been reported to be more effective at inhibiting RNR than free 3-AP (Shao et al., 2006), consistent with the notion of Y• reduction by FeII-(3-AP) as the main mechanism of rapid RNR inhibition by 3-AP.
Conclusions
RNR is uniquely positioned as the rate-limiting step in de novo dNTP biosynthesis required for DNA replication and repair and hence an important therapeutic target. RNR level and activity is tightly regulated both during the cell cycle progression in proliferating cells and in response to genotoxic stress in all cell types. The molecular details of RNR regulation, which involves transcription, protein degradation and subcellular localization, have emerged from both biochemical and genetic studies in model organisms and cell cultures. A much less understood mode of RNR regulation is at the level of its metallocofactor assembly, a process that requires sources of bioavailable iron and reducing equivalents. Recent studies in S. cerevisiae have identified the monothiol glutaiedxoins Grx3/4 and the Fe-S cluster containing protein Dre2 as potential sources of iron and electrons, respectively, in RNR cofactor assembly. Delineation of the steps involved in RNR cofactor biosynthesis demands future biochemical and genetics studies. The inherent instability of the Y• of eukaryotic RNR suggests the existence of a maintenance pathway that sustains RNR activity to provide adequate dNTP pools for completion of DNA replication during each cell cycle. Several classes of RNR inhibitors targeting different components of the holoenzyme have been in clinical use and trials. A detailed mechanistic understanding of RNR cofactor assembly may provide critical insights into future development and investigation of RNR-targeting therapeutics with higher specificity and selectivity.
Acknowledgements
This work was supported by National Institutes of Health (CA125574 and GM81393).
Footnotes
Compliance with ethics guidelines The authors declare no conflict of interest.
This article does not contain any studies with human or animal as subjects performed by any of the authors.
References
- Allen JB, Zhou Z, Siede W, Friedberg EC, Elledge SJ. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8(20):2401–2415. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- Atkin CL, Thelander L, Reichard P, Lang G. Iron and free radical in ribonucleotide reductase. Exchange of iron and Mössbauer spectroscopy of the protein B2 subunit of the Escherichia coli enzyme. J Biol Chem. 1973;248(21):7464–7472. [PubMed] [Google Scholar]
- Aye Y, Brignole EJ, Long MJ, Chittuluiu J, Drennan CL, Asturias FJ, Stubbe J. Clofarabine targets the large subunit (α) of human ribonucleotide reductase in live cells by assembly into persistent hexamers. Chem Biol. 2012a;19(7):799–805. doi: 10.1016/j.chembiol.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aye Y, Long MJ, Stubbe J. Mechanistic studies of semicarbazone triapine targeting human ribonucleotide reductase in vitro and in mammalian cells: tyrosyl radical quenching not involving reactive oxygen species. J Biol Chem. 2012b;287(42):35768–35778. doi: 10.1074/jbc.M112.396911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow T, Eliasson R, Platz A, Reichard P, Sjöberg BM. Enzymic modification of a tyrosine residue to a stable free radical in ribonucleotide reductase. Proc Natl Acad Sci USA. 1983;80(6):1492–1495. doi: 10.1073/pnas.80.6.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi V, Pontis E, Reichard P. Changes of deoxyribonucleoside triphosphate pools induced by hydroxyurea and their relation to DNA synthesis. J Biol Chem. 1986;261(34):16037–16042. [PubMed] [Google Scholar]
- Björklund S, Skog S, Tribukait B, Thelander L. S-phase-specific expression of mammalian ribonucleotide reductase R1 and R2 subunit mRNAs. Biochemistry. 1990;29(23):5452–5458. doi: 10.1021/bi00475a007. [DOI] [PubMed] [Google Scholar]
- Boehmer PE, Lehman IR. Herpes simplex virus DNA replication. Annu Rev Biochem. 1997;66(1):347–384. doi: 10.1146/annurev.biochem.66.1.347. [DOI] [PubMed] [Google Scholar]
- Bollinger JM, Jr, Edmondson DE, Huynh BH, Filley J, Norton JR, Stubbe J. Mechanism of assembly of the tyrosyl radical-dinuclear iron cluster cofactor of ribonucleotide reductase. Science. 1991;253(5017):292–298. doi: 10.1126/science.1650033. [DOI] [PubMed] [Google Scholar]
- Bollinger JM, Jr, Jiang W, Green MT, Krebs C. The manganese (IV)/iron(III) cofactor of Chlamydia trachomatis ribonucleotide reductase: structure, assembly, radical initiation, and evolution. Cutr Opin Struct Biol. 2008;18(6):650–657. doi: 10.1016/j.sbi.2008.11.007. [DOI] [PubMed] [Google Scholar]
- Bollinger JM, Jr, Tong WH, Ravi N, Huynh BH, Edmondson DE, Stubbe JA. Use of rapid kinetics methods to study the assembly of the diferric-tyrosyl radical cofactor of E. coli ribonucleotide reductase. Methods Enzymol. 1995;258:278–303. doi: 10.1016/0076-6879(95)58052-2. [DOI] [PubMed] [Google Scholar]
- Brockman RW, Shaddix S, Laster WR, Jr, Schabel FM., Jr Inhibition of ribonucleotide reductase, DNA synthesis, and L1210 leukemia by guanazole. Cancer Res. 1970;30(9):2358–2368. [PubMed] [Google Scholar]
- Burrell RA, McClelland SE, Endesfelder D, Groth P, Weller MC, Shaikh N, Domingo E, Kanu N, Dewhurst SM, Gronroos E, Chew SK, Rowan AJ, Schenk A, Sheffer M, Howell M, Kschischo M, Behrens A, Helleday T, Bartek J, Tomlinson IP, Swanton C. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494(7438):492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss JL, Torti FM, Torti SV. The role of iron chelation in cancer therapy. Curr Med Chem. 2003;10(12):1021–1034. doi: 10.2174/0929867033457638. [DOI] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Campestre C, Agamennone M, Tortorella P, Preziuso S, Biasone A, Gavuzzo E, Pochetti G, Mazza F, Hiller O, Tschesche H, Consalvi V, Gallina C. N-Hydroxyurea as zinc binding group in matrix metalloproteinase inhibition: mode of binding in a complex with MMP-8. Bioorg Med Chem Lett. 2006;16(1):20–24. doi: 10.1016/j.bmcl.2005.09.057. [DOI] [PubMed] [Google Scholar]
- Cerqueira NM, Fernandes PA, Ramos MJ. Ribonucleotide reductase: a critical enzyme for cancer chemotherapy and antiviral agents. Recent Patents Anticancer Drug Discov. 2007;2(1):11–29. doi: 10.2174/157489207779561408. [DOI] [PubMed] [Google Scholar]
- Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 2003a;112(3):391–401. doi: 10.1016/s0092-8674(03)00075-8. [DOI] [PubMed] [Google Scholar]
- Chabes A, Thelander L. DNA building blocks at the foundation of better survival. Cell Cycle. 2003;2(3):171–173. doi: 10.4161/cc.2.3.354. [DOI] [PubMed] [Google Scholar]
- Chabes AL, Pfleger CM, Kirschner MW, Thelander L. Mouse ribonucleotide reductase R2 protein: a new target for anaphase-promoting complex-Cdh1-mediated proteolysis. Proc Natl Acad Sci USA. 2003b;100(7):3925–3929. doi: 10.1073/pnas.0330774100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabouté ME, Clément B, Philipps G. S phase and meristem-specific expression of the tobacco RNR1b gene is mediated by an E2F element located in the 5′ leader sequence. J Biol Chem. 2002;277(20):17845–17851. doi: 10.1074/jbc.M200959200. [DOI] [PubMed] [Google Scholar]
- Chabouté ME, Clément B, Sekine M, Philipps G, Chaubet-Gigot N. Cell cycle regulation of the tobacco ribonucleotide reductase small subunit gene is mediated by E2F-like elements. Plant Cell. 2000;12(10):1987–2000. [PMC free article] [PubMed] [Google Scholar]
- Chaston TB, Lovejoy DB, Watts RN, Richardson DR. Examination of the antiproliferative activity of iron chelators: multiple cellular targets and the different mechanism of action of triapine compared with desferrioxamine and the potent pyridoxal isonicotinoyl hydrazone analogue 311. Clin Cancer Res. 2003;9(1):402–414. [PubMed] [Google Scholar]
- Chaston TB, Richardson DR. Iron chelators for the treatment of iron overload disease: relationship between structure, redox activity, and toxicity. Am J Hematol. 2003;73(3):200–210. doi: 10.1002/ajh.10348. [DOI] [PubMed] [Google Scholar]
- Climent I, Sjöberg BM, Huang CY. Carboxyl-terminal peptides as probes for Escherichia coli ribonucleotide reductase subunit interaction: kinetic analysis of inhibition studies. Biochemistry. 1991;30(21):5164–5171. doi: 10.1021/bi00235a008. [DOI] [PubMed] [Google Scholar]
- Cotruvo JA, Jr, Stubbe J. An active dimanganese(III)-tyrosyl radical cofactor in Escherichia coli class Ib ribonucleotide reductase. Biochemistry. 2010;49(6):1297–1309. doi: 10.1021/bi902106n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotruvo JA, Jr, Stubbe J. Class I ribonucleotide reductases: metallocofactor assembly and repair in vitro and in vivo. Annu Rev Biochem. 2011;80(1):733–767. doi: 10.1146/annurev-biochem-061408-095817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coves J, Zeghouf M, Macherel D, Guigliarelli B, Asso M, Fontecave M. Flavin mononucleotide-binding domain of the flavoprotein component of the sulfite reductase from Escherichia coli. Biochemistry. 1997;36(19):5921–5928. doi: 10.1021/bi9623744. [DOI] [PubMed] [Google Scholar]
- D’Angiolella V, Donato V, Forrester FM, Jeong YT, Pellacani C, Kudo Y, Saraf A, Florens L, Washburn MP, Pagano M. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell. 2012;149(5):1023–1034. doi: 10.1016/j.cell.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BW, Kohanski MA, Simmons LA, Winkler JA, Collins JJ, Walker GC. Hydroxyurea induces hydroxyl radical-mediated cell death in Escherichia coli. Mol Cell. 2009;36(5):845–860. doi: 10.1016/j.molcel.2009.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayani PN, Bishop MC, Black K, Zeltzer PM. Desferoxamine (DFO)—mediated iron chelation: rationale for a novel approach to therapy for brain cancer. J Neurooncol. 2004;67(3):367–377. doi: 10.1023/b:neon.0000024238.21349.37. [DOI] [PubMed] [Google Scholar]
- Donehower RC. An overview of the clinical experience with hydroxyurea. Semin Oncol. 1992;19(3 Suppl 9):11–19. [PubMed] [Google Scholar]
- Eklund H, Uhlin U, Fämegårdh M, Logan DT, Nordlund P. Structure and function of the radical enzyme ribonucleotide reductase. Prog Biophys Mol Biol. 2001;77(3):177–268. doi: 10.1016/s0079-6107(01)00014-1. [DOI] [PubMed] [Google Scholar]
- Elledge SJ, Zhou Z, Allen JB, Navas TA. DNA damage and cell cycle regulation of ribonucleotide reductase. BioEssays. 1993;15:333–339. doi: 10.1002/bies.950150507. [DOI] [PubMed] [Google Scholar]
- Enyedy EA, Primik MF, Kowol CR, Arion VB, Kiss T, Keppler BK. Interaction of Triapine and related thiosemicarbazones with iron(III)/(II) and gallium(III): a comparative solution equilibrium study. Dalton Trans. 2011;40(22):5895–5905. doi: 10.1039/c0dt01835j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Villegas C, Huang A, Wright JA. The mammalian ribonucleotide reductase R2 component cooperates with a variety of oncogenes in mechanisms of cellular transformation. Cancer Res. 1998;58(8):1650–1653. [PubMed] [Google Scholar]
- Finch RA, Liu MC, Cory AH, Cory JG, Sartorelli AC. Triapine (3-aminopyridine-2-carboxaldehyde thiosemicarbazone; 3-AP): an inhibitor of ribonucleotide reductase with antineoplastic activity. Adv Enzyme Regul. 1999;39(1):3–12. doi: 10.1016/s0065-2571(98)00017-x. [DOI] [PubMed] [Google Scholar]
- Fishbein WN, Carbone PP. Hydroxyurea: Mechanism of Action. Science. 1963;142(3595):1069–1070. doi: 10.1126/science.142.3595.1069. [DOI] [PubMed] [Google Scholar]
- Fontecave M, Eliasson R, Reichard P. NAD(P)H:flavin oxidoreductase of Escherichia coli. A ferric iron reductase participating in the generation of the free radical of ribonucleotide reductase. J Biol Chem. 1987;262(25):12325–12331. [PubMed] [Google Scholar]
- Fontecave M, Eliasson R, Reichard P. Enzymatic regulation of the radical content of the small subunit of Escherichia coli ribonucleotide reductase involving reduction of its redox centers. J Biol Chem. 1989;264(16):9164–9170. [PubMed] [Google Scholar]
- Fontecave M, Mulliez E, Logan DT. Deoxyribonucleotide synthesis in anaerobic microorganisms: the class III ribonucleotide reductase. Prog Nucleic Acid Res Mol Biol. 2002;72:95–127. doi: 10.1016/s0079-6603(02)72068-0. [DOI] [PubMed] [Google Scholar]
- Gwilt PR, Tracewell WG. Pharmacokinetics and pharmacodynamics of hydroxyurea. Clin Pharmacokinet. 1998;34(5):347–358. doi: 10.2165/00003088-199834050-00002. [DOI] [PubMed] [Google Scholar]
- Halimi Y, Dessau M, Poliak S, Ast T, Erez T, Livnat-Levanon N, Kamiol B, Hirsch JA, Chamovitz DA. COP9 signalosome subunit 7 from Arabidopsis interacts with and regulates the small subunit of ribonucleotide reductase (RNR2) Plant Mol Biol. 2011;77(1-2):77–89. doi: 10.1007/s11103-011-9795-8. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hao Z, Qiao T, Jin X, Li X, Gao J, Fan D. Preparation and characterization of a specific monoclonal antibody against CIAPIN1. Hybridoma (Larchmt) 2005;24(3):141–145. doi: 10.1089/hyb.2005.24.141. [DOI] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol CeU. 2007;28(5):739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Haunhorst P, Hanschmann EM, Bräutigam L, Stehling O, Hoffinann B, Mühlenholf U, Lill R, Bemdt C, Lillig CH. Crucial function of vertebrate ghitaredoxin 3 (PICOT) in iron homeostasis and hemoglobin maturation. Mol Biol Cell. 2013;24(12):1895–1903. doi: 10.1091/mbc.E12-09-0648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer A, Crona M, Logan DT, Sjöberg BM. DNA building blocks: keeping control of manufacture. Crit Rev Biochem Mol Biol. 2012;47(1):50–63. doi: 10.3109/10409238.2011.630372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hristova D, Wu CH, Jiang W, Krebs C, Stubbe J. Importance of the maintenance pathway in the regulation of the activity of Escherichia coli ribonucleotide reductase. Biochemistry. 2008;47(13):3989–3999. doi: 10.1021/bi702408k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CM, Yeh MT, Tsao N, Chen CW, Gao QZ, Chang CY, Lee MH, Fang JM, Sheu SY, Lin CJ, Tseng MC, Chen YJ, Chang ZF. Tumor cells require thymidylate kinase to prevent dUTP incorporation during DNA repair. Cancer Cell. 2012;22(1):36–50. doi: 10.1016/j.ccr.2012.04.038. [DOI] [PubMed] [Google Scholar]
- Huang M, Elledge SJ. Identification of RNR4, encoding a second essential small subunit of ribonucleotide reductase in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17(10):6105–6113. doi: 10.1128/mcb.17.10.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Yun D, Saleh L, Ban EW, Xing G, Hoffart LM, Maslak MA, Krebs C, Bollinger JM., Jr A manganese(IV)/iron(III) cofactor in Chlamydia trachomatis ribonucleotide reductase. Science. 2007;316(5828):1188–1191. doi: 10.1126/science.1141179. [DOI] [PubMed] [Google Scholar]
- Johns DG, Gao WY. Selective depletion of DNA precursors: an evolving strategy for potentiation of dideoxynucleoside activity against human immunodeficiency virus. Biochem Pharmacol. 1998;55(10):1551–1556. doi: 10.1016/s0006-2952(97)00664-3. [DOI] [PubMed] [Google Scholar]
- Jordan A, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 1998;67(1):71–98. doi: 10.1146/annurev.biochem.67.1.71. [DOI] [PubMed] [Google Scholar]
- Kaplan J, McVey Ward D, Crisp RJ, Philpott CC. Iron-dependent metabolic remodeling in S. cerevisiae. Biochim Biophys Acta. 2006;1763(7):646–651. doi: 10.1016/j.bbamcr.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Kashlan OB, Cooperman BS. Comprehensive model for allosteric regulation of mammalian ribonucleotide reductase: refinements and consequences. Biochemistry. 2003;42(6):1696–1706. doi: 10.1021/bi020634d. [DOI] [PubMed] [Google Scholar]
- Ke PY, Kuo YY, Hu CM, Chang ZF. Control of dTTP pool size by anaphase promoting complex/cyclosome is essential for the maintenance of genetic stability. Genes Dev. 2005;19(16):1920–1933. doi: 10.1101/gad.1322905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BJ. The evolution of hydroxyurea therapy in chronic myelogenous leukemia. Semin Oncol. 1992;19(3 Suppl 9):21–26. [PubMed] [Google Scholar]
- Kolberg M, Strand KR, Graff P, Andersson KK. Structure, fonction, and mechanism of ribonucleotide reductases. Biochim Biophys Acta. 2004;1699(1-2):1–34. doi: 10.1016/j.bbapap.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Kumar D, Viberg J, Nilsson AK, Chabes A. Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res. 2010;38(12):3975–3983. doi: 10.1093/nar/gkq128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Silber JR, Loeb LA. The mutagenic effect of deoxynucleotide substrate imbalances during DNA synthesis with mammalian DNA polymerases. Mutat Res. 1982;94(2):413–419. doi: 10.1016/0027-5107(82)90304-9. [DOI] [PubMed] [Google Scholar]
- Kunos C, Radivoyevitch T, Abdul-Karim FW, Fanning J, Abulafia O, Bonebrake AJ, Usha L. Ribonucleotide reductase inhibition restores platinum-sensitivity in platinum-resistant ovarian cancer: a Gynecologic Oncology Group Study. J Transl Med. 2012;10(1):79. doi: 10.1186/1479-5876-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita J, Nakajima K. Megalocytiviruses. Viruses. 2012;4(4):521–538. doi: 10.3390/v4040521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann G, Thelander L, Gräslund A. EPR stopped-flow studies of the reaction of the tyrosyl radical of protein R2 from ribonucleotide reductase with hydroxyurea. Biochem Biophys Res Commun. 1992;188(2):879–887. doi: 10.1016/0006-291x(92)91138-g. [DOI] [PubMed] [Google Scholar]
- Lee YD, Wang J, Stubbe J, Elledge SJ. Dif1 is a DNA-damage-regulated facilitator of nuclear import for ribonucleotide reductase. Mol CeU. 2008;32(1):70–80. doi: 10.1016/j.molcel.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R, Hoffmann B, Molik S, Pierik AJ, Rietzschel N, Stehling O, Uzarska MA, Webert H, Wilbrecht C, Mühlenhoff U. The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism. Biochim Biophys Acta. 2012;1823(9):1491–1508. doi: 10.1016/j.bbamcr.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Liu C, Powell KA, Mundt K, Wu L, Carr AM, Caspari T. Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and-independent mechanisms. Genes Dev. 2003;17(9):1130–1140. doi: 10.1101/gad.1090803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhou B, Xue L, Yen F, Chu P, Un F, Yen Y. Ribonucleotide reductase subunits M2 and p53R2 are potential biomarkers for metastasis of colon cancer. Clin Colorectal Cancer. 2007;6(5):374–381. doi: 10.3816/CCC.2007.n.007. [DOI] [PubMed] [Google Scholar]
- Loeb LA, Kunkel TA. Fidelity of DNA synthesis. Annu Rev Biochem. 1982;51(1):429–157. doi: 10.1146/annurev.bi.51.070182.002241. [DOI] [PubMed] [Google Scholar]
- Logan DT, Mulliez E, Larsson KM, Bodevin S, Atta M, Garnaud PE, Sjoberg BM, Fontecave M. A metal-binding site in the catalytic subunit of anaerobic ribonucleotide reductase. Proc Natl Acad Sci USA. 2003;100(7):3826–3831. doi: 10.1073/pnas.0736456100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy DB, Richardson DR. Iron chelators as anti-neoplastic agents: current developments and promise of the PIH class of chelators. Curr Med Chem. 2003;10(12):1035–1049. doi: 10.2174/0929867033457557. [DOI] [PubMed] [Google Scholar]
- Madaan K, Kaushik D, Verma T. Hydroxyurea: a key player in cancer chemotherapy. Expert Rev Anticancer Ther. 2012;12(1):19–29. doi: 10.1586/era.11.175. [DOI] [PubMed] [Google Scholar]
- Mathews CK. DNA precursor metabolism and genomic stability. FASEB J. 2006;20:1300–1314. doi: 10.1096/fj.06-5730rev. [DOI] [PubMed] [Google Scholar]
- Mayhew CN, Sumpter R, Inayat M, Cibull M, Phillips JD, Elford HL, Gallicchio VS. Combination of inhibitors of lymphocyte activation (hydroxyurea, trimidox, and didox) and reverse transcriptase (didanosine) suppresses development of murine retrovirus-induced lymphoproliferative disease. Antiviral Res. 2005;65(1):13–22. doi: 10.1016/j.antiviral.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Minnihan EC, Ando N, Brignole EJ, Olshansky L, Chittuluru J, Asturias FJ, Drennan CL, Nocera DG, Stubbe J. Generation of a stable, aminotyrosyl radical-induced α2β2 complex of Escherichia coli class Ia ribonucleotide reductase. Proc Natl Acad Sci USA. 2013a;110(10):3835–3840. doi: 10.1073/pnas.1220691110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnihan EC, Nocera DG, Stubbe J. Reversible, long-range radical transfer in E. coli class Ia ribonucleotide reductase. Acc Chem Res. 2013b;46(11):2524–2535. doi: 10.1021/ar4000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima H, Takahashi Y, Kamata T, Shimizu H, Sakai N, Gitlin JD. Use of desferrioxamine in the treatment of aceruloplasminemia. Ann Neurol. 1997;41(3):404–407. doi: 10.1002/ana.410410318. [DOI] [PubMed] [Google Scholar]
- Mowa MB, Warner DF, Kaplan G, Kana BD, Mizrahi V. Function and regulation of class I ribonucleotide reductase-encoding genes in mycobacteria. J Bacteriol. 2009;191(3):985–995. doi: 10.1128/JB.01409-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlenhoff U, Molik S, Godoy JR, Uzarska MA, Richter N, Seubert A, Zhang Y, Stubbe J, Pierrel F, Herrero E, Lillig CH, Lill R. Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab. 2010;12(4):373–385. doi: 10.1016/j.cmet.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Bálint E, Ashcroft M, Vousden KH. A ribonucleotide reductase gene is a transcriptional target of p53 and p73. Oncogene. 2000;19(37):4283–4289. doi: 10.1038/sj.onc.1203774. [DOI] [PubMed] [Google Scholar]
- Netz DJ, Stümpfig M, Doré C, Mühlenhoff U, Pierik AJ, Lill R. Tah18 transfers electrons to Dre2 in cytosolic iron-sulfur protein biogenesis. Nat Chem Biol. 2010;6(10):758–765. doi: 10.1038/nchembio.432. [DOI] [PubMed] [Google Scholar]
- Niida H, Katsuno Y, Sengoku M, Shimada M, Yukawa M, Ikura M, Ikura T, Kohno K, Shima H, Suzuki H, Tashiro S, Nakanishi M. Essential role of Tip60-dependent recruitment of ribonucleotide reductase at DNA damage sites in DNA repair during G1 phase. Genes Dev. 2010;24(4):333–338. doi: 10.1101/gad.1863810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75(1):681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- Nyholm S, Thelander L, Gräslund A. Reduction and loss of the iron center in the reaction of the small subunit of mouse ribonucleotide reductase with hydroxyurea. Biochemistry. 1993;32(43):11569–11574. doi: 10.1021/bi00094a013. [DOI] [PubMed] [Google Scholar]
- Offenbacher AR, Vassiliev IR, Seyedsayamdost MR, Stubbe J, Barry BA. Redox-linked structural changes in ribonucleotide reductase. J Am Chem Soc. 2009;131(22):7496–7497. doi: 10.1021/ja901908j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormö M, deMaré F, Regnström K, Abeig A, Sahlin M, Ling J, Loehr TM, Sanders-Loehr J, Sjöberg BM. Engineering ofthe iron site in ribonucleotide reductase to a self-hydroxylating monooxygenase. J Biol Chem. 1992;267(13):8711–8714. [PubMed] [Google Scholar]
- Ortigosa AD, Hristova D, Perlstein DL, Zhang Z, Huang M, Stubbe J. Determination of the in vivo stoichiometiy of tyrosyl radical per betabeta’ in Saccharomyces cerevisiae ribonucleotide reductase. Biochemistry. 2006;45(40):12282–12294. doi: 10.1021/bi0610404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollycove M, Maqsood M. Existence of an erythropoietic labile iron pool in animals. Nature. 1962;194(4824):152–154. doi: 10.1038/194152a0. [DOI] [PubMed] [Google Scholar]
- Pujol-Carrion N, Belli G, Herrero E, Nogues A, de Ia Torre-Ruiz MA. Glutaredoxins Grx3 and Grx4 regulate nuclear localisation of Aft1 and the oxidative stress response in Saccharomyces cerevisiae. J Cell Sci. 2006;119(Pt 21):4554–564. doi: 10.1242/jcs.03229. [DOI] [PubMed] [Google Scholar]
- Reece SY, Hodgkiss JM, Stubbe J, Nocera DG. Proton-coupled electron transfer: the mechanistic underpinning for radical transport and catalysis in biology. Philos Trans R Soc Lond B Biol Sci. 2006;361(1472):1351–1364. doi: 10.1098/rstb.2006.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichard P. Interactions between deoxyribonucleotide and DNA synthesis. Annu Rev Biochem. 1988;57(1):349–374. doi: 10.1146/annurev.bi.57.070188.002025. [DOI] [PubMed] [Google Scholar]
- Reichard P. From RNA to DNA, why so many ribonucleotide reductases? Science. 1993;260(5115):1773–1777. doi: 10.1126/science.8511586. [DOI] [PubMed] [Google Scholar]
- Ren S, Wang R, Komatsu K, Bonaz-Krause P, Zyrianov Y, McKenna CE, Csipke C, Tokes ZA, Lien EJ. Synthesis, biological evaluation, and quantitative structure-activity relationship analysis of new Schiff bases of hydroxysemicarbazide as potential antitumor agents. J Med Chem. 2002;45(2):410–419. doi: 10.1021/jm010252q. [DOI] [PubMed] [Google Scholar]
- Richardson DR, Kalinowski DS, Richardson V, Sharpe PC, Lovejoy DB, Islam M, Bernhardt PV. 2-Acetylpyridine thiosemicaiba-zones are potent iron chelators and antiproliferative agents: redox activity, iron complexation and characterization of their antitumor activity. J Med Chem. 2009;52(5):1459–1470. doi: 10.1021/jm801585u. [DOI] [PubMed] [Google Scholar]
- Rofougaran R, Vodnala M, Hofer A. Enzymatically active mammalian ribonucleotide reductase exists primarily as an alpha6-beta2 octamer. J Biol Chem. 2006;281(38):27705–27711. doi: 10.1074/jbc.M605573200. [DOI] [PubMed] [Google Scholar]
- Rouault T, Klausner R. Regulation of iron metabolism in eukaryotes. Curr Top Cell Regul. 1997;35:1–19. doi: 10.1016/s0070-2137(97)80001-5. [DOI] [PubMed] [Google Scholar]
- Saito Y, Shibayama H, Tanaka H, Tanimura A, Matsumura I, Kanakura Y. PICOT is a molecule which binds to anamorein. Biochem Biophys Res Commun. 2011;408(2):329–333. doi: 10.1016/j.bbrc.2011.04.033. [DOI] [PubMed] [Google Scholar]
- Saldivar JC, Miuma S, Bene J, Hosseini SA, Shibata H, Sun J, Wheeler LJ, Mathews CK, Huebner K. Initiation of genome instability and preneoplastic processes through loss of Fhit expressioa. PLoS Genet. 2012;8(11):el003077. doi: 10.1371/journal.pgen.1003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salowe S, Bollinger JM, Jr, Ator M, Stubbe J, McCraken J, Peisach J, Samano MC, Robins MJ. Alternative model for mechanism-based inhibition of Escherichia coli ribonucleotide reductase by 2′-azido-2′-deoxyuridine 5′-diphosphate. Biochemistry. 1993;32(47):12749–12760. doi: 10.1021/bi00210a026. [DOI] [PubMed] [Google Scholar]
- Sanvisens N, de Llanos R, Puig S. Function and regulation of yeast ribonucleotide reductase: cell cycle, genotoxic stress, and iron bioavailability. Biomed J. 2013;36:51–58. doi: 10.4103/2319-4170.110398. [DOI] [PubMed] [Google Scholar]
- Scozzafava A, Supuran CT. Hydroxyurea is a carbonic anhydrase inhibitor. Bioorg Med Chem. 2003;11(10):2241–2246. doi: 10.1016/s0968-0896(03)00112-3. [DOI] [PubMed] [Google Scholar]
- Shao J, Liu X, Zhu L, Yen Y. Targeting ribonucleotide reductase for cancer therapy. Expert Opin Ther Targets. 2013;17(12):1423–1437. doi: 10.1517/14728222.2013.840293. [DOI] [PubMed] [Google Scholar]
- Shao J, Zhou B, Di Bilio AJ, Zhu L, Wang T, Qi C, Shih J, Yen Y. A Ferrous-Triapine complex mediates formation of reactive oxygen species that inactivate human ribonucleotide reductase. Mol Cancer Ther. 2006;5(3):586–592. doi: 10.1158/1535-7163.MCT-05-0384. [DOI] [PubMed] [Google Scholar]
- Shao J, Zhou B, Zhu L, Qiu W, Yuan YC, Xi B, Yen Y. In vitro characterization of enzymatic properties and inhibition of the p53R2 subunit of human ribonucleotide reductase. Cancer Res. 2004;64(1):1–6. doi: 10.1158/0008-5472.can-03-3048. [DOI] [PubMed] [Google Scholar]
- Shibayama H, Takai E, Matsumura I, Kouno M, Morii E, Kitamura Y, Takeda J, Kanakura Y. Identification of a cytokine-induced antiapoptotic molecule anamorsin essential for definitive hematopoi-esis. J Exp Med. 2004;199(4):581–592. doi: 10.1084/jem.20031858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöberg BM, Reichard P. Nature of the free radical in ribonucleotide reductase from Escherichia coli. J Biol Chem. 1977;252(2):536–541. [PubMed] [Google Scholar]
- Song S, Pursell ZF, Copeland WC, Longley MJ, Kunkel TA, Mathews CK. DNA precursor asymmetries in mammalian tissue mitochondria and possible contribution to mutagenesis through reduced replication fidelity. Proc Natl Acad Sci USA. 2005;102(14):49904995. doi: 10.1073/pnas.0500253102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souglakos J, Boukovinas I, Taron M, Mendez P, Mavroudis D, Tripaki M, Hatzidaki D, Koutsopoulos A, Stathopoulos E, Georgoulias V, Rosell R. Ribonucleotide reductase subunits M1 and M2 mRNA expression levels and clinical outcome of lung adenocarcinoma patients treated with docetaxel/gemc itabine. Br J Cancer. 2008;98(10):1710–1715. doi: 10.1038/sj.bjc.6604344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbe J. Di-iron-tyrosyl radical ribonucleotide reductases. Curr Opin Chem Biol. 2003;7(2):183–188. doi: 10.1016/s1367-5931(03)00025-5. [DOI] [PubMed] [Google Scholar]
- Stubbe J, Cotruvo JA., Jr Control of metallation and active cofactor assembly in the class Ia and Ib ribonucleotide reductases: diiron or dimanganese? Curr Opin Chem Biol. 2011;15(2):284–290. doi: 10.1016/j.cbpa.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbe J, Ge J, Yee CS. The evolution of ribonucleotide reduction revisited. Trends Biochem Sci. 2001;26(2):93–99. doi: 10.1016/s0968-0004(00)01764-3. [DOI] [PubMed] [Google Scholar]
- Stubbe J, van Der Donk WA. Protein Radicals in Enzyme Catalysis. Chem Rev. 1998;98(2):705–762. doi: 10.1021/cr9400875. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404(6773):42–9. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- Thelander L, Gräslund A, Thelander M. Continual presence of oxygen and iron required for mammalian ribonucleotide reduction: possible regulation mechanism. Biochem Biophys Res Commun. 1983;110(3):859–865. doi: 10.1016/0006-291x(83)91040-9. [DOI] [PubMed] [Google Scholar]
- Wang J, Lohman GJ, Stubbe J. Enhanced subunit interactions with gemcitabine-5′-diphosphate inhibit ribonucleotide reductases. Proc Natl Acad Sci USA. 2007;104(36):14324–14329. doi: 10.1073/pnas.0706803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MF, Dillingham MS. Iron-sulphur clusters in nucleic acid processing enzymes. Curr Opin Struct Biol. 2012;22(1):94–100. doi: 10.1016/j.sbi.2011.11.004. [DOI] [PubMed] [Google Scholar]
- Willing A, Follmann H, Auling G. Ribonucleotide reductase of Brevibacterium ammoniagenes is a manganese enzyme. Eur J Bio. 1988;170:603–611. doi: 10.1111/j.1432-1033.1988.tb13740.x. [DOI] [PubMed] [Google Scholar]
- Wu CH, Jiang W, Krebs C, Stubbe J. YfaE, a ferredoxin involved in diferric-tyrosyl radical maintenance in Escherichia coli ribonucleotide reductase. Biochemistry. 2007;46(41):11577–11588. doi: 10.1021/bi7012454. [DOI] [PubMed] [Google Scholar]
- Wu X, Huang M. Dif1 controls subcellular localization of ribonucleotide reductase by mediating nuclear import of the R2 subunit. Mol Cell Biol. 2008;28(23):7156–7167. doi: 10.1128/MCB.01388-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L, Zhou B, Liu X, Qiu W, Tin Z, Yen Y. Wild-type p53 regulates human ribonucleotide reductase by protein-protein interaction with p53R2 as well as hRRM2 subunits. Cancer Res. 2003;63(5):980–986. [PubMed] [Google Scholar]
- Yang FD, Spanevello RA, Celiker I, Hirschmann R, Rubin H, Cooperman BS. The carboxyl terminus heptapeptide of the R2 subunit of mammalian ribonucleotide reductase inhibits enzyme activity and can be used to purify the R1 subunit. FEBS Lett. 1990;272(1-2):61–64. doi: 10.1016/0014-5793(90)80449-s. [DOI] [PubMed] [Google Scholar]
- Yao R, Zhang Z, An X, Bucci B, Perlstein DL, Stubbe J, Huang M. Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways. Proc Natl Acad Sci USA. 2003;100(11):6628–6633. doi: 10.1073/pnas.1131932100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Rouault TA. Human iron-sulfur cluster assembly, cellular iron homeostasis, and disease. Biochemistry. 2010;49(24):4945–4956. doi: 10.1021/bi1004798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen Y, Grill SP, Dutschman GE, Chang CN, Zhou BS, Cheng YC. Characterization of a hydroxyurea-resistant human KB cell line with supersensitivity to 6-thioguanine. Cancer Res. 1994;54(14):3686–3691. [PubMed] [Google Scholar]
- Yu Y, Wong J, Lovejoy DB, Kalinowski DS, Richardson DR. Chelators at the cancer coalface: desferrioxamine to Triapine and beyond. Clin Cancer Res. 2006;12(23):6876–6883. doi: 10.1158/1078-0432.CCR-06-1954. [DOI] [PubMed] [Google Scholar]
- Zeidner JF, Karp JE, Blackford AL, Smith BD, Gojo I, Gore SD, Levis MJ, Carraway HE, Greer JM, Ivy SP, Pratz KW, McDevitt MA. Phase II trial of sequential ribonucleotide reductase inhibition in aggressive myeloproliferative neoplasms. Haematologica. 2013 doi: 10.3324/haematol.2013.097246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu L, Wu X, An X, Stubbe J, Huang M. Investigation of in vivo diferric tyrosyl radical formation in Saccharomyces cerevisiae Rnr2 protein: requirement of Rnr4 and contribution of Grx3/4 AND Dre2 proteins. J Biol Chem. 2011;286(48):41499–41509. doi: 10.1074/jbc.M111.294074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lyver ER, Nakamaru-Ogiso E, Yoon H, Amutha B, Lee DW, Bi E, Ohnishi T, Daldal F, Pain D, Dancis A. Dre2, a conserved eukaryotic Fe/S cluster protein, functions in cytosolic Fe/S protein biogenesis. Mol Cell Biol. 2008;28(18):5569–5582. doi: 10.1128/MCB.00642-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 2001;20(13):3544–3553. doi: 10.1093/emboj/20.13.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerman JE, Hsueh T, Koya RC, Davis ME, Ribas A. siRNA knockdown of ribonucleotide reductase inhibits melanoma cell line proliferation alone or synergistically with temozolomide. J Invest Dermatol. 2011;131(2):453–460. doi: 10.1038/jid.2010.310. [DOI] [PubMed] [Google Scholar]