Abstract
Examination of gene functions in specific tumor types improves insight in tumorigenesis and helps design better treatments. Due to the rarity of histiocytic/dendritic cell sarcoma in humans, it is difficult to accrue such knowledge. Therefore, comparative research of these cancers in predisposed dog breeds, such as the Flatcoated retriever, can be of value. Histiocytic sarcoma in the dog can be grouped into a soft tissue- and visceral form. The soft tissue form at first is localized, while the visceral form progresses more quickly to a terminal state, which might be related to variations in gene expression. Microarray analyses were performed on fresh-frozen tissue from Flatcoated retrievers with either soft tissue- or visceral histiocytic sarcoma. Expression differences of ten most significantly differentially expressed genes were validated with quantitative real-time PCR (q PCR) analyses. Q PCR analyses confirmed the significantly aberrant expression of three of the selected genes: C6 was up-regulated; CLEC12A and CCL5 were down-regulated in the visceral histiocytic sarcoma compared to the soft tissue form. The findings of our study indicate that these two forms of histiocytic sarcoma in the dog display a variation in gene expression and warrant analysis of functional changes in the expression of those genes in these rare sarcomas in man.
Introduction
Histiocytic malignancies include dendritic cell- and histiocytic sarcomas and disseminated Langerhans cell histiocytosis [1]. In humans, the frequency of this group of diseases is very low [2]. This hampers an evidence based therapeutic approach for instance based on selective inhibition of specific signal transduction pathways. Canine histiocytic sarcoma (HS) resemble the human histiocytic malignancies [3]–[6]. As for such human cancers, canine HS involves the proliferation of members of both histiocytic lineages; dendritic cells and macrophages [5], [6]. The dog is genetically closer related to man than mice [7], [8] and the study of genetic changes of spontaneous cancers in the dog therefore has high comparative value [3], [9], in particular for cancers that are rare in the human such as histiocytic malignancies [10], [11]. Research could lead to more insight in the pathogenesis of this disease and could facilitate the identification of therapeutic targets valuable for both species [4], [7], [12]–[15].
In the dog, the limited genetic flow within breeds is responsible for specific breed traits including disease predispositions. However, the disease predispositions resulting from selective inbreeding can be studied for the benefit of affected breeds as well as for humans in which the rarity of diseases such as histiocytic malignancies hampers scientific progress. The Flatcoated retriever (FCR) has a strongly increased risk for HS development [16], [17]. In dogs [3], [4], [16] and humans [10], [11], histiocytic malignancies are grave conditions though the prognosis varies between subtypes [1]. HS has two common subtypes. Localized, soft tissue histiocytic sarcoma (STHS), manifests itself as a tumor arising in the deeply seated soft tissues of limbs, often in association with joints. In this form, chemotherapy as an adjunct to tumor resection can improve survival in some dogs that suffer from STHS [18]. The prognosis for the second form; visceral histiocytic sarcoma (VHS), which is a multifocal and disseminated form that is manifested in internal organs, uniformly is very poor [3], [16], [17], [19], [20]. There is presently no immunohistochemical method available to definitively differentiate between STHS and VHS and some have stated that VHS and STHS represent two different stages along a continuum of the same disease [5], [21].
Our previous research of HS in Flatcoated retrievers has provided evidence that the expression of nine common genes is altered when comparing both STHS and VHS with normal spleen; indicating a common ground for the general development of HS [15]. As a next step we compared the two forms of HS with one another. In this additional study we provide evidence that, in addition to common changes in STHS and VHS compared to healthy tissues described previously, there are some marked genetic differences between these two common forms of histiocytic malignancies.
Materials and Methods
The experimental protocol (ID 2007.III.08.110) was peer-reviewed by the scientific committee of the Department of Animals in Science & Society, Utrecht University, The Netherlands, and approved by the Animal Experiments Committee of the Academic Biomedical Centre, Utrecht, The Netherlands. The Animal Experiments Committee based its decision on ‘De Wet op de Dierproeven’ (The Dutch ‘Experiments on Animals Act’, 1996) and on the ‘Dierproevenbesluit’ (the Dutch ‘animal experiments decree’, 1996). Both documents are available online at http://wetten.overheid.nl.
Case recruitment and histopathological evaluation
As mentioned in the earlier study [15], all tumor samples were confirmed as being spontaneously occurring histiocytic malignancies obtained from (previously untreated) Dutch family-owned FCR. All samples were obtained with informed owner consent. Case selection and obtainment of tissues were similar as mentioned in the previous study. As a common reference pool a multitude of canine organs (testis, liver, spleen, prostate, duodenum, lung, kidney and brain) from healthy crossbreeds (n = 8) was used [15].
Patient details are listed in Table 1.
Table 1. Patient details.
| Name | Sex | Pathology | AO (yrs) | Site(s) | Microarray/PCR |
| D1, TJ | MN | STHS | 7.7 | shoulder | Y/Y |
| D3, UH | FN | STHS | 8 | shoulder | Y/Y |
| D4, BS | MN | STHS | 6.5 | elbow | Y/Y |
| D5, BaS | M | STHS | 7.6 | knee | Y/Y |
| D6, TV | MN | STHS | 8.1 | elbow | Y/Y |
| D7, DV | MN | STHS | 11 | shoulder | Y/Y |
| DX, YM | MN | STHS | 9.7 | shoulder | N/Y |
| D2, BE | M | VHS | 9.4 | liver/spleen/lnn abd | Y/Y |
| D8, DW | M | VHS | 9.5 | spleen/lnn abd | Y/N |
| D9, JV | FN | VHS | 8.9 | lung/lnn mediast | Y/Y |
| D11, BT | F | VHS | 8.5 | lung | Y/Y |
| D12, AG | M | VHS | 7.3 | lung/spleen/kidney | Y/Y |
| D13, TR | FN | VHS | 7.9 | lung | Y/Y |
| D14, SG | F | VHS | 4.1 | lung/lnn mediast | Y/Y |
| DX, SC | MG | VHS | 10 | liver/spleen | N/Y |
AO: Age of onset, M: male, MN: male neutered, F: female, FN: female neutered, STHS: soft tissue (localized) histiocytic sarcoma, VHS: visceral (disseminated) histiocytic sarcoma, lnn abd: abdominal lymphnodes, lnn mediast: mediastinal lymph nodes. Note: For cases with VHS the site sampled for gene expression is indicated in bold letters N = no Y = yes
RNA isolation
RNA isolation was performed as described previously [15]. In short, approximately 30 mg of frozen tumor was disrupted/lysed and homogenized and total RNA was isolated and treated with DNase using the RNeasy mini kit (Qiagen, The Netherlands) according to the manufacturer's protocol. Quantity and integrity were assessed with the Bioanalyzer Agilent BioAnalyzer-2100 (Bioanalyzer, Agilent Technologies, Santa Clara, CA) in combination with an RNA 6000 Pico-LabChip. The average RNA integrity number 8.5 (range: 7.2–9.8) was found to be appropriate [22]. RNA concentration was quantified using a NanoDrop ND-1000 (Isogen Life Science) spectrophotometer.
Expression profiling
Expression profiling was performed as described previously [15]. In short, RNA was labeled twice and hybridized against the common reference RNA on dual channel arrays, with RNA amplifications and labeling being performed on an automated system (Caliper Life Sciences NV/SA, Belgium) as described previously [23]. Dye swap of Cy3 and Cy5 was performed to reduce dye bias. Hybridizations were done on a HS4800PRO system supplemented with QuadChambers (Tecan Benelux B.V.B.A.) using 500–1000 ng labeled cRNA per channel as described [24]. After automated data extraction using Imagene 8.0 (BioDiscovery), printtip Loess normalization was performed [25] on mean spot-intensities. Dye-bias was corrected based on a within-set estimate as described [26]. Data were analyzed using ANOVA [27] (R version 2.2.1/MAANOVA version 0.98–7) (http://www.r-project.org/). Briefly, both tumourgroups were compared through the common reference channel. P-values were determined by a permutation F2-test, in which residuals were shuffled 5000 times globally. Thus analyzed 191 gene probes with P<0.05 after family wise error correction (FWER) were considered significantly changed.
Functional annotation
To examine whether certain pathways are over- or under-represented in the gene list, all genes differentially expressed between STHS and VHS, were introduced into DAVID (http://david.abcc.ncifcrf.gov/).
Quantitative real time PCR; gene selection
Following the outcome of the microarray expression profiling, 11 genes were selected; namely C-type lectin domain family 12, member A (CLEC12A); C-C motif chemokine 5 Precursor (Small-inducible cytokine A5) (CCL5_CANFA); Asporin (ASPN); CD9 molecule (CD9); Transketolase-like 1, transcript variant 1 (TKTL1); Complement component 6, transcript variant 1(C6); S100 calcium binding protein A12 (S100A12); Immunoglobulin J polypeptide (IGJ); S100 calcium binding protein A8 (S100A8); Phytanoyl-CoA dioxygenase, peroxisomal like (PHYH). Selection of these genes was based on the ones most significantly differently expressed on the micro-array chip and their fold changes.
Details of the qPCR reactions and primer sequences are depicted in Table 2. Delta Ct method, using efficiencies between 95.0 and 104.8%, was used for both the reference- as well as the target genes.
Table 2. QPCR primers for genes of interest (based on Microarray analyses) and reference genes (efficiencies varied between 95.0 and 104.8%).
| Gene name | ENSID | Primer sequence | Forward/reverse | Amplicon size (bp) | AnnealingT (°C) |
| CLEC12A | ENSCAFG00000025113 | AAATGCCAGCCTGTTGAC | F | 110 | 61 |
| TGGTAATCTCTGTCATACTTGGG | R | ||||
| CCL5_CANFA | ENSCAFG00000018171 | TATGCCTCAGACACCACAC | F | 119 | 63 |
| GACAAAGACGACTGCTGG | R | ||||
| ASPN | ENSCAFG00000002307 | CCACGAGTCAGAGAAATACAC | F | 135 | 59 |
| GGCAGAAGTCATTCACTCC | R | ||||
| CD9 | ENSCAFG00000015172 | TGTGCTGTCATCCATCAC | F | 118 | 57 |
| TGCCAAATATCATCACTACGG | R | ||||
| TKTL1 | ENSCAFG00000019451 | ATGAGATACAAACAGGAGGAC | F | 136 | 61 |
| CCAGTATATGCCATCCCAC | R | ||||
| C6 | ENSCAFG00000018598 | CCGTTGTGATTGACTTTGAG | F | 88 | 61 |
| CTTTCTGAGGTTGTTCCGT | R | ||||
| S100A12 | ENSCAFG00000023324 | AAAGGGTGAGATGAAGCAG | F | 156 | 61 |
| CACAACAGAAACCAGGGA | R | ||||
| IGJ | ENSCAFG00000002911 | CCTTCTCCCGATGATCCT | F | 120 | 63 |
| GGTACACAAATTTCGTTCTCAC | R | ||||
| S100A8 | ENSCAFG00000017557 | GTTTACCACAAGTACTCCCTG | F | 148 | 63 |
| CCATCGCTATTGACATCCA | R | ||||
| PHYH | ENSCAFG00000023349 | CTGAAGCCACACGATTATCC | F | 112 | 58 |
| TCTCCTTTCTCCATCACGA | R | ||||
| HPRT | ENSCAFG00000018870 | AGCTTGCTGGTGAAAAGGAC | F | 104 | 56 |
| TTATAGTCAAGGGCATATCC | R | ||||
| RPS19 | ENSCAFG00000028485 | CCTTCCTCAAAAAGTCTGGG | F | 95 | 61 |
| GTTCTCATCGTAGGGAGCAAG | R | ||||
| RPL8 | ENSCAFG00000001677 | CCATGAATCCTGTGGAGC | F | 64 | 55 |
| GTAGAGGGTTTGCCGATG | R | ||||
| SRPR | ENSCAFG00000010474 | GCTTCAGGATCTGGACTGC | F | 81 | 61 |
| GTTCCCTTGGTAGCACTGG | R | ||||
| RPL13 | ENSCAFG00000019840 | GCCGGAAGGTTGTAGTCGT | F | 87 | 61 |
| GGAGGAAGGCCAGGTAATTC | R | ||||
| GUSB | ENSCAFG00000010193 | AGACGCTTCCAAGTACCCC | F | 103 | 62 |
| AGGTGTGGTGTAGAGGAGCAC | R | ||||
| GAPDH | ENSCAFG00000024323 | TGTCCCCACCCCCAATGTATC | F | 100 | 58 |
| CTCCGATGCCTGCTTCACTACCTT | R | ||||
| B2MG | ENSCAFG00000013633 | TCCTCATCCTCCTCGCT | F | 85 | 61 |
| TTCTCTGCTGGGTGTCG | R | ||||
| RPS5 | ENSCAFG00000002366 | TCACTGGTGAGAACCCCCT | F | 141 | 62.5 |
| CCTGATTCACACGGCGTAG | R |
Genes identified using microarray as being significantly different comparing Soft Tissue Histiocytic Sarcoma (STHS) and Visceral Histiocytic Sarcoma (VHS): C-type lectin domain family 12, member A (CLEC12A); C-C motif chemokine 5 Precursor (Small-inducible cytokine A5) (CCL5_CANFA); Asporin (ASPN); CD9 molecule (CD9);Transketolase-like 1, transcript variant 1 (TKTL1); Complement component 6, transcript variant 1(C6); S100 calcium binding protein A12 (S100A12); Immunoglobulin J polypeptide (IGJ); S100 calcium binding protein A8 (S100A8); Phytanoyl-CoA dioxygenase, peroxisomal like (PHYH) Reference genes primers for q PCR: Hypoxanthine phosphoribosyltransferase (HPRT), Ribosomal protein S19 (RPS19) ribosomalprotein L8 (RPL8), Signal recognition particle receptor (SRPR), Ribosomal protein L13, (RPL13), glucuronidase, beta (GUSB), Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH), Beta-2-Microglobulin (B2MG), 40S ribosomal protein S5 (RPS5), Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-2-microglobulin (B2MG), Ribosomal protein S5 (RPS5).
RNA isolation and cDNA synthesis
Besides tissues from all but one patient (of which the insufficient tissue remained for the validating qPCR experiment) used in the microarray experiment (six soft tissue HS and seven visceral HS), two additional samples (one soft tissue HS and one visceral HS that met the inclusion criteria) were added. Total RNA from these samples was isolated. After isolation, total RNA was treated with DNase using the RNeasy mini kit (Qiagen, The Netherlands) according to the manufacturer's protocol.
Reverse transcription (RT) was performed of all 20 samples in a 80 µl reaction using 2000 ng total RNA, 16 µl iScript Reaction mix and 4 µl iScript Reverse Transcriptase (iScript cDNA Synthesis kit, Bio Rad, Veenendaal, The Netherlands). The mixture, contain both random hexamers and oligo-dT primers,was incubated 5 min. at 25°C, 30 min. at 42°C and followed by 5 min. at 85°C. Minus RT controls were prepared from 500 ng of the same RNA under the same conditions, but without addition of reverse transcriptase.
Reference genes and primer development
Nine reference genes were used as the non-regulated reference genes for normalization, based on their stable expression in canine tissue [28], [29] namely ribosomal protein S19 (RPS19) hypoxanthine phosphoribosyltransferase (HPRT), ribosomal protein L8 (RPL8), signal recognition particle receptor (SRPR), and ribosomal protein L1 (RPL13), glucuronidase, beta (GUSB), Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH), Beta-2-Microglobulin (B2MG), 40S ribosomal protein S5 (RPS5). Primers for reference genes, including their optimum annealing temperature are listed in Table 2.
Using Ensembl (Ensembl 70; CanFam3.1), through annotated transcripts, PCR primers were designed using the Perl Primer software (version 2.0.0.7) according to the parameters outlined in the Bio-Rad i-cycler manual. The specificity of each primer pair was confirmed by sequencing its product and also in qPCR by checking the meltcurve and reaction efficiency. GeNorm [30] was used to establish expression stability. Amplicon sequence-reactions were performed using BigDye v3.1 according to the manufacturer's (Life Technologies, Bleiswijk, The Netherlands) instructions on an ABI3130XL and analyzed in Lasergene (version 9.1 DNASTAR) and confirmed the specificity of each amplicon.
Quantitative PCR
Published guidelines for the qPCR experiment were followed according to the MIQE guidelines [31]–[33]. For qPCR, the CFX detection system (Bio-Rad.) with SYBR green fluorophore was used. Reactions were performed in a total volume of 10 µl containing 5 µl of 2× SYBR green super mixes (Bio-Rad Laboratories Ltd.), 0.5 µl of each primer at 400 nM concentration, 0.8 µl of cDNA and 3.2 µl RNase and DNase free water as previously described [15], [34], [35]. Expression analysis was performed on sample duplicates in duplicate. A minus RT sample and a no template control were performed as control. Expression levels were based on Ct values normalized using the mean of seven of the nine reference genes. GAPDH and B2MG did not behave fully stable according to GeNorm [28].
A Wilcoxon rank sum test was performed to determine the significance of differential gene expression. All results were Bonferroni corrected.
Results and Discussion
The Microarray enabled analysis of the expression of 42,034 features; however, since only 21,682 (51%) were annotated (CanFam 2.0), it is possible that important genes are missed. Gene expression profiles of the two HS forms were compared with each other to identify genes that are specific for each manifestation.
When comparing VHS and STHS, 191 probes were significantly differentially expressed, listed in Additional Table S1. From the selection of most significant (P<0.003), unique probes with a threshold of log2 fold change of 1.5 (n = 19), eight were excluded for either comprising chromosomal regions (n = 3) or as clones that did not align (n = 5). The eleven genes that remained, are visualized in a heatmap (Figure 1). Their potential involvement in tumor development or behavior was considered by a literature research. All microarray gene expression data were deposited in the public data repository GEO (accession number GSE45832).
Figure 1. Microarray- based heatmap of 11 genes.
C-type lectin domain family 12, member A (CLEC12A); C-C motif chemokine 5 Precursor (Small-inducible cytokine A5) (CCL5_CANFA); Asporin (ASPN); CD9 molecule (CD9);Transketolase-like 1, transcript variant 1 (TKTL1); Complement component 6, transcript variant 1(C6); S100 calcium binding protein A12 (S100A12); Immunoglobulin J polypeptide (IGJ); S100 calcium binding protein A8 (S100A8); Phytanoyl-CoA dioxygenase, peroxisomal like (PHYH).
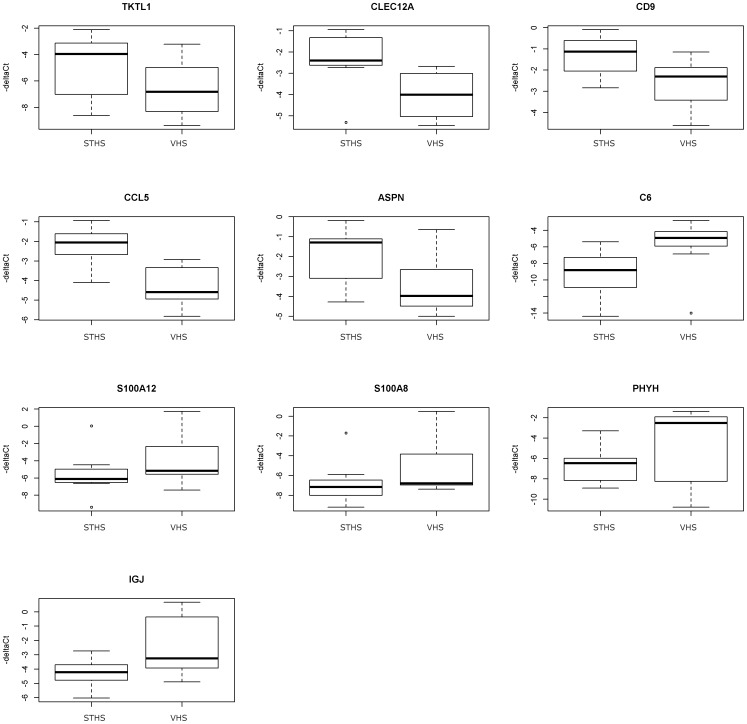
QPCR confirmed and quantified the differential expression of the 11 genes selected from the micro-array data. As a result, C6, S100A12, S100A8, PHYH and IGJ were up-regulated and TKTL1, CLEC12A, CD9, CCL5 and ASPN were down-regulated in VHS compared to STHS. Only for three gene products was the difference in expression significant: C6 (P = 0.038), CLEC12A (P = 0.026) and CCL5 (P = 0.0069) (Figure 2).
Figure 2. Quantitative PCR results.
The upregulation or downregulation of selected genes in STHS (Soft Tissue Histiocytic Sarcoma) and VHS (Visceral Histiocytic Sarcoma). The thick black line represents the median (50th percentile) and also the first and third quartile (25th and 75th percentile respectively) are displayed.Three genes are significantly differentially expressed: C6 was up-regulated when comparing the more aggressive visceral histiocytic sarcoma to the soft tissue form, and CLEC12A and CCL5 were down-regulated when comparing the more aggressive visceral histiocytic sarcoma to the soft tissue histiocytic sarcoma. Abbreviations: C-type lectin domain family 12, member A (CLEC12A); C-C motif chemokine 5 Precursor (Small-inducible cytokine A5) (CCL5_CANFA); Asporin (ASPN); CD9 molecule (CD9);Transketolase-like 1, transcript variant 1 (TKTL1); Complement component 6, transcript variant 1(C6); S100 calcium binding protein A12 (S100A12); Immunoglobulin J polypeptide (IGJ); S100 calcium binding protein A8 (S100A8); Phytanoyl-CoA dioxygenase, peroxisomal like (PHYH).
For technical reasons, no qPCR data could be obtained for CAMK2A.
Our observations of CLEC12A and CCL5 and those made in several human cancer types, make it conceivable that these gene products play a role in HS. In humans, C chemokine ligand 5 (CCL5) functions as one of the natural ligands for the CC Chemokine Receptor 5 (CCR5). It mediates chemotactic activity in immune cells including monocytes and dendritic cells [36] CCL5 [37], [38] and CCR5 [39] promote breast cancer invasiveness and metastatic potential, while CCR5 inhibition abrogates this [39]. For inflammatory breast cancer, CCL5 is considered to constitute a prominent part of a poor prognosis signature [40]. Also in human colorectal carcinoma CCL5 appears to stimulate cancer progression [41]. Furthermore, the CCL5/CCR5 axis has been shown to promote cell motility in human osteosarcoma [42]. Thus, for these human cancers CCL5 appears to be associated with a metastatic phenotype. Yet, in our study, CCL5 was expressed at lower level in VHS than in STHS. Considering the more rapid metastatic nature of VHS, this is surprising, and this finding needs confirmation (also at protein level) in future studies. Still, a decrease in CCL5 expression has been described in other human malignancies such as colon carcinoma when compared to normal tissue [43]. An alternative explanation for a reduction in CCL5 expression in visceral as compared to localized HS could be that a reduction in expression of CCL5 could protect against immunosurveillance [44] and hence be related to more aggressive behavior of HS.
CLEC12A expression was found to be significantly lowered in VHS compared to STHS. CLEC12A (or MICL) is considered a negative regulator of granulocyte and monocyte function [45]. Activation of myeloid cells and recruitment to sites of inflammation – but not increase or decrease in the level of differentiation – was accompanied by reduced expression [46]. Whereas normal lymphocytes have no or low MICL expression, a relatively high expression of this gene in acute lymphoblastic leukemia was found to be associated with prolonged relapse-free survival [47]. How the reduced expression in VHS as compared to STHS relates to these findings remains to be determined, but a resulting increased migratory capacity as present in VHS could be an explanation.
Complement component 6 (C6) gene expression was significantly increased in VHS as compared to STHS. In acute leukemia's an increase in circulating complement is common. As one early study has demonstrated increased expression of complement in monocytes by conditioned media of leukemic cells [48]. At present, a straight forward hypothesis on the functional consequences of the variation in C6 expression in the different forms of HS is not easily postulated but it might be associated with a reaction of the innate immune system to the neoplasm and not a direct effect of the neoplastic cell population.
The variation in expression of C6, CCL5 and CLEC12A, all three members of the immune response, is worthy of follow up investigations, with focus upon their character as deranged histiocytes and may include comparisons with the canine reactive histiocytic diseases [1].
When comparing the results of qPCR and microarray the difference in the expression of some genes between STHS and VHS did not attain statistical significance. The variation between the two methods relates to the fact that a microarray experiment is a semi-quantitative screening method and the qPCR quantitative. Many methodological factors can lead to a lack of correlation between array results and qPCR measurements [49]. The expression of CD9 (synonym MRP-1; motility related protein 1) in VHS was suppressed as compared to STHS at a level trending towards significance (P = 0.07) and further investigation of this gene in these sarcomas seems warranted in view of associations with reduced expression of this gene with tumor aggressiveness such as reported for other cancers. A lower expression of CD9 was found to be associated with the formation of bony metastases in studies using breast cancer cell lines [50], [51]. Similarly, in human colon cancer patients, patients that lacked CD9 mRNA expression, had a worse prognosis than the cases that did express CD9 mRNA [52].
DAVID pathway analyses of the significantly differentially expressed genes in the micro array did not lead to the detection of altered expression of whole pathways. Also the genes chosen for qPCR confirmation did not appear to be related at the level of regulation.
The alterations in gene function as detected in the current analyses, need follow up by subsequent investigations by use of antibodies - most still need to be developed and validated for use in the dog to examine an altered expression at protein level.
When looking at tumor conditions in the human that share features with canine HS, none of the eleven genes for which altered function was observed in the present study, have been recognized as aberrantly expressed in micro-array/qPCR studies in Langerhans cell histiocytosis [53].
Conclusions
As a valuable addition to our previous study, in which we were able to provide evidence for involvement of several genes in the development of HS, irrespective of form, this study provides the most comprehensive database to date of genetic variations in the two most common forms of HS, namely VHS and STHS. Using fold-change analysis, it reveals genetic variations not previously associated with these two forms.
On the basis of quantitative differences in expression of C6 (up-regulated in VHS versus STHS), CLEC12A and CCL5 (down-regulated in VHS versus STHS) were associated with each subtype. Down-regulation of CLEC12A in VHS, the more aggressive form of HS, is in line with previously published observations that such reduced function facilitates migratory capacity of myeloid cells in humans [46]. Down-regulation of CCL5 is in line with several studies in human cancers [37], [47], however contradictory to others [43]. Further investigations should focus on changes of gene function at protein level and a comparison of these histiocytic malignancies in dog and human.
Supporting Information
All 191 probes significantly differentially expressed. Of four of the 191 probes, the annotation could not be traced back. Of the remaining probes, 28 have a ‘genomic’ location for which no gene could be mapped at this moment. 159 Probes have a gene-name, of which 142 are unique. From the selection of most significant (P<0.003), unique probes with a threshold of log2 fold change of 1.5 (n = 19), eight were excluded; either for comprising chromosomal regions (n = 3) or clones that did not align (n = 5, marked with *). The eleven genes that remained, are written in bold and italic.
(DOCX)
Funding Statement
This study was partly funded by the European Commission (LUPA-GA-201370). No external financers contributed to this experiment. Intervet International BV, that in later stage became part of Merck Sharp Dohme, provided a gift to this study in order to help finance the purchase of micro-arrays. The donation was provided without any obligation by authors to this company. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Moore PF (2014) A review of histiocytic diseases of dogs and cats. Vet Pathol 51: 167–184 10.1177/0300985813510413 ; 10.1177/0300985813510413 10.1177/0300985813510413; 10.1177/0300985813510413 [DOI] [PubMed] [Google Scholar]
- 2.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, et al.. (2008) WHO classification of tumours of haematopoietic and lymphoid tissues, fourth edition. Lyon: IARC Press. [Google Scholar]
- 3. Hedan B, Thomas R, Motsinger-Reif A, Abadie J, Andre C, et al. (2011) Molecular cytogenetic characterization of canine histiocytic sarcoma: A spontaneous model for human histiocytic cancer identifies deletion of tumor suppressor genes and highlights influence of genetic background on tumor behavior. BMC Cancer 11: 201 10.1186/1471-2407-11-201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abadie J, Hedan B, Cadieu E, De Brito C, Devauchelle P, et al. (2009) Epidemiology, pathology, and genetics of histiocytic sarcoma in the bernese mountain dog breed. J Hered 100 Suppl 1S19–27 10.1093/jhered/esp039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Affolter VK, Moore PF (2002) Localized and disseminated histiocytic sarcoma of dendritic cell origin in dogs. Vet Pathol 39: 74–83. [DOI] [PubMed] [Google Scholar]
- 6. Moore PF, Affolter VK, Vernau W (2006) Canine hemophagocytic histiocytic sarcoma: A proliferative disorder of CD11d+ macrophages. Vet Pathol 43: 632–645 10.1354/vp.43-5-632 [DOI] [PubMed] [Google Scholar]
- 7. Paoloni M, Khanna C (2008) Translation of new cancer treatments from pet dogs to humans. Nat Rev Cancer 8: 147–156 10.1038/nrc2273 [DOI] [PubMed] [Google Scholar]
- 8. Hoffman MM, Birney E (2007) Estimating the neutral rate of nucleotide substitution using introns. Mol Biol Evol 24: 522–531 10.1093/molbev/msl179 [DOI] [PubMed] [Google Scholar]
- 9. Breen M, Modiano JF (2008) Evolutionarily conserved cytogenetic changes in hematological malignancies of dogs and humans—man and his best friend share more than companionship. Chromosome Res 16: 145–154 10.1007/s10577-007-1212-4 [DOI] [PubMed] [Google Scholar]
- 10. Beverley PC, Egeler RM, Arceci RJ, Pritchard J (2005) The nikolas symposia and histiocytosis. Nat Rev Cancer 5: 488–494 10.1038/nrc1632 [DOI] [PubMed] [Google Scholar]
- 11. Pileri SA, Grogan TM, Harris NL, Banks P, Campo E, et al. (2002) Tumours of histiocytes and accessory dendritic cells: An immunohistochemical approach to classification from the international lymphoma study group based on 61 cases. Histopathology 41: 1–29. [DOI] [PubMed] [Google Scholar]
- 12.Shearin AL, Hedan B, Cadieu E, Erich SA, Schmidt EV, et al.. (2012) The MTAP-CDKN2A locus confers susceptibility to a naturally occurring canine cancer. Cancer Epidemiol Biomarkers Prev. 10.1158/1055-9965. EPI-12-0190-T. [DOI] [PMC free article] [PubMed]
- 13. Tamburini BA, Trapp S, Phang TL, Schappa JT, Hunter LE, et al. (2009) Gene expression profiles of sporadic canine hemangiosarcoma are uniquely associated with breed. PLoS One 4: e5549 10.1371/journal.pone.0005549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tamburini BA, Phang TL, Fosmire SP, Scott MC, Trapp SC, et al. (2010) Gene expression profiling identifies inflammation and angiogenesis as distinguishing features of canine hemangiosarcoma. BMC Cancer 10: 619 10.1186/1471-2407-10-619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boerkamp KM, van der Kooij M, van Steenbeek FG, van Wolferen ME, Groot Koerkamp MJA, et al. (2013) Gene expression profiling of histiocytic sarcomas in a canine model: The predisposed flatcoated retriever dog. PLoS One 8(8): e71094; 0071094/journal.pone [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dobson J, Hoather T, McKinley TJ, Wood JL (2009) Mortality in a cohort of flat-coated retrievers in the UK. Vet Comp Oncol 7: 115–121 10.1111/j.1476-5829.2009.00181 [DOI] [PubMed] [Google Scholar]
- 17. Fidel J, Schiller I, Hauser B, Jausi Y, Rohrer-Bley C, et al. (2006) Histiocytic sarcomas in flat-coated retrievers: A summary of 37 cases (november 1998-march 2005). Vet Comp Oncol 4: 63–74 10.1111/j.1476-5810.2006.00090 [DOI] [PubMed] [Google Scholar]
- 18. Skorupski KA, Rodriguez CO, Krick EL, Clifford CA, Ward R, et al. (2009) Long-term survival in dogs with localized histiocytic sarcoma treated with CCNU as an adjuvant to local therapy. Veterinary and comparative oncology 7: 139–144 10.1111/j.1476-5829.2009.00186 [DOI] [PubMed] [Google Scholar]
- 19.Meuten DJ (ed) (2002) Tumors in domestic animals. Fourth Edition, Iowa State Press, Ames, Iowa, USA. 788 p.doi: 10.1002/9780470376928.fmatter. [Google Scholar]
- 20. Constantino-Casas F, Mayhew D, Hoather TM, Dobson JM (2011) The clinical presentation and histopathologic-immunohistochemical classification of histiocytic sarcomas in the flat coated retriever. Vet Pathol 48: 764–771 10.1177/0300985810385153 [DOI] [PubMed] [Google Scholar]
- 21. Fulmer AK, Mauldin GE (2007) Canine histiocytic neoplasia: An overview. Can Vet J 48: 1041–3, 1046–50. [PMC free article] [PubMed] [Google Scholar]
- 22. Opitz L, Salinas-Riester G, Grade M, Jung K, Jo P, et al. (2010) Impact of RNA degradation on gene expression profiling. BMC Med Genomics 3: 36 10.1186/1755-8794-3-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roepman P, Wessels LF, Kettelarij N, Kemmeren P, Miles AJ, et al. (2005) An expression profile for diagnosis of lymph node metastases from primary head and neck squamous cell carcinomas. Nat Genet 37: 182–186 10.1038/ng1502 [DOI] [PubMed] [Google Scholar]
- 24. van de Peppel J, Kemmeren P, van Bakel H, Radonjic M, van Leenen D, et al. (2003) Monitoring global messenger RNA changes in externally controlled microarray experiments. EMBO Rep 4: 387–393 10.1038/sj.embor.embor798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang YH, Dudoit S, Luu P, Lin DM, Peng V, et al. (2002) Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Margaritis T, Lijnzaad P, van Leenen D, Bouwmeester D, Kemmeren P, et al. (2009) Adaptable gene-specific dye bias correction for two-channel DNA microarrays. Mol Syst Biol 5: 266 10.1038/msb.2009.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu H, Kerr M, Cui X, Churchill G (2003) MAANOVA: A software package for the analysis of spotted cDNA microarray experiments. In: Anonymous The Analysis of Gene Expression Data. pp.313–341. [Google Scholar]
- 28. Schlotter YM, Veenhof EZ, Brinkhof B, Rutten VP, Spee B, et al. (2009) A GeNorm algorithm-based selection of reference genes for quantitative real-time PCR in skin biopsies of healthy dogs and dogs with atopic dermatitis. Vet Immunol Immunopathol 129: 115–118 10.1016/j.vetimm.2008.12.004 [DOI] [PubMed] [Google Scholar]
- 29. Brinkhof B, Spee B, Rothuizen J, Penning LC (2006) Development and evaluation of canine reference genes for accurate quantification of gene expression. Anal Biochem 356: 36–43 10.1016/j.ab.2006.06.001 [DOI] [PubMed] [Google Scholar]
- 30. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: Research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, et al. (2009) The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55: 611–622 10.1373/clinchem.2008.112797 ; 10.1373/clinchem.2008.112797 10.1373/clinchem.2008.112797; 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 32. Bustin SA, Beaulieu JF, Huggett J, Jaggi R, Kibenge FS, et al. (2010) MIQE precis: Practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC Mol Biol 11: 74–2199-11-74 10.1186/1471-2199-11-74 ; 10.1186/1471-2199-11-74 10.1186/1471-2199-11-74; 10.1186/1471-2199-11-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huggett JF, Foy CA, Benes V, Emslie K, Garson JA, et al. (2013) The digital MIQE guidelines: Minimum information for publication of quantitative digital PCR experiments. Clin Chem 59: 892–902 10.1373/clinchem.2013.206375 ; 10.1373/clinchem.2013.206375 10.1373/clinchem.2013.206375; 10.1373/clinchem.2013.206375 [DOI] [PubMed] [Google Scholar]
- 34. van Steenbeek FG, Van den Bossche L, Grinwis GC, Kummeling A, van Gils IH, et al. (2013) Aberrant gene expression in dogs with portosystemic shunts. PLoS One 8: e57662 10.1371/journal.pone.0057662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van Steenbeek FG, Spee B, Penning LC, Kummeling A, van Gils IH, et al. (2013) Altered subcellular localization of heat shock protein 90 is associated with impaired expression of the aryl hydrocarbon receptor pathway in dogs. PLoS One 8: e57973 10.1371/journal.pone.0057973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. de la Rosa G, Longo N, Rodríguez-Fernández JL, Puig-Kroger A, Pineda A, et al. (2003) Migration of human blood dendritic cells across endothelial cell monolayers: Adhesion molecules and chemokines involved in subset-specific transmigration. J Leukoc Biol 73: 639–649. [DOI] [PubMed] [Google Scholar]
- 37. Swamydas M, Ricci K, Rego S, Dreau D (2013) Mesenchymal stem cell-derived CCL-9 and CCL-5 promote mammary tumor cell invasion and the activation of matrix metalloproteinases. Cell adhesion migration 7: 315–324 10.4161/cam.25138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, et al. (2007) Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature 449: 557–563 10.1038/nature06188 [DOI] [PubMed] [Google Scholar]
- 39. Velasco Velázquez MP, Pestell RG (2013) The CCL5/CCR5 axis promotes metastasis in basal breast cancer. Oncoimmunology 2: e23660–e23660 10.4161/onci.23660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bieche I, Lerebours F, Tozlu S, Espie M, Marty M, et al. (2004) Molecular profiling of inflammatory breast cancer: Identification of a poor-prognosis gene expression signature. Clinical cancer research 10: 6789–6795 10.1158/1078-0432.CCR-04-0306 [DOI] [PubMed] [Google Scholar]
- 41. Cambien B, Richard-Fiardo P, Karimdjee BF, Martini V, Ferrua B, et al. (2011) CCL5 neutralization restricts cancer growth and potentiates the targeting of PDGFRbeta in colorectal carcinoma. PLoS One 6: e28842 10.1371/journal.pone.0028842; 10.1371/journal.pone.0028842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang SW, Wu HH, Wang PC, Ou WC, Chou WY, et al. (2012) CCL5 and CCR5 interaction promotes cell motility in human osteosarcoma. PLoS ONE 7: e35101–e35101 10.1371/journal.pone.0035101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Baier PK, Eggstein S, Wolff Vorbeck G, Baumgartner U, Hopt UT (2005) Chemokines in human colorectal carcinoma. Anticancer Res 25: 3581–3584. [PubMed] [Google Scholar]
- 44. Qian X, Zhang J, Liu J (2011) Tumor-secreted PGE2 inhibits CCL5 production in activated macrophages through cAMP/PKA signaling pathway. J Biol Chem 286: 2111–2120 10.1074/jbc.M110.154971; 10.1074/jbc.M110.154971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Marshall AS, Willment JA, Lin HH, Williams DL, Gordon S, et al. (2004) Identification and characterization of a novel human myeloid inhibitory C-type lectin-like receptor (MICL) that is predominantly expressed on granulocytes and monocytes. J Biol Chem 279: 14792–14802 10.1074/jbc.M313127200 [DOI] [PubMed] [Google Scholar]
- 46. Marshall AS, Willment JA, Pyz E, Dennehy KM, Reid DM, et al. (2006) Human MICL (CLEC12A) is differentially glycosylated and is down-regulated following cellular activation. Eur J Immunol 36: 2159–2169 10.1002/eji.200535628 [DOI] [PubMed] [Google Scholar]
- 47. Harvey RC, Mullighan CG, Wang X, Dobbin KK, Davidson GS, et al. (2010) Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: Correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood 116: 4874–4884 10.1182/blood-2009-08-239681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gyapay G, Schmidt B, Válay M, Falus A, Anh Tuan N, et al. (1991) Effect of conditioned media of acute myeloid leukemia blast cells on complement synthesis by cultured human cells of monocyte and hepatocyte origin. Complement Inflamm 8: 370–377. [DOI] [PubMed] [Google Scholar]
- 49. Chuaqui RF, Bonner RF, Best CJ, Gillespie JW, Flaig MJ, et al. (2002) Post-analysis follow-up and validation of microarray experiments. Nat Genet 32 Suppl: 509–514 10.1038/ng1034 [DOI] [PubMed] [Google Scholar]
- 50. Kischel P, Bellahcene A, Deux B, Lamour V, Dobson R, et al. (2012) Overexpression of CD9 in human breast cancer cells promotes the development of bone metastases. Anticancer Res 32: 5211–5220. [PubMed] [Google Scholar]
- 51. Mimori K, Kataoka A, Yoshinaga K, Ohta M, Sagara Y, et al. (2005) Identification of molecular markers for metastasis-related genes in primary breast cancer cells. Clinical experimental metastasis 22: 59–67 10.1007/s10585-005-4417-y [DOI] [PubMed] [Google Scholar]
- 52. Mori M, Mimori K, Shiraishi T, Haraguchi M, Ueo H, et al. (1998) Motility related protein 1 (MRP1/CD9) expression in colon cancer. Clinical cancer research 4: 1507–1510. [PubMed] [Google Scholar]
- 53. Rust R, Kluiver J, Visser L, Harms G, Blokzijl T, et al. (2006) Gene expression analysis of dendritic/Langerhans cells and langerhans cell histiocytosis. J Pathol 209: 474–483 10.1002/path.2003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All 191 probes significantly differentially expressed. Of four of the 191 probes, the annotation could not be traced back. Of the remaining probes, 28 have a ‘genomic’ location for which no gene could be mapped at this moment. 159 Probes have a gene-name, of which 142 are unique. From the selection of most significant (P<0.003), unique probes with a threshold of log2 fold change of 1.5 (n = 19), eight were excluded; either for comprising chromosomal regions (n = 3) or clones that did not align (n = 5, marked with *). The eleven genes that remained, are written in bold and italic.
(DOCX)