Abstract
The filamentous fungus Penicillium chrysogenum harbors an astonishing variety of nonribosomal peptide synthetase genes, which encode proteins known to produce complex bioactive metabolites from simple building blocks. Here we report a novel non-canonical tetra-modular nonribosomal peptide synthetase (NRPS) with microheterogenicity of all involved adenylation domains towards their respective substrates. By deleting the putative gene in combination with comparative metabolite profiling various unique cyclic and derived linear tetrapeptides were identified which were associated with this NRPS, including fungisporin. In combination with substrate predictions for each module, we propose a mechanism for a ‘trans-acting’ adenylation domain.
Introduction
Fungal non-ribosomal peptides contribute a large variety of secondary metabolites with remarkable properties such as antibacterial, antifungal, antiparasitic, anticancer and immunosuppressive activities. These metabolites are produced by large, multifunctional protein complexes, called nonribosomal peptide synthetases (NRPS). These enzymes catalyze the stepwise condensation of simple amino acid building blocks to complex molecules. NRPSs have a modular organization, with each module responsible for one discrete chain-elongation step. Every single module can be subdivided into domains that carry all essential information for recognition, activation and modification of the corresponding substrate. At a minimum, a typical NRPS module consists of an adenylation (A) domain, responsible for amino acid activation, a thiolation domain, also known as peptidyl carrier protein (PCP), which binds the activated amino acid and a condensation (C) domain that catalyzes peptide-bond formation. The common arrangements of these domains follow a (C-A-PCP)n organization. Additionally, a variety of optional domains have been described such as methyltransferase (MT) and epimerization (E) domains [1].
The number of modules and their domain organization within NRPS enzymes controls the structures of the final product(s) [1]–[3]. Thus, the order of modules usually corresponds to the sequence of amino acids in the peptide. Many NRPS systems adhere to this mechanistic paradigm, which is often referred to as the “co-linearity rule” [4]. Also exceptions to this rule have been discovered, including iterative NRPSs, which incorporate multiple residues of the same amino acid iteratively into the peptide structure and the so called nonlinear NRPSs, which deviate completely from the standard domain organization leading to unexpected products [3], [5], [6].
The impact of non-ribosomal peptide metabolites on the quality of human life raised the interest of pharmaceutical industries to invest in identification, engineering and heterologous expression of NRPS genes and pathways to ensure the rational production of novel compounds [7]–[9]. To understand the basic mechanisms of the biosynthesis of these complex NRPSs, detailed studies have been performed during the past few decades. These included the structural analysis of adenylation domains, mutational analysis of substrate specificity of these modules, the fusion of unrelated modules to produce new products and the identification of helper proteins for optimal activation of adenylation domains [10]–[12]. Although this has led to detailed insights into catalytic mechanisms, so far a structure of a complete NRPS is lacking that would reveal how modules cooperate to facilitate product formation. The availability of genome sequencing data and sophisticated bioinformatics analysis of various fungi revealed the presence of many NRPS genes that have not been associated with known secondary metabolites [13]–[15]. Moreover, most of these genes are not expressed when the fungi are grown under laboratory conditions, implying that many more secondary metabolites await discovery.
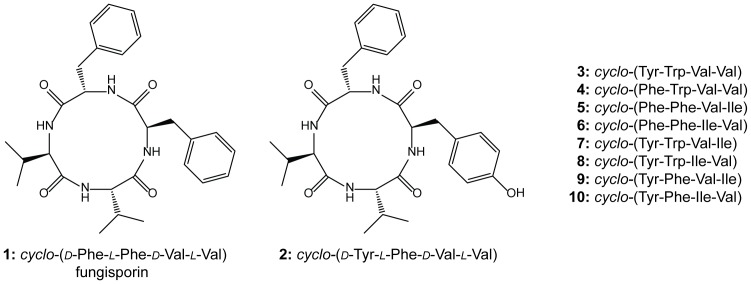
The filamentous fungus Penicillium chrysogenum is well known for the production of the antibiotic penicillin G that is synthesized by the tri-modular NRPS δ-(L-α-aminoadipyl)-L-cysteinyl-D-valine synthetase. In addition, other NRPS derived secondary metabolites like the roquefortines and meleagrin have been reported from P. chrysogenum [16]–[18]. Here, we describe the identification and structural characterization of cyclic tetrapeptides (Figure 1), including the previously identified metabolite fungisporin [19], [20], and the discovery of a tetra-modular NRPS with an unusual domain organization with adenylation domains showing microheterogenicity. It is proposed to term this NRPS HcpA (CAP93139.1) based on the produced Hydrophobic cyclic peptides.
Figure 1. Identified secondary metabolites.
Structures of cyclic tetrapeptides identified in P. chrysogenum.
Materials and Methods
A. Chemicals
6-aminoquinolyl-N-hydroxysuccinimidyl carbamate (AQC) reagent and borate buffer were obtained as part of AccQ Tag Reagent Kit from Waters (Waters, Milford, MA, USA). cyclo-(d-Tyr-l-Phe-d-Val-l-Val) was obtained from Celtek Peptides (Nashville, TN).
B. Host strains, media, grown condition and plasmid construction
Deletion of the hcpA gene was carried out in P. chrysogenum strain DS54555, which lacks penicillin cluster genes and the ku70 gene [18]. This strain was kindly provided by the DSM Biotechnology Center (Delft, Netherlands). A deletion plasmid was constructed by amplifying the flanking regions of the targeted gene with the Multisite Gateway®Three-Fragment Vector Construction Kit according to the procedure described by Invitrogen using pDEST R4-R3p as template. Primers used for the construction of the deletion plasmid pDEST R4-R3p PcHcpA (Figure S1) are listed in Table S1. Escherichia coli DH5α (F– Φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK–, mK+) phoA supE44 λ– thi-1 gyrA96 relA1) was used as host strain for high frequency transformation and plasmid DNA amplification [21]. All the strains were grown on yeast nitrogen base-glucose-yeast extract (YGG)-medium for protoplasts formation and transformation [22]. Both mutant and host strains of P. chrysogenum were grown on secondary metabolite production medium as described previously [18].
C. Transformation procedure
The deletion plasmid pDes R4-R3p PcHcpA was transformed to the protoplasts of P. chrysogenum DS54555 [23] yielding the ΔhcpA derivative of strain DS54555 The phleomycin resistance gene was used as selection marker for the deletion of the HcpA gene [22], [24].
D. Genomic DNA extraction, total RNA extraction, cDNA amplification and qPCR analysis
Genomic DNA (gDNA) was isolated after 96 hours of growth on SMP medium (secondary metabolite production medium) using the modified yeast gDNA isolation protocol [25] in which the fungal mycelium is broken in a FastPrep FP120 system (Qbiogene). Isolated gDNA was measured using a NanoDrop ND-1000. gDNA of the host and various deletion strains was isolated using the E.Z.N.A. Fungal DNA kit (Omega Bio-tek). Total RNA of the host strain was isolated after 48 hours of growth in SMP medium for the first time and then with the interval of 24 hours up to 216 hours of growth using the Trizol reagent (Invitrogen), with additional DNase treatment using the Turbo DNA-free kit (Ambion). Total RNA was measured with the NanoDrop ND-1000 and a concentration of 500 ng per cDNA reaction was used. cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad) in a 10 µl end volume. The primers used to analyze the expression of the hcpA gene were designed around an intron to avoid amplification on gDNA (Table S2). For expression analyses, the γ-actin gene was used as a control for normalization (Table S2). A negative reverse transcriptase (RT) control was used to determine the gDNA contamination in isolated total RNA. The expression levels were determined as described [18].
E. Southern blotting
Southern blotting was carried out by digesting gDNA (5 µg) with the indicated restriction enzymes. Digested DNA fragments were separated on a 0.8 % agarose gel, blotted onto a Zeta-Probe membrane (Biorad) as described earlier [26], and hybridized with the indicated DIG labeled probes.
F. Metabolite profiling and analysis
1. Sample preparation
Host and deletion strains of P. chrysogenum strains used for gene assignments were grown in quintuplicate according to the procedure described above. Samples for acquisition of the metabolite profiles from the growth curves were from five replicates. Metabolite profiling was carried out with modifications as described earlier [18]. Briefly, 4 µl of internal standard mixture (855 nmol/mL ranitidine, 657 nmol/mL reserpine and 114 nmol/mL ampicillin) was added to 100 µl fermentation broth followed by the addition of 400 µl methanol for protein precipitation. The samples were vortexed and spun down at 14,000 g for 10 minutes. 300 µL supernatant was evaporated for 30 minutes in a speedvac (Thermo Scientific, San Jose, CA) and re-dissolved in 100 µL water. LC-UV-MS analysis was performed on an Agilent 1200 Capillary pump (Agilent, Santa Clara, CA) coupled in-line to a Surveyor PDA detector (Thermo Scientific, San Jose, CA) and LTQ-FT mass spectrometer (Thermo Scientific, San Jose, CA) using electrospray ionization and operated in a scan range between m/z 110 and m/z 2000 in positive/negative ion switching mode. Separation was performed on a Waters Atlantis T3 column (2.1×100 mm, 3 µm) (Waters, Milford, MA) starting with 98 % of solvent A (1 % acetonitrile and 0.1 % formic acid in water) and 2 % solvent B (1 % water and 0.1 % formic acid in acetonitrile) for 1.5 minutes at a flow rate of 300 µL/min. 40 % B were reached after 22 minutes and 100 % B at 25 minutes. The column was flushed with 100 % B and re-equilibrated to initial conditions. Peak detection and integration were performed using an in-house tool followed by statistical tests to discover significant different features. Finally, discovered features were integrated using LCquan v.26 (Thermo Scientific, San Jose, CA). The non-related non-endogenous compound reserpine was used as internal standard.
2. Identification of cyclic tetrapeptides
The identity of cyclic tetrapeptides was determined using samples from liquid cultures of P. chrysogenum and a crude spontaneous precipitate obtained from P. chrysogenum cultures, containing primarily 1 and 2 next to various minor abundant cyclic tetrapeptides in relative concentrations of 70 and 15 %, respectively. LC-MSn experiments for the determination of consecutive amino acid losses were performed according the metabolite profiling section with normalized collision energies of 35 %, an isolation width of 1 amu and an activation Q of 0.30. NMR spectra were recorded on a Bruker Avance III 700 MHz NMR spectrometer (Bruker, Billerica, MA), equipped with a 5 mm TCI probe. 2 mg of each sample was dissolved in 0.6 mL anhydrous DMSO. NMR spectra were acquired at 340 K.
3. Identification of linear tetrapeptides
Linear tetrapeptides were identified according their multiple-stage fragmentation after AQC derivatization [27]. Methanol (400 µL) was added to an aliquot of 100 µL fermentation broth for protein precipitation. Samples were vortexed for 10 minutes, spun down for 10 minutes and 300 µL of the supernatant was evaporated to dryness in a speedvac (Thermo Scientific, San Jose, CA). Derivatization was done according to the supplier's procedure by re-dissolving the sample in 40 µL water, 40 µL borate buffer (pH 8.5) and 20 µL AQC solution. The mixture was vortexed for 10 minutes and heated for 10 minutes at 55°C.
LC-MSn experiments were conducted on an Agilent 1200 Capillary pump (Agilent, Santa Clara, CA) coupled to a LTQ-FT mass spectrometer (Thermo Scientific, San Jose, CA) using electrospray ionization. Separation was performed on a Waters Atlantis T3 column (2.1×100 mm, 3 µm) (Waters, Milford, MA) starting with 72 % of solvent A (1 % acetonitrile and 0.1 % formic acid in water) and 28 % of solvent B (1 % water and 0.1 % formic acid in acetonitrile) for 1.5 minutes at a flow rate of 300 µL/min. After 8 minutes the gradient reached 60 % of solvent B. Subsequently, the column was flushed with 100 % B before it was re-equilibrated to initial conditions. The peptide sequences were elucidated using multiple-stage collision-induced dissociation (CID) of the protonated molecule following consecutive cleavages of amino acids residues, starting from the C-terminus of the derivatized linear tetrapeptide. CID was performed with normalized collision energies of 35 %, an isolation width of 1 amu and an activation Q of 0.30.
Results
Bioinformatic analysis of a tetrapeptide NRPS
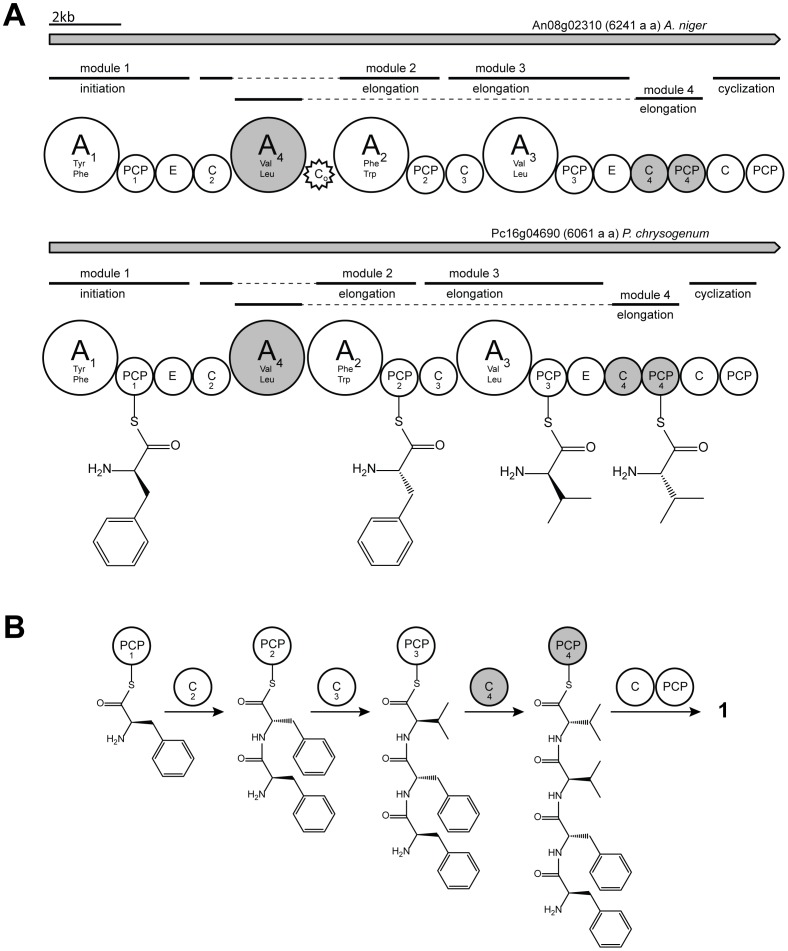
Genome sequencing revealed that P. chrysogenum encodes 11 NRPS genes [15]. Microarray expression analysis under glucose-limited chemostat culture conditions as well as quantitative PCR under shake flask culture condition showed that Pc16g04690 (genbank protein identifier CAP93139.1) (hcpA) is highly expressed (Figure 2A) [15]. The hcpA gene encodes a large multimodular non-ribosomal peptide synthetase enzyme (Figure 3) with 6064 amino acids and a calculated molecular mass of about 670 kDa. HcpA, which shows 54% sequence identity to the orthologous An08g02310 in A. niger, has the domain architecture A1-PCP1-E-C2-A4-A2-PCP2-C3-A3-PCP3-E-C4-PCP4-C-PCP (A = adenylation, C = condensation, PCP = thiolation, and E = epimerization) (Figure 3A) [15]. A similar domain architecture was deduced for the orthologous protein of A. niger except for an insertion of a 177 amino acid long sequence between the adenylation domain A4 and A2 which shows homology to conserved motifs of an incomplete condensation domain (Co) (Figure 3A). To predict the substrate specificity of the four adenylation domains of the two HcpA proteins, NRPSPredictor2 was used [28]. This program extracted the active site amino acid motifs DAACVAGVAK and DAVIIAAVAK as the signature sequences for the A1 domain in the P. chrysogenum and A. nidulans HcpA proteins. This motif exhibits similarity with the signature sequence of the adenylation domain of a bacitracine-producing NRPS that activates phenylalanine as a substrate. The signature sequences for the A4 domain (DAVSAGVAAK and DMQSAWFICK in the P. chrysogenum and A. nidulans HcpA, respectively) shows homology with the valine-activating adenylation domain of the gramicidin synthetase, whereas the A2 (DAMTVGGVFK and DVLSTGAICK for the P. chrysogenum and A. nidulans HcpA, respectively) and A3 (DAMFVGGVFK and DAMFVGGIFK for the P. chrysogenum and A. nidulans HcpA, respectively) domains have predicted specificities towards phenylalanine and valine, respectively. The overall architecture of both synthetases is unusual, as the A2 and A4 domains occur adjacent to each other, flanked by a single C and PCP domain in a C2-A4-A2-PCP2 pattern. On the other hand, an incomplete module (C4-PCP4) without an adjacent A domain is found at the N-termini of these NRPSs (Figure 3A).
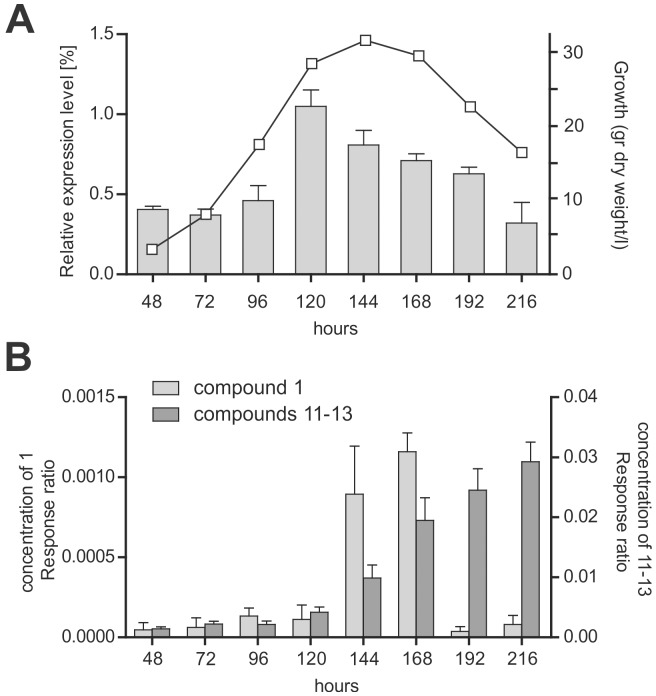
Figure 2. Correlation between the expression level of HcpA and metabolite formation during growth.
A: Time dependent expression level of hcpA as determined by quantitative RT-PCR during growth of P. chrysogenum as monitored by biomass formation. B: Internal standard corrected concentration of the cyclic peptide 1 and its degradation products 11–13 present in the growth media. The concentration of peptides was determined by HPLC-UV-MS.
Figure 3. Hypothetical model for the biosynthesis of compound 1 in P. chrysogenum and A. niger by the HcpA NRPS.
A: Binding of the monomers on the carrier protein domains. Both isolated condensation and thiolation domains C4 and T4, missing a preceding adenylation domain, are assumed to correspond to the adenylation domain A4, located upstream. B: Proposed assembly of compound 1 including condensation domains presumed for catalyzing the formation of the peptide bond.
Genetic deletion of the tetrapeptide NRPS and secondary metabolite identification
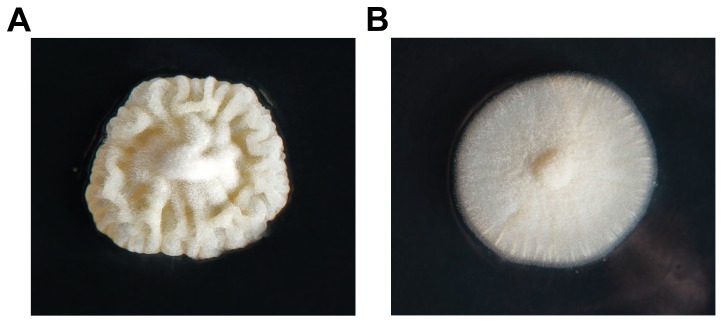
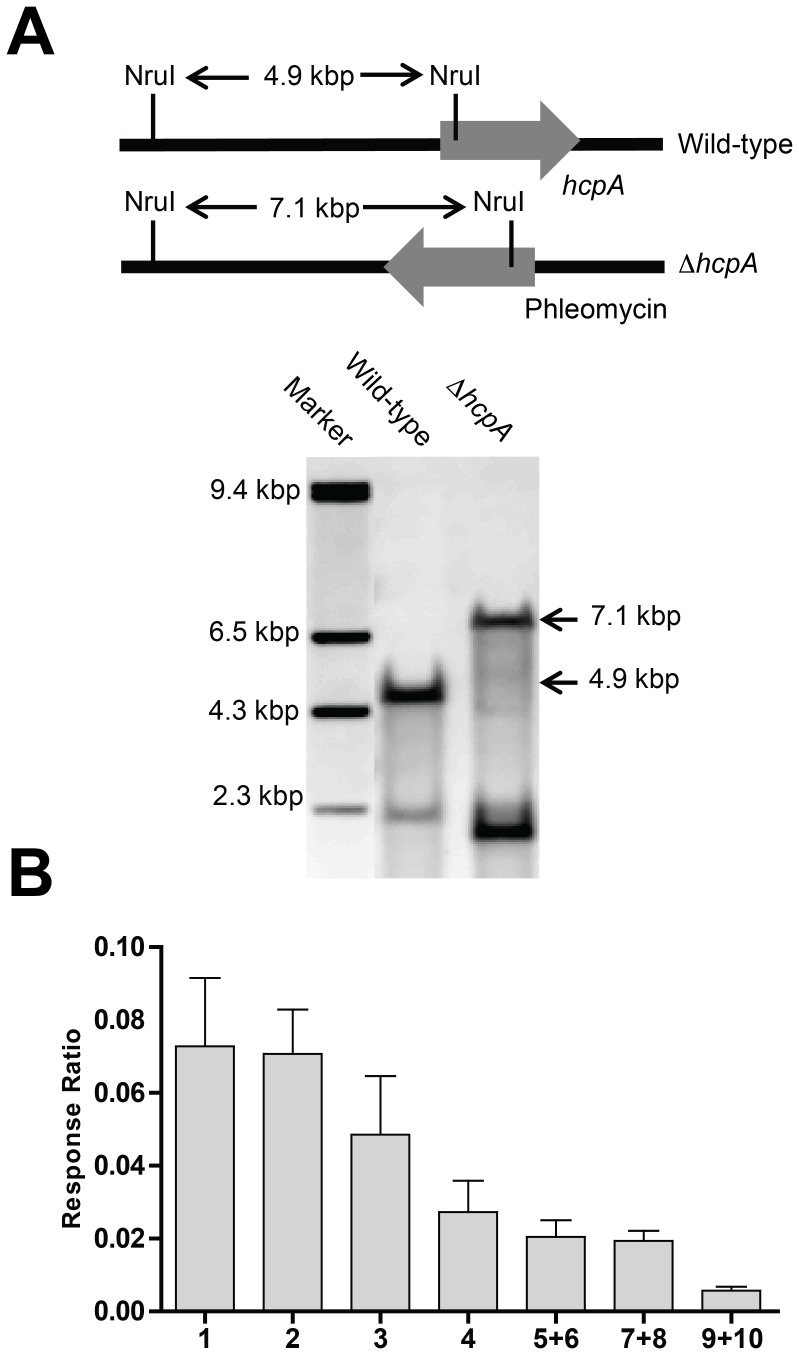
In order to identify the secondary metabolites synthetized by HcpA, the corresponding gene was deleted and comparative metabolite profiling was performed on the culture supernatant of the host and deletion strain. As host strain, P. chrysogenum DS54555 was used, which is derived from the industrial DS17690 strain and that lacks the ku70 gene to make it competent for homologous recombination. The DS54555 strain also lacks the multiple penicillin biosynthetic genes clusters in order to facilitate the detection of unknown secondary metabolites in the culture broth as the profile is no longer dominated by β-lactams. The hcpA gene was resequenced from the genome of P. chrysogenum DS54555 and the nucleotide sequence of the open reading frame (genbank KJ679502) including the promotor region was found to be identical to the hcpA gene present in the sequenced genome of P. chrysogenum Wisconsin54-1255 [15]. The gene deleted by homologous recombination using the deletion plasmid pDEST R4-R3p (Figure S1) containing the flanking regions of hcpA and the phleomycin resistance gene. Colonies were selected on phleomycin containing agar plates where the mutant colonies showed smooth phenotypic characteristics compared to the wrinkled surface of the colonies of the parental strain (Figure 4). The deletion of the hcpA gene was confirmed by southern blot hybridization (Figure 5A).
Figure 4. Colonies of P. chrysogenum strain DS54555.
A: Colony of the wild type strain showing a wrinkled surface. B: Colony of the ΔhcpA strain with a smooth surface.
Figure 5. Southern blot analysis for the hcpA deletion and concentration of cyclic tetrapeptides in culture broth of host strain.
A: Southern blot hybridization validating the complete gene deletion of hcpA. B: Internal standard corrected concentrations of the cyclic tetrapeptides 1–10 present in the culture broth of the host strain of P. chrysogenum grown for 168 hours. Isomers are presented together as no chromatographic separation was obtained during profiling. No cyclic or linear tetrapeptides could be found in the deletion strain.
The host and ΔhcpA strain were grown for 168 hours in SMP Medium followed by comparative metabolite analysis using HPLC-UV-MS. Several secondary metabolites were found to be present in the host but absent in the deletion strain (Figure 6, Table S3). These compounds could be classified into two groups according their chemical structure. The first group consists of ten cyclic tetrapeptides, which were identified using HPLC-MSn, NMR and a synthetic standard (Figure 1). Upon excitation, cyclic tetrapeptides undergo ring opening in the mass spectrometer resulting in four linear tetrapeptides, which can be sequenced in a similar fashion as linear peptides. By following the sequential loss of amino acids from b-ions of each generated linear tetrapeptide, the sequence of their cyclic origin could be determined (Figure S2). The identities of the involved amino acids, corresponding to the losses in the mass spectrometer, as well as their sequence were additionally confirmed by 1H-NMR and 13C-NMR experiments for the compounds 1 and 2 (Table S4 and S5). To discriminate the amino acid isoleucine from its isomer leucine, which is represented by a loss of 113 Da (C6H11ON) in the mass spectra of compounds 5–10, 1H-, 13C- and various 2D-NMR experiments were conducted (Figure S3, Table S6). Overall, cyclic tetrapeptides obtained from metabolic profiling contain various combinations of the five amino acids valine, isoleucine, phenylalanine, tyrosine and tryptophan making them extremely hydrophobic. They are arranged in a common sequence in which two aromatic amino acids are followed by two aliphatic amino acids. The absolute stereochemistry of compound 2 was confirmed by spiking its synthetic standard to a natural extract, which did not lead to additional signals in the 1H-NMR spectra, whereas the intensity of the main signals increased as compared to the impurities (Figure S4A and B). In addition, superimposing the HMBC spectra of the natural and the synthetic sample of peptide 2 illustrated identical correlations and shifts (Figure S5). Furthermore, retention time and MS2 fragmentation did not show differences between the extracted compound and the synthesized standard. This leads to the conclusion that not only the sequence of amino acids is identical in the synthetic and natural peptide, but also the chirality of the individual amino acids. Therefore, the cyclic tetrapeptide 2 has the same stereochemistry as observed for 1 [19] with the first amino acid of an aliphatic and aromatic pair in D- and the second amino acid in L-form. Due to the low concentration of cyclic tetrapeptides 3–10, reliable stereochemical information could not be obtained. However, as these peptides originate from the same NRPS as 1–2 and share the same sequence of aliphatic and aromatic amino acids, identical stereochemistry is expected.
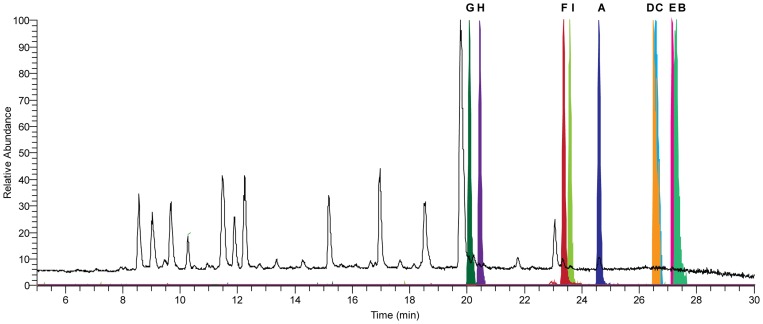
Figure 6. HPLC-MS elution profiles of the internal standard reserpine and the highest abundant identified peptides.
Total ion chromatogram (TIC, black) and normalized extracted ion chromatograms (EIC, colored) of the protonated molecule ([M+H]+ with 5 ppm accuracy) of the used internal standard reserpine (A), the four highest abundant cyclic tetrapeptides 1 (B), 2 (C), 3 (D), 4 (E) and linear tetrapeptides 11–13 (F), 14–16 (G), 17–19 (H), 20–22 (I) present in the culture broth of P. chrysogenum.
The second category of identified compounds consists of 18 linear tetrapeptides which are comprised of the same five amino acids as their cyclic analogues (Table S3 and S7). Similar to the cyclic peptides, each linear tetrapeptide contains two aliphatic and two aromatic amino acids in different arrangements yielding various isomeric structures. Due to similar chromatographic properties and an identical mass-over-charge ratio, these isomers are represented as a group in data obtained from metabolic profiling (Figure S6A and B). Their structure elucidation is challenging as minor fragments can be attributed to a fragmentation of low abundant linear tetrapeptides as well as to possible sequence scrambling of major linear tetrapeptides which was reported for similar linear peptides [29]. To separate and sequence these isomeric tetrapeptides and to prevent possible sequence scrambling, their N-terminus was derivatized using 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate (AQC) (Figure S6C and D) [30]. De novo peptide sequencing was performed by following the consecutive amino acid losses of various b-ions using multiple-stage fragmentation mass spectrometry (Figure S6E and Table S7). Similar to the cyclic peptides, each linear tetrapeptide incorporates two consecutive aromatic and/or two consecutive aliphatic amino acids.
In conclusion, several linear and cyclic tetrapeptides with similar structural features were found to be present in the host but absent in the deletion strain which shows that they originate from one single NRPS, namely HcpA.
Expression of the hcpA gene and secondary metabolite production
To examine the expression of the hcpA gene, the host strain was grown for up to 216 hours in SMP medium. Samples were collected for total mRNA extraction and extracellular metabolites analysis during growth. Metabolite concentrations were determined by HPLC-UV-MS, while transcript levels were determined by quantitative PCR using γ-actin as a reference gene. A high expression level of the hcpA gene was observed between 120 and 168 hours of growth, which was paralleled by a 12 times increase in the concentration of cyclic tetrapeptide 1 (Figure 2A and B) in the medium. In general, the concentration of cyclic tetrapeptides was exceptionally high around 168 hours of growth, while the concentration of the linear tetrapeptides increased with time. These data suggest that the linear tetrapeptides are derived from the cyclic tetrapeptides by degradation.
Discussion
Here we have demonstrated that the NRPS HcpA is responsible for the biosynthesis of cyclic hydrophobic tetrapeptides. Bioinformatics analysis of HcpA shows an unusual domain architecture in comparison to classical linear NRPSs with no products associated so far. However, through the deletion of the hcpA gene and comparative metabolite profiling, ten cyclic tetrapeptides were identified, including the previously described secondary metabolite fungisporin 1 [19]. According to the non-ribosomal code in combination with discovered cyclic products, the first NRPS module is specific for phenylalanine. Due to the adjacent E domain, responsible for epimerization of the activated amino acid, a D-configuration is expected as observed for product 1 (Figure 1). The next two modules of the NRPS contain an unusual architecture in which PCP2 and C3 are flanked by two neighboring adenylation domains A2 and A4. As adenylation domain A4 shows high specificity towards valine and adenylation domain A2 shows high homology towards A1, both adenylation domains activate phenylalanine as supported by the structure of 1. Due to the lack of an adjacent PCP domain for A4 and a missing C domain for A2, which are necessary for loading and condensation of the substrate, no module activity is expected according to the classical C-A-PCP geometry (Figure 3A). Surprisingly, both products 1 and 2 show the incorporation of L-phenylalanine at the second position. As module 2 is predicted to be the only module capable to catalyze the incorporation of L-phenylalanine, A2 indeed must be active. Consequently, it seems likely that A4 is skipped, leaving the N-terminal C2 domain of module 2 to catalyze the condensation of the first amino acid from module 1 to the second amino acid from module 2, as observed for the products 1 and 2. The third module of HcpA contains the domains C3, A3, PCP3 and E arranged in a common linear order. The A3 domain is predicted to activate valine, which agrees with the peptide sequence of 1 and 2, as D-valine is their third amino acid. The fourth module of the NRPS is an incomplete module consisting of C4 and PCP4. Due to a missing preceding A domain no activity is expected. However, the chemical structures of both cyclic tetrapeptides 1 and 2 show the incorporation of L-valine as fourth amino acid in their peptide sequence. As A4 is the only domain predicted to be specific to valine without an adjacent epimerization domain, it is very likely that this domain is a ‘trans-acting’ A domain that interacts with C4 and PCP4 to add the last amino acid to the tetrapeptide. A similar architectural flexibility has been observed in the biosynthesis of yersiniabactin, in which one A domain, located in HMWP2, loads three PCPs located on different modules [31]–[33]. As the linear domain organization of HcpA does not reflect a linear assembly of substrate incorporation into the final product, non-linear interactions are deduced. Although A4, C4 and PCP4 are not in a consecutive sequence on a genomic level, they might still be closely arranged in the final three-dimensional enzymatic structure. Structural characterization would be necessary to determine spatial proximity. Finally, after the incorporation of L-valine into the peptide chain, the C and PCP domain of the last module catalyze the cyclization of the peptide leading to the final cyclic structure, as previously observed in other NRPS systems [34], [35]. It should be stressed that non-linear NRPS organizations are a very heterogenous group of NRPS systems which deviate from the colinearity rule thus showing various unusual mechanisms [6]. In an alternative model A4 might be non-functional, leaving A3 loading two PCPs, namely PCP3 and PCP4, similar to the cysteine-specific A domain of HMWP2 in yersiniabactin biosynthesis [3]. However, more detailed biochemical studies are required to fully understand the interplay between these enzymes and to confirm the exact biosynthetic mechanisms involved.
Next to the production of 1 and 2, eight additional lower abundant cyclic tetrapeptides were identified to be present in the host strain and absent in the deletion strain (Figure 1). They show a similar peptide sequence as 1, containing two aromatic amino acids followed by two aliphatic amino acids. Although stereochemical information is only available for the compounds 1 and 2, it can be assumed that each of the cyclic tetrapeptides contains an aliphatic and aromatic amino acid in the D configuration, more specifically at the first and third position. These assumptions in combination with the stereochemical structure of 1 and 2 lead to the conclusion, that each adenylation domain of HcpA shows specificity towards more than one precursor amino acid with A1 being specific towards phenylalanine and tyrosine and A2 being specific towards phenylalanine and to a lesser extent to tryptophan (Table S8) reminiscent of microheterogenicity. Together with the two aliphatic amino acid selecting adenylation domains A3 and A4, which preferably activate valine before isoleucine, 16 cyclic tetrapeptide combinations are theoretically possible. However, only ten of these were detected in the fermentation broth of P. chrysogenum confirming a different degree of specificity towards their precursors. Based on a similar chemical scaffold of identified compounds 1–10 to the tetrapeptides cyclo-(N-MePhe-Ile)2, cyclo-(N-MePhe-Val)2 and cyclo-(N-MePhe-Val-N-MePhe-Ile) reported from Onychocola sclerotic, cardiac channel blocking activities can be expected for the hydrophobic cyclic peptides presented here [36]. In addition, the colonies of the ΔhcpA strain lost the ability to produce a wrinkled surface leading to a rather smooth appearance (Figure 4). As this change is attributed to the deletion of the hcpA gene, the hydrophobic cyclic peptides 1–10 need to be involved. Possibly, these molecules function analogous to hydrophobins in altering the surface properties and influencing aerial growth. The exact function is, however, still unclear.
In addition to the cyclic tetrapeptides several highly abundant linear tetrapeptides could be observed in the cultural broth of the host strain that were absent in the deletion strain. To each of the cyclic tetrapeptides, several linear tetrapeptides with the same sequence were present. For instance, for the cyclic tetrapeptide 1 with the sequence cyclo-(Phe-Phe-Val-Val), three linear tetrapeptides with the sequences Phe-Val-Val-Phe, Val-Phe-Phe-Val and Phe-Phe-Val-Val could be found at different ratios. Their concentration increased over time in the media while their cyclic counterpart decreased after 168 hours (Figure 2B). This leads to the conclusion that the linear peptides originate from the degradation of their cyclic counterparts by hydrolysis of their peptide bonds, which was observed exclusively between two aromatic, two aliphatic or an aliphatic followed by an aromatic amino acid (Table S3 and S7). Linear tetrapeptides with a N-terminal aliphatic amino acid and a C-terminal aromatic amino acid were not detected, leading to the conclusion that cleavage of this bond is not favorable. As cyclic tetrapeptides are relatively stable towards chemical and thermal degradation, enzymatic hydrolysis might be most probable.
P. chrysogenum contains a second NRPS that could potentially be involved in tetrapeptide formation, i.e., Pc13g14330. This protein has a linear organization and thus differs from the HcpA protein. Pc13g14330 is hardly expressed under batch culture conditions as employed in this study. Moreover, overexpression of Pc13g14330 under control of the strong pcbC promoter did not lead to any novel detectable metabolite in the growth medium, nor did the deletion of the Pc13g14330 gene affect the cyclic tetrapeptide production or result in a loss of other metabolites (unpublished data). Therefore, we conclude that Pc13g14330 is unrelated to HcpA, and not responsible for cyclic tetrapeptide formation. HcpA shows 54% amino acid sequence identity with the orthologous protein from A. niger with exactly the same module organization (Figure 3A). Furthermore, all cyclic products 1–10 present in P. chrysogenum could also be found in the supernatant of an A. niger strain while they were absent in the HcpA deletion strain (unpublished data). Therefore, it is concluded that HcpA is involved in production of all cyclic metabolites (1–10) in A. niger. A small difference exists in the organization of both HcpA proteins with a short additional amino acid sequence present between domains A4 and A2 in the A. nidulans enzyme. This sequence showed limited homology to a condensation domain and appears as an incomplete condensation domain. Hence, one may deduce that this non-canonical situation might have evolved quite recently. Perhaps, a complete C-A-PCP-C-A-PCP module structure was present before, but degenerated after new interactions between the domains evolved.
Conclusions
Cyclic tetrapeptides are a structurally interesting group of peptides produced by fungi that have attracted much interest because of their cardiac ion-channel blocking properties. The complete biosynthetic mechanism of these metabolites has been deduced by gene deletion experiments in P. chrysogenum in combination with comparative metabolite profiling and consecutive structure elucidation. By this analysis, trans-aminoacylation for chain elongation in NRPS has been found. Furthermore, a distinct microheterogenicity of each adenylation domain towards different amino acid building blocks resulted in a range of cyclic tetrapeptides as produced by a single NRP synthetase.
Supporting Information
Plasmid used for the deletion of the hcpA gene in P. chrysogenum .
(TIF)
Multiple-stage fragmentation for de novo sequencing of compound 2 based on sequential amino acid losses. A: MS2 fragmentation spectra of the cyclic tetrapeptide 2 cyclo-(d-Tyr-l-Phe-d-Val-l-Val). Due to ring opening of the cyclic peptide in the mass spectrometer at different positions, three different amino acid losses occurred, yielding different b3-ions. B-D: MS3 fragmentation spectra obtained by further fragmenting b3-ions from MS2 showing b2-ions used for peptide sequencing. The cyclic tetrapeptides 1 and 3–10 were identified accordingly.
(TIF)
HSQC spectrum of isolated mixture of cyclic tetrapeptides in DMSO. The spectra were used for the identification of isoleucine present in the minor abundant products 5 and 9. Signals corresponding to isoleucine are indicated with circles. Conducted TOCSY, COSY and HMBC experiments further confirm this conclusion (data not shown). 1H and 13C chemical shifts are shown in Table S6.
(TIF)
1H-NMR spectra of mixtures of cyclic tetrapeptides. A: 1H-NMR spectrum of a precipitate of various cyclic tetrapeptides containing primarily peptide 1 and 2 (bottom). Synthetic compound 2 spiked to the precipitated mix of various cyclic tetrapeptides in DMSO at 340 K (top). Signals corresponding to 2 were increased as compared to the impurities whereas additional signals did not appear. B: Zoomed regions of 1H-NMR spectrum of natural precipitate (bottom) and precipitate spiked with compound 2 (top). Signals which increased after spiking are indicated (*).
(TIF)
Superimposed HMBC spectra of synthetic compound 2 (red) and an isolated mixture of cyclic tetrapeptides containing naturally produced compound 2 (black). Correlations between NH and CO are shown which indicate identical shifts for both samples. Assignments can be found in Table S4.
(TIF)
Sequencing of the linear isomers 11 (Phe-Phe-Val-Val), 12 (Val-Phe-Phe-Val) and 13 (Phe-Val-Val-Phe). A: Extracted ion chromatogram (EIC) of the linear tetrapeptides 11, 12 and 13 using the profiling method. No chromatographic separation could be achieved. B: MS2 fragmentation spectrum of unseparated linear tetrapeptides 11–13. Although the spectrum is dominated by fragments originating from 11, several lower abundant fragments can be found originating from a fragmentation of 12 and 13 or possible sequence scrambling of 11. C: Sequence of linear tetrapeptides after N-terminal AQC derivatization to achieve better chromatographic separation and to prevent sequence scrambling. D: Normalized total ion chromatogram (TIC) of AQC derivatized linear peptides 11–13 after first C-terminal amino acid loss showing chromatographic separation and allowing peptide sequencing. E: Individual MS3 fragmentation spectra of chromatographically separated derivatized peptides 11, 12 and 13 showing b2 and b1 ions used for peptide sequencing. The linear peptides 14–28 were identified accordingly.
(TIF)
Primers designed for making the hcpA deletion construct in P. chrysogenum .
(DOCX)
Primers designed for the expression analysis of the hcpA and actin gene.
(DOCX)
Retention time, formula and acquired m/z of cyclic and linear tetrapeptides obtained from metabolic profiling. Isomers have the same retention time as they could not be chromatographically separated during profiling.
(DOCX)
1H and 13C-NMR chemical shifts of synthetically produced compound 2 with sequence cyclo -( d -Tyr- l -Phe- d -Val- l -Val) in DMSO at 340 K. As synthetically and naturally produced 2 show identical NMR spectra, only chemical shifts for the synthetically produced compound are shown. δDMSO (1H/13C) = (2.55/40.50), (δ in ppm).
(DOCX)
1H and 13C chemical shifts of naturally produced compound 1 with sequence cyclo -( d -Phe- l -Phe- d -Val- l -Val) present in a mix of various cyclic tetrapeptides in DMSO acquired at 340 K. Signals overlapping with the highest abundant compound 2 are indicated (*). δDMSO (1H/13C) = (2.55/40.50), (δ in ppm).
(DOCX)
1H and 13C chemical shifts of isoleucine in compound 5 with sequence cyclo- (Phe-Phe-Val-Ile) and compound 9 with sequence cyclo- (Phe-Phe-Val-Ile) present in a extracted mix of cyclic tetrapeptides. NMR signals corresponding to C = O and NH as well as remaining amino acid signals are not observed (n.o.) due to overlap with the main constituents. δDMSO (1H/13C) = (2.55/40.50).
(DOCX)
Sequencing data leading to the identification of linear tetrapeptides after AQC derivatization. Multiple-stage fragmentation was used for determination of b-ions for de-novo peptide sequencing.
(DOCX)
Identified and proposed cyclic and linear tetrapeptides in respect to the adenylation domain specificity in the NRPS HcpA.
(DOCX)
Acknowledgments
We thank Oleksandr Salo for processing of the hcpA gene sequence.
Funding Statement
HA and MR were supported by STW (Stichting Technische Wetenschappen) in the framework of the Genbiotics Perspectief Programme. HA was also supported by Higher education commission Pakistan (HEC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Schwarzer D, Finking R, Marahiel MA (2003) Nonribosomal peptides: from genes to products. Nat Prod Rep 20: 275–287. [DOI] [PubMed] [Google Scholar]
- 2. Grunewald J, Marahiel MA (2006) Chemoenzymatic and template-directed synthesis of bioactive macrocyclic peptides. Microbiol Mol Biol Rev 70: 121–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mootz HD, Schwarzer D, Marahiel MA (2002) Ways of assembling complex natural products on modular nonribosomal peptide synthetases. Chembiochem 3: 490–504. [DOI] [PubMed] [Google Scholar]
- 4. Fischbach MA, Walsh CT (2006) Assembly-line enzymology for polyketide and nonribosomal Peptide antibiotics: logic, machinery, and mechanisms. Chem Rev 106: 3468–3496. [DOI] [PubMed] [Google Scholar]
- 5. Shaw-Reid CA, Kelleher NL, Losey HC, Gehring AM, Berg C, et al. (1999) Assembly line enzymology by multimodular nonribosomal peptide synthetases: the thioesterase domain of E. coli EntF catalyzes both elongation and cyclolactonization. Chem Biol 6: 385–400. [DOI] [PubMed] [Google Scholar]
- 6. Sussmuth R, Muller J, von Dohren H, Molnar I (2011) Fungal cyclooligomer depsipeptides: from classical biochemistry to combinatorial biosynthesis. Nat Prod Rep 28: 99–124. [DOI] [PubMed] [Google Scholar]
- 7. Stevens BW, Lilien RH, Georgiev I, Donald BR, Anderson AC (2006) Redesigning the PheA domain of gramicidin synthetase leads to a new understanding of the enzyme's mechanism and selectivity. Biochemistry 45: 15495–15504. [DOI] [PubMed] [Google Scholar]
- 8. Wyatt MA, Mok MC, Junop M, Magarvey NA (2012) Heterologous expression and structural characterisation of a pyrazinone natural product assembly line. Chembiochem 13: 2408–2415. [DOI] [PubMed] [Google Scholar]
- 9. Zhang K, Nelson KM, Bhuripanyo K, Grimes KD, Zhao B, et al. (2013) Engineering the substrate specificity of the DhbE adenylation domain by yeast cell surface display. Chem Biol 20: 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baltz RH (2011) Function of MbtH homologs in nonribosomal peptide biosynthesis and applications in secondary metabolite discovery. J Ind Microbiol Biotechnol 38: 1747–1760. [DOI] [PubMed] [Google Scholar]
- 11. Conti E, Stachelhaus T, Marahiel MA, Brick P (1997) Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J 16: 4174–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Doekel S, Coeffet-Le Gal MF, Gu JQ, Chu M, Baltz RH, et al. (2008) Non-ribosomal peptide synthetase module fusions to produce derivatives of daptomycin in Streptomyces roseosporus. Microbiology 154: 2872–2880. [DOI] [PubMed] [Google Scholar]
- 13. Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, et al. (2010) SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol 47: 736–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, et al. (2007) Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol 25: 221–231. [DOI] [PubMed] [Google Scholar]
- 15. van den Berg MA, Albang R, Albermann K, Badger JH, Daran JM, et al. (2008) Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat Biotechnol 26: 1161–1168. [DOI] [PubMed] [Google Scholar]
- 16. Frisvad JC, Smedsgaard J, Larsen TO, Samson RA (2004) Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Studies in Mycology: 201–241. [Google Scholar]
- 17. Garcia-Estrada C, Ullan RV, Albillos SM, Fernandez-Bodega MA, Durek P, et al. (2011) A single cluster of coregulated genes encodes the biosynthesis of the mycotoxins roquefortine C and meleagrin in Penicillium chrysogenum. Chem Biol 18: 1499–1512. [DOI] [PubMed] [Google Scholar]
- 18. Ali H, Ries MI, Nijland JG, Lankhorst PP, Hankemeier T, et al. (2013) A branched biosynthetic pathway is involved in production of roquefortine and related compounds in Penicillium chrysogenum. PLoS One 8: e65328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Studer RO (1969) Synthesis and structure of fungisporin. Experientia 25: 899. [DOI] [PubMed] [Google Scholar]
- 20. Miyao K (1960) The structure of fungisporin (Studies on fungisporin III). Bull Agr Chem Soc Japan 24: 23–30. [Google Scholar]
- 21.Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press 2nd ed.
- 22. Kovalchuk A, Weber SS, Nijland JG, Bovenberg RA, Driessen AJ (2012) Fungal ABC transporter deletion and localization analysis. Methods Mol Biol 835: 1–16. [DOI] [PubMed] [Google Scholar]
- 23. Alvarez E, Cantoral JM, Barredo JL, Diez B, Martin JF (1987) Purification to homogeneity and characterization of acyl coenzyme A:6-aminopenicillanic acid acyltransferase of Penicillium chrysogenum. Antimicrob Agents Chemother 31: 1675–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kolar M, Punt PJ, van den Hondel CA, Schwab H (1988) Transformation of Penicillium chrysogenum using dominant selection markers and expression of an Escherichia coli lacZ fusion gene. Gene 62: 127–134. [DOI] [PubMed] [Google Scholar]
- 25. Harju S, Fedosyuk H, Peterson KR (2004) Rapid isolation of yeast genomic DNA: Bust n' Grab. BMC Biotechnol 4: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nijland JG, Kovalchuk A, van den Berg MA, Bovenberg RA, Driessen AJ (2008) Expression of the transporter encoded by the cefT gene of Acremonium chrysogenum increases cephalosporin production in Penicillium chrysogenum. Fungal Genet Biol 45: 1415–1421. [DOI] [PubMed] [Google Scholar]
- 27. Noga MJ, Dane A, Shi S, Attali A, van Aken H, et al. (2012) Metabolomics of cerebrospinal fluid reveals changes in the central nervous system metabolism in a rat model of multiple sclerosis. Metabolomics 8: 253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rottig M, Medema MH, Blin K, Weber T, Rausch C, et al. (2011) NRPSpredictor2–a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res 39: W362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bleiholder C, Osburn S, Williams TD, Suhai S, Van Stipdonk M, et al. (2008) Sequence-scrambling fragmentation pathways of protonated peptides. J Am Chem Soc 130: 17774–17789. [DOI] [PubMed] [Google Scholar]
- 30. Cohen SA, Michaud DP (1993) Synthesis of a fluorescent derivatizing reagent, 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate, and its application for the analysis of hydrolysate amino acids via high-performance liquid chromatography. Anal Biochem 211: 279–287. [DOI] [PubMed] [Google Scholar]
- 31. Gehring AM, DeMoll E, Fetherston JD, Mori I, Mayhew GF, et al. (1998) Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem Biol 5: 573–586. [DOI] [PubMed] [Google Scholar]
- 32. Gehring AM, Mori I, Perry RD, Walsh CT (1998) The nonribosomal peptide synthetase HMWP2 forms a thiazoline ring during biogenesis of yersiniabactin, an iron-chelating virulence factor of Yersinia pestis. Biochemistry 37: 11637–11650. [DOI] [PubMed] [Google Scholar]
- 33. Suo Z, Tseng CC, Walsh CT (2001) Purification, priming, and catalytic acylation of carrier protein domains in the polyketide synthase and nonribosomal peptidyl synthetase modules of the HMWP1 subunit of yersiniabactin synthetase. Proc Natl Acad Sci U S A 98: 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gao X, Haynes SW, Ames BD, Wang P, Vien LP, et al. (2012) Cyclization of fungal nonribosomal peptides by a terminal condensation-like domain. Nat Chem Biol 8: 823–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Keating TA, Ehmann DE, Kohli RM, Marshall CG, Trauger JW, et al. (2001) Chain termination steps in nonribosomal peptide synthetase assembly lines: directed acyl-S-enzyme breakdown in antibiotic and siderophore biosynthesis. Chembiochem 2: 99–107. [DOI] [PubMed] [Google Scholar]
- 36. Perez-Victoria I, Martin J, Gonzalez-Menendez V, de Pedro N, El Aouad N, et al. (2012) Isolation and structural elucidation of cyclic tetrapeptides from Onychocola sclerotica. J Nat Prod 75: 1210–1214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Plasmid used for the deletion of the hcpA gene in P. chrysogenum .
(TIF)
Multiple-stage fragmentation for de novo sequencing of compound 2 based on sequential amino acid losses. A: MS2 fragmentation spectra of the cyclic tetrapeptide 2 cyclo-(d-Tyr-l-Phe-d-Val-l-Val). Due to ring opening of the cyclic peptide in the mass spectrometer at different positions, three different amino acid losses occurred, yielding different b3-ions. B-D: MS3 fragmentation spectra obtained by further fragmenting b3-ions from MS2 showing b2-ions used for peptide sequencing. The cyclic tetrapeptides 1 and 3–10 were identified accordingly.
(TIF)
HSQC spectrum of isolated mixture of cyclic tetrapeptides in DMSO. The spectra were used for the identification of isoleucine present in the minor abundant products 5 and 9. Signals corresponding to isoleucine are indicated with circles. Conducted TOCSY, COSY and HMBC experiments further confirm this conclusion (data not shown). 1H and 13C chemical shifts are shown in Table S6.
(TIF)
1H-NMR spectra of mixtures of cyclic tetrapeptides. A: 1H-NMR spectrum of a precipitate of various cyclic tetrapeptides containing primarily peptide 1 and 2 (bottom). Synthetic compound 2 spiked to the precipitated mix of various cyclic tetrapeptides in DMSO at 340 K (top). Signals corresponding to 2 were increased as compared to the impurities whereas additional signals did not appear. B: Zoomed regions of 1H-NMR spectrum of natural precipitate (bottom) and precipitate spiked with compound 2 (top). Signals which increased after spiking are indicated (*).
(TIF)
Superimposed HMBC spectra of synthetic compound 2 (red) and an isolated mixture of cyclic tetrapeptides containing naturally produced compound 2 (black). Correlations between NH and CO are shown which indicate identical shifts for both samples. Assignments can be found in Table S4.
(TIF)
Sequencing of the linear isomers 11 (Phe-Phe-Val-Val), 12 (Val-Phe-Phe-Val) and 13 (Phe-Val-Val-Phe). A: Extracted ion chromatogram (EIC) of the linear tetrapeptides 11, 12 and 13 using the profiling method. No chromatographic separation could be achieved. B: MS2 fragmentation spectrum of unseparated linear tetrapeptides 11–13. Although the spectrum is dominated by fragments originating from 11, several lower abundant fragments can be found originating from a fragmentation of 12 and 13 or possible sequence scrambling of 11. C: Sequence of linear tetrapeptides after N-terminal AQC derivatization to achieve better chromatographic separation and to prevent sequence scrambling. D: Normalized total ion chromatogram (TIC) of AQC derivatized linear peptides 11–13 after first C-terminal amino acid loss showing chromatographic separation and allowing peptide sequencing. E: Individual MS3 fragmentation spectra of chromatographically separated derivatized peptides 11, 12 and 13 showing b2 and b1 ions used for peptide sequencing. The linear peptides 14–28 were identified accordingly.
(TIF)
Primers designed for making the hcpA deletion construct in P. chrysogenum .
(DOCX)
Primers designed for the expression analysis of the hcpA and actin gene.
(DOCX)
Retention time, formula and acquired m/z of cyclic and linear tetrapeptides obtained from metabolic profiling. Isomers have the same retention time as they could not be chromatographically separated during profiling.
(DOCX)
1H and 13C-NMR chemical shifts of synthetically produced compound 2 with sequence cyclo -( d -Tyr- l -Phe- d -Val- l -Val) in DMSO at 340 K. As synthetically and naturally produced 2 show identical NMR spectra, only chemical shifts for the synthetically produced compound are shown. δDMSO (1H/13C) = (2.55/40.50), (δ in ppm).
(DOCX)
1H and 13C chemical shifts of naturally produced compound 1 with sequence cyclo -( d -Phe- l -Phe- d -Val- l -Val) present in a mix of various cyclic tetrapeptides in DMSO acquired at 340 K. Signals overlapping with the highest abundant compound 2 are indicated (*). δDMSO (1H/13C) = (2.55/40.50), (δ in ppm).
(DOCX)
1H and 13C chemical shifts of isoleucine in compound 5 with sequence cyclo- (Phe-Phe-Val-Ile) and compound 9 with sequence cyclo- (Phe-Phe-Val-Ile) present in a extracted mix of cyclic tetrapeptides. NMR signals corresponding to C = O and NH as well as remaining amino acid signals are not observed (n.o.) due to overlap with the main constituents. δDMSO (1H/13C) = (2.55/40.50).
(DOCX)
Sequencing data leading to the identification of linear tetrapeptides after AQC derivatization. Multiple-stage fragmentation was used for determination of b-ions for de-novo peptide sequencing.
(DOCX)
Identified and proposed cyclic and linear tetrapeptides in respect to the adenylation domain specificity in the NRPS HcpA.
(DOCX)