Abstract
Pseudomonas extremaustralis produces mainly polyhydroxybutyrate (PHB), a short chain length polyhydroxyalkanoate (sclPHA) infrequently found in Pseudomonas species. Previous studies with this strain demonstrated that PHB genes are located in a genomic island. In this work, the analysis of the genome of P. extremaustralis revealed the presence of another PHB cluster phbFPX, with high similarity to genes belonging to Burkholderiales, and also a cluster, phaC1ZC2D, coding for medium chain length PHA production (mclPHA). All mclPHA genes showed high similarity to genes from Pseudomonas species and interestingly, this cluster also showed a natural insertion of seven ORFs not related to mclPHA metabolism. Besides PHB, P. extremaustralis is able to produce mclPHA although in minor amounts. Complementation analysis demonstrated that both mclPHA synthases, PhaC1 and PhaC2, were functional. RT-qPCR analysis showed different levels of expression for the PHB synthase, phbC, and the mclPHA synthases. The expression level of phbC, was significantly higher than the obtained for phaC1 and phaC2, in late exponential phase cultures. The analysis of the proteins bound to the PHA granules showed the presence of PhbC and PhaC1, whilst PhaC2 could not be detected. In addition, two phasin like proteins (PhbP and PhaI) associated with the production of scl and mcl PHAs, respectively, were detected. The results of this work show the high efficiency of a foreign gene (phbC) in comparison with the mclPHA core genome genes (phaC1 and phaC2) indicating that the ability of P. extremaustralis to produce high amounts of PHB could be explained by the different expression levels of the genes encoding the scl and mcl PHA synthases.
Introduction
Polyhydroxyalkanoates (PHAs) are reserve polymers produced by a wide range of bacteria accumulated in intracellular granules as carbon and energy storage material under unbalanced growth conditions. These polymers exhibit thermoplastic and elastomeric features in addition to other interesting properties, such us biodegradability and biocompatibility, which make them promising industrial materials [1]. The physical and chemical properties of these bioplastics are directly related to the monomer composition of the polymer. According to the number of carbons in the monomer units, PHAs can be classified into two classes: short-chain-length PHAs (sclPHAs) and medium-chain-length PHAs (mclPHAs). The sclPHAs, composed of C3 to C5 monomer units, are thermoplastic in nature, hard and brittle, meanwhile mclPHAs, which are formed of C6 to C14 monomer units, are elastomeric in nature.
The gene cluster involved in polyhydroxybutyrate (PHB) production, the most widely produced sclPHA by bacteria, includes three structural genes named phaA or phbA, phaB or phbB and phaC or phbC, encoding a β-ketothiolase, a (R)-specific acetoacetyl-CoA reductase and a class I synthase, respectively. Class I synthase is the key enzyme for PHB biosynthesis.
The mclPHA cluster (phaC1ZC2DFI) is well conserved among the mclPHA producers bacteria. This cluster encode two class II synthase genes (phaC1 and phaC2) separated by a gene encoding a PHA depolymerase (phaZ); a transcriptional activator (phaD) and two genes (phaFI) encoding phasin-like proteins [2]. In most Pseudomonas, β-oxidation and fatty acid de novo synthesis are used to convert fatty acid or carbohydrate intermediates, respectively into (R)-3-hydroxyacyl-CoAs, which are used as substrates by the Class II PHA synthases [3].
Synthesis of mcl-PHAs has been observed in fluorescent and non-fluorescent Pseudomonas species, whereas PHB production is not a common characteristic of Pseudomonas spp. [4]. PHB production has been reported in some Pseudomonas strains, such as those included in the “P. oleovorans group” [5].
Pseudomonas extremaustralis is a highly stress resistant bacterium isolated from Antarctica, able to accumulate large quantities of PHB [6]. Previous studies in this strain have allowed the identification of a PHB gene cluster (phaRBAC) located within a genomic island [7], [8]. The PHB gene cluster of P. extremaustralis showed a mosaic structure with genes of different origin, indicating their acquisition by horizontal transfer [8]. Recently, we have obtained the complete genome of P. extremaustralis 14-3b (DSM 25547), a natural derivative of the type strain (14-3T) [9]. This strain accumulates mainly PHB and minor amounts of mclPHAs when grown with octanoate or glucose as carbon source.
Based on this evidence, we hypothesized that genes involved in mclPHA production should be also present in the genome of P. extremaustralis. In the present work, we identified and analyzed the genes involved in mclPHA metabolism in P. extremaustralis. Besides, the PHB and mclPHA content as well as the expression of the genes encoding the corresponding synthases were compared under different growth conditions. Finally, the PHA granules of P. extremaustralis were isolated in order to identify the proteins bound to PHA in this bacterium.
Materials and Methods
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria belonging to the Pseudomonas genus were cultured at 30°C in 0.5NE2 medium [10] supplemented with sodium octanoate (0.25%) or glucose (0.27 or 3%) as carbon source. The 0.5NE2 medium has reduced nitrogen content that favors PHA accumulation [10]. Escherichia coli was cultured at 37°C in Luria Broth (LB). When necessary, antibiotics were added at the following concentrations: 50 µg.ml−1 kanamycin (Km), 100 µg.ml−1 ampicillin (Amp), 10 µg.ml−1 gentamicin (Gm) and 10 µg.ml−1 tetracycline (Tet).
Table 1. Bacterial strains and plasmids used in this study.
| Strains and plasmids | Relevant characteristics | Source or references |
| Bacterial strains | ||
| P. extremaustralis DSM 25547 (14-3b) | Wild type PHB producer | [9] |
| P. putida GPp104 | PHA negative mutant | [10] |
| Escherichia coli DH5α | fhuA2 Δ(argF-lacZ)U169 phoA glnV44 Φ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17− | [11] |
| E. coli S17-1 | Conjugative. recA, tra genes | [12] |
| Plasmids | ||
| pGEM-T Easy | Cloning vector | Promega |
| pBBR1MCS-5 | GmR; medium-copy number, conjugative | [13] |
| pBBR1MCS-2 | KmR; medium-copy number, conjugative | [13] |
| pSJ33 | KmR; low-copy number, conjugative | [14] |
| pMHZ59 | pBBR1MCS-5 derivative, containing phaC1 | This work |
| pMMic2 | pBBR1MCS-5 derivative, containing phaC2 | This work |
| pM-phbX | pBBR1MCS-2 derivative, containing phbX | This work |
| pPSJ-phbX | pSJ33 derivative, containing phbX | This work |
Screening of genes involved in PHA metabolism
The search of genes responsible of mclPHA production in the genome of P. extremaustralis was first performed using PCR amplification strategies. Degenerate primers were designed to amplify the mclPHA genes of P. extremaustralis, using the sequences of mclPHA genes of other Pseudomonas species available in the databases (Table 2). PCR amplification reactions were performed in a final volume of 50 µl containing 0.3 µM of each primer, 0.2 mM of dNTPs, and 2.5U of Go Taq polymerase (Promega). The reaction mixtures were subjected to 30 cycles of amplification in a Techne Termocycler with the following conditions: 94°C for 1 min, 50/60°C for 30 s, 72°C for 1 min and a final extension of 10 min at 72°C. Amplification products were sequenced by Macrogen Inc. (Korea). Sequences were aligned, assembled and analyzed using the following programs available online: BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi), CAP3 Sequence Assembly Program (http://pbil.univ-lyon1.fr/cap3.php), ORFfinder (http://www.ncbi.nlm.nih.gov/projects/gorf/), and ClustalW (http://www.ebi.ac.uk/Tools/clustalw/). Promoters were predicted using Softberry program (http://linux1.softberry.com/berry.phtml).
Table 2. Oligonucleotides used in this study.
| Gene | Sequence primer 5′ to 3′ forward (up) | Sequence primer 5′ to 3′ reverse (low) |
| phaC1 | TGTTCACYTGGGAVTACC | CGGGTAATCAGGAACAGA |
| phaZ | CAAGCACACCGATTCCTGGTG | TTGAGVAGGGTGTTGGAA |
| phaC2 | TGATCAACATGCGCMTGCT | GGATCCTGAACAAATACAGGGCACAT |
| phaD | CGCTTCGTGCTGTCCAAC | GCYACCAGCATCATSAYCTGRTA |
| phbX | GCGTGGAGATGAGCCAGGAAAGTTTTTTTGCGA | CGCTCGAGTCATTCGGCGTTGCCTCCCTT |
| rtphaC1 | CGATGACTTGAAACGCCA | ACGGTTGCTTGATGGCTT |
| rtphaC2 | GAGAAAGACCCGTGACGAAC | GCGTCCCATCTGACCGCCCAGG |
| rtphbC | CTTCGTCCTCGGATGTTCTG | ATCGACCCACCAACTCCTG |
| rt16S | CAGCTCGTGTCGTGAGATGT | AAGGGCCATGATGACTTGAC |
Subsequently, the availability of the genome of P. extremaustralis allowed us to look for other genes involved in PHA metabolism and to determine their location using the bioinformatic tools included in the RAST server (http://rast.nmpdr.org/). Analysis of genomic islands was performed using the Island Viewer software [15].
Complementation assays
The genes encoding the PHA synthases (phaC1 and phaC2) of P. extremaustralis were amplified by PCR. Fragments of 2.9 and 2.1 kb containing phaC1 and phaC2 genes with their own promoter sequences, respectively, were cloned into the pGEM-T Easy Plasmid (Promega) and subsequently subcloned into the vector pBBR1MCS-5.
The vectors carrying the corresponding genes were introduced by conjugation into the PHA negative strain P. putida GPp104, following a modified protocol from Friedrich et al. [16]. Briefly, cell cultures were grown until exponential phase. Recipient and donor cells were mixed on a 1:1 ratio. Afterwards 1 ml of the mixture was centrifuged; the pellet was resuspended in 100 µl of 10 mM MgSO4 and put onto a sterile filter (0.22 µm, Millipore) over a LB agar plate at 30°C for 18 h. Subsequently, cells were washed off of the filter with 1 ml of 10 mM MgSO4, collected by centrifugation and plated on 0.5NE2 supplemented with 0.25% sodium octanoate and the adequate antibiotic concentration.
Cells from the transconjugant colonies were subjected to Nile Blue staining [17] in order to qualitatively analyze their ability to produced PHA. Quantitative determinations of PHA were also performed from lyophilized cells subjected to methanolysis and hot chloroform extraction. Methyl ester derivatives were analyzed by gas chromatography as previously described [18]. The PHA content was expressed as percentage of cell dry weight (CDW).
Quantitative Real Time RT-PCR
Total RNA was extracted from pellets of late exponential phase cultures (OD600≈1.8) grown under PHA accumulation conditions as described earlier, using the Total RNA extraction kit (Real Biotech Corporation) according to manufacturer's indications. The RNA samples were subsequently treated with DNase I and the cDNA was synthesized using random hexamers (Promega) and AMV retrotranscriptase according to the manufacturer's instructions. Total RNA was quantified using a Nanodrop 2000 Spectrophotometer (Thermo Scientific). RT qPCR amplification of the cDNAs was performed in a LightCycler (DNA Engine M.J. Research) using Hot Firepol EvaGreen qPCR Mix Plus (no ROX, Solis Biodyne), plus 0.25 µM of the following forward and reverse primers corresponding to phbC, phaC1 and phaC2, respectively: rtphbCup/low, rtphaC1up/low and rtphaC2up/low, (Table 2). In addition, primers rt16Sup/low were used for amplification of 16S rRNA gene, that was used for normalization of the pha genes expression levels. The cycling parameters used were: initial denaturation at 95°C for 5 min; 95°C for 15 s, 40 cycles of 56.4°C for 15 s, and a final extension step at 72°C for 5 min. The cDNA from three independent bacterial cultures grown with 0.25% sodium octanoate and 0.27% of glucose as carbon source were used for amplification.
Quantification of the fluorescence values obtained after amplification of the cDNAs was performed using a calibration curve as previously described [19]. Serial dilutions of genomic DNA of P. extremaustralis (0.014 ng to 1.65 ng) were used as template. Genomic DNA extraction was performed with the DNeasy Tissue Kit (Qiagen) following the manufacturer's instructions.
Isolation of native PHA granules
Cultures of P. extremaustralis grown at 30°C in 0.5NE2 medium containing sodium octanoate were used for granule isolation. Exponential and stationary phase cultures were collected after 8 and 24 h of incubation, respectively. In addition, for carbon starvation, 24 h cultures were centrifuged, washed, resuspended in the same culture medium without carbon sources, and incubated for 5 h at 30°C. PHA granules were isolated as previously described in several works [20]. Cell extracts, obtained after disruption of cells through French press, were loaded into a glycerol gradient (87% v/v and 50% v/v). After ultracentrifugation, the granule layer found between the two glycerol solutions was reloaded on top of a second glycerol gradient (87%v/v, 80%v/v, 60% v/v and 40%v/v). Finally, the layer between the 60% and the 80% glycerol solutions was collected and subjected to overnight dialysis in 50 mM cold Tris-HCl buffer. The protein concentration was determined as described by Lowry [21]. The native granule extracts were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) [22] and stained with Coomasie Blue R-250 [23]. The major bands were excised from the gel and identified by high resolution mass spectrometry at the Laboratory of Proteomics (Institute of Biotechnology, UNAM, Mexico). Briefly, samples were reduced, alkylated, digested with trypsin and applied into a LC- MS system consisting of a micro flow Accela (Thermo - Ficher Co.) with “spliter” (1/20) coupled with a mass spectrometer LTQ- Orbitrap Velos (Thermo - Ficher Co.) with nano- electrospray ionization (ESI) source. Database search was carried out using Mascot and Protein Prospector programs.
Sequences accession number
The sequences have been deposited in EMBL database under accession number AHIP01000001-AHIP01000135.
Statistical analysis
For statistical analysis, three parallel experiments were conducted and data represent mean ± SD. Student's two-tailed t-test with confidence level >95% (P<0.05) was performed to determine the significance of differences.
Results
Characterization of genes involved in PHA metabolism in Pseudomonas extremaustralis
Pseudomonas extremaustralis is able to accumulate PHB (Table 3). Previous studies allowed us the identification of the gene cluster (phaRBAC), involved in PHB production, located within a genomic island [8]. Analysis of PHA production in P. extremaustralis showed that this bacterium is also able to produce low quantities of mclPHA during growth with 0.25% sodium octanoate or 3% glucose as carbon sources (Table 3). Since most of the bacteria belonging to the Pseudomonas genus have a conserved cluster of genes involved in mclPHA production, we searched for this cluster of genes in the genome sequence of P. extremaustralis. We found a mclPHA gene cluster comprised by two different genes encoding class II synthases, phaC1 and phaC2 (locus_tag: PE143B_0104515 and PE143B_0104505), which were separated by the phaZ gene (locus_tag: PE143B_0104510), encoding a putative intracellular PHA depolymerase. Inspection of the intergenic region between phaC1 and phaZ revealed the presence of a hairpin structure of 18 bp (ΔG = -37kcal), consisting of a palindromic sequence that displays dyad symmetry. This sequence was similar to the previously found in P. corrugata 388 and P. putida GPo1, which acts as a transcription terminator in both species [24], [25], suggesting a similar role in P. extremaustralis. A putative transcriptional regulator, phaD (locus_tag: PE143B_0104500), was found downstream of phaC2.
Table 3. Production of PHA in P. extremaustralis and recombinants P. putida GPp104.
| Strain | Carbon source | Time of culture (h) | PHB content (wt% of CDW) | PHA content (PHHX+ PHO) (wt% of CDW) |
| P. extremaustralis 14-3b | 0.25% octanoate | 8 | 13.60±1.40 | 3.40±0.30 |
| 0.25% octanoate | 24 | 35.80±3.60 | 1.05±0.10 | |
| 0.27% glucose | 8 | 0.37±0.04 | ND | |
| 0.27% glucose | 24 | 1.30±0.10 | ND | |
| 3% glucose | 24 | 12.20±1.00 | 0.26±0.02 | |
| P. putida GPp104 pMHZ59 | 0.25% octanoate | 24 | 2.60±0.30 | 28.00±0.80 |
| P. putida GPp104 pMMic2 | 0.25% octanoate | 24 | 0.10±0.01 | 9.60±1.00 |
| P. putida GPp104 pMHZ59 | 2% glucose | 24 | 0.60±0.01 | 6.10±0.82 |
| P. putida GPp104 pMMic2 | 2% glucose | 24 | 0.27±0.03 | 2.66±0.33 |
| P. putida GPp104 pBBR1MCS-5 | 0.25% octanoate | 24 | ND | ND |
| P. putida GPp104 pBBR1MCS-5 | 2% glucose | 24 | ND | ND |
Cultures were grown in 0.5NE2 with the indicate carbon source.
ND = no detected, PHHX: polyhydroxyhexanoate, PHO: polyhydroxyoctanoate.
The availability of the genome sequence of P. extremaustralis allowed us to perform a more extensive analysis of the genes involved in mclPHA metabolism. In addition to the phaC1ZC2D gene cluster, two genes, phaF and phaI (locus_tag: PE143B_0104460 and PE143B_0104455), encoding phasin like proteins were identified. These genes were not located immediately downstream phaD, as observed in other Pseudomonas species [26], due to the presence of a natural insertion containing seven ORFs (locus_tag: PE143B_0104495, PE143B_0104490, PE143B_0104485, PE143B_0104480, PE143B_0104475, PE143B_0104470, PE143B_0104465) not related to PHA metabolism (Fig. 1). Some of these ORFs encode putative proteins related to pili and fimbriae similar to those belonging to β-Proteobacteria, like the P pilus assembly protein pilin FimA, a bacterial pili assembly chaperone and the outer membrane usher protein FimD. The deduced amino acid sequences of PhaC1, PhaZ, PhaC2, PhaD, PhaF and PhaI of P. extremaustralis showed a conserved size and high similarity with the corresponding proteins of other Pseudomonas species. The highest similarity was found with P. fluorescens SBW25 [27] sharing 98%, 78%, 96%, 95%, 94% and 75% of identity with PhaC1, PhaZ, PhaC2, PhaD, PhaF and PhaI, respectively. The aminoacids involved in the catalytic activity (Cys 296, Asp 452 and His 480) of the class II synthases [28] and other conserved amino acid (Trp 398 and Cys 431) were present in PhaC1 and PhaC2 of P. extremaustralis.
Figure 1. Genetic organization of the mclPHA gene cluster in P. extremaustralis.
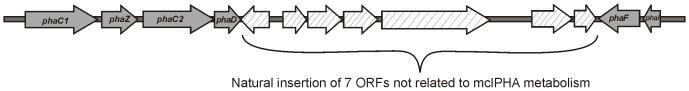
mclPHA genes belonging to γ Proteobacteria (smooth arrows). Natural insertion containing 7 ORFs related to β Proteobacteria between the genes phaD and phaF (broken arrows). Arrows indicate the direction of gene transcription and the relative size of each ORF. From left to right: phaC1(1680 bp), phaZ (846 bp), phaC2 (1683 bp), phaD (621 bp), followed by genes encoding a LuxR family DNA binding response regulator (642 bp), a putative fimbrial subunit (579 bp), a bacterial pili assembly chaperone (780 bp), a pili assembly chaperone (759 bp), an outer membrane usher protein FimD (2505 bp), a putative fimbrial protein (945 bp) and a Pi fimbriae major subunit (528 bp), not related to PHA metabolism, and phaF (927 bp) and phaI (423 bp).
Complementation analyses of the genes encoding the class II PHA synthase genes were performed in order to determine if the identified phaC1 and phaC2 were functionally active. Plasmids pMHZ59 and pMMic2 containing phaC1 and phaC2, respectively, restored PHA production in P. putida GPp104 (a derivative of P. putida KT2442 unable to produce PHAs), during growth with sodium octanoate or glucose as carbon sources (Table 3). Both class II PHA synthases were able to synthesize mainly mclPHA, however PhaC1 was also able to produce low quantities of PHB (around 3% of the cell dry weight) when the bacterium was grown with sodium octanoate as carbon source (Table 3). According to these results, both class II PHA synthases are functionally active in P. extremaustralis.
Further bioinformatic analysis of the genome of P. extremaustralis, allowed the identification of a cluster of 4078 bp, that was named phbFPX, presumably associated with PHB metabolism. The phbFPX cluster was located in the proximity (4000 bp of distance) of the previously identified phaRBAC cluster [8] involved in PHB biosynthesis. The genes included in the phbFPX cluster showed high similarity with sequences belonging to β-Proteobacteria, indicating that they were probably acquired by horizontal gene transfer. Prediction of genomic islands using the Island Viewer Program indicates that both clusters (phaRBAC and phbFPX) could belong to the same genomic island. The bioinformatic analysis of the cluster phbFPX (locus_tag: PE143B_0105770, PE143B_0105765, PE143B_0105760, respectively) revealed the presence of two genes encoding granule associated proteins (GAPs), named phbF and phbP, as well as an ORF that was named phbX. The deduced aminoacid sequences of PhbF and PhbP showed 100% identity with proteins of Pseudomonas stutzeri A1501 and between 60 and 76% identity with GAPs (phasin like or regulatory proteins) belonging to Azotobacter spp. The protein PhbX showed 97% identity with a protein annotated as PHA synthase in the P. stutzeri A1501 genome [29] and 62% identity with a putative PHA depolymerase of Azotobacter vinelandii DJ [30]. PhbX of P. extremaustralis showed an alfa-beta hydrolase domain as described for PHA synthases and depolymerases. However, the key amino acids involved in the functionality of PHA synthases could not be found in this protein and we were not able to complement the PHA negative strain P. putida GPp104 with a plasmid carrying phbX (data not shown). Likewise, the lipase box (G-X1-S-X2-G) present in many PHA depolymerases [31], could not be found in PhbX.
Based on the fact that several gene clusters involved in PHAs metabolism are present in the genome of P. extremaustralis, and in order to avoid confusion, the cluster involved in PHB metabolism that was previously named phaRBAC [7], [8] is now renamed phbRBAC.
Expression of genes encoding PHA synthases
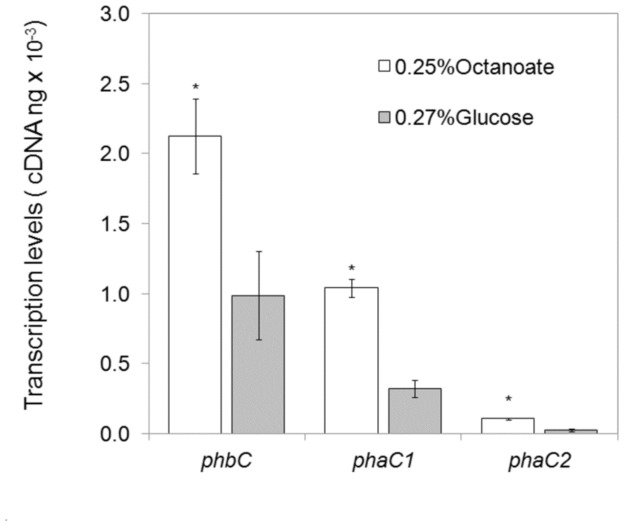
In order to understand why P. extremaustralis produces mainly PHB in spite of having genes encoding functional mclPHA synthases, we analyzed the expression of the three different PHA synthases present in the genome of this strain. The expression levels of the three PHA synthases genes were compared using RNA extracted from late exponential phase cultures (8h of growth, OD600≈1.8) grown under two different PHA accumulation conditions: 0.25% sodium octanoate (high PHA accumulation) or 0.27% glucose (low PHA accumulation). Cultures grown with sodium octanoate accumulated around 14% of PHB, while cells grown with glucose accumulated less than 1% of PHB. Levels of mclPHAs, polyhydroxyoctanoate (PHO) plus polyhydroxyhexanoate (PHHx) were less than 4% or undetectable, when bacteria were grown with sodium octanoate or glucose, respectively (Table 3). The expression of all the PHA synthases genes, phbC, phaC1 and phaC2, was higher in cultures supplemented with octanoate. The level of expression of phbC compared with phaC1 and phaC2, was significantly higher in both culture conditions (P<0.005, Fig. 2). The transcript levels of the class I PHA synthase, phbC, were 2 and 3 times higher than the transcript levels of phaC1, and 19 and 35 times higher than phaC2 when the cells were cultivated with octanoate and glucose, respectively (Fig. 2). Finally, the expression level of phaC1 was 9 and 11 times higher than the expression level of phaC2 when bacteria were grown with octanoate and glucose, respectively.
Figure 2. Analysis of the expression of phbC, phaC1 and phaC2.
qRT-PCR assays were performed with cells grown on 0.5NE2 supplemented with 0.25% sodium octanoate or 0.27% glucose until late exponential phase. Results are the average ± standard deviations from three independent cultures.
Analysis of the proteins associated to the PHA granules
To further investigate the expression and functionality of P. extremaustralis PHB and PHA synthases and to identify the proteins bound to the PHA granules in this bacterium, we carried out the isolation of the PHA granules and the analysis of the major proteins bound to these granules from cultures grown until exponential or stationary phase under different polymer accumulation conditions, using sodium octanoate or glucose as carbon sources. The composition of the proteins bound to the PHA granules was also analyzed from cells subjected to 5h of carbon starvation. SDS-PAGE of PHA granules, extracted from cells grown under different conditions, displayed a similar qualitative pattern of proteins (data not shown). For that reason, only the proteins obtained from granules extracted from 24 h-cultures, grown with sodium octanoate, were identified. Based on the molecular weight of the proteins normally found in PHA granules, and their relative amount in the SDS-PAGE (Fig. 3), nine bands were excised for the gel and subsequent identified by peptide fingerprint analysis (Table 4). Several proteins directly involved in PHA metabolism, such as the synthases PhbC and PhaC1, the phasin like protein PhaI and the phasin like protein PhbP, were identified among the granule bound proteins. PhbP represented the major band (Fig 3). In addition, PhbX, encoded by an ORF located downstream of phbP, was also found in the granules. Another four proteins, not known to be related to PHA metabolism, were also found: the cytosolic aminopeptidase PepA, a possible secretion protein, the heat shock protein Hsp20 and the DNA binding protein HU (Table 4).
Figure 3. Proteins associated to the PHA granules of P. extremaustralis.
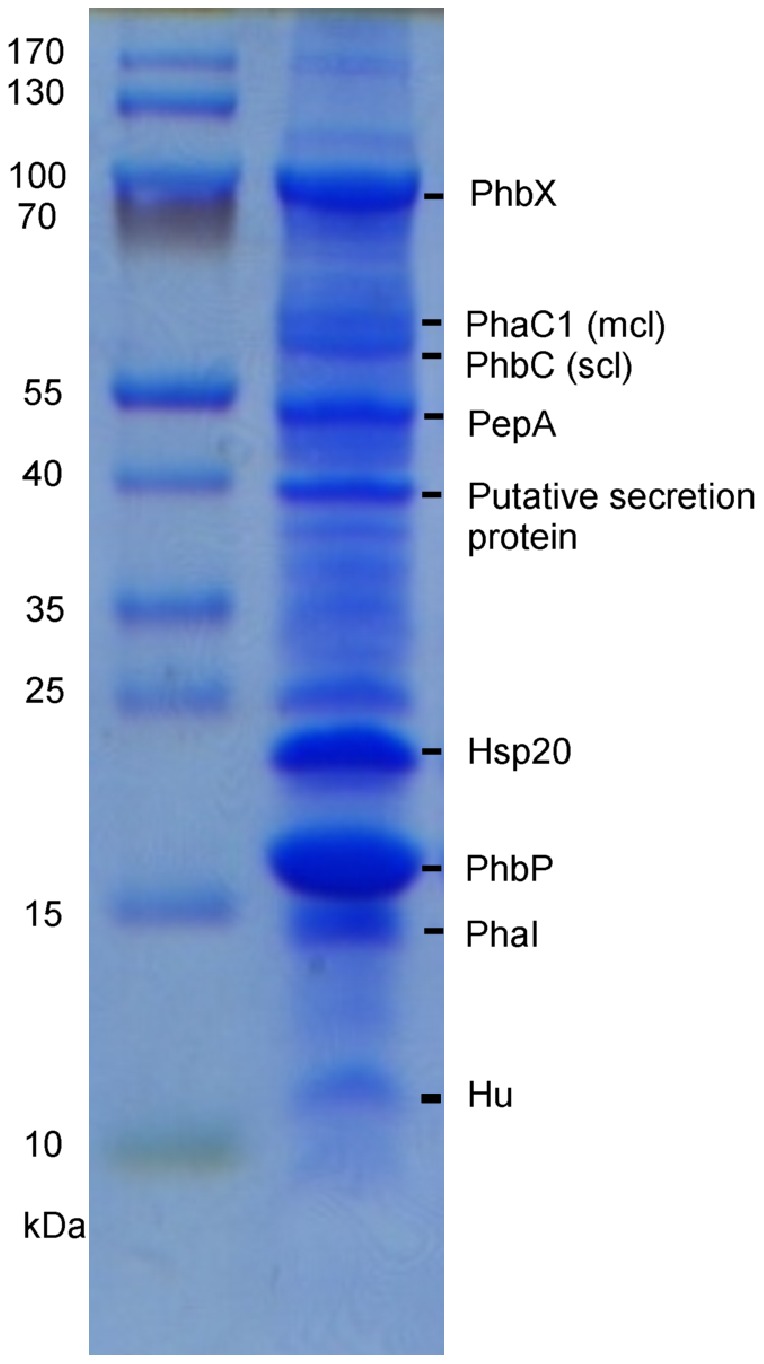
Granules were obtained from cells grown under PHA accumulation conditions. Proteins were separated by SDS-PAGE, stained with Coomassie brilliant blue and subsequently identified by peptide fingerprint analysis as described in Materials and Methods. M, molecular mass standard.
Table 4. Proteins detected in PHA granules of P. extremaustralis by proteome analysis.
| Protein | MW (kDa) | Locus_tag | Annotated function |
| PhbX | 93.50 | PE143B_0105760 | Poly(3-hydroxyalkanoate) synthase |
| PhbC | 64.15 | PE143B_0105720 | Poly(R)-hydroxyalkanoic acid synthase |
| PhaC1 | 62.58 | PE143B_0104515 | Poly(R)-hydroxyalkanoic acid synthase |
| PhbP | 19.88 | PE143B_0105765 | Phasin |
| PhaI | 15.31 | PE143B_0104455 | Putative polyhydroxyalkanoate biosynthesis-like protein |
| Aminopeptidase | 52.42 | PE143B_0109130 | Cytosol aminopeptidase. COG0260 Leucyl aminopeptidase |
| Secretion protein | 38.36 | PE143B_0105715 | Glycoside hydrolase family 43. Multidrug resistance efflux pump. COG0845 Membrane-fusion protein |
| Heat-shock protein | 20.45 | PE143B_0103035 | Molecular chaperone Hsp20. COG0071 Molecular chaperone (small heat shock protein) |
| DNA binding protein HU | 9.80 | PE143B_0124435 | Integration host factor. COG0776 Bacterial nucleoid DNA-binding protein |
The annotated functions of the identified proteins, the locus tags of the coding genes and the calculated molecular masses (MW) are shown.
Discussion
The analysis of the genome of Pseudomonas extremaustralis revealed the presence of different clusters of genes involved in PHA metabolism. Previous studies allowed us to determine that the cluster phbRBAC, involved in PHB metabolism, was located within a genomic island in P. extremaustralis [7], [8]. The genes belonging to this cluster have different origins, indicating that they were acquired by horizontal transfer, probably derived from Burkholderiales [8]. It has also been demonstrated that these genes confer adaptability to different stress conditions [32], [33].
In this work, an additional gene cluster, phbFPX, was found located 4000 bp upstream of the previously described phbRBAC [7]. This gene cluster showed similarity with phb genes found in diverse Proteobacteria, including Burkholderiales species, Azotobacter vinelandii and P. stutzeri A1501, suggesting the acquisition of these genes by horizontal transfer events. Interestingly, the region that includes both PHB gene clusters of P. extremaustralis showed high similarity with a genomic island with similar characteristics present in P. stutzeri A1501.
Besides, phbFPX and phbRBAC, the gene cluster phaC1ZC2D, identified as belonging to the Pseudomonas core genome, was found in P. extremaustralis. In several studied Pseudomonas spp., the phasin encoding genes, phaF and phaI, are located immediately downstream of phaD [24], [34], [35]. However, in P. extremaustralis, seven ORFs encoding proteins involved in pili and fimbriae assembly with similarity to β-Proteobacteria were found between phaD and phaF, suggesting the occurrence of an insertion in this region. This finding provides further evidence about the existence of genetic elements of diverse origin in the genome of P. extremaustralis.
PHA synthases are the enzymes responsible for PHA biosynthesis. Both class II synthases, PhaC1 and PhaC2, were able to restore the ability to synthesize mclPHA in a PHA minus strain of P. putida. PhbX, a putative synthase encoded in the phbFPX cluster, did not confer to P. putida GPp104 the capacity to synthesize PHAs, thus its role in PHA metabolism remains to be investigated.
The analysis of the expression of the genes phbC (encoding the PHB synthase) and phaC1 and phaC2 (encoding mclPHA synthases) of P. extremaustralis demonstrated that these three PHA synthases are expressed, but at different levels. When P. extremaustralis was cultivated with sodium octanoate as carbon source, the expression levels of phbC, phaC1 and phaC2 were significantly higher that when the cells were grown with glucose. The observed differences in the expression levels of the genes encoding PHA synthases correlated with a higher polymer production when P. extremaustralis was cultivated with sodium octanoate. In agreement with our results, RT-qPCR studies and analysis of the activity of the promoter regions of phaC1 and phaC2 conducted with different strains of P. corrugata and P. putida KT2442 showed higher expression levels for phaC1 in comparison with phaC2 when the cells were grown with octanoate as carbon source [36], [19]. The higher expression level of phbC in comparison with phaC1 and phaC2 could account for the higher percentage of sclPHA in comparison with mclPHA in P. extremaustralis. The type strain of P. extremaustralis, 14-3T, is a natural phaA mutant able to produce PHB from octanoate but not from glucose [6], [7] showing the capability of class I synthase to use monomers derived from fatty acids metabolism. In line with these results, strain 14-3b showed higher PHB production when grown in octanoate compared to glucose, indicating a high ability to use substrates derived from fatty acids for PHB biosynthesis. This fact, added to the higher expression of class I synthase, could result in a strong competence for substrate availability between PHB and mclPHA synthases.
The RT-qPCR assays of pha genes in P. extremaustralis agreed with the results obtained by the analysis of the granule associated proteins, in which PHA synthases involved in the synthesis of scl and mclPHAs, PhbC and PhaC1, were detected. In concordance with the low expression levels of phaC2 in the RT-qPCR experiments, this protein was not detected in the PHA granules. In line with these results, PhaC2 was also not detected in PHA granules of P. putida GPo1, while a band of 60 kDa, corresponding to PhaC1 was identified [37]. The analysis of the proteins bound to the PHA granule in P. extremaustralis allowed us to identify a major band belonging to PhbP, protein directly associated with PHB production as the main phasin-like-protein, as well as the phasin PhaI, involved in mclPHA production. Analysis of the phasin-like-proteins bound to PHB granules showed that PhaP1 in Ralstonia eutropha and Herbaspirillum seropedicae, as well as PhaP in Azotobacter sp. FA8, proteins homologous to PhbP of P. extremaustralis, were found as the main GAPs [38], [9], [40]; whilst in mclPHA granules produced by P. oleovorans GPo1 the main GAP was PhaI [24]. In addition, we also found several proteins not directly related to PHA metabolism associated to the PHA granules, such as a cytosolic aminopeptidase. In PHA granules extracted from P. putida GPo1 a leucine aminopeptidase, with a putative role in the turnover of granule-associated proteins, was also detected [41]. This leucine aminopeptidase is similar to PepA of E. coli and PhpA of P. aeruginosa. The cytosolic aminopeptidase found in the PHA granules of P. extremaustralis showed 55% and 81% similarity to PepA of E. coli and PhpA of P. aeruginosa, respectively. In addition, in the PHB producers R. eutropha and H. seropedicae a leucyl aminopeptidase was also found [42], [39]. The DNA binding protein HU was also found associated to the PHA granules in P. extremaustralis. The role of this protein in the granules is not clear, but the presence of other histone like proteins, not directly related to metabolism, associated to the granules has been previously reported [39]. Nucleoid DNA-binding proteins involved in PHA metabolism have been described in both scl and mcl PHA producers. The proteins PhaM of the PHB producer R. eutropha, and PhaF found in the mcl-PHA-accumulating P. putida, interact with the DNA and the nucleoid ensuring equitable segregation of granules in daughter cells during cell division [43], [35].
The heat shock protein Hsp20, has also been found attached to PHA granules in P. extremaustralis. No heat shock proteins have been found in granules of native PHA producers, but in recombinant strains of E. coli carrying the genes for PHB production, an increase in the heat shock proteins DnaK and IbpAB has been observed associated with polymer granules [44], [45]. Moreover, DnaK and GroEL were detected in PhaP1 mutants of R. eutropha [46]. A putative secretion protein belonging to the HlyD family, with high percentage of identity to Type I antifreeze proteins, was also found attached to the PHA granules of P. extremaustralis. This protein presents a high content of alanine residues, characteristic of this kind of proteins [8]. The gene encoding this secretion protein is located next to the phbC gene, encoding the PHB synthase. A role of this protein in PHB metabolism remains to be investigated.
Due to the presence of the machinery to produce both scl and mclPHA in P. extremaustralis, we cannot determine if the identified granule-bound proteins proceed from a mixture of two types of granules composed only by one kind of polymer (PHB or mclPHA), or a single type of granule containing a mix of PHB and mcl-PHA. However, according to the polymer content data, the composition of the granules was mainly PHB.
Metabolism of mclPHAs in other Pseudomonas spp. has a complex regulation, which is controlled at the enzymatic level by cofactor inhibition and metabolite availability; at transcriptional level by specific and global transcriptional regulatory factors; and at translational level by global post-transcriptional regulators [47]. Up to now, only few reports have shown the presence of genes associated with the synthesis of scl and mclPHAs in the same organism [48], [49]. For this reason, the physiological and regulatory mechanisms that govern the production of both types of polymers in the same bacterium are still poorly understood. In this paper, we propose that the differential expression of PHB and PHA synthases may account for the high production of PHB in P. extremaustralis. An interesting finding of this work is that the PHB synthase, phbC, presumably acquired by horizontal transfer, showed higher expression in comparison with the Pseudomonads own mclPHA synthases, conferring the ability to produce high amounts of PHB in P. extremaustralis.
Finally, the study of the genes involved in PHA metabolism in P. extremaustralis could be useful for biotechnological purposes in order to achieve the production of different PHAs with varied elastomeric properties.
Acknowledgments
We thank Dr. Beatriz S. Mendez and Dr. M. Julia Pettinari for the critical reading of the manuscript.
Funding Statement
This work was supported by grants from Universidad de Buenos Aires (UBACyT 2011-2014, N° 20020100100854), Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, PIP 2011, N° 0338) and Agencia Nacional de Promoción Científica (ANPCyT: PICT 2007, N° 0639), Argentina and from Consejo Nacional de Ciencia y Tecnología (CONACyT, grant 127979), México. J.A.R. and N.I.L. are career investigators from CONICET, and M.V.C. has received a graduate student fellowship from ANPCyT and financial support from the program Ciencia y Tecnología para el Desarrollo (CYTED). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hazer B, Steinbüchel A (2007) Increased diversification of polyhydroxyalkanoates by modification reactions for industrial and medical applications. Appl Microbiol Biotechnol 74: 1–12. [DOI] [PubMed] [Google Scholar]
- 2. de Eugenio LI, Escapa IF, Morales V, Dinjaski N, Galán B, et al. (2010) The turnover of medium-chain-length polyhydroxyalkanoates in Pseudomonas putida KT2442 and the fundamental role of PhaZ depolymerase for the metabolic balance. Environ Microbiol 12: 207–221. [DOI] [PubMed] [Google Scholar]
- 3. Huijberts GN, de Rijk TC, de Waard P, Eggink G (1994) 13C nuclear magnetic resonance studies of Pseudomonas putida fatty acid metabolic routes involved in poly(3-hydroxyalkanoate) synthesis. J Bacteriol 176: 1661–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kessler B, Palleroni NJ (2000) Taxonomic implications of synthesis of poly-beta-hydroxybutyrato and other poly-beta-hydroxyalkanoates by aerobic Pseudomonas . Int J Sys Evol Microbiol 50: 711–713. [DOI] [PubMed] [Google Scholar]
- 5. Diard S, Carlier JP, Ageron E, Grimont PA, Langlois V, et al. (2002) Accumulation of poly(3-hydroxybutyrate) from octanoate in different Pseudomonas belonging to the rRNA homology group I. Syst Appl Microbiol 25: 183–188. [DOI] [PubMed] [Google Scholar]
- 6. López NI, Pettinari MJ, Stackebrandt E, Tribelli PM, Potter M, et al. (2009) Pseudomonas extremaustralis sp. nova poly(3-hydroxybutyrate) producer isolated from an Antarctic environment. Curr Microbiol 59: 514–519. [DOI] [PubMed] [Google Scholar]
- 7. Ayub ND, Pettinari MJ, Ruiz JA, López NI (2006) Impaired polyhydroxybutyrate biosynthesis from glucose in Pseudomonas sp. 14-3 is due to a defective β-Ketothiolase gene. FEMS Microbiol Lett 264: 125–131. [DOI] [PubMed] [Google Scholar]
- 8. Ayub ND, Pettinari MJ, Méndez BS, López NI (2007) The polyhydroxyalkanoate genes of a stress resistant Antarctic Pseudomonas are situated within a genomic island. Plasmid 58: 240–248. [DOI] [PubMed] [Google Scholar]
- 9. Tribelli PM, Raiger Iustman LJ, Catone MV, Di Martino C, Revale S, et al. (2012) Genome sequence of the polyhydroxybutyrate producer Pseudomonas extremaustralis, a highly stress-resistant Antarctic bacterium. J Bacteriol 194: 2381–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huisman GW, Wonink E, Meima R, Kazemier B, Terpstra P (1991) Metabolism of poly(3-hydroxyalkanoates) (PHAs) by Pseudomonas oleovorans: identification and sequences of genes and function of the encoded proteins in the synthesis and degradation of PHA. J Biol Chem 266: 2191–2198. [PubMed] [Google Scholar]
- 11. Meselson M, Yuan R (1968) DNA restriction enzyme from E. coli . Nature 217: 1110–1114. [DOI] [PubMed] [Google Scholar]
- 12. Simon R, Priefer UU, Pühler A (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1: 784–791. [Google Scholar]
- 13. Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, et al. (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166: 175–176. [DOI] [PubMed] [Google Scholar]
- 14. Jaenecke S, Díaz E (1999) Construction of plasmid vectors bearing a NotI-expression cassette based on the lac promoter. Int Microbiol 2: 29–31. [PubMed] [Google Scholar]
- 15. Langille MGI, Brinkman FSL (2009) IslandViewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics 25: 664–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Friedrich B, Hogrefe C, Schlegel HG (1981) Naturally occurring genetic transfer of hydrogen-oxidizing ability between strains of Alcaligenes eutrophus . J Bacteriol 174: 198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ostle A, Holt JG (1982) Nile blue A as a fluorescent stain for poly-β-hydroxybutyrate. Appl Environ Microbiol 44: 238–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Braunegg G, Sonnleitner B, Lafferty RM (1978) A rapid gas chromatographic method for the determination of poly-β-hydroxybutyric acid in microbial biomass. Eur J Appl Microbiol Biotechnol 6: 29–37. [Google Scholar]
- 19. de Eugenio LI, Galán B, Escapa IF, Maestro B, Sanz JM, et al. (2010) The PhaD regulator controls the simultaneous expression of the pha genes involved in polyhydroxyalkanoate metabolism and turnover in Pseudomonas putida KT2442. Env Microbiol 12: 1591–1603. [DOI] [PubMed] [Google Scholar]
- 20. Jendrossek D (2007) Peculiarities of PHA granules preparation and PHA depolymerase activity determination. Appl Microbiol Biotech 74: 1186–1196. [DOI] [PubMed] [Google Scholar]
- 21. Lowry OH, Rosebrough NJ, Far AL, Randall RJ (1951) Protein measure with the Folin-phenol reagent. J Biol Chem 193: 265–267. [PubMed] [Google Scholar]
- 22. Shagger H, Von Jagow G (1987) Tricine-Sodium Dodecyl sulfate-polyacrylamide gel electrophoresis for separation of protein in the range from 1 to 100 kDa. Anal Biochem 166: 386–379. [DOI] [PubMed] [Google Scholar]
- 23. Weber K, Osborn M (1969) The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem 244: 4406–4412. [PubMed] [Google Scholar]
- 24. Prieto MA, Buhler B, Jung K, Witholt B, Kessler B (1999) PhaF, a polyhydroxyalkanoate-granule-associated protein of Pseudomonas oleovorans GPo1 involved in the regulatory expression system for pha genes. J Bacteriol 181: 858–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Solaiman DKY, Ashby RD, Licciardello G, Catara V (2008) Genetic organization of pha gene locus affects phaC expression, poly(hydroxyalkanoate) composition and granule morphology in Pseudomonas corrugata . J Ind Microbiol Biotech 35: 111–120. [DOI] [PubMed] [Google Scholar]
- 26. Hoffmann N, Rehm BHA (2004) Regulation of polyhydroxyalkanoate biosynthesis in Pseudomonas putida and Pseudomonas aeruginosa . FEMS Microbiol Lett 237: 1–7. [DOI] [PubMed] [Google Scholar]
- 27. Silby MW, Cerdeño-Tárraga AM, Vernikos GS, Giddens SR, Jackson RW, et al. (2009) Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens . Genome Biol 10: R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rehm BH (2003) Polyester synthases: natural catalysts for plastics. Biochem J 15: 15–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yan Y, Yang J, Dou Y, Chen M, Ping S, et al. (2008) Nitrogen fixation island and rhizosphere competence traits in the genome of root-associated Pseudomonas stutzeri A1501. Proc Natl Acad Sci USA 105: 7564–7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Setubal JC, dos Santos P, Goldman BS, Ertesvag H, Espin G, et al. (2009) Genome sequence of Azotobacter vinelandii, an obligate aerobe specialized to support diverse anaerobic metabolic processes. J Bacteriol 191: 4534–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Knoll M, Hamm TM, Wagner F, Martinez V, Pleiss J (2009) The PHA Depolymerase Engineering Database: A systematic analysis tool for the diverse family of polyhydroxyalkanoate (PHA) depolymerases. BMC Bioinformatics 8: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ayub ND, Tribelli PM, López NI (2009) Polyhydroxyalkanoates are essential for maintenance of redox state in the Antartic bacterium Pseudomonas sp. 14-3 during low temperature adaptation. Extremophiles 13: 59–66. [DOI] [PubMed] [Google Scholar]
- 33. Tribelli PM, López NI (2011) Poly(3-hydroxybutyrate) influences biofilm formation and motility in the novel Antarctic species Pseudomonas extremaustralis under cold conditions. Extremophiles 15: 541–547. [DOI] [PubMed] [Google Scholar]
- 34. Timm A, Steinbüchel A (1992) Cloning and molecular analysis of the poly(3-hydroxyalkanoic acid) gene locus of Pseudomonas aeruginosa PAO1. J Biol Chem. 209: 15–30. [DOI] [PubMed] [Google Scholar]
- 35. Galán B, Dinjaski N, Maestro B, de Eugenio LI, Escapa IF, et al. (2011) Nucleoid-associated PhaF phasin drives intracellular location and segregation of polyhydroxyalkanoate granules in Pseudomonas putida KT2442. Molecular Microbiol 79: 402–418. [DOI] [PubMed] [Google Scholar]
- 36. Conte E, Catara V, Greco S, Russo M, Alicata R, et al. (2006) Regulation of polyhydroxyalkanoate synthases (phaC1 and phaC2) gene expression in Pseudomonas corrugata . Appl Microbiol Biotechnol 72: 1054–1062. [DOI] [PubMed] [Google Scholar]
- 37. de Roo G, Ren Q, Witholt B, Kessler B (2000) Development of an improved in vitro activity assay for medium chain length PHA polymerases based on coenzymeA release measurements. J Microbiol Methods 41: 1–8. [DOI] [PubMed] [Google Scholar]
- 38. Pötter M, Müller H, Reinecke F, Wieczorek R, Fricke F, et al. (2004) The complex structure of polyhydroxybutyrate (PHB) granule: four orthologous and paralogous phasins occur in Ralstonia eutropha . Microbiology 150: 2301–2311. [DOI] [PubMed] [Google Scholar]
- 39. Tirapelle EF, Muller-Santos M, Tadra-Sfeir MZ, Kadowaki MAS, Steffens MBR, et al. (2013) Identification of proteins associated with polyhydroxybutyrate granules from Herbaspirillum seropedicae SmR1 - Old partners, New players. PLoS ONE 8(9): e75066 doi:10.1371/journal.pone.0075066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pettinari MJ, Chaneton L, Vazquez G, Steinbüchel A, Mendez BS (2003) Insertion sequence-like elements associated with putative polyhydroxybutyrate regulatory genes in Azotobacter sp. FA8. Plasmid 50: 36–44. [DOI] [PubMed] [Google Scholar]
- 41. Ren Q, de Roo G, Ruth K, Witholt B, Zinn M, et al. (2009) Simultaneous accumulation and degradation of polyhydroxyalkanoates: futile cycle or clever regulation? Biomacromolecules 10: 916–922. [DOI] [PubMed] [Google Scholar]
- 42.Jendrossek D, Pfeiffer D (2014) New insights in the formation of polyhydroxyalkanoate granules (carbonosomes) and novel functions of poly(3-hydroxybutyrate). Environ Microbiol doi:10.1111/1462-2920.12356. [DOI] [PubMed]
- 43. Pfeiffer D, Wahl A, Jendrossek D (2011) Identification of a multifunctional protein, PhaM, that determines number, surface to volume ratio, subcellular localization and distribution to daughter cells of poly(3-hydroxybutyrate), PHB, granules in Ralstonia eutropha H16. Mol Microbiol 82: 936–951. [DOI] [PubMed] [Google Scholar]
- 44. Han MJ, Park SJ, Lee JW, Min BH, Lee SY, et al. (2006) Analysis of poly(3-hydroxybutyrate) granule-associated proteome in recombinant Escherichia coli . J Microbiol Biotechnol 16: 901–910. [Google Scholar]
- 45. de Almeida A, Catone MV, Rhodius V, Gross C, Pettinari MJ (2011) Unexpected stress-reducing effect of PhaP, a poly(3-hydroxybutyrate) granule-associated protein, in Escherichia coli . Appl Environ Microbiol 77: 6622–6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pötter M, Müller H, Reinecke F, Wieczorek R, Fricke F, et al. (2004) The complex structure of polyhydroxybutyrate (PHB) granules: four orthologous and paralogous phasins occur in Ralstonia eutropha. . Microbiology 150: 2301–2311. [DOI] [PubMed] [Google Scholar]
- 47. Tortajada M, Ferreira da Silva L, Prieto MA (2013) Second-generation functionalized medium chain-length polyhydroxyalkanoates: the gateway to high-value bioplastic applications. Int Microbiol 16: 1–15. [DOI] [PubMed] [Google Scholar]
- 48. Matsusaki H, Manji S, Taguchi K, Kato M, Fukui T, et al. (1998) Cloning and molecular analysis of the poly(3-hydroxybutyrate) and poly(3-hydroxybutyrate-co-3-hydroxyalkanoate) biosynthesis genes in Pseudomonas sp. strain 61-3. J Bacteriol 180: 6459–6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hu F, You S (2007) Inactivation of type I polyhydroxyalkanoate synthase in Aeromonas hydrophila resulted in discovery of another potential PHA synthase. J Ind Microbiol Biotechnol 34: 255–260. [DOI] [PubMed] [Google Scholar]