Abstract
Pet dogs very frequently develop spontaneous mammary tumors and have been suggested as a good model organism for breast cancer research. In order to obtain an insight into underlying signaling mechanisms during canine mammary tumorigenesis, in this study we assessed the incidence and the mechanism of canonical Wnt activation in a panel of 12 canine mammary tumor cell lines. We show that a subset of canine mammary cell lines exhibit a moderate canonical Wnt activity that is dependent on Wnt ligands, similar to what has been described in human breast cancer cell lines. In addition, three of the tested canine mammary cell lines have a high canonical Wnt activity that is not responsive to inhibitors of Wnt ligand secretion. Tumor cell lines with highly active canonical Wnt signaling often carry mutations in key members of the Wnt signaling cascade. These cell lines, however, carry no mutations in the coding regions of intracellular Wnt pathway components (APC, β-catenin, GSK3β, CK1α and Axin1) and have a functional β-catenin destruction complex. Interestingly, however, the cell lines with high canonical Wnt activity specifically overexpress LEF1 mRNA and the knock-down of LEF1 significantly inhibits TCF-reporter activity. In addition, LEF1 is overexpressed in a subset of canine mammary carcinomas, implicating LEF1 in ligand-independent activation of canonical Wnt signaling in canine mammary tumors. We conclude that canonical Wnt activation may be a frequent event in canine mammary tumors both through Wnt ligand-dependent and novel ligand–independent mechanisms.
Introduction
The Wnt signaling is one of the key players during normal mammary gland development as well as during mammary tumorigenesis [1]. The canonical, β-catenin-mediated Wnt signaling is activated by secreted Wnt ligands through activation of transmembrane frizzled (Fzd) receptors and LDL-receptor related protein 5 or 6 (Lpr5/6) co-receptors. This triggers Dishevelled-dependent disruption of the β-catenin-destruction complex in the cytoplasm that is composed of multiple proteins including, glycogen synthase kinase 3β (GSK3β), adenomatous polyposis coli (APC), Axin1 (or Axin2) and casein kinase 1 (CK1). Consequently, stabilized β-catenin translocates to the nucleus where it can associate with the T-cell factor (TCF)/Lymphoid enhancer-binding factor (LEF)-family of transcription factors to regulate the expression of specific target genes. The Wnt signal strength can be negatively regulated by secreted factors that competitively bind Wnt ligands (e.g. secreted Fzd related protein (sFRP)) or by factors that disrupt the assembly of Fzd/Lpr5/6 co-receptor complex (e.g. dikkopf-1) [2].
Many studies have documented active Wnt signaling in mammary tissue based on presence of stabilized β-catenin protein and often in combination with aberrant expression of target genes (e.g. Axin2). Based on these criteria, over 50% of human breast tumor tissue samples assessed, showed signs of aberrant canonical Wnt activity (reviewed in [3]). In contrast, reports on the signaling activity in cultured human breast cancer cells are somewhat contradicting. A number of studies have used the presence of active (phosphorylated) or uncomplexed β-catenin in cell lysates as an indicator of active canonical Wnt signaling (Table 1 and references therein). Other studies have applied a more quantitative manner to assess Wnt activity in cultured cells using a TCF-reporter assay. In this assay, the ratio of luciferase signal from reporters containing a promoter with either functional of mutated TCF response elements can be taken as a measure of canonical Wnt activity, with ratios higher than 1.0 indicating an active signaling. Although not always consistent between different studies, a subset of human breast cancer cell lines was found to have moderate TCF-reporter activity (Table 1 and references therein). In addition, a number of studies showed that the effect of canonical Wnt signaling could be attenuated by Wnt inhibitors sFRP1 or Dkk1 [4], [5] or by blocking the receptor Fzd7 [6]. This indicates a ligand-dependent mechanism of canonical Wnt pathway activation in human breast cancer. In concordance, mutations in downstream signaling components (e.g. APC and β-catenin) are rarely found in human primary mammary tumors. Similarly, in cultured cells only one out of 24 screened human breast cancer cell lines (DU4475) had a truncating mutation in APC and none in β-catenin [7]. In contrast, epigenetic silencing of APC and Wnt ligand inhibitors (sFRP1 and Wif1) have often been reported in primary human mammary tumors and in human breast cancer cell lines [8]–[10]. The ligand-dependent nature of canonical Wnt activation in human breast cancer and benign breast lesions is further supported by frequent overexpression of Wnt ligands [3].
Table 1. Assessment of canonical Wnt activity in human mammary cell lines.
| Cell line | Active or uncomplexed β-catenin | TCF-reporter activity (ratio >1.0) | Reference |
| AB589 | no | [4] | |
| BC3 | yes | [4] | |
| BT20 | no | [40] | |
| BT474 | yes | [41] | |
| BT483 | no | [42] | |
| BT549 | no | [40], [42] | |
| DU4475 | yes | [40] | |
| EVSA-T | no | [40] | |
| HCC1187 | yes | [25] | |
| HCC1395 | yes | [25] | |
| HCC1937 | no | [42] | |
| Hs578T | no | [40] | |
| MCF7 | no/yes | [4]/[41], [43] | |
| no/yes | [40]/[24] | ||
| MDA-MB-134 | no | [4] | |
| MDA-MB-134VI | no | [40] | |
| MDA-MB-157 | yes | [4] | |
| MDA-MB-175 | no | [4] | |
| MDA-MB-231 | no/yes | [41]/[4], [43] | |
| no/yes | [25], [40], [44]/[5] | ||
| MDA-MB-361 | no | [4] | |
| no | [42] | ||
| MDA-MB-415 | no | [4] | |
| no | [42] | ||
| MDA-MB-435 | no | [4] | |
| MDA-MB-453 | no | [40] | |
| no | [4], [43] | ||
| MDA-MB-468 | no | [4] | |
| no | [42] | ||
| JIMT-1 | yes | [41] | |
| OCUB-F | no | [40] | |
| SkBr3 | no | [41] | |
| SK-BR-3 | no | [40] | |
| SK-BR-5 | no | [40] | |
| SK-BR-7 | no | [42] | |
| SUM159 | no | [42] | |
| SUM185 | no | [42] | |
| T47D | yes | [41], [43] | |
| no | [40], [44] | ||
| UACC893 | no | [42] | |
| ZR-75-1 | yes | [41] | |
| no | [42] |
Pet dogs have been suggested as a valuable breast cancer model for preclinical research due to the high incidence and spontaneous nature of the tumor development, shared environmental risk factors, strong genetic similarity with humans and shared aspects of mammary tumor biology [11], [12]. Moreover, development of canine mammary tumors is highly dependent on steroid hormone exposure, with progesterone being the main risk factor [13], [14].
The activity of canonical Wnt signaling in canine mammary tumorigenesis has not been quantitatively assessed so far. Previous studies have only addressed the expression of β-catenin protein in spontaneous canine mammary tumors in relation to E-cadherin and/or APC [15]–[17]. However, comparative gene expression profiling of human and canine mammary tumors has implicated a significant similarity in deregulation of multiple cancer-related pathways, including Wnt signaling [18]. In this study we aimed to assess the activation of canonical Wnt signaling in canine mammary tumors using a panel of canine mammary cell lines. We report that subsets of canine mammary tumor cell lines exhibit moderate, ligand-dependent-, and high, ligand-independent-mechanisms of canonical Wnt activation. Moreover, we show that the ligand-independent activation of canonical Wnt signaling is coupled to the overexpression of LEF1.
Materials and Methods
Canine mammary cell lines and tissue
Canine mammary tumor cell lines used in this study were established from primary tumors diagnosed as carcinoma (CMT1, CMT-U27, CMT9, P114, CHMp, CNMp and CIPp) or its metastasis (CHMm, CNMm and CIPm), benign mixed tumor (CMT-U229) and osteosarcoma-like tumor (CMT-U335) [19]–[21]. All cell lines were cultured in DMEM/F12 (Invitrogen, Bleiswijk, The Netherlands) supplemented with 10% fetal bovine serum (FBS) (FBS Gold, PAA, Cölbe, Germany). Canine mammary tissue used in this study originates from privately owned dogs that were referred to clinics of Veterinary Faculty in Zagreb, Croatia. Canine mammary surgery was performed as a part of a necessary medical treatment due to the presence of mammary tumor. This was done under the common rules for veterinary surgery for which owners asked for medical treatment of their pets. In contrast to medical intervention in laboratory animals no external permission was necessary other than that the surgery is done by qualified veterinary surgeons. The dog's owners were informed and gave their consent that the collected tissues can be used for research purposes. Histopathology of all tumor and paired normal tissue was evaluated by Prof. E. Hellmen (Table 2). Pictures of cell morphology were captured using an Olympus microscope (Zoeterwoude, The Netherlands) with 10×10 magnification.
Table 2. Information about histopathology and RNA quality of canine mammary tumor tissue.
| Sample ID | Tumor histopathology | Normal (RIN) | Tumor (RIN) |
| 2 | Benign mixed tumor | 8.8 | 8.1 |
| 3 | Complex adenoma | 7.7 | 8.4 |
| 7 | Complex adenoma | 8.3 | 8.6 |
| 14 | Carcinosarcoma (combined osteosarcoma and ductular carcinoma) | 7.9 | 8.3 |
| 20 | In situ carcinoma | 7.6 | 7.9 |
| 25 | Atypical sclerosing adenosis and purulent inflammation | 7.8 | 9.2 |
| 26 | Simple solid carcinoma | 8.9 | 9.0 |
| 1 | Simple ductal carcinoma | 7.1 | 9.3 |
| 5 | Simple carcinoma | 6.5 | 8.0 |
| 24 | Complex carcinoma | 8.4 | 9.8 |
| 31 | Probable fibrosarcoma/complex carcinoma | 8.6 | 7.1 |
TCF-reporter assay
Transfection was performed in FBS-free medium using 3 µl Lipofectamine 2000 (Invitrogen), 800 ng pTOPFLASH (TOP) or pFOPFLASH (FOP) (gift from Dr. Marc van de Wetering, Hubrecht Institute, The Netherlands) and 0.5 ng human β-actin-promoter renilla construct [22] as an internal control. Cells were seeded 48 h before transfection at a density optimal for transfection, according to the manufacturer's protocol in a 24 wells plate (Primaria, BD Biosciences, Breda, The Netherlands). In case of Wnt3a cotransfection, 10 ng mouse pcDNA4-Wnt3a construct (gift from Dr. Wim de Lau, Hubrecht Institute, The Netherlands) was used. Transfection was stopped after 5 h and cells were left to recover for 24 h on DMEM/F12 supplemented with 10% FBS. Cells were then treated with increasing concentrations of IWP-2 (Stemgent, Cambridge, UK) or 5 mM LiCl for 24 h. Control DMSO concentration reflected the DMSO concentration in the 10 µM IWP-2 solution. The firefly and renilla luciferase activities were measured using a Dual-Luciferase Assay System (Promega, Leiden, The Netherlands) in a Centro LB 960 luminometer (Berthold Technologies, Vilvoorde, Belgium). Differences in pTOPFLASH/pFOPFLASH were statistically assessed using unpaired, two tailed Student's t test in Microsoft Office Excel. All transfection experiments were performed using three replicate samples and each experiment was independently repeated 2–4 times.
siRNA
Canine sequence-specific LEF1 (synonym: TCF1-alpha, Genbank: XM_003434032) and β-catenin (CTNNB1, Genbank: NM_001137652) siRNA was designed on the website http://www.dharmacon.com/designcenter/designcenterpage.aspx (DharmaconRNAi technologies, ThermoScientific, USA). Universal MOCKsiRNA (ON-TARGET plus non-targeting pool species H, M, R) was used as the negative control for siRNA experiments. There was no cross-silencing of non-target genes checked by blasting the siRNA designed sequences against the canine genome database. The sequence of the LEF1 siRNA duplex is as follows: sense GAAGAGGAAGAGAGAGAAAUU and antisense UUUCUCUCUCUUCCUCUUCUU, and for β-catenin sense GAACGAAGGUGUAGCAACAUU and antisense UGUUGCUACACCUUCGUUCUU. Cell transfections were first optimized with siGLO (Dharmacon, Colorado) (data not shown). 80.000 CMT-U27 cells were transfected with 1 µl DharmaFECT Duo as transfection reagent (Dharmacon, Colorado), 50 nM siRNA and 0.5 µg DNA (TOP or FOP) in 24 wells plates (Primaria, BD, The Netherlands). After 24 h and 48 h incubation in DMEM:F12 and 10% FCS cells were harvested for RNA isolation or TCF-reporter assay.
RNA isolation, cDNA synthesis, sequencing and quantitative RT-PCR
From all cell lines, total RNA was isolated from two different passages. From canine mammary tissue and from the mammary cell lines total RNA was isolated and treated with deoxyribonuclease using the RNeasy mini kit (Qiagen, Venlo, The Netherlands) according to the manufacturer's protocol. Quality of mRNA from tissue samples were assessed using a 2100 Bioanalyzer (Agilent Technologies, Amstelveen, The Netherlands) and RNA integrity number (RIN) of each sample is presented in Table 2. cDNA synthesis was performed using iScript kit (Bio-Rad Laboratories) according to manufacturer's protocol. Specific primer sets were used to amplify gene products for quantitative RT-PCR (Table 3) and sequencing (Table 4). Quantitative RT-PCR was performed using Bio Rad MyIQ detection system (Bio-RAD Laboratories) with SYBR Green Fluorophore. Relative target gene expression was normalized to that of the reference gene RPS19 using a delta Ct method [23], and relative induction of gene expression was statistically assessed using paired, two tailed Student's t test in Microsoft Office Excel. For comparison of relative gene expression between three sets of cell line, REST-MCS beta software was used (http://www.gene-quantification.de/rest-mcs.html). For the sequence reactions we used a standard amplification with Phusion-Hot Start Taq (Finnzymes, Espoo, Finland) according to manufacturer's protocol. DNA sequence reactions were performed using BigDye v3.1 according to the manufacturer's protocol (Applied Biosystems, Foster City, CA). All amplifications were performed on an ABI 3130XL (Applied Biosystems, Foster City, CA) and analyzed in Lasergene (version 9.1 DNASTAR). The obtained sequences were compared with DNA sequences in databases using BLASTn (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Table 3. Information about primers used to assess gene expression.
| Gene name | Forward primer | Reverse primer | Annealing T (°C) |
| SFRP1 | AGCGAGTTTGCATTGAGGAT | TCTTGATGGGTCCCAACTTC | 60 |
| APC | AGTCCCAAGCAACAGAAGC | GCAGTTGAACCCTGAGCA | 63 |
| β-catenin (CTNNB1) | ATGGGTAGGGCAAATCAGTAAGAGGT | AAGCATCGTATCACAGCAGGTTAC | 64 |
| E-cadherin (CDH1) | CAGGAAGCTCTCCACCAGAG | CTGGGAAATGTGAGCACCTC | 58 |
| LEF1 | AGACATCCTCCAGCTCCTGA | GATGGATAGGGTTGCCTGAA | 60 |
| TCF1 (TCF7) | CTACTCCGCCTTCAATCTGC | AGAGAGTTGTGGGACGCTGT | 60 |
| TCF3 (TCF7L1) | CCTGGAGCTGTTGGACAAAT | AAACCAGGCTGGACATTGAG | 60 |
| TCF4 (TCF7L2) | CGAGTGCACGTTGAAAGAAA | ATGTGAAGCTGTCGCTCCTT | 60 |
| WNT1 | CTGGCACGTTGACTCAGAGA | AAGAGCTGCATAGCCACCAC | 63 |
| WNT2 | GACAGGGATCACAGCCTCTT | TGGTGATGGCAAACACAACT | 63 |
| WNT3 | ATGAACAAGCACAACAACGAG | TTGAGGAAGTCGCCGATAG | 61.5 |
| WNT4 | CGAGGAGTGCCAGTACCAGT | AGAGATGGCGTACACGAAGG | 61,6 |
| WNT5B | CCCTGTACAGAGACCCGAGA | ACAACTGGCACAGCTTCCTC | 61.5 |
| WNT7A | GCCTCGACGAGTGTCAGTTT | GATGATGGCGTAGGTGAAGG | 60 |
| RPS19 | CCTTCCTCAAAAAGTCTGGG | GTTCTCATCGTAGGGAGCAAG | 61 |
| AXIN2 | GGACAAATGCGTGGATACCT | TGCTTGGAGACAATGCTGTT | 60 |
Table 4. Information about primers used to sequence gene coding regions.
| gene name | Forward primer | Location | Reverse primer | Location |
| β-catenin | AAGCACACCATACAACGG | F4 | CCTAAACCACTCCCACCCT | R2494 |
| (CTNNB1) | GGCTGCTATGTTCCCTGAGA | F327 | CCACCTGGTCCTCATCATTA | R541 |
| GGGACCTTGCACAATCTTTCTC | F688 | |||
| AATGCAGGCTTTAGGGCTTCA | F1128 | |||
| CCTGCCATCTGTGCTCTTCGTC | F1429 | |||
| TCACAACCGAATCGTAATCAGA | F1794 | |||
| APC | AGAGGCAGACTCAGCACCAT | F3728 | GGGGCTTATAATGCCACTCA | R4311 |
| GGCATTATAAGCCCCAGTGA | F4297 | ACAGGGGGAGGTAATTTTGG | R4865 | |
| AAAGAGCCCGAAAAGCCTAC | F4702 | ACACGGAAAGGCTTGTGACT | R5279 | |
| GCCCAAAGGAAAAAGTCACA | F5247 | CGATTTACGGGGTGTTTTGT | R5810 | |
| CCAGGGAAAAGGCTGAATTA | F5675 | ACTCCTGCAACAGGTCGTCT | R6178 | |
| AGACGACCTGTTGCAGGAGT | F6159 | GGGCTGTTTCATTTGGCTTA | R6702 | |
| AGCAAACATGCCTTCGATCT | F6708 | CCTTTGGAGGCAGACTCACT | R7253 | |
| ACGTCTCCAGGCAGACAGAT | F7153 | GAATGGGAGCGTGCAATATC | R7652 | |
| CACGCTCCCATTCTGAAAGT | F7640 | CCGTTACCCACACTTGGTTT | R8180 | |
| GAGATCCCCAACAGGAAACA | F8070 | CACACGGATGTCACGAGGTA | R8585 | |
| GSK3B | GAGGGTGATTCGGGAAGAG | F1005 | TAGGCTAAACTTCGGAACAG | R1542 |
| CGGAAACAGTATACAGAGTTGC | F1437 | AAGTAACTGGTGGTTCTTCCTG | R2338 | |
| TTCCCTCAAATTAAGCAC | F1907 | |||
| CK1 | GAGCGGCGGCGATCAGGTTCC | F217 | ATACCCATTAGGAAGTTATCTGGT | R835 |
| (CSNK1A1) | TACACAGAGACATTAAACCAGATA | F796 | ATCTGCTCTGCTTCTTCTGTTC | R1446 |
| ATAAAAATCTCACTGGCACTG | F1016 | |||
| AXIN1 | CTTCGTTCAGAGTGGGCAGGTAGC | R730 | ||
| GTTTGACCAGGCACAGACGGAGAT | F543 | CGGTAAGTGCGAGGAATGTGAGGT | R1127 | |
| ACCGACAGCAGCGTGGAT | F1006 | ACTGTGGTGACTGTGGTGGTG | R1626 | |
| CCGGCCATCGTTCCCCTGACAAT | F1466 | GCGGTGCCTGCTGATCTCCTTCTC | R1941 | |
| TCGGAGGACACAGACAAGAAC | F1867 | GAACCTCCTCGAACACCACTC | R2536 | |
| GCGGAGTGGTGTTCGAGGAGGTTC | F2513 | |||
| E-cadherin | TCTGGTTTATGAAGTTGTAGAGGC | R399 | ||
| (CDH1) | CAGCCTATGTTTCTGATGAC | F312 | ACAGATCCTTGGAAGACTGC | R921 |
| AGTCTTCCAAGGATCTGTCAC | F904 | CAGTCACGTACAAGACATACTG | R1475 | |
| TTTGTTGTCACCACAGACCC | F1376 | CAATGATGTTGATGACATGAGG | R1955 | |
| ACCTCGAAATATGGACTTCTG | F1897 | CAACTGGCTCAAGTCAAAGTC | R2428 | |
| CTCGCTCTACTAATCCTGATTCTG | F2264 | CCATTCATTCAGGTAGTCATAGTC | R2707 | |
| TTCTCCAGAAATTCCTCTCAG | R2861 | |||
| LEF1 | GAGCGGAGATTGCAGAGC | F615 | CGTTGGGAATGAGTTTCGTT | R1875 |
Protein extraction and Western blot
For whole cell lysis cells were washed with cold HANK's balanced salt solution and scraped in cold RIPA buffer (6.5 mM Na2HPO4, 1.5 mM KH2PO4, 137 mM NaCl, and 2.7 mM KCl (pH 7.4); 1% sodium dodecyl sulfate (SDS) (vol/vol), 1% Igepal (vol/vol), 0.5% Na-deoxycholate (wt/vol), 1 mM phenylmethylsulfonylfluoride,1 mM Na-orthovanadate and 1 µg/ml aprotinin). After 20 min incubation on ice, samples were centrifuged for 15 min at 16,000 g at 4°C. Protein concentration was determined using Bio-Rad Dc Protein Assay (Bio-Rad Laboratories). Fifty microgram protein of total cell lysates was subjected to SDS-PAGE and analyzed by Western blot. For extraction of cytoplasmic and nuclear protein fractions NE-PER Reagent kit (Thermo Scientific, Breda, The Netherlands) was used according to the manufacturer's protocol. Ten µg protein was subsequently subjected to western blot analysis. Primary antibodies used in this study were directed against APC (AB-1) Mouse (FE9) (OP44 1∶1000), (Calbiochem, Merck, Amsterdam, The Netherlands), β-catenin (Ab6302 1∶4000)(Abcam, Cambridge, UK), human E-cadherin (610181, 1∶2000 BD Biosciences, Breda, The Netherlands), GAPDH (ab9485 1∶2000) (Abcam) and β-actin pan Ab-5 (MS-1295-P1 1∶2000)(Thermo Scientific) as a reference protein. And as secondary antibody goat anti-mouse HRP-conjugated (HAF007, R&D Systems, Abingdon, UK) was used. HRP was visualized using Advance TM_Enhancedchemiluminescence (ECL, Amersham, GE Healthcare, Eindhoven, The Netherlands) and analyzed using GelDoc2000 (BioRad).
Results and Discussion
Canonical Wnt signaling is aberrantly active in a subset of canine mammary tumor cell lines
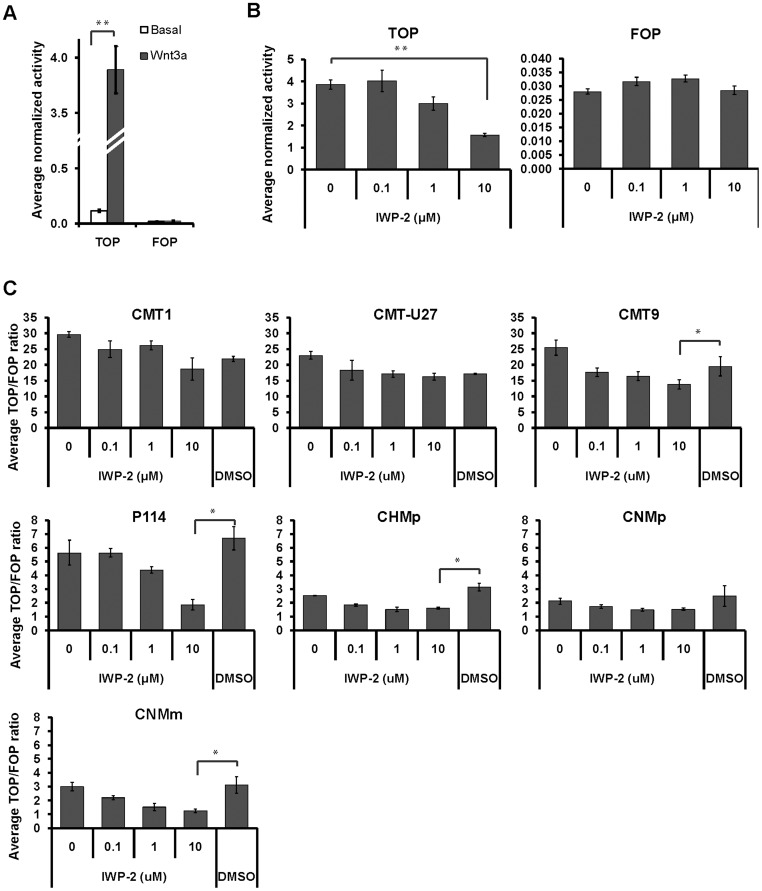
To quantitatively assess canonical Wnt activity we tested a panel of 12 canine mammary cell lines using a TCF-reporter assay (Fig. 1). Three cell lines (CMT1, CMT-U27 and CMT9) showed high TCF-reporter activity. Four cell lines (P114, CHMp, CNMp and CNMm) showed moderate reporter activity comparable to previously reported activity in human mammary cell lines [24], [25]. The remaining five cell lines (CMT-U229, CMT-U335, CHMm, CIPp and CIPm) with the TOP/FOP ratio around 1, lacked canonical Wnt activity.
Figure 1. TCF-reporter activity in canine mammary tumor cell lines.
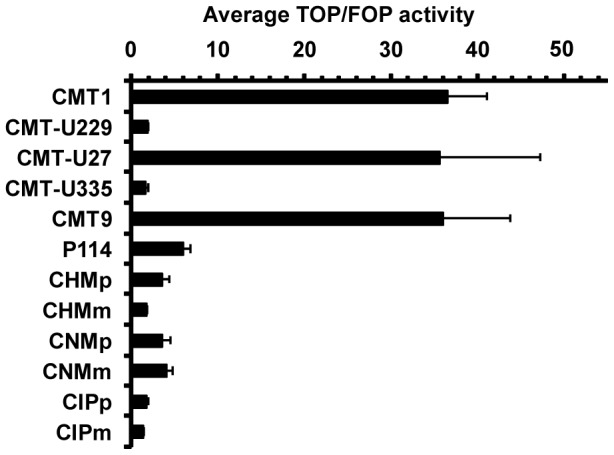
Average TOP/FOP ratio (+/− s.e.m.) in canine mammary cell lines from 3–4 independent experiments.
Ligand-dependent and -independent mechanism of canonical Wnt activation in canine mammary cell lines
IWP-2 is a small molecule inhibitor that impairs Wnt ligand palmitoylation and secretion [26] and could, therefore, be used to discriminate between ligand-dependent and -independent mechanisms of canonical Wnt activation in cells. To assess the activity of IWP-2 in canine cells, P114 cell line was transiently co-transfected with canonical Wnt ligand Wnt3a (Fig. 2A) and treated with increasing concentration of IWP-2. IWP-2 effectively inhibited Wnt-3a-dependent TOP-flash activity, but not the FOP-flash activity (Fig. 2B), confirming its specificity as canonical Wnt inhibitor in canine cells. The effect of IWP-2 treatment on the basal canonical Wnt activity was subsequently evaluated in all cell lines with active Wnt signaling. In cell lines with moderate basal canonical Wnt activity (i.e. P114, CHMp, CNMp and CNMm), IWP-2 was able to efficiently inhibit the TCF-reporter activity. Treatment with 10 µM IWP-2 resulted in TOP/FOP ratios around 1, suggesting a full ligand-dependency in the cell lines with moderately activate Wnt signaling (Fig. 2C). Moreover, we have assessed the expression of several Wnt ligands previously reported as activators of canonical signaling and/or being expressed in mammary tissue and cell lines [27], [28]. Ligand-dependent activation of the pathway in these cell lines is further supported by the high expression of multiple Wnt ligands (especially Wnt5b and Wnt7a) and undetectable levels of the inhibitor sFRP1 (Fig. 3). IWP-2 treatment in CMT1, CMT-U27 and CMT9 cells, however, had no or only a minor effect on the TCF-reporter activity (Fig. 2C). These three cell lines are therefore expected to have a ligand-independent component for canonical Wnt activation. Recently, it has been reported that conditioned medium of tumor-associated macrophages or co-culture with macrophages mediate a switch from canonical to non-canonical Wnt signaling in multiple canine mammary cell lines, including P114 and CMT-U27 [29]. Inhibition of canonical Wnt signaling (demonstrated by downregulation of cytoplasmic and nuclear β-catenin protein levels) was associated with exposure of cells to increased levels of non-canonical Wnt ligands and canonical Wnt inhibitor Dkk-1. Our data supports the responsiveness of P114 cell line to Wnt ligands and inhibitors. However, the insensitivity of basal TCF-reporter activity in CMT-U27 cells to treatment with IWP-2 suggests that the reported inhibition of canonical Wnt signaling [29] is not mediated by altered Wnt ligand or inhibitor expression, but most probably is caused by other mechanisms.
Figure 2. Inhibition of canonical Wnt activity using porcupine inhibitor IWP-2.
(A) Effect of transient Wnt3a co-transfection on TOP and FOP activities in P114 cells. (B) Effect of IWP-2 treatment on TOP and FOP activities in P114 cells co-transfected with Wnt-3a. (C) Effect of IWP-2 treatment on basal TOP/FOP ratio in CMT1, CMT-U27, CMT9, P114, CHMp, CNMp and CNMm cell lines. TOP/FOP ratio after treatment with 10 µM IWP-2 was tested against the control DMSO treatment. * indicates p<0.05 and **p<0.01.
Figure 3. Expression of multiple Wnt ligand- and inhibitor Sfrp1-mRNA.
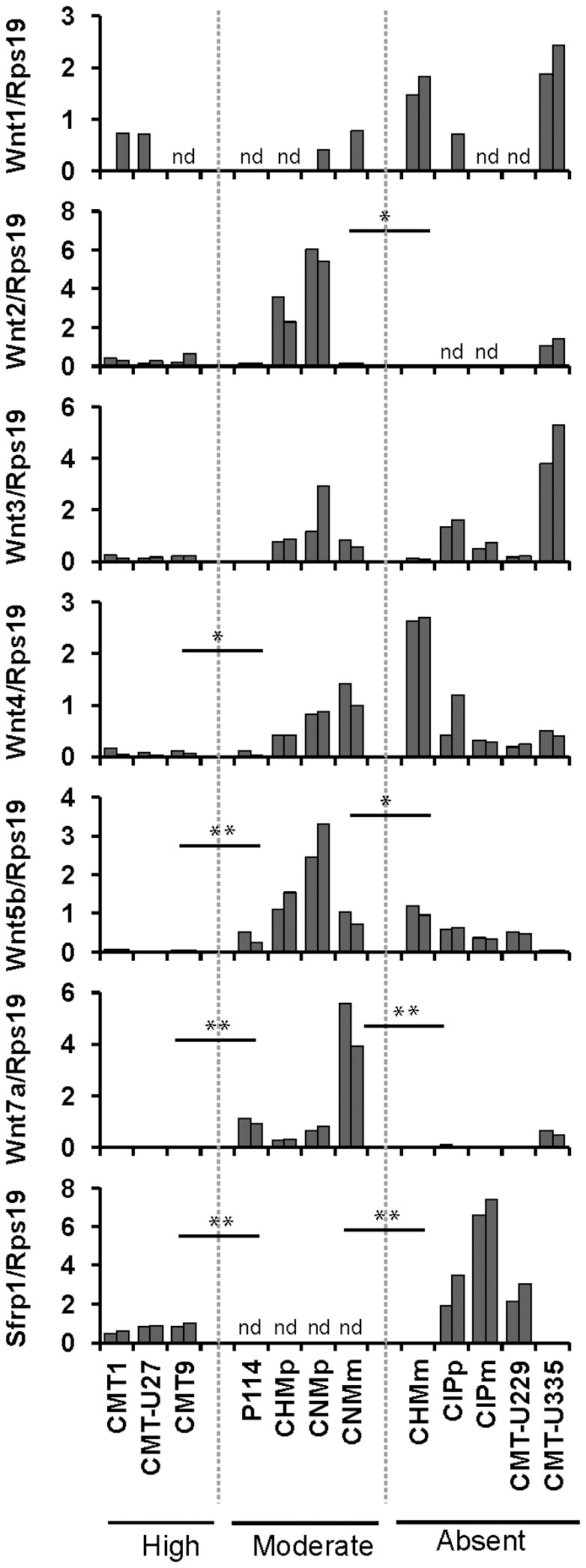
mRNA expression of Wnt1, Wnt2, Wnt3, Wnt4, Wnt5b, Wnt7a and Sfrp1 in two different passages of cell lines. Target gene expression was normalized to that of a reference gene Rps19. Cell lines were divided in three groups (from left to right): cell lines with high, moderate or absent canonical Wnt activity. * indicates p<0.05, **p<0.01 and nd stands for non-detectable.
High canonical Wnt activity is not associated with a lack of functional E-cadherin
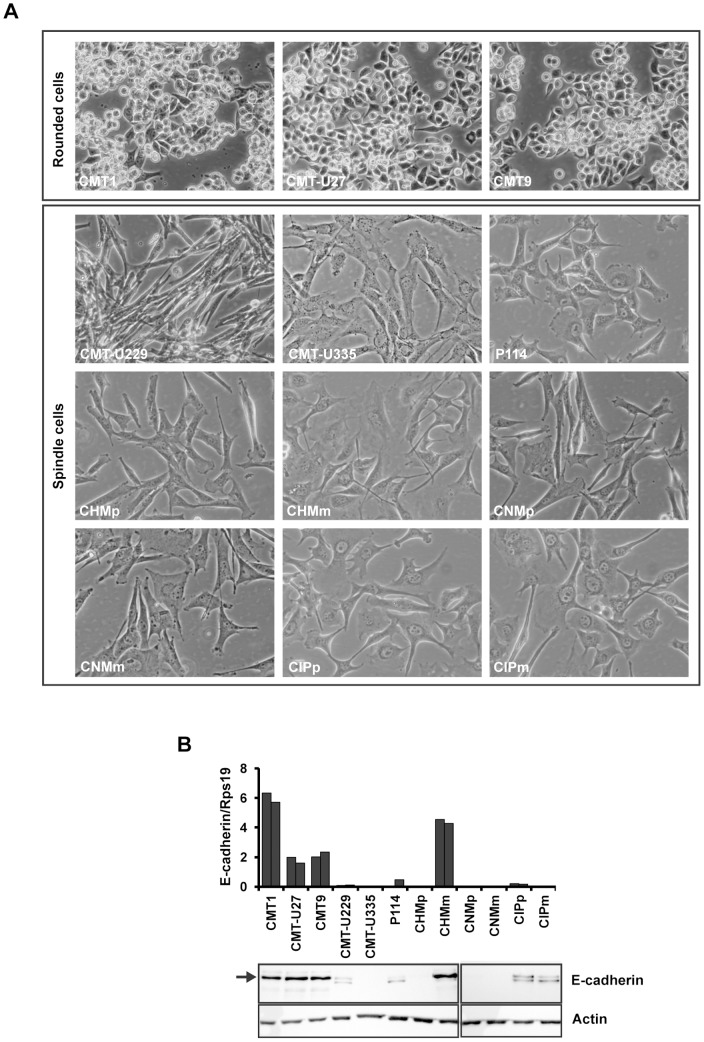
The majority of the canine mammary cell lines used in this study had spindle-cell-like morphology, except for CMT1, CMT-U27 and CMT9, which grow as attached cells but also as partially rounded cells (Fig. 4A). A partially rounded morphology has been associated with E-cadherin mutations in human breast cancer cell lines [30]. As the loss of E-cadherin protein can stimulate canonical Wnt activity [31], we analyzed its coding sequence and mRNA and protein levels in all 12 canine mammary cell lines. Sequencing analysis of the whole CDH1 coding region revealed no mutations in any of the cell lines (Table S1). Moreover, CMT1, CMT-U27 and CMT9 highly expressed mRNA and mature protein of E-cadherin (Fig. 4B), suggesting a different mechanism of canonical Wnt activation in these cell lines.
Figure 4. Cell line morphology and E-cadherin expression.
(A) Canine mammary cell lines grouped based on their morphology as (partially) rounded cells or spindle cells. (B) Expression of E-cadherin at mRNA (top) and protein (bottom) level. Rps19 and actin expression served as reference mRNA and protein, respectively. Arrow indicates the position of full-length mature E-cadherin protein. Additional E-cadherin protein band present in some of the cell lines represents the unprocessed form of the protein.
High canonical Wnt activity is not associated with defects in β-catenin destruction complex
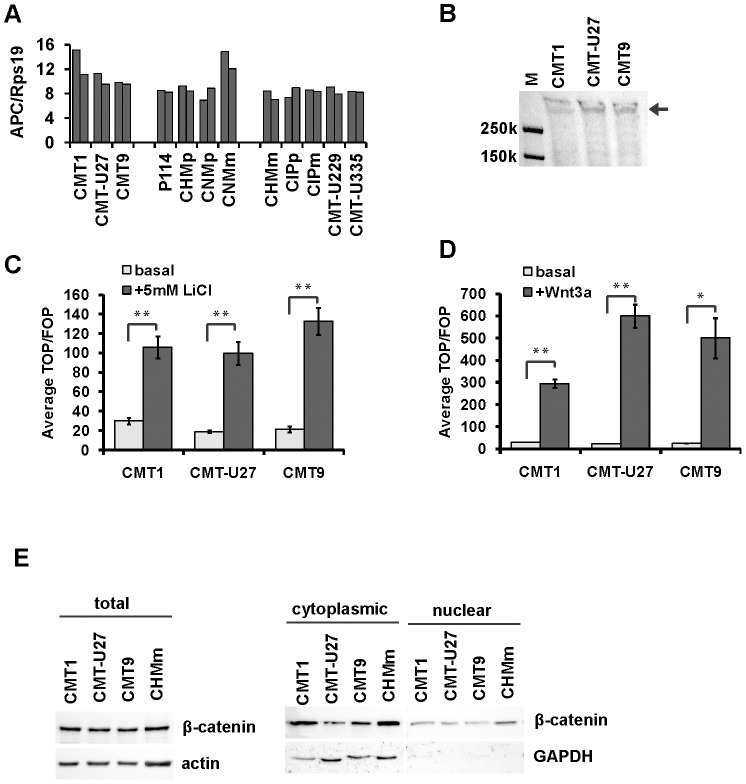
In multiple tumors, elevated canonical Wnt activity has been shown to result from mutations in components of β-catenin destruction complex [32]. Mutational analysis of coding sequences of APC, β-catenin, GSK3β, CK1α and Axin1 in canine mammary cell lines revealed, however, no mutations that were restricted to cell lines with the active canonical Wnt signaling (Table S1). As APC is also known to be epigenetically silenced or proteolytically cleaved in tumors [33], [34], its mRNA and protein expression were additionally assessed. All 12 canine mammary cell lines expressed comparable levels of APC mRNA (Fig. 5A). Analysis of protein expression in CMT1, CMT-U27 and CMT9 revealed that APC was expressed as a full-length protein (Fig. 5B). We next asked whether high canonical Wnt activity in these cells is a consequence of a defect at the level of β-catenin destruction complex. Canonical Wnt signaling in cells in which β-catenin destruction complex function is fully impaired is expected to be insensitive to further stimulation of the pathway by Wnt ligands or to treatment with GSK3β inhibitors [35]. CMT1, CMT-U27 and CMT9 cells, however, responded potently to GSK3β inhibitor, LiCl (Fig. 5C) as well as to Wnt3a transfection (Fig. 5D). To determine whether the high Wnt activity is associated with increased stabilization of β-catenin protein we assessed total, cytoplasmic and nuclear levels of β-catenin by western blot. GAPDH was used as a marker of cytoplasmic proteins to assess the purity of extracts from different cell fractions. When compared to a cell line lacking canonical Wnt activity (CHMm), CMT1, CMT-U27 and CMT9 cells did not show evidence of marked cytoplasmic or nuclear β-catenin protein stabilization (Fig. 5E), implying no major defect in the β-catenin destruction complex function.
Figure 5. Assessment of defects at the level of β-catenin destruction complex.
(A) mRNA expression of APC in two different passages of canine mammary cell lines normalized to the expression of Rps19. (B) APC protein expression in CMT1, CMT-U27 and CMT9. M indicates loading marker with reference molecular weight bands. (C) Effect of treatment with 5 mM LiCl or co-transfection with Wnt-3a (D) on TCF-reporter activity in CMT1, CMT-U27 and CMT9. * indicates p<0.05 and **p<0.01. (E) β-catenin protein expression in total cell lysates (total) and the cytoplasmic and nuclear fractions.
Ectopic LEF1 expression contributes to the high canonical Wnt activity in CMT1, CMT-U27 and CMT9
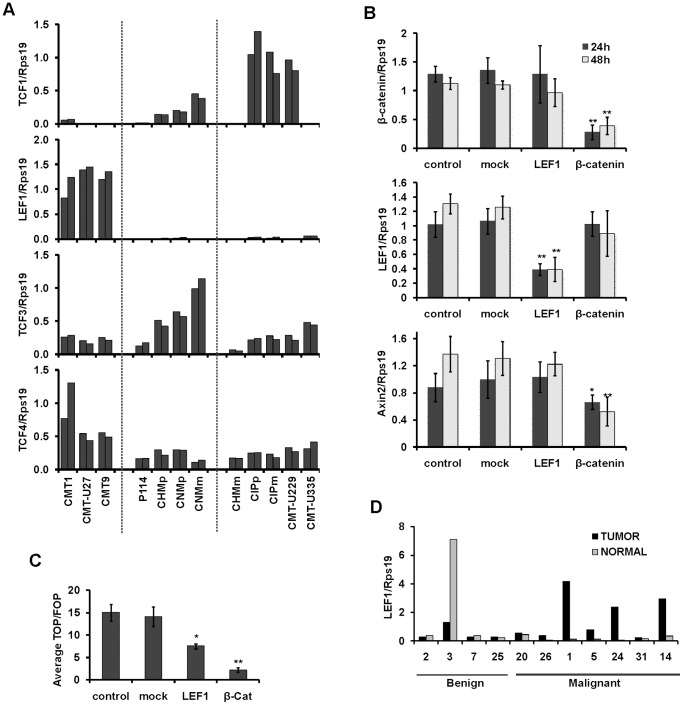
Upon stabilization, β-catenin translocates to the nucleus and interacts with members of TCF/LEF family of transcription factors (TCF1, LEF1, TCF2 and TCF4). Interestingly, although all TCF/LEF family members were expressed in canine mammary cell lines, LEF1 showed high mRNA expression specifically in CMT1, CMT-U27 and CMT9 (Fig. 6A). Different LEF1 isoforms have been described, resulting from alternative splicing of LEF1 transcript [36]. Sequencing of LEF1 coding region in CMT1, CMT-U27, CMT9 and CIPp showed that the first three cell lines express LEF1 transcript lacking exon 6. Lack of exon 6 in Xenopus LEF1 has been shown to lower its transcriptional potential on TOPFlash reporter in HEK293 cells [37]. Lack of exon 6 in LEF1 in CMT1, CMT-U27 and CMT9 can therefore not be attributing to the high canonical Wnt activity in these cell lines. As LEF1 is known to be a direct target gene of canonical Wnt signaling [38] we asked whether high expression of LEF1 in these three cell lines could be a cause or a consequence of high canonical Wnt activity. For this purpose β-catenin and LEF1 knock down was performed in CMT-U27 cells. Knock-down of β-catenin resulted in a potent inhibition of TCF-reporter activity (Fig. 6C) and down regulation of Axin2 target gene (Fig. 6B), but it had no effect on the expression of LEF1 (Fig. 6B). Knock-down of LEF1 did not affect β-catenin expression (Fig. 6B) but was able to significantly inhibit TCF-reporter activity (Fig. 6C). Altogether, this suggests that the ectopic expression of LEF1 in CMT1, CMT-U27 and CMT9 is not a consequence but rather a contributing factor to the high canonical Wnt activity in these cell lines. LEF1 was recently shown to affect the viability, invasion and migration of breast cancer cells [39]. The correlation between LEF1 overexpression and high canonical Wnt activity in canine mammary tumor cell lines prompted us, therefore, to assess LEF1 expression in spontaneous canine mammary tumors. To assess the tumor-specific overexpression of LEF1, each canine mammary tumor was compared to the corresponding normal tissue from the same patient. Interestingly, five out of seven malignant tumor samples showed overexpression of LEF1 (Fig. 6D). However, the sample size will need to be increased in order to test the statistical significance of these results. A challenging task remains to assess whether LEF1 overexpression in canine mammary tumors is associated with high canonical Wnt activity. Considering that canine mammary tumor cell lines with high LEF1 expression do not seem to show marked overstabilization of β-catenin protein (Fig. 5E), an alternative marker for canonical Wnt activity in tissue samples is needed. In this regard, Axin2 mRNA levels were suggested to correlate with mutations in the Wnt signaling pathway in a panel of human cancer cell lines [32]. However, in canine mammary cell lines, basal Axin2 mRNA levels do not correlate with the canonical Wnt activity (Fig. S1), implicating that Axin2 expression is also not a reliable canonical Wnt activity marker in canine mammary tumors. On a further note, LEF1 knock-down was not able to fully inhibit TCF-reporter activity in canine mammary cell lines. This may be a consequence of insufficient knock-down of LEF1 mRNA but it may also argue for involvement of additional canonical Wnt activating factors. To test whether other TCFs may be compensating for the knock-down of LEF1, the levels of all TCF-family members were assessed 24 h after knock-down of LEF1 (Fig. S2). Expression of neither of TCFs showed, however, signs of compensation. Lastly, the use of transient transfection system prevented us from investigating a relationship between LEF1 overexpression and the cellular morphology. For this purpose stable transfection of inducible LEF1 knock-down system should be employed.
Figure 6. Association between high canonical Wnt activity and LEF1 expression.
(A) mRNA expression of TCF1, LEF1, TCF3 and TCF4 in two different passages of canine mammary cell lines normalized to the expression of Rps19. (B) β-catenin, LEF1 and Axin2 mRNA expression in CMT-U27 cells that were either non-transfected (control) or transfected with mock control (mock), LEF1 siRNA (LEF1) or β-catenin siRNA (β-catenin). mRNA expression was analyzed 24 h and 48 h post-transfection. (C) Average TOP/FOP ratio in CMT-U27 cells as described in (B). * indicates p<0.05 and **p<0.01 compared to the mock control. (D) Rps19 normalized mRNA expression of LEF1 in a panel of canine mammary tumors (tumor) and normal mammary tissue (normal) from the same dog.
Conclusions
Altogether, this study provides evidence for moderate, ligand-dependent canonical Wnt activation in canine mammary tumors that is comparable to human breast cancer. In addition, we report a novel ligand-independent mechanism involving LEF1 overexpression, which results in high canonical Wnt activity. Our further studies aim to explore this ligand-independent mechanism extensively and to identify the underlying gene mutations.
Supporting Information
Axin2 mRNA expression. Rps19 normalized Axin2 mRNA expression in two different passages of canine mammary cell lines. Cell lines were divided in three groups (from left to right): cell lines with high, moderate or absent canonical Wnt activity.
(TIF)
Expression of TCF-family members upon LEF1 knock-down. Relative Rps19 normalized mRNA expression of LEF1, TCF1, TCF3 and TCF4 24 h after LEF1 knock-down in CMT-U27 cells. Average expression of control conditions for each target gene is set to 100.
(TIF)
Sequencing results of target gene coding regions in canine mammary tumor cell lines.
(DOCX)
Funding Statement
This work was supported by the Mozaiek Grant 017.004.081 (to A.G.) from the Dutch Society for Scientific Research (NWO). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hatsell S, Rowlands T, Hiremath M, Cowin P (2003) Beta-catenin and tcfs in mammary development and cancer. J Mammary Gland Biol Neoplasia 8: 145–158. [DOI] [PubMed] [Google Scholar]
- 2. Clevers H, Nusse R (2012) Wnt/beta-catenin signaling and disease. Cell 149: 1192–1205 10.1016/j.cell.2012.05.012; 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 3. Howe LR, Brown AM (2004) Wnt signaling and breast cancer. Cancer Biol Ther 3: 36–41. [DOI] [PubMed] [Google Scholar]
- 4. Bafico A, Liu G, Goldin L, Harris V, Aaronson SA (2004) An autocrine mechanism for constitutive wnt pathway activation in human cancer cells. Cancer Cell 6: 497–506 10.1016/j.ccr.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 5. Matsuda Y, Schlange T, Oakeley EJ, Boulay A, Hynes NE (2009) WNT signaling enhances breast cancer cell motility and blockade of the WNT pathway by sFRP1 suppresses MDA-MB-231 xenograft growth. Breast Cancer Res 11: R32 10.1186/bcr2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang L, Wu X, Wang Y, Zhang K, Wu J, et al. (2011) FZD7 has a critical role in cell proliferation in triple negative breast cancer. Oncogene 30: 4437–4446 10.1038/onc.2011.145; 10.1038/onc.2011.145. [DOI] [PubMed] [Google Scholar]
- 7. Schlosshauer PW, Brown SA, Eisinger K, Yan Q, Guglielminetti ER, et al. (2000) APC truncation and increased beta-catenin levels in a human breast cancer cell line. Carcinogenesis 21: 1453–1456. [PubMed] [Google Scholar]
- 8. Ai L, Tao Q, Zhong S, Fields CR, Kim WJ, et al. (2006) Inactivation of wnt inhibitory factor-1 (WIF1) expression by epigenetic silencing is a common event in breast cancer. Carcinogenesis 27: 1341–1348 10.1093/carcin/bgi379. [DOI] [PubMed] [Google Scholar]
- 9. Veeck J, Niederacher D, An H, Klopocki E, Wiesmann F, et al. (2006) Aberrant methylation of the wnt antagonist SFRP1 in breast cancer is associated with unfavourable prognosis. Oncogene 25: 3479–3488 10.1038/sj.onc.1209386. [DOI] [PubMed] [Google Scholar]
- 10. Virmani AK, Rathi A, Sathyanarayana UG, Padar A, Huang CX, et al. (2001) Aberrant methylation of the adenomatous polyposis coli (APC) gene promoter 1A in breast and lung carcinomas. Clin Cancer Res 7: 1998–2004. [PubMed] [Google Scholar]
- 11. Hansen K, Khanna C (2004) Spontaneous and genetically engineered animal models; use in preclinical cancer drug development. Eur J Cancer 40: 858–880 10.1016/j.ejca.2003.11.031. [DOI] [PubMed] [Google Scholar]
- 12. Pinho SS, Carvalho S, Cabral J, Reis CA, Gartner F (2012) Canine tumors: A spontaneous animal model of human carcinogenesis. Transl Res 159: 165–172 10.1016/j.trsl.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 13. Misdorp W (1988) Canine mammary tumours: Protective effect of late ovariectomy and stimulating effect of progestins. Vet Q 10: 26–33. [DOI] [PubMed] [Google Scholar]
- 14. Schneider R, Dorn CR, Taylor DO (1969) Factors influencing canine mammary cancer development and postsurgical survival. J Natl Cancer Inst 43: 1249–1261. [PubMed] [Google Scholar]
- 15. Restucci B, Maiolino P, Martano M, Esposito G, De Filippis D, et al. (2007) Expression of beta-catenin, E-cadherin and APC in canine mammary tumors. Anticancer Res 27: 3083–3089. [PubMed] [Google Scholar]
- 16. De Matos AJ, Lopes CC, Faustino AM, Carvalheira JG, Rutteman GR, et al. (2007) E-cadherin, beta-catenin, invasion and lymph node metastases in canine malignant mammary tumours. APMIS 115: 327–334 10.1111/j.1600-0463.2007.apm_544.x. [DOI] [PubMed] [Google Scholar]
- 17. Gama A, Paredes J, Gartner F, Alves A, Schmitt F (2008) Expression of E-cadherin, P-cadherin and beta-catenin in canine malignant mammary tumours in relation to clinicopathological parameters, proliferation and survival. Vet J 177: 45–53 10.1016/j.tvjl.2007.05.024. [DOI] [PubMed] [Google Scholar]
- 18. Uva P, Aurisicchio L, Watters J, Loboda A, Kulkarni A, et al. (2009) Comparative expression pathway analysis of human and canine mammary tumors. BMC Genomics 10: 135 10.1186/1471-2164-10-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hellmen E (1992) Characterization of four in vitro established canine mammary carcinoma and one atypical benign mixed tumor cell lines. In Vitro Cell Dev Biol 28A: 309–319. [DOI] [PubMed] [Google Scholar]
- 20. Van Leeuwen IS, Hellmen E, Cornelisse CJ, Van den Burgh B, Rutteman GR (1996) P53 mutations in mammary tumor cell lines and corresponding tumor tissues in the dog. Anticancer Res 16: 3737–3744. [PubMed] [Google Scholar]
- 21. Uyama R, Nakagawa T, Hong SH, Mochizuki M, Nishimura R, et al. (2006) Establishment of four pairs of canine mammary tumour cell lines derived from primary and metastatic origin and their E-cadherin expression. Vet Comp Oncol 4: 104–113 10.1111/j.1476-5810.2006.00098.x. [DOI] [PubMed] [Google Scholar]
- 22. Doleschall M, Mayer B, Cervenak J, Cervenak L, Kacskovics I (2007) Cloning, expression and characterization of the bovine p65 subunit of NFkappaB. Dev Comp Immunol 31: 945–961 10.1016/j.dci.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 23. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25: 402–408 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24. Bjorklund P, Svedlund J, Olsson AK, Akerstrom G, Westin G (2009) The internally truncated LRP5 receptor presents a therapeutic target in breast cancer. PLoS One 4: e4243 10.1371/journal.pone.0004243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rieger ME, Sims AH, Coats ER, Clarke RB, Briegel KJ (2010) The embryonic transcription cofactor LBH is a direct target of the wnt signaling pathway in epithelial development and in aggressive basal subtype breast cancers. Mol Cell Biol 30: 4267–4279 10.1128/MCB.01418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen B, Dodge ME, Tang W, Lu J, Ma Z, et al. (2009) Small molecule-mediated disruption of wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol 5: 100–107 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carmon KS, Loose DS (2008) Secreted frizzled-related protein 4 regulates two Wnt7a signaling pathways and inhibits proliferation in endometrial cancer cells. Mol Cancer Res 6: 1017–1028 10.1158/1541-7786.MCR-08-0039; 10.1158/1541-7786.MCR-08-0039. [DOI] [PubMed] [Google Scholar]
- 28. Benhaj K, Akcali KC, Ozturk M (2006) Redundant expression of canonical wnt ligands in human breast cancer cell lines. Oncol Rep 15: 701–707. [PubMed] [Google Scholar]
- 29. Krol M, Mucha J, Majchrzak K, Homa A, Bulkowska M, et al. (2014) Macrophages mediate a switch between canonical and non-canonical wnt pathways in canine mammary tumors. PLoS One 9: e83995 10.1371/journal.pone.0083995; 10.1371/journal.pone.0083995. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30. Hollestelle A, Elstrodt F, Timmermans M, Sieuwerts AM, Klijn JG, et al. (2010) Four human breast cancer cell lines with biallelic inactivating alpha-catenin gene mutations. Breast Cancer Res Treat 122: 125–133 10.1007/s10549-009-0545-4. [DOI] [PubMed] [Google Scholar]
- 31. Kuphal F, Behrens J (2006) E-cadherin modulates wnt-dependent transcription in colorectal cancer cells but does not alter wnt-independent gene expression in fibroblasts. Exp Cell Res 312: 457–467 10.1016/j.yexcr.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 32. Polakis P (2012) Drugging wnt signalling in cancer. EMBO J 31: 2737–2746 10.1038/emboj.2012.126; 10.1038/emboj.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Browne SJ, MacFarlane M, Cohen GM, Paraskeva C (1998) The adenomatous polyposis coli protein and retinoblastoma protein are cleaved early in apoptosis and are potential substrates for caspases. Cell Death Differ 5: 206–213 10.1038/sj.cdd.4400331. [DOI] [PubMed] [Google Scholar]
- 34. Klarmann GJ, Decker A, Farrar WL (2008) Epigenetic gene silencing in the wnt pathway in breast cancer. Epigenetics 3: 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Easwaran V, Song V, Polakis P, Byers S (1999) The ubiquitin-proteasome pathway and serine kinase activity modulate adenomatous polyposis coli protein-mediated regulation of beta-catenin-lymphocyte enhancer-binding factor signaling. J Biol Chem 274: 16641–16645. [DOI] [PubMed] [Google Scholar]
- 36. Hovanes K, Li TW, Waterman ML (2000) The human LEF-1 gene contains a promoter preferentially active in lymphocytes and encodes multiple isoforms derived from alternative splicing. Nucleic Acids Res 28: 1994–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ghogomu SM, van Venrooy S, Ritthaler M, Wedlich D, Gradl D (2006) HIC-5 is a novel repressor of lymphoid enhancer factor/T-cell factor-driven transcription. J Biol Chem 281: 1755–1764 10.1074/jbc.M505869200. [DOI] [PubMed] [Google Scholar]
- 38. Filali M, Cheng N, Abbott D, Leontiev V, Engelhardt JF (2002) Wnt-3A/beta-catenin signaling induces transcription from the LEF-1 promoter. J Biol Chem 277: 33398–33410 10.1074/jbc.M107977200. [DOI] [PubMed] [Google Scholar]
- 39. Hsieh TH, Tsai CF, Hsu CY, Kuo PL, Hsi E, et al. (2012) n-butyl benzyl phthalate promotes breast cancer progression by inducing expression of lymphoid enhancer factor 1. PLoS One 7: e42750 10.1371/journal.pone.0042750; 10.1371/journal.pone.0042750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van de Wetering M, Barker N, Harkes IC, van der Heyden M, Dijk NJ, et al. (2001) Mutant E-cadherin breast cancer cells do not display constitutive wnt signaling. Cancer Res 61: 278–284. [PubMed] [Google Scholar]
- 41. Schlange T, Matsuda Y, Lienhard S, Huber A, Hynes NE (2007) Autocrine WNT signaling contributes to breast cancer cell proliferation via the canonical WNT pathway and EGFR transactivation. Breast Cancer Res 9: R63 10.1186/bcr1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ng SS, Mahmoudi T, Danenberg E, Bejaoui I, de Lau W, et al. (2009) Phosphatidylinositol 3-kinase signaling does not activate the wnt cascade. J Biol Chem 284: 35308–35313 10.1074/jbc.M109.078261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim SY, Dunn IF, Firestein R, Gupta P, Wardwell L, et al. (2010) CK1epsilon is required for breast cancers dependent on beta-catenin activity. PLoS One 5: e8979 10.1371/journal.pone.0008979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mikheev AM, Mikheeva SA, Maxwell JP, Rivo JV, Rostomily R, et al. (2008) Dickkopf-1 mediated tumor suppression in human breast carcinoma cells. Breast Cancer Res Treat 112: 263–273 10.1007/s10549-007-9867-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Axin2 mRNA expression. Rps19 normalized Axin2 mRNA expression in two different passages of canine mammary cell lines. Cell lines were divided in three groups (from left to right): cell lines with high, moderate or absent canonical Wnt activity.
(TIF)
Expression of TCF-family members upon LEF1 knock-down. Relative Rps19 normalized mRNA expression of LEF1, TCF1, TCF3 and TCF4 24 h after LEF1 knock-down in CMT-U27 cells. Average expression of control conditions for each target gene is set to 100.
(TIF)
Sequencing results of target gene coding regions in canine mammary tumor cell lines.
(DOCX)