Abstract
Excitable tissues rely on junctional membrane complexes to couple cell surface signals to intracellular channels. The junctophilins have emerged as a family of proteins critical in coordinating the maturation and maintenance of this cellular ultrastructure. Within skeletal and cardiac muscle, junctophilin-1 and junctophilin-2, respectively, couple sarcolemmal and intracellular calcium channels. In neuronal tissue, junctophilin 3 and junctophilin 4 may have an emerging role in coupling membrane neurotransmitter receptors and intracellular calcium channels. These important physiologic roles are highlighted by the pathophysiology which results when these proteins are perturbed and a growing body of literature has associated junctophilins with the pathogenesis of human disease.
Keywords: calcium, excitation contraction coupling, JPH, junctophilin, mutation
AN EMERGING ROLE IN THE PATHOPHYSIOLOGY OF EXCITABLE CELLS
Since the sentinel discovery of the junctophilin (JPH) family of proteins a decade ago[1], a rapidly progressing body of literature has linked these proteins to critical roles in all excitable cells with implications for human physiology and pathophysiology. Through maintaining critical subcellular architecture, and an emerging function as a direct regulator of calcium-handling proteins, impaired JPHs have been implicated in a number of human diseases. Investigations into the role of JPHs in cellular physiology have elucidated novel mechanisms of skeletal muscle myopathy, cardiomyopathy, heart failure, arrhythmia, behavioral and learning deficits, motor control, and Huntington Disease-like pathology. Future investigations hold the promise of novel therapeutic targets and molecular interventions for a number of these diseases.
PROTEIN PHYLOGENY AND STRUCTURE
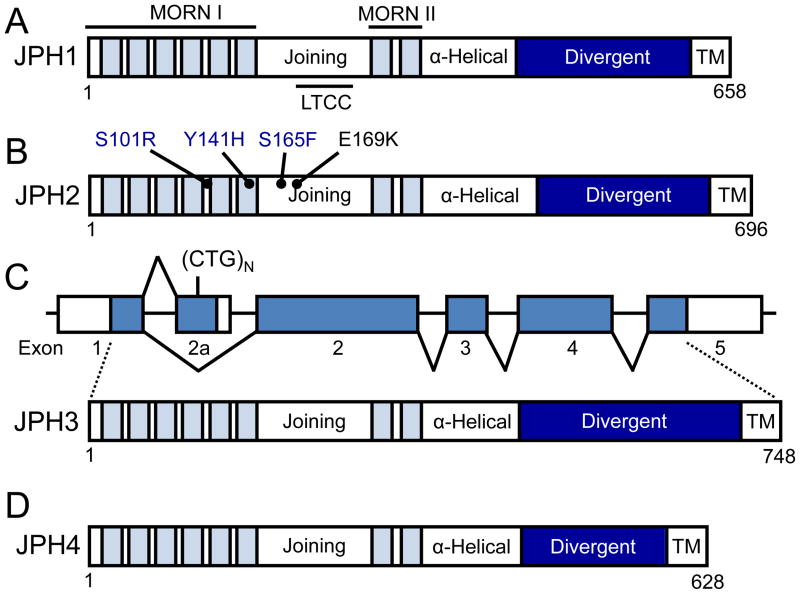
Junctophilins are members of a family of junctional membrane complex (JMC)-associated proteins found in all excitable cells from striated muscle to neurons. JPHs were originally discovered using an immuno-proteomic monoclonal antibody library. This library was created from mice immunized with rabbit skeletal muscle membranes taken from transverse tubules (T-tubules) and muscle triads[2]. JPHs contain specific protein domains which define them as a family, including eight amino-terminal membrane occupation and recognition nexus (MORN) motifs separated by a joining region, followed by an alpha helix, divergent, and a C-terminal transmembrane motif that anchors the protein into the endoplasmic reticulum (ER) or sarcoplasmic reticulum (SR)[1, 3]. The protein topology for each JPH isoform is depicted in Figure 1.
Figure 1. Protein topology of the four junctophilin isoforms expressed in humans.
The JPH amino terminus has 8 MORN motifs (light blue fill) distributed across two MORN domains separated by a joining region (white fill). The carboxy terminus contains a transmembrane (TM) domain (white fill) which embeds this end of the protein into intracellular membranes such as the sarcoplasmic reticulum in striated muscle. An alpha-helical domain (white fill) and a less evolutionarily conserved divergent region (dark blue fill) join these two termini. The isoforms vary in length from 628 (JPH4) to 758 (JPH3) residues. (A) Protein topology of JPH1, which is found predominantly in skeletal muscle. JPH1 binds to the LTCC via a binding domain within the joining region of the protein. (B) Protein topology of JPH2, the major cardiac isoform. Three JPH2 mutations have been linked with the development of hypertrophic cardiomyopathy (blue text) and one to arrhythmogenesis (black text). (C) Schematic of the two alternatively spliced transcripts of JPH3 including the untranslated regions (white fill) and coding exons (blue fill). The full length transcript consisting of 5 exons which encode JPH3 is depicted as well as the 2 exon alternative transcript containing the CTG trinucleotide repeat expansion in the alternate exon 2 (2a). Below is the protein topology of JPH3. (D) Protein topology of JPH4. Abbreviations: JPH, junctophilin; LTCC, L-type calcium channel binding domain; MORN, membrane occupation and recognition nexi; TM, transmembrane domain.
MORN motifs have been found to be highly conserved across all JPH isoforms, as well as across various species, and demonstrate a consensus sequence of YxGxWxxGxRHGYG[4]. Initially identified by BLAST analysis, the eight MORN motifs in JPH are separated by a ‘joining region’ which links MORN motifs I–VI to VII and VIII. As a whole, the MORN motifs are thought to traffic and bind JPHs to the plasma membrane. This hypothesis was based on early experiments which truncated the carboxy-terminus of the protein and resulted in localization of the MORN motif-containing amino terminus strictly to the plasma membrane[1]. Further, homologous MORN motifs in plants, such as A. thaliana, have high affinity for membrane phospholipids such as phosphatidic acid and somewhat lower affinity for membrane phosphatidylinositol (PI)4P and PI(4,5)P2[5]. In addition to JPHs, multiple MORN motifs are found in several other classes of proteins including histone-lysine N-methyltransferase and phosphatidylinositol-4-phosphate 5-kinase proteins[1, 6].
The carboxy-terminal membrane-spanning domain of JPH is believed to embed the protein into intracellular membranes such as the ER or SR[1, 3]. This domain is highly conserved across species and isoforms, and in silico secondary structure analysis as well as hydrophobicity analysis predict the presence of this domain[1, 4]. Linking the MORN motifs and the membrane-spanning domain are two domains, the alpha-helical domain and the divergent region. Highly conserved across JPH isoforms, in silico analysis predict multiple alpha-helical stretches within the primary sequence of this domain. Spanning approximately one hundred amino acids, it is thought to bridge the gap between the plasmalemma/sarcolemma and the ER/SR[1, 3, 7]. Given the possible alpha-helical structure, this domain may provide mechanical elasticity to regulate dyad distance[1, 4]. Conversely, the divergent region derives its name from the relatively low primary sequence conservation between the various JPH isoforms, and its role is unclear at present[4]. Overall, these presumed functional domains allow for JPHs to play critical roles in the maintenance of cellular ultrastructure, particularly, the interface between intracellular membranes such as the ER/SR and the plasmalemma/sarcolemma.
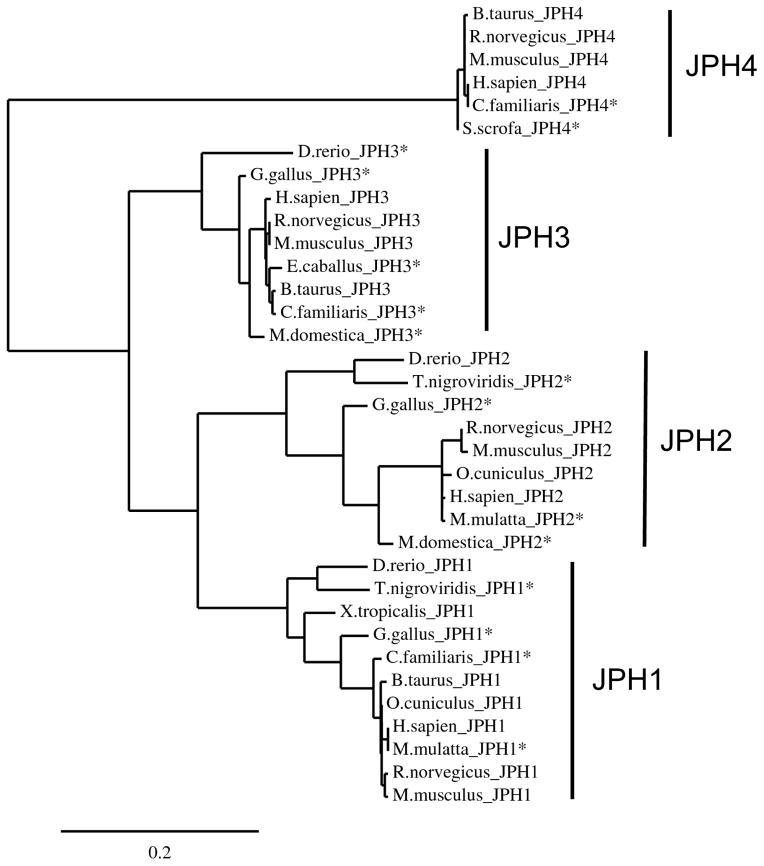
Initial studies in rodent models as well as in-depth bioinformatic and phylogenic analysis have identified four major JPH family members in higher chordate organisms, including humans[4]. Each is expressed preferentially in various excitable tissues and include skeletal muscle JPH1-encoded junctophilin type 1 (JPH1), cardiac muscle JPH2-encoded JPH2, and neuronal tissue JHP3-encoded JPH3, and JPH4-encoded JPH4. A phylogenic tree generated from multiple JPH isoforms across a diverse group of species is depicted in Figure 2.
Figure 2. Junctophilin phylogenic tree.
A phylogenic tree comprised of the primary sequences of 35 JPH isoforms derived from multiple species created using MUSCLE alignment and PhyML for phylogeny and TreeDyn tree rendering[4, 80–82]. Clades containing each of the isoforms are indicated. * indicate that the protein sequence is a putative JPH isoform that has not been experimentally validated. Abbreviations: JPH, junctophilin.
CALCIUM SIGNALING IN STRIATED MUSCLE
A major role of Ca2+ within muscle is the initiation and coordination of myofilament contraction[8–11]. In cardiac myocytes, Ca2+-mediated contraction signaling begins with the opening of voltage-gated L-type Ca2+ channels (LTCC) at the sarcolemma in response to membrane excitation. LTCC Ca2+ influx down its electrochemical gradient triggers a larger Ca2+ release from the SR via the intracellular Ca2+ release channel ryanodine receptor type 2 (RyR2) in a process known as Ca2+-induced Ca2+-release (CICR, see Glossary). In this manner, Ca2+ traveling across the cardiac dyad triggers the opening of RyR2 and release of SR Ca2+ without direct coupling between the LTCC and RyR2. This process serves as the molecular initiator for ATP-expending myofilament-based mechanical contraction of the heart. Contraction is terminated by active removal of Ca2+ from the cytosol into the SR via the action of the sarcoplasmic reticulum calcium ATPase (SERCA2a) or removal from the cell by the sodium-calcium exchanger (NCX1). While skeletal muscle is analogous in many ways, store Ca2+ release is directly triggered by alterations in sarcolemmal potential, a process known as voltage-dependent calcium-release[12]. During this process, voltage changes directly trigger release of store Ca2+ through direct coupling of the LTCC and RyR1[13, 14].
JUNCTOPHILIN TYPE 1 AND SKELETAL MUSCLE
JPH1-encoded JPH1 is the major JPH family member expressed in skeletal muscle but also demonstrates a low level of expression in the heart. JPH1 contains 5 coding exons localizing to 8q21 and encodes a 658 amino acid 90 kDa protein in humans[15]. JPH2-encoded JPH2 is also expressed in skeletal muscle and is the predominant cardiac isoform[16]. JPH1 maintains the critical ultrastructural geometry of the skeletal muscle triad which is comprised of three independent membranous structures including the T-tubular invagination of the sarcolemma flanked on two sides by cytosolic SR[17]. Loss of JPH1 expression has been shown to reduce the number of intact triads and to deform intact triads[18]. A detailed explanation of skeletal muscle triads, cardiac dyads, and the junctional membrane complex can be found in Box 1.
Box 1. Subcellular Architecture and the Junctional Membrane Complex.
Precise subcellular architecture of intracellular membrane and associated proteins is required for effective transmission of membrane excitation to mechanical contraction of skeletal and cardiac muscle. To facilitate this, the sarcolemma of striated muscle cells has deep, finger-like invaginations that penetrate the interior of the myocyte. Running parallel along these T-tubules are portions of the SR which are so-called junctional SR. In skeletal muscle, the SR runs alongside a shared portion of the T-tubule on two opposing sides which create a triad of membranes observed on microscopy. In cardiac muscle, a single junctional SR runs alongside the T-tubule over a given portion of the T-tubule appearing as a dyad. The distance of the triad or dyad is carefully controlled and is critical for efficient movement and signaling of ions, particularly Ca2+. In the cardiac cell, it is movement of extracellular Ca2+ across the sarcolemma and the cardiac dyad which triggers CICR and allows for efficient EC coupling. A host of proteins, including JPHs, are crucial to the development and maintenance of this architecture. Further, the proteins which localize to the interface of these membranes and are responsible for Ca2+ cross talk are referred to as the JMC.
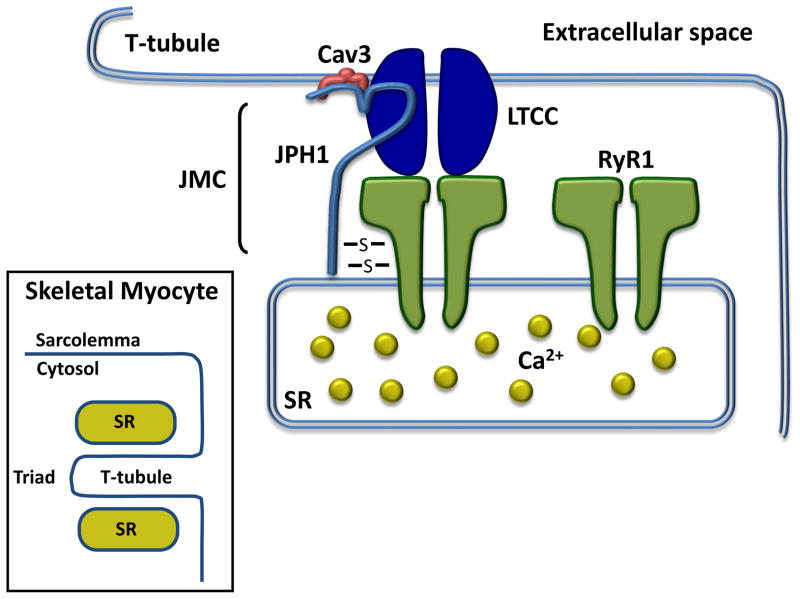
In addition to a structural role, JPH1 directly binds and regulates a number of proteins within the skeletal muscle JMC. JPH1 directly interacts with the sarcolemmal LTCC through a binding domain on JPH1 spanning from amino acids 232–369 within the joining region domain of the protein. In addition, JPH1 directly interacts with sarcolemmal CAV3-encoded caveolin type 3 (Cav3)[19]. JPH1 also directly interacts with the skeletal muscle specific, RYR1-encoded ryanodine receptor type 1 (RyR1) which mediates release of Ca2+ stored within the SR. This interaction is mediated through highly reactive thiol groups which are sensitive to oxidative insult, and changes in this interaction can alter gating of RyR1. Overall, these interactions suggest a mechanism through which release of SR-stored Ca2+ via RyR1 is mediated by a direct interaction with JPH1 in an oxidation-dependent fashion[20]. A schematic depicting JPH1 within the skeletal muscle triad is depicted in Figure 3.
Figure 3. JPH1 and the skeletal muscle triad.
The N-terminus of JPH1 localizes to the plasma membrane of the skeletal myocyte T-tubule while simultaneously approximating the sarcolemmal via a C-terminal transmembrane domain which allows it to approximate the triad subcellular membrane architecture. The N-terminal aspect of JPH1 (light blue) binds plasmalemmal-limited Cav3 (red) and the LTCC (dark blue) while the C-terminal aspect directly binds RyR1 (green) within the SR. The JPH1-RyR1 interaction is mediated by reactive thiols interactions (depicted by S) which regulate RyR1 channel gating. Together, these proteins, along with other neighboring proteins, comprise the JMC. Inset, a schematic depicting the ultrastructure of the triad within skeletal myocytes. Abbreviations: Cav3, caveolin-3; JMC, junctional membrane complex; JPH1, junctophilin-1; LTCC, L-type calcium channel; RyR1, ryanodine receptor 1; SR, sarcoplasmic reticulum.
In vitro, colocalization of LTCC and RyR1 has been demonstrated by immunofluorescence into discrete areas along C2C12 murine skeletal myoblasts. This localization is disrupted following expression silencing of JPH1 and is associated with impaired LTCC Ca2+ influx[19]. Further, JPH1 expression silencing in vitro decreases store-operated Ca2+ entry (SOCE) which balances net Ca2+ loss from intracellular Ca2+ stores[18]. This results in reduced SR-stored Ca2+ release via RyR1[18]. Follow-up studies suggest that impaired SOCE may occur through disruption of stromal interaction molecule 1 (STIM1) and calcium release-activated calcium modulator 1 (ORAI1)[21]. In this model, depletion of SR-store Ca2+ allows for STIM1 oligimerization within the SR and interacts with ORAI1, which allows for SOCE through the sarcolemma. This interaction is disrupted with loss of JPH1 expression impairing the ability of the myocyte to increase sarcolemmal Ca2+ influx to balance SR Ca2+ depletion. RyR1 function is also regulated by the transient receptor potential channel canonical type 3 (TRPC3)[22]. Knockout of TRPC3 in primary skeletal myotubes demonstrate impaired RyR1 EC coupling gain and SR Ca2+ release with intact SR store Ca2+ levels. Taken together, these results support the role of JPH1 in regulating the skeletal muscle triad ultrastructure and the JMC. This role is critical to maintain efficient EC coupling gain, SR store Ca2+ release, and SOCE.
While the precise mechanism of SOCE remains controversial, a body of evidence suggests that SOCE plays a minimal role, if any, during physiologic muscle contraction. As the majority of cytosolic Ca2+ is removed from the cytosol by SERCA into the SR, a relatively small amount of Ca2+ is extruded from the myocyte[23]. Conversely, during non-physiologic experimental conditions, or under states of muscle fatigue, SOCE plays a critical role in maintaining store Ca2+ homeostasis[24, 25]. Further, since there is minimal net extravasation of cytosolic Ca2+ from intact skeletal myocytes during relaxation, SOCE is seen more prominently with in vitro myoblasts, such as C2C12 cells [26]. Thus, additional studies are needed to further elucidate the role of JPH1 in regulating SOCE utilizing in vivo models of intact, adult skeletal myocytes.
Skeletal Muscle Myopathy and Damage
Mice lacking JPH1 die shortly after birth due to defective suckling. This early mortality cannot be rescued by placement of an orogastric tube for feeding, suggesting that milk aspiration or respiratory failure, likely due to pharyngeal muscle and diaphragmatic dysfunction, contributes to pup death[27]. Skeletal myocytes isolated from these mice demonstrate fewer apparent JMCs, reduced contractile force, and reduced Ca2+-mediated twitch tension[27]. In addition, this perinatal mortality is associated with failure of normal triad development as well as disruption and vacuolization of normal SR structure[28]. These studies suggest that JPH1 plays a key role in skeletal muscle EC coupling and that disruption or impairment of JPH1 function is associated with poor skeletal muscle function through a loss of cellular ultrastructure and triad development.
JPH1 expression has been shown to decrease after skeletal muscle damage from eccentric contraction, a type of contraction in which the muscle elongates under tension due to an opposing force which is greater than the traditional contractile force generated by the muscle[29]. This loss of expression coincides with loss of EC coupling, and gradual return of JPH1 protein levels correlates with recovery of EC coupling and force generation. Further investigation found that increased skeletal muscle contraction, as well as directly increasing cytosolic Ca2+ levels to supra-physiologic levels, result in calpain-mediated proteolysis of JPH1[30]. Specifically, JPH1 is cleaved proximal to the C-terminus creating a 75 kDa and a 17 kDa fragment with a loss of expression of full-length protein. Interestingly, aberrant JPH1 proteolysis was identified in a murine mouse model of muscular dystrophy providing an association with the development of primary muscle disease[30]. These findings suggest that eccentric contraction, prolonged muscle contraction, and other mechanical stress may develop impaired contractile force and EC coupling secondary to loss of JPH1 function.
JUNCTOPHILIN TYPE 2 AND CARDIAC MUSCLE
The JPH2 gene contains five coding exons localizing to 20q13.12 and encodes a 696 amino acid 74kDa protein JPH2 in humans[15]. JPH2 is the major JPH family member expressed in cardiac muscle and shares significant primary sequence identity and functional parallels with JPH1 in skeletal muscle. In contrast to skeletal muscle, which demonstrates triadic ultrastructure, cardiac muscle characteristically has dyadic ultrastructure whereby a span of junctional SR runs parallel to one sarcolemmal t-tubular membrane at a given location[31, 32]. JPH2 plays a critical role in maintaining this dyadic structure and associated effective Ca2+ signaling. For a schematic depicting JPH2 within the cardiac dyad see Figure 4.
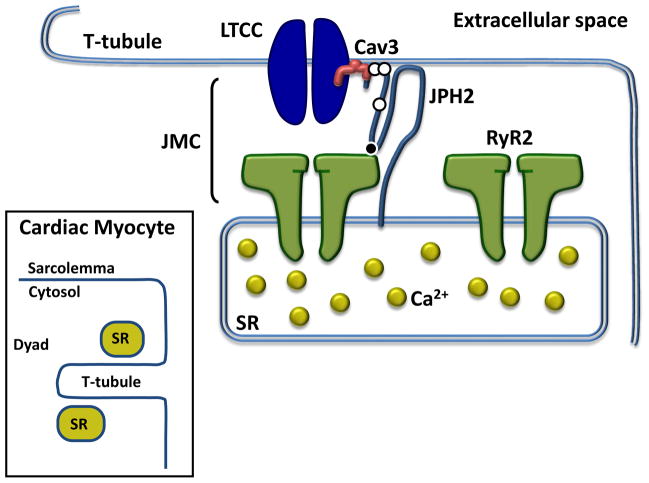
Figure 4. JPH2 and the cardiac muscle dyad.
The N-terminus of JPH2 (light blue) localizes to the plasma membrane of the cardiomyocyte, binds plasmalemmal-limited Cav3 (red) and directly interacts with RyR2 (green) via the MORN motif joining region and possibly the C-terminal aspect of the protein. This interaction regulates RyR2 gating and is responsible for pathologic Ca2+ (yellow) release in some JPH2 mutations. Together, these proteins along with other neighboring proteins, comprise the JMC. The white circles on JPH2 indicate the position of the three mutations associated with hypertrophic cardiomyopathy while the black circle denotes the location of the mutation associated with arrhythmia. Inset, a schematic depicting the ultrastructure of the dyad within cardiac myocytes. Abbreviations: Cav3, caveolin-3; JMC, junctional membrane complex; JPH2, junctophilin-2; LTCC, L-type calcium channel; RyR2, ryanodine receptor 2; SR, sarcoplasmic reticulum.
JPH2 null mice, while embryonic lethal, demonstrate a 90% increase in the distance of the cardiac dyad as well as vacuolization of the SR[1]. This disruption in cellular ultrastructure was associated with random cardiomyocyte contraction and irregular Ca2+ transients which occurred spontaneously without Ca2+ influx through the LTCC. Further, Ca2+ transients in JPH2 null mice are significantly reduced in amplitude perhaps due to the disruption of the SR ultrastructure[1]. From these early studies, JPH2 was originally viewed as a structural protein serving as a molecular tether which maintained the cardiac dyad ultrastructure required for effective CICR and Ca2+ signaling. Recent studies have added complexity to the function of this protein.
Development of the Junctional Membrane Complex and the T-Tubule
JPH2 plays a key role in cardiomyocyte development and differentiation. While it is possible that the amino-terminal MORN motifs of JPH2 might directly interact with the inner leaflet of the sarcolemma, JPH2 has also been shown to bind Cav3, a protein which localizes to the sarcolemma[33]. Cav3 is necessary for the formation of “little cave”-like invaginations of the cardiomyocyte sarcolemma called caveolae as well as regulation of several cardiac ion channels including the LTCC, and various K+ channels[34, 35]. Immunofluorescent localization of the LTCC, RyR2, Cav3, and JPH2 in rat myocytes demonstrates a temporal correlation between development of influx of Ca2+ from the LTCCa, CICR, and T-tubule formation[36]. While expression of Cav3 and RyR2 was observed early in development, JPH2 expression was not seen until day 15 and was associated with formation of the T-tubule and maturation of CICR. Further, embryonic stem cell-derived cardiomyocytes with reduced JPH2 expression demonstrate perturbed cardiac development and decreased Ca2+ transients that are sporadic and irregular. These cells also demonstrated disordered myofilament production, perturbed energy-generating capacity, and Ca2+ dysregulation, which culminated in fewer spontaneously contractile myocytes[37].
These studies in developing cardiomyocytes correlate well with studies in differentiated cardiac cells. JPH2 expression silencing in cardiac cells in vitro has been shown to impair CICR with decreased Ca2+ transient amplitude and blunting of spontaneous generation of Ca2+ transients[38]. These findings were seen with unchanged expression of the LTCC, RyR2, NCX, SERCA, and phospholamban and suggest altered function of RyR2 rather than altered protein expression. Induced JPH2 knock-down in adult mice demonstrated RyR2 hyperactivity with increased spark frequency despite impaired CICR and EC coupling gain[7]. Co-immunoprecipitation showed RyR2 directly binds JPH2 suggesting a role in regulation of RyR2 gating. Further, single molecule super-resolution microscopy in rat ventricular myocytes identified RyR2 clusters in close proximity to similarly shaped clusters of JPH2 with approximately ~80% colocalization[39]. These studies suggest that direct binding of JPH2 to RyR2 is necessary for proper RyR2 gating and loss of JPH2 expression results in reduced EC coupling gain during CICR and increased RyR2 Ca2+ leakage during diastole.
In addition to Ca2+ signaling, JPH2 plays a key role in the development and stability of myocyte ultrastructure. Mice with decreased levels of cardiac JPH2 showed defective postnatal T-tubule maturation, while mice over-expressing JPH2 had accelerated T-tubule maturation by postnatal day 8[40]. Further, independent studies utilizing acute RNAi-mediated knockdown of JPH2 in cultured cardiomyocytes have shown T-tubule disorganization[41]. Taken together, these studies suggest that JPH2 plays both a structural as well as protein-regulatory role within the cardiomyocyte. Reflecting this critical role, perturbations in JPH2 expression or mutations result in cardiac pathology.
Cardiomyopathy and Arrhythmogenesis
Primary myocardial diseases, such as cardiomyopathies, are a common cause of morbidity and mortality worldwide[42]. JPH2 has been linked to the development of various types of myocardial disease in both rodent models as well as humans. Murine genetic models of dilated cardiomyopathy (DCM) and hypertrophic cardiomyopathy (HCM), demonstrate reduced protein expression[33]. In addition, a rat model of aortic-banding pressure-induced hypertrophy similarly demonstrated reduced JPH2 expression and was associated with disruption of the cardiac dyad and junctional SR surface[43]. In silico modeling of the effect of this disrupted junctional interaction between LTCC and RyR on CICR predicts slow Ca2+ transient generation and reduced transient amplitude[44]. These studies suggest that loss of JPH2 expression, and the resulting disruption in Ca2+ transient generation, are associated with early molecular remodeling that may precede pathologic cellular remodeling.
Based on these findings, a series of studies were conducted to determine whether loss of JPH2 expression could drive cardiomyopathic disease. Knock-down of JPH2 in immortalized cardiac cells induced cellular hypertrophy, up-regulation of several hypertrophy markers including skeletal actin, atrial natriuretic factor, brain natriuretic factor, myosin heavy chain, regulator of calcineurin 1-exon 4 splice isoform, and was associated with reduced transient amplitude[38]. In vivo, conditional cardiac knockdown of JPH2 in mice rapidly leads to heart failure and early mortality[7]. These mice demonstrated increased cardiac mass with dilated ventricles and reduced fractional shortening as well as reduced CICR due to reduced EC coupling gain[7]. These murine model findings were supported by complementary human clinical studies where myocardium taken from surgical resection of HCM patients with left ventricular outflow tract obstruction demonstrated reduced, and in some cases undetectable, levels of JPH2[38]. In addition, cardiomyocytes isolated from patients with DCM and ischemic cardiomyopathy were observed to have reduced and mislocalized T-tubule-SR junctions with reduced JPH2 expression[44].
Based on the observation that altered JPH2 expression is associated with cardiomyopathic disease and heart failure, several studies have sought to identify whether mutations in the gene encoding JPH2 might be similarly associated with disease. The sentinel study in this regard identified three mutations localizing to the amino terminus first MORN motif domain and the linker domain of JPH2[45]. These mutations include JPH2–S101R, Y141H, and S165F. Each mutation was identified in a Caucasian individual who demonstrated clear clinical disease yet did not have a mutation in a gene traditionally associated with the development of HCM[45–47]. As with reduced JPH2 expression, these mutations were found to reduce CICR amplitude and disrupt cellular ultrastructure resulting in cellular hypertrophy when over-expressed[45]. Two additional genetic variants localizing to the divergent domain, JPH2-R436C and G505S, were reported in a small cohort of Japanese patients with HCM[48]. Further genetic analyses have identified each of these mutations in control cohorts of ostensibly healthy individuals without HCM, thus it is likely these variants are polymorphisms without clinical significance[49]. Recently, a JPH2 mutation has been identified in a small family with HCM and prominent clinical arrhythmia[50]. The JPH2-E169K mutation was associated with development of supraventricular arrhythmias in a small multi-generational family. A murine model of this mutation exhibited a higher incidence of pacing-induced atrial fibrillation secondary to abnormal spontaneous Ca2+ waves and increased spark frequency resulting from decreased RyR2-JPH2 interaction[50]. These genetic studies suggest that JPH2 may function beyond JMC structural integrity and may also have a regulatory role on RyR2 and perhaps other JMC-associated proteins. Finally, while the frequency of JPH2 mutations among patients with HCM is rare (<1%), loss of JPH2 expression may demonstrate a common feature of hypertrophic and arrhythmic remodeling[49].
Skeletal muscle myopathy linked to JPH2 mutations
While JPH2 mutations have been associated with cardiomyopathy and arrhythmias, a small number of studies have linked these mutations to perturbed skeletal muscle function in vitro. Over-expression of the JPH2-S165F mutation in primary murine skeletal myotubes resulted in hypertrophy, reduced CICR, and reduced SR Ca2+ release via RyR1[51]. Immunoprecipitation assays suggest that the S165F mutation impairs protein kinase C-mediated phosphorylation resulting in impaired binding to TRPC3[51]. TRPC3 had been previously found to modulate the function of RyR1 through a direct interaction with JPH2 in skeletal muscle and binds JPH2 near the joining region[52, 53]. Thus, the S165F mutation is believed to lead to skeletal myocyte hypertrophy through impaired regulation of RyR1 function. JPH2-Y141H has also been linked to skeletal muscle hypertrophy and altered Ca2+ signaling[54]. Over-expression of JPH2-Y141H in primary skeletal myotubes demonstrated increase in cellular diameter and reduced EC coupling; however, unlike S165F, impairment of RyR1-mediated SR Ca2+ release was not observed. Rather, increased SOCE via Orai1 and increased resting Ca2+ levels was observed. These findings suggest that the JPH2-Y141H mutation is associated with skeletal muscle hypertrophy through a mechanism independent of RyR1 impairment. Interestingly, the initial probands hosting the JPH2-S165F and JPH2-Y141H mutations demonstrated no clinical evidence of skeletal muscle myopathy such as muscular dystrophy. Further, these studied have not been replicated using in vivo models and the clinical relevance of these mutations on skeletal muscle remains unclear at present.
JUNCTOPHILIN TYPE 3 AND 4 AND NEURONAL TISSUE
JPH3 (also referred to as JP3 elsewhere in the literature) and JPH4 are primarily expressed within the neurons of the brain. While the precise role of these proteins remains enigmatic, an emerging body of evidence suggests that neuronal JPHs may play roles in mediating balance and motor control through maintenance of efficient Ca2+ signaling. JPH3 knock-out mice demonstrate reduced balance and impaired motor coordination at 3 months of age without overt alterations in brain morphology or significant defects in molecular signaling[55]. While JPH1 and JPH2 knock-out mice demonstrated disrupted ultrastructure of the skeletal myocyte triad and cardiomyocyte dyad, respectively, JPH3 knockout mice had no discernible disruption in neuronal cellular architecture. In addition, these mice demonstrated no apparent difference in purkinje cell action potentials and normal synaptic function[55]. A follow-up study utilizing JPH3 knock-out and hemizygous knock-out mice aged to 6 and 9 months of age, identified progressive defects in neuromuscular strength, coordination, and balance which was greater in the knock-out mouse and progressive over time[56]. JPH4 localizes to spatially discrete areas in the brain compared to JPH3; however, the knockout mouse has no discernible phenotype[57, 58]. Detailed intracellular Ca2+ signaling studies in these mice have not been done to date, nor has a comprehensive analysis of all major functional regions of the brain been done to determine whether a more robust phenotype may be identifiable. Despite these shortcomings, these results suggest that JPH3 and 4 are not required for cellular architecture stabilization and brain development individually which raises the possibility that the function of these isoforms may be redundant.
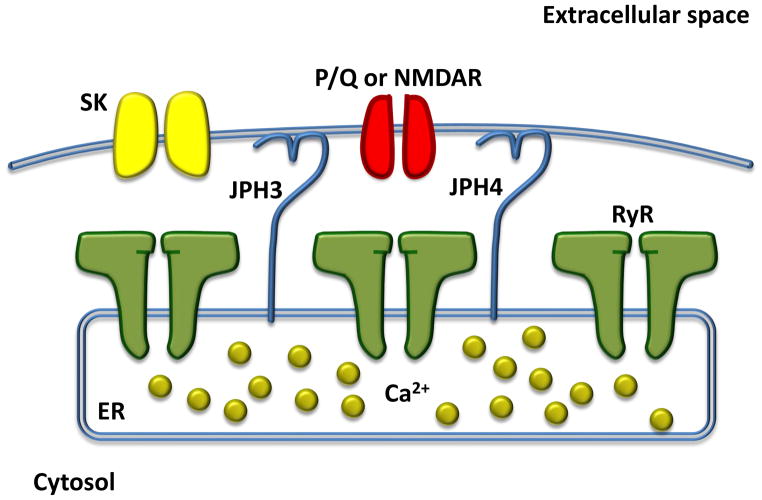
Supporting this hypothesis, JPH3/4 double knock-out mice demonstrate disrupted intracellular Ca2+ signaling[58]. Indeed, disrupted communication between plasmalemmal Ca2+ entry via the N-methyl D-aspartate (NMDA) glutamate receptors and intracellular neuronal RyRs as well as the small-conductance Ca2+-activated potassium (SK) channels was noted in hippocampal neurons[58]. Under normal physiologic conditions, these plasmalemmal and intracellular channels are necessary for intracellular Ca2+ signaling and proper neuronal function[58]. In addition to this molecular defect in hippocampal neurons, JPH3/4 double knock-out mice also demonstrate a similar impairment in Ca2+ signaling of purkinje cells[59]. Specifically, purkinje cells obtained from double knock-out mice have impaired Ca2+ crosstalk between plasmalemmal P/Q-type voltage gated channels and intracellular RyR channels impairing SK channel-mediated after-hyperpolarization[59]. These mice demonstrate reduced memory and exploratory activity, as well as irregular hind limb reflexes ultimately culminating in death of young mice within five weeks of birth[58]. While the full mechanistic implications of this defective Ca2+ signaling has yet to be clarified, these early studies raise the possibility that defective store Ca2+ release by RyR isoforms may be associated with neuronal JPH loss. While little is known about the role of RyR within neuronal tissue, RyR3 has canonically been viewed as the major isoform of the brain while RyR1 and RyR2 are also expressed[60, 61]. While the specific RyR isoform responsible for ER Ca2+ release has not been clarified for the numerous neuronal cell subtypes in the brain, it is hypothesized that RyR isoforms may by neuronal subtype-specific[62, 63]. In addition, recent evidence has suggested that neuronal LTCCs may couple with RyR1 in a manner analogous to voltage triggered-RyR1 opening in skeletal muscle. Indeed, heterozygous knock-in mice harboring a known RyR1-I4895T mutation were noted to have significantly impaired voltage-induced Ca2+ release of store Ca2+ in addition to skeletal muscle myopathy[64]. These recent studies raise the possibility that molecular uncoupling of voltage-sensitive Ca2+ channels from neuronal RyRs in JPH3/4 deficient mice may mirror to the uncoupling of SOCE from RyR1 in skeletal muscle with loss of JPH1. JPH3 and 4 within neurons are depicted in Figure 5.
Figure 5. JPH3/4 and neuronal ultrastructure.
In purkinje cells, the two JPH3 and 4 isoforms (light blue) may be responsible for maintain appropriate spacing between neuronal plasma membrane and the ER. This is critical for effective Ca2+ signaling from the P/Q-type channel (red), RyR (green), and SK channels (yellow). In other neuronal subtypes, such as hippocampal neurons, the NMDA receptor has been shown to mediate influx of Ca2+. There is no clear evidence for direct binding of JPH3 or 4 with other members of the neuronal JMC to date. While it is hypothesized that particular RyR isoforms, including RyR1, 2, and 3, may have important roles in ER Ca2+ release in neuronal type-specific manner, this has yet to be clearly demonstrated Abbreviations: ER, endoplasmic reticulum; JPH3, junctophilin-3; JPH4, junctophilin-4; NMDAR, N-methyl D-aspartate glutamate receptor; P/Q, P/Q-type voltage gated channel; RyR, ryanodine receptor; SK, small-conductance Ca2+-activated potassium channel.
Huntington Disease-Like Syndrome
JPH3 was the first member of the junctophilin family to be implicated in the pathogenesis of human disease. An insidiously progressive neurodegenerative disease, Huntington’s disease (HD) is associated with development of involuntary and uncoordinated muscle movements (chorea) and subcortical dementia[65]. The etiology of this disease is traditionally believed to be a CAG trinucleotide repeat expansion within the gene HTT which encodes the huntingtin protein[66]. This trinucleotide repeat results in the insertion of a polyglutamine tract within the huntingtin protein. While this mutation represents the classic cause of HD, a subset of patients are negative for mutations in this gene and yet are clinically indistinguishable from patients with mutations in HTT. These mutation-negative individuals are considered Huntington’s disease-like syndrome phenocopies. Some patients with HD-like syndrome were found to host octapeptide repeat expansion in the PRNP-encoded prion protein and have been designated HDL1[67]. In 2001, a trinucleotide CAG/CTG repeat expansion was identified in a family with HD-like syndrome that was negative for mutations in HTT that localized to an alternatively-spliced exon of JPH3[68, 69]. This has led to the labeling of JPH3 as the second HD-like associated gene (HDL2). While large studies estimating the frequency of JPH3 mutations in HD or HD-like syndromes have not been done, a study examining nine independent series of patients with HD-like syndrome estimated that 0–15% of patients who are negative for mutations in HTT will host mutations in JPH3[70]. A recent study of a cohort of Venezuelan subjects with HD-like syndrome found that approximately 25% (4/16) hosted JPH3 expansion mutations[71]. These numbers represent early estimates derived from small studies; however, it is generally accepted that JPH3 mutations are a relatively rare cause of HD-like syndrome.
While mutations in JPH3 underlie a relatively small proportion of individuals with HD-like syndrome, multiple independent cohorts have identified this disease association particularly in individuals of African ancestry[70, 72–74]. Individuals with HDL2 demonstrate a triad of symptoms marked by abnormal movements (most often chorea), dementia, and psychiatric disturbances, and the number of trinucleotide repeats is inversely proportional to the age of symptom onset[70]. Recent familial studies have added additional complexity to the clinical phenotype associated with JPH3 mutations. Several large, multi-generational families hosting JPH3 trinucleotide repeat expansions demonstrated severe akinetic-rigid syndrome with a relatively minor degree of chorea[71, 75]. Despite the possible emergence of distinct clinical presentations of individuals with HDL2, there has been no reproducible genotype-phenotype correlation which might explain why a given family demonstrates hyperkinetic chorea symptoms while another develops akinetic rigid syndrome symptoms.
While the precise mechanism of disease pathogenesis in HDL2 remains unknown, some progress has been in made in clarifying the pathogenic mechanism of the JPH3 trinucleotide repeat expansion. Fluorescent in situ hybridization (FISH) analysis of neuronal specimens obtained from HDL2 patients at autopsy demonstrated RNA foci within the neurons which were absent in traditional HD patients[76]. FISH for CAG/CTG repeat expansion of the mutated JPH3 colocalize to these areas of RNA foci. Further, over-expression of the alternatively spliced exon 2A containing the repeat expansion resulted in cellular toxicity in vitro[76]. These studies suggest that the trinucleotide repeat expansion of JPH3 may result in aberrant RNA transcription and formation of toxic RNA foci. This possibility is supported by murine models over-expressing the trinucleotide repeat that demonstrate accelerated development of motor deficits as well as neurodegeneration[77]. Recent in vitro studies have raised the possibility that additional anti-sense transcription in an open reading frame within the JPH3 gene locus may also play a role in neuronal toxicity[56]. While this possible anti-sense transcript has yet to be identified in vivo, RNA toxicity remains an interesting possible mechanism of neuronal dysfunction. Taken together, these studies demonstrate that neuronal toxicity and death may be occurring through multiple mechanisms including loss of JPH3 expression or generation of toxic RNA.
CONCLUDING REMARKS
The JPH family of proteins is critical for proper Ca2+ signaling within excitable cells, and pertubation results in cellular dysfunction that can manifest in clinical disease. While initially thought to be structural proteins holding together ultrastructural cellular architecture, it has become clear that striated muscle JPHs also modulate the function of Ca2+-handling proteins and are needed for physiologic Ca2+ signaling. Perturbations in JPH expression, or function-impairing mutations, can alter intracellular Ca2+ signaling resulting in myopathic disease secondary to pathologic cellular remodeling as well as arrhythmogenesis. An emerging body of literature raises the possibility of a similar role in neuronal tissue such as the brain, and mutations in neuronal-specific JPHs have been linked to neurodegenerative disease. While the number of patients with clearly defined mutations in JPH genes remains small, understanding this class of proteins may provide significant understanding to the way in which excitable cells function and how to intervene when they fail. A list of outstanding questions and possible future directions for investigation is located in Box 2.
Box 2. Outstanding Questions.
Do JPH isoforms undergo post-translational modifications such as phosphorylation, and if so, what is the effect of these modifications on Ca2+ signaling within the cell?
What is the role of JPH1 mutations, if any, in the pathogenesis of skeletal muscle myopathies?
Are there additional binding partners of striated muscle-specific JPH1 and JPH2 which may play roles in the development of myopathic disease?
Are there additional binding partners of cardiac JPH2 which may play roles in arrhythmic disease?
How do mutations in various functional domains of JPH1 and JPH2 create differing disease phenotypes?
What are physiologic and pathophysiologic triggers for proteolytic degradation of JPH1 and 2, and can this form of post-translational modification be extended to JPH3 and 4?
Can over-expression of JPH2 in cardiac cells, or JPH3/4 in neuronal cells, be protective against cardiomyopathic and neurodegenerative processes, respectively?
What is the mechanism of disease pathogenesis triggered by the JPH3 trinucleotide expansion in patients with Huntington’s Disease-Like syndrome?
What is the role of mutations in JPH4, if any, in the pathogenesis of neurodegenerative disease?
Are there other brain or neuronal diseases that can be linked to perturbed JPH expression or genetic mutations?
Do JPH3 and/or 4 play a role in learning and memory?
HIGHLIGHTS.
Junctophilins are a conserved family of proteins found in all excitable cells
Junctophilins maintain subcellular architecture and calcium signaling
Mutations have been linked to cardiomyopathy, arrhythmia and neurodegeneration
Acknowledgments
D.B is supported by AHA Predoctoral Fellowships and by NIH grant T32HL007676-21A1. X.H.T.W. is an Established Investigator of the American Heart Association (AHA; 13EIA14560061), a W.M. Keck Foundation distinguished young scholar, and is supported by the Muscular Dystrophy Association, and NIH grants HL089598, HL091947 and HL117641, and the Juanita P. Quigley Endowed Chair in Cardiology. This work was also supported in part by Fondation Leducq (‘Alliance for CaMKII Signaling in Heart’) to X.H.T.W.
GLOSSARY
- Akinetic-rigid syndromes
A constellation of symptoms characterized by a paucity of voluntary movements which, when present, are slow, and associated with increased muscle tone/rigidity
- Alternative splicing
A molecular process during which the messenger RNA transcribed from a single gene can be processed differently to create various mature mRNA sequences which are then ultimately translated into different proteins
- Calcium-induced calcium-release (CICR)
Release of a relatively large amount of stored intracellular calcium is triggered by a relatively small amount of calcium, such as from extracellular calcium moving intracellularly
- Chorea
A neurologic disorder characterized by abnormal involuntary movements which are generally abrupt, irregular, and non-stereotyped in nature. It is derived from the Greek word meaning “dance”
- Diastole
Relaxation of the heart or cardiac myocyte
- Dilated cardiomyopathy (DCM)
Enlargement and dilation of the heart associated with impaired function and weak contraction
- Excitation-contraction coupling (EC coupling)
Mechanical cellular contraction, such as in a striated muscle cell, is linked to cellular excitations, such as change in the sarcolemmal membrane potential
- Hypertrophic cardiomyopathy (HCM)
Asymmetric hypertrophy of predominantly the left ventricle of the heart in the absence of a clinically identifiable cause such as co-morbid hypertension
- Huntington’s disease-like syndrome (HD-like)
A clinical entity mimicking the clinical presentation of Huntington’s disease, including progressive involuntary and uncoordinated muscle movements (chorea) and subcortical dementia in a patient without an identifiable trinucleotide expansion mutation in the HTT-encoding huntingtin protein
- Junctional membrane complex (JMC)
A complex formed by a plasma membrane and a juxtaposed intracellular membrane such as the endoplasmic reticulum or sarcoplasmic reticulum which hosts a number of proteins required for intracellular ion signaling
- Purkinje cells
A type of neuron located in the cerebellum of the brain
- Store-operated calcium entry (SOCE)
A cellular process to replace a depleted store of intracellular calcium, such as the sarcoplasmic reticulum, which results in net influx of calcium across the plasma membrane to replenish the store
- Transient
A rapid increase in cytosolic calcium resulting from calcium-induced calcium-release
- Transverse tubule (T-tubule)
A finger-like invagination of the sarcolemma of striated muscle cells which allows penetration of a membrane potential into the myocyte
- Supraventricular tachycardia
An arrhythmia of the heart with rapid heart rate with an arrhythmic origin above the ventricles
- Systole
Contraction of the heart or cardiac myocyte
Footnotes
CONFLICTS OF INTEREST
X.H.T.W. is a founding partner of Elex Biotech, a company that develops new drugs targeting intracellular calcium leak.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Takeshima H, et al. Junctophilins: a novel family of junctional membrane complex proteins. Mol Cell. 2000;6:11–22. doi: 10.1016/s1097-2765(00)00003-4. [DOI] [PubMed] [Google Scholar]
- 2.Weisleder N, et al. Immuno-proteomic approach to excitation-contraction coupling in skeletal and cardiac muscle: Molecular insights revealed by the mitsugumins. Cell Calcium. 2008;43:1–8. doi: 10.1016/j.ceca.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishi M, et al. Characterization of human junctophilin subtype genes. Biochem Biophys Res Commun. 2000;273:920–927. doi: 10.1006/bbrc.2000.3011. [DOI] [PubMed] [Google Scholar]
- 4.Garbino A, et al. Molecular evolution of the junctophilin gene family. Physiol Genomics. 2009;37:175–186. doi: 10.1152/physiolgenomics.00017.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma H, et al. MORN motifs in plant PIPKs are involved in the regulation of subcellular localization and phospholipid binding. Cell Res. 2006;16:466–478. doi: 10.1038/sj.cr.7310058. [DOI] [PubMed] [Google Scholar]
- 6.Im YJ, et al. The N-terminal membrane occupation and recognition nexus domain of Arabidopsis phosphatidylinositol phosphate kinase 1 regulates enzyme activity. J Biol Chem. 2007;282:5443–5452. doi: 10.1074/jbc.M611342200. [DOI] [PubMed] [Google Scholar]
- 7.van Oort R, et al. Disrupted junctional membrane complexes and hyperactive ryanodine receptors after acute junctophilin knockdown in mice. Circulation. 2011;123:979–988. doi: 10.1161/CIRCULATIONAHA.110.006437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bers D. Excitation-Contraction Coupling and Cardiac Contractile Force. Boston: Kluwer Academic Publisher; 2001. [Google Scholar]
- 9.Bers DM. Cardiac excitation-contraction coupling. Nature. 2002;415:198–205. doi: 10.1038/415198a. [DOI] [PubMed] [Google Scholar]
- 10.Catterall WA. Excitation-contraction coupling in vertebrate skeletal muscle: A tale of two calcium channels. Cell. 1991;64:871–874. doi: 10.1016/0092-8674(91)90309-m. [DOI] [PubMed] [Google Scholar]
- 11.Endo M. Calcium-induced calcium release in skeletal muscle. Physiol Rev. 2009;89:1153–1176. doi: 10.1152/physrev.00040.2008. [DOI] [PubMed] [Google Scholar]
- 12.Hodgkin AL, Horowicz P. Potassium contractures in single muscle fibres. J Physiol. 1960;153:386–403. doi: 10.1113/jphysiol.1960.sp006541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider M, Chandler W. Voltage dependent charge movement in skeletal muscle: a possible step in excitation-contraction coupling. Nature. 1973;242:244–246. doi: 10.1038/242244a0. [DOI] [PubMed] [Google Scholar]
- 14.Curtis BM, Catterall WA. Purification of the calcium antagonist receptor of the voltage-sensitive calcium channel from skeletal muscle transverse tubules. Biochemistry. 1984;23:2113–2118. doi: 10.1021/bi00305a001. [DOI] [PubMed] [Google Scholar]
- 15.Hoffmann R, Valencia A. Implementing the iHOP concept for navigation of biomedical literature. Bioinformatics. 2005;21:ii252–ii258. doi: 10.1093/bioinformatics/bti1142. [DOI] [PubMed] [Google Scholar]
- 16.Nishi M, et al. Characterization of human junctophilin subtype genes. Biochem Biophys Res Commun. 2000;273:920–927. doi: 10.1006/bbrc.2000.3011. [DOI] [PubMed] [Google Scholar]
- 17.Block B, et al. Structural evidence for direct interaction between the molecular components of the transverse tubule/sarcoplasmic reticulum junction in skeletal muscle. J Cell Biol. 1988;107:2587–2600. doi: 10.1083/jcb.107.6.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirata Y, et al. Uncoupling store-operated Ca2+ entry and altered Ca2+ release from sarcoplasmic reticulum through silencing of junctophilin genes. Biophys J. 2006;90:4418–4427. doi: 10.1529/biophysj.105.076570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Golini L, et al. Junctophilin 1 and 2 proteins interact with the L-type Ca2+ channel dihydropyridine receptors (DHPRs) in skeletal muscle. J Biol Chem. 2011;286:43717–43725. doi: 10.1074/jbc.M111.292755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phimister AJ, et al. Conformation-dependent stability of junctophilin 1 (JP1) and ryanodine receptor type 1 (RyR1) channel complex is mediated by their hyper-reactive thiols. J Biol Chem. 2007:8667–8677. doi: 10.1074/jbc.M609936200. [DOI] [PubMed] [Google Scholar]
- 21.Li H, et al. Impaired orai1-mediated resting Ca2+ entry reduces the cytosolic [Ca2+] and sarcoplasmic reticulum Ca2+ loading in quiescent junctophilin 1 knock-out myotubes. J Biol Chem. 2010;285:39171–39179. doi: 10.1074/jbc.M110.149690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee EH, et al. Functional coupling between TRPC3 and RyR1 regulates the expressions of key triadic proteins. J Biol Chem. 2006;281:10042–10048. doi: 10.1074/jbc.M600981200. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez-Serratos H, et al. Na-Ca exchange studies in sarcolemmal skeletal muscle. Ann NY Acad Sci. 1996;779:556–560. doi: 10.1111/j.1749-6632.1996.tb44837.x. [DOI] [PubMed] [Google Scholar]
- 24.Cully TR, Launikonis BS. Store-operated Ca2+ entry is not required for store refilling in skeletal muscle. Clin Exp Pharmacol Physiol. 2013;40:338–344. doi: 10.1111/1440-1681.12078. [DOI] [PubMed] [Google Scholar]
- 25.Launikonis B, et al. Toward the roles of store-operated Ca2+ entry in skeletal muscle. Pflugers Arch. 2010;460:813–823. doi: 10.1007/s00424-010-0856-7. [DOI] [PubMed] [Google Scholar]
- 26.Stiber JA, Rosenberg PB. The role of store-operated calcium influx in skeletal muscle signaling. Cell Calcium. 2011;49:341–349. doi: 10.1016/j.ceca.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito K, et al. Deficiency of triad junction and contraction in mutant skeletal muscle lacking junctophilin type 1. J Cell Biol. 2001;154:1059–1068. doi: 10.1083/jcb.200105040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komazaki S, et al. Deficiency of triad formation in developing skeletal muscle cells lacking junctophilin type 1. FEBS Letters. 2002;524:225–229. doi: 10.1016/s0014-5793(02)03042-9. [DOI] [PubMed] [Google Scholar]
- 29.Corona BT, et al. Junctophilin damage contributes to early strength deficits and EC coupling failure after eccentric contractions. Am J Physiol Cell Physiol. 2010;298:C365–376. doi: 10.1152/ajpcell.00365.2009. [DOI] [PubMed] [Google Scholar]
- 30.Murphy RM, et al. Ca2+-dependent proteolysis of junctophilin-1 and junctophilin-2 in skeletal and cardiac muscle. J Physiol. 2013;591:719–729. doi: 10.1113/jphysiol.2012.243279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rayns DG, et al. Surface features of striated muscle: I. guinea-pig cardiac muscle. J Cell Sci. 1968;3:467–474. doi: 10.1242/jcs.3.4.467. [DOI] [PubMed] [Google Scholar]
- 32.Fawcett DW, McNutt NS. The ultrastructure of the cat myocardium: I. ventricular papillary muscle. J Cell Biol. 1969;42:1–45. doi: 10.1083/jcb.42.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minamisawa S, et al. Junctophilin type 2 is associated with caveolin-3 and is down-regulated in the hypertrophic and dilated cardiomyopathies. Biochem Biophys Res Commun. 2004;325:852–856. doi: 10.1016/j.bbrc.2004.10.107. [DOI] [PubMed] [Google Scholar]
- 34.Galbiati F, et al. Caveolin-3 null mice show a loss of caveolae, changes in the microdomain distribution of the dystrophin-glycoprotein complex, and t-tubule abnormalities. J Biol Chem. 2001;276:21425–21433. doi: 10.1074/jbc.M100828200. [DOI] [PubMed] [Google Scholar]
- 35.Balijepalli RC, Kamp TJ. Caveolae, ion channels and cardiac arrhythmias. Prog Biophys Mol Biol. 2008;98:149–160. doi: 10.1016/j.pbiomolbio.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ziman AP, et al. Excitation-contraction coupling changes during postnatal cardiac development. J Mol Cell Cardiol. 2010;48:379–386. doi: 10.1016/j.yjmcc.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang X, et al. Junctophilin 2 knockdown interfere with mitochondrium status in ESC-CMs and cardiogenesis of ES cells. J Cell Biochem. 2012;113:2884–2894. doi: 10.1002/jcb.24164. [DOI] [PubMed] [Google Scholar]
- 38.Landstrom A, et al. Junctophilin-2 expression silencing causes cardiocyte hypertrophy and abnormal intracellular calcium-handling. Circ Heart Fail. 2011;4:214–223. doi: 10.1161/CIRCHEARTFAILURE.110.958694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jayasinghe Isuru D, et al. Nanoscale organization of junctophilin-2 and ryanodine receptors within peripheral couplings of rat ventricular cardiomyocytes. Biophys J. 2012;102:L19–L21. doi: 10.1016/j.bpj.2012.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reynolds JO, et al. Junctophilin-2 is necessary for T-tubule maturation during mouse heart development. Cardiovasc Res. 2013;100:44–53. doi: 10.1093/cvr/cvt133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han J, et al. Morphogenesis of t-tubules in heart cells: the role of junctophilin-2. Sci China Life Sci. 2013;56:647–652. doi: 10.1007/s11427-013-4490-4. [DOI] [PubMed] [Google Scholar]
- 42.Go AS, et al. Heart disease and stroke statistics 2013 update: A report from the American Heart Association. Circulation. 2013;127:e6–e245. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu M, et al. Intermolecular failure of l-type Ca2+ channel and ryanodine receptor signaling in hypertrophy. PLoS Biology. 2007;5:e21. doi: 10.1371/journal.pbio.0050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu H-D, et al. Ultrastructural remodelling of Ca2+ signalling apparatus in failing heart cells. Cardiovasc Res. 2012;95:430–438. doi: 10.1093/cvr/cvs195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Landstrom AP, et al. Mutations in JPH2-encoded junctophilin-2 associated with hypertrophic cardiomyopathy in humans. J Mol Cell Cardiol. 2007;42:1026–1035. doi: 10.1016/j.yjmcc.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Landstrom AP, Ackerman MJ. GWAS or gee whiz, PSAS or pshaw: elucidating the biologic and clinical significance of genetic variation in cardiovascular disease. Heart Rhythm. 2009;6:1751–1753. doi: 10.1016/j.hrthm.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Landstrom AP, Ackerman MJ. The Achilles’ heel of cardiovascular genetic testing: distinguishing pathogenic mutations from background genetic noise. Clin Pharmacol Ther. 2011;90:496. doi: 10.1038/clpt.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsushita Y, et al. Mutation of junctophilin type 2 associated with hypertrophic cardiomyopathy. J Hum Genet. 2007;52:543–548. doi: 10.1007/s10038-007-0149-y. [DOI] [PubMed] [Google Scholar]
- 49.Landstrom AP, Ackerman MJ. Beyond the cardiac myofilament: Hypertrophic cardiomyopathy-associated mutations in genes that encode calcium-handling proteins. Curr Mol Med. 2012;12:507–518. doi: 10.2174/156652412800620020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beavers DL, et al. Mutation E169K in junctophilin-2 causes atrial fibrillation due to impaired RyR2 stabilization. J Am Coll Cardiol. 2013;62:2010–2019. doi: 10.1016/j.jacc.2013.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woo JS, et al. S165F mutation of junctophilin 2 affects Ca2+ signalling in skeletal muscle. Biochem J. 2010;427:125–134. doi: 10.1042/BJ20091225. [DOI] [PubMed] [Google Scholar]
- 52.Woo JS, et al. Glutamate at position 227 of junctophilin-2 is involved in binding to TRPC3. Mol Cell Biochem. 2009;328:25–32. doi: 10.1007/s11010-009-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woo JS, et al. TRPC3-interacting triadic proteins in skeletal muscle. Biochemical Journal. 2008;411:399–405. doi: 10.1042/bj20071504. [DOI] [PubMed] [Google Scholar]
- 54.Woo JS, et al. Hypertrophy in skeletal myotubes induced by junctophilin-2 mutant, Y141H, involves an increase in store-operated Ca2+ entry via orai1. J Biol Chem. 2012;287:14336–14348. doi: 10.1074/jbc.M111.304808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nishi M, et al. Motor discoordination in mutant mice lacking junctophilin type 3. Biochem Biophys Res Commun. 2002;292:318–324. doi: 10.1006/bbrc.2002.6649. [DOI] [PubMed] [Google Scholar]
- 56.Seixas AI, et al. Loss of junctophilin-3 contributes to huntington disease-like 2 pathogenesis. Ann Neurol. 2012;71:245–257. doi: 10.1002/ana.22598. [DOI] [PubMed] [Google Scholar]
- 57.Nishi M, et al. Coexpression of junctophilin type 3 and type 4 in brain. Mol Brain Res. 2003;118:102–110. doi: 10.1016/s0169-328x(03)00341-3. [DOI] [PubMed] [Google Scholar]
- 58.Moriguchi S, et al. Functional uncoupling between Ca2+ release and after hyperpolarization in mutant hippocampal neurons lacking junctophilins. Proc Natl Acad Sci USA. 2006;103:10811–10816. doi: 10.1073/pnas.0509863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kakizawa S, et al. Functional crosstalk between cell-surface and intracellular channels mediated by junctophilins essential for neuronal functions. Cerebellum. 2008;7:385–391. doi: 10.1007/s12311-008-0040-1. [DOI] [PubMed] [Google Scholar]
- 60.Mori F, et al. Developmental changes in expression of the three ryanodine receptor mRNAs in the mouse brain. Neurosci Lett. 2000;285:57–60. doi: 10.1016/s0304-3940(00)01046-6. [DOI] [PubMed] [Google Scholar]
- 61.Balschun D, et al. Deletion of the ryanodine receptor type 3 (RyR3) impairs forms of synaptic plasticity and spatial learning. EMBO J. 1999;18:5264–5273. doi: 10.1093/emboj/18.19.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shimuta M, et al. Postsynaptic modulation of AMPA receptor-mediated synaptic responses and LTP by the type 3 ryanodine receptor. Mol Cell Neurosci. 2001;17:921–930. doi: 10.1006/mcne.2001.0981. [DOI] [PubMed] [Google Scholar]
- 63.Kushnir A, et al. Ryanodine receptor studies using genetically engineered mice. FEBS Lett. 2010;584:1956–1965. doi: 10.1016/j.febslet.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.De Crescenzo V, et al. Type 1 ryanodine receptor knock-in mutation causing central core disease of skeletal muscle also displays a neuronal phenotype. Proc Natl Acad Sci U S A. 2012;109:610–615. doi: 10.1073/pnas.1115111108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walker FO. Huntington’s disease. Lancet. 2007;369:218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- 66.Gusella J, et al. A polymorphic DNA marker genetically linked to Huntington’s disease. Nature. 1983;306:234–238. doi: 10.1038/306234a0. [DOI] [PubMed] [Google Scholar]
- 67.Moore RC, et al. Huntington disease phenocopy is a familial prion disease. Am J Hum Gen. 2001;69:1385–1388. doi: 10.1086/324414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Margolis R, et al. A disorder similar to Huntington’s disease is associated with a novel CAG repeat expansion. Ann Neurol. 2001;50:373–380. doi: 10.1002/ana.1312. [DOI] [PubMed] [Google Scholar]
- 69.Holmes SE, et al. A repeat expansion in the gene encoding junctophilin-3 is associated with Huntington disease-like 2. Nat Genet. 2001;29:377–378. doi: 10.1038/ng760. [DOI] [PubMed] [Google Scholar]
- 70.Margolis R, et al. Huntington’s disease-like 2 (HDL2) in North America and Japan. Ann Neurol. 2004;56:670–674. doi: 10.1002/ana.20248. [DOI] [PubMed] [Google Scholar]
- 71.Paradisi I, et al. Huntington disease-like 2 (HDL2) in Venezuela: frequency and ethnic origin. J Hum Genet. 2013;58:3–6. doi: 10.1038/jhg.2012.111. [DOI] [PubMed] [Google Scholar]
- 72.Stevanin G, et al. CAG/CTG repeat expansions at the Huntington’s disease-like 2 locus are rare in Huntington’s disease patients. Neurology. 2002;58:965–967. doi: 10.1212/wnl.58.6.965. [DOI] [PubMed] [Google Scholar]
- 73.Santos C, et al. Huntington disease-like 2: the first patient with apparent European ancestry. Clin Genet. 2008;73:480–485. doi: 10.1111/j.1399-0004.2008.00981.x. [DOI] [PubMed] [Google Scholar]
- 74.Rodrigues G, et al. Huntington’s disease-like 2 in Brazil--report of 4 patients. Mov Disord. 2008;23:2244–2247. doi: 10.1002/mds.22223. [DOI] [PubMed] [Google Scholar]
- 75.Schneider S, et al. JPH3 repeat expansions cause a progressive akinetic-rigid syndrome with severe dementia and putaminal rim in a five-generation African-American family. Neurogenetics. 2012;13:133–140. doi: 10.1007/s10048-012-0318-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rudnicki DD, et al. Huntington’s disease–like 2 is associated with CUG repeat-containing RNA foci. Ann Neurol. 2007;61:272–282. doi: 10.1002/ana.21081. [DOI] [PubMed] [Google Scholar]
- 77.Wilburn B, et al. An antisense CAG repeat transcript at JPH3 locus mediates expanded polyglutamine protein toxicity in huntington’s disease-like 2 mice. Neuron. 2011;70:427–440. doi: 10.1016/j.neuron.2011.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matlin AJ, et al. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 79.Pan Q, et al. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 80.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dereeper A, et al. Phylogeny.fr: robust phylogenetic analysis for the nonspecialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chevenet F, et al. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]