Abstract
The origin of NO signaling can be traceable back to the origin of life with the large scale of parallel evolution of NO synthases (NOSs). Inducible-like NOSs may be the most basal prototype of all NOSs and that neuronal-like NOS might have evolved several times from this prototype. Other enzymatic and non-enzymatic pathways for NO synthesis have been discovered using reduction of nitrites, an alternative source of NO. Diverse synthetic mechanisms can co-exist within the same cell providing a complex NO-oxygen microenvironment tightly coupled with cellular energetics. The dissection of multiple sources of NO formation is crucial in analysis of complex biological processes such as neuronal integration and learning mechanisms when NO can act as a volume transmitter within memory-forming circuits. In particular, the molecular analysis of learning mechanisms (most notably in insects and gastropod molluscs) opens conceptually different perspectives to understand the logic of recruiting evolutionarily conserved pathways for novel functions. Giant uniquely identified cells from Aplysia and related species precent unuque opportunities for integrative analysis of NO signaling at the single cell level.
Keywords: Nitric Oxide Synthase, Evolution, Vertebrates, Invertebrates, Protozoa, Plants, Fungi, Nitrites, Cnidaria, Molluscs, Bacteria, Non-Enzymatic Synthesis, Neurons, Nervous Systems, Aplysia, Learning and Memory, Review
2. INTRODUCTION
2.1. Toxic gases as endogenous messengers in multicellular organisms
At least six gaseous molecules can formally fulfill the criteria of being intra- and intercellular messengers. All of them can be endogenously synthesized, released and specifically effect cellular processes and homeostasis both in plants and animals as well as in unicellular eukaryotes, bacteria and archaea. If we set aside the obvious regulatory functions of oxygen and carbon dioxide, four other gaseous molecules have been extensively studied as “true” signal molecules in the last decades: ethylene, carbon monoxide, hydrogen sulfide and nitric oxide (NO).
2.1.1. Ethylene (C2H4)
Historically, the concept that gases can be endogenous intercellular messengers in multicellular organisms can be traced back to the 1900s. It was first shown that gaseous ethylene inhibits both growth and geotropism in plants, and that ethylene is endogenously produced by fruits (see reviews of (1, 2)). Further observations both confirmed the crucial role of ethylene in plant development and revealed its signal transduction mechanisms (3-10). Unexpectedly, the endogenous synthesis of ethylene was also reported in sponges, a basal animal group (11), where ethylene is involved in the regulation of calcium homeostasis (12, 13) and can interact with NO-dependent pathways (14). Apparently, hormonal and signaling functions of ethylene might not be restricted to the plant kingdom. It would be intriguing to evaluate the role of ethylene in other basal metazoans such as Placozoa and Ctenophores as well as its potential signaling function as a mediator of symbiotic relationships between algae and Cnidarians.
2.1.2. Carbon monoxide (CO)
In 1991 the toxic gas CO was proposed as a signal molecule in animal tissues (15) and a neurotransmitter in particular (16, 17). Indeed, CO is enzymatically produced from heme by two isoforms of heme oxygenase (HO): (i) the constitutively expressed enzyme HO-2, known to be abundant in the mammalian brain and activated by calcium-calmodulin (18)); and (ii) the inducible enzyme HO-1 found in some peripheral tissues. In addition, CO is formed as a by-product of lipid peroxidation (e.g. following redox stress and injury). Endogenously released CO might activate soluble guanylyl cyclase and therefore act as an extracellular messenger. Therefore its functions in neuronal and circulatory systems, as well as other peripheral tissues, may be similar but not identical to NO because of different regulatory mechanisms and a different biological half-life (see recent reviews in (19-32).
2.1.3. Hydrogen sulfide (H2S)
The second toxic gas, hydrogen sulfide, joined the club of gaseous messengers in 1996 (33). In mammalian tissues H2S is produced from L-cysteine metabolism mainly by three enzymes: cystathionine beta-synthetase (CBS), cystathionine gamma-lyase (CSE) and 3-mercaptosulfurtransferase (MST) (22, 34-39). H2S was first reported as an endogenous modulator involved in hippocampal long-term potentiation and other neural functions (33, 35, 40-43). Later it was shown that H2S can act as a physiological vasorelaxant (44), a modulator within the gastrointestinal tract and liver, and be involved in pathogenesis of various cardiovascular diseases and pain (22, 35, 45-53). Its functions are now the subject of many research projects and we might expect to see a growth of comparative studies describing signaling in a variety of invertebrates, expecially those living in hypoxic conditioins.
Nevertheless, we know little about physiological concentrations of CO and H2S in different tissues and cells. Many mechanisms of their action are still under intense investigation, and their interactions with other regulatory systems remain open for future exploration.
2.1.4. Nitric oxide (NO)
In contrast to other gaseous messengers, the information related to NO signaling is overwhelming with more than 120,000 publications by the end of 2009. Such an unprecedented burst of activity started immediately after the 1986-1987 discovery of NO as an endogenous vasoactive molecule, and was followed by the 1998 Nobel Prize for three pioneers in the field: Robert F. Fuschgott, Louis J. Ignarro, and Ferid Murad. It might appear that NO is the foremost member of the “class” of gaseous messengers in animals and even one of the best characterized neurotransmitters in the brain. Yet we have very modest knowledge about its specific functions in neural circuits, neurodevelopment and generation of specific behaviors. Major challenges include the radical (and reactive) nature of NO, difficulties in direct measurements of NO production and complex redox chemistry which can modify practically all classes of biomolecules intra- or extracellularly. It is not surprising that NO can be implicated in almost all biological processes of an entire organism but it is tricky to integrate its molecular functions in specific cells or complex neural assembles. Important directions in the field will include NO analysis in relatively large and accessible cells, simpler nervous systems, or organs of invertebrates (54-65) to study intercellular NO signaling. Use of unicellular organisms may help clarify the system biology of NO.
One of the current obstacles is that most of the data within this rapidly growing field focus on mammals and were obtained using highly heterogeneous and complex tissues or brain structures. As a result, certain aspects of synthesis, regulation and even classification of NO-dependent mechanisms in specialized cells or neurons, although generalized from a few selected models, might only have a limited application if other animal groups are to be considered. On the other hand, a number of widely distributed mechanisms described in various bacteria, plants and invertebrate animals (e.g. both non-enzymatic and enzymatic NO synthesis from nitrites) received relatively little attention in mammalian oriented studies. Furthermore, there is a tendency for a quite biased approach in analysis of NO signaling (e.g. limited to the use of standard mammalian NOS inhibitors) within complex systems such as nervous system functions or animal development.
The goal of this paper is to review comparative aspects of NO synthesis. Specifically, we would like to stress and illuminate the enormous diversity of NO synthases, as well as non-enzymatic mechanisms of NO production. We also will discuss NO mediated signaling with a focus on the distribution and function of NO in invertebrate animals and non-animal groups.
Historically, studies of relatively simple organisms have revealed fundamental principles that apply to more complex systems. It should be true for NO signaling as well. NO-related regulatory mechanisms may be as old as cellular organization itself (66-69), so that ‘ancestral’ functions of NO in prokaryotes and basal eukaryotes are likely well preserved across billions of years of biological evolution and can be essential for biomedical studies and clinical applications.
3. THE CONCEPT OF THE NO MICROENVIRONMENT AND VOLUME SIGNALING IN ANIMAL PHYSIOLOGY
From a historical point of view, the 1986-1987 discovery that the radical NO is the major dilatory agent in the circulatory system of humans (70-72) immediately generated a burst of interest in the whole concept that living systems are able to produce and utilize gaseous species as genuine intra- and extracellular signal molecules. It should be noted, however, that in many physiologically oriented studies the term of “NO” is used to refer to three forms: (i) the nitrosyl radical (.NO itself) and (ii) its nitroxyl (NO-) or (iii) nitrosonium (NO+) ions (see these reviews for further details (73-76)).
Animal physiologists immediately recognized that NO is a new and ubiquitous messenger molecule with signaling mechanisms dissimilar to those found for classical hormones, neuropeptides and other transmitters (19, 54, 63, 67, 71, 74, 77-83). In particular, the synthesis of NO does not require any special storage machinery or specialized cargo delivery or release mechanisms. NO can act directly at the place of its initial synthesis or it can diffuse across membrane barriers in 3D space, potentially affecting targets that are far from its origin (84-88).
NO primarily exerts its effects through direct covalent binding to target molecules (75, 89-96) resulting in their direct chemical modification (e.g. forming SNitrosothiols or nitrosotyrosine residues in target proteins (97-101). As a result of its distinct physical and chemical properties, the concept of a NO microenvironment with transient NO gradients is a more accurate way to describe the actual situation in living tissues. Finally, being a radical NO is chemically very active; NO gradients are not constant. They can be both dynamic and restricted to highly localized compartments even within small cells. Endogenous NO concentrations (ranging from 10-12 to 10-6 M) are dramatically affected by both the intracellular and extracellular redox state. Therefore, the biological half-life of NO can be extremely variable (from milliseconds to minutes and even hours) depending upon many environmental factors (such as the NO concentration itself, pO2, the presence of thiols, heme groups, or various endogenous scavengers, etc.).
For example, NO oxidation in aqueous solution is not linear with respect to NO concentration and can be summarized by the following reaction, with NO-2 as the predominant reaction product (101-104):
The loss of NO from the reaction is described as –d[NO]/dt = 4k[NO]2[O2]; k=2×106 M-2s-1 (i.e. this reaction is second order in NO concentration and first order in O2 concentration). For example, with [O2] = 200 mkM it will take about 1 min for 10 mkM NO (a concentration which can be reached locally following iNOS activation in macrophages) to degrade to 5 mkM (or half of the starting level). If the initial concentration of NO is 10 nM (a level that can be achieved following activation of constitutive NOSs) it will take over 70 hours to degrade to 5 nM (101). Maximal NO concentration in solution in equilibrium with headspace gas of pure NO (at 1 atm pressure and 25° C) will be about 1.9mM and its degradation to half of its original concentration will take seconds (105). Consequently, physiologically relevant NO concentrations within the range of 100 pM to 1nM will be very little effected by direct oxidation in solutions and can be maintained for many days.
In addition, the solubility of NO in hydrophobic solvents is ~9 times greater than in water (105) and O2 is approximately threefold greater. As a result, NO might be nine times more concentrated in cell membranes, lipid inclusions and lipoprotein complexes. Therefore, the calculated rate of NO autoxidation in the membrane will be 243 times faster than in the aqueous phase because of the concentration effect of the reactants (106, 107). As a second outcome of this process, NO reacts with various lipid radicals in membranes and can act as a potential inhibitor of lipid peroxidation and low-density lipoprotein oxidation. Surprisingly, nitrated fatty acids not only represent one of the largest pools of nitrogen oxides in vasculature (108-110); they are also recognized as a novel class of bioactive cell-signaling molecules (110-114).
It is evident that in biological tissues NO will react reasonably well with many other biomolecules such as thiols, metal adducts in heme-containing proteins and complexes that can act both as a “NO sink” and/or as NO carriers depending on cellular microenvironments (100). Furthermore, the reactivity of NO with radicals such as O-2 is a diffusion limited process. Additional details about diffusion range and modeling of NO gradients and concentrations is well described elsewhere (84-87). Importantly, even ubiquitous products of NO-dependent posttranslational modifications (via N-nitrosation, hemenitrosylation and S-nitrosation) are also highly dynamic. They have fairly short lifetimes and are linked to tissue oxygenation and redox state; for example, hypoxia induces profound changes in nitrosylation within 1-5 minutes (115). In other words, the complexity of NO gradients in living cells and tissues might look intractable and unpredictable because of our limited knowledge about chemical microenvironments in cells and cell compartments. Nevertheless, some of the most intriguing relationships were found between NO signaling and oxygen gradients.
Both on the long time-scale of biological evolution in ecosystems and in the relatively short-term events in an organism, tissue, cell or cell compartment, there is always a strong, reciprocal, concentration-dependent influence of O2 and NO on each other's physiological actions. As indicated by Carl Nathan, “NO exerts more control when the concentration of oxygen ([O2]) falls”(116). As a result, NO and reactive nitrogen intermediates (RNI, such as nitrites, .NO2, N2O3, N2O4, S-nitrothiols, peroxynitrite (OONO-), and dinitrosyl-iron complexes) with their reciprocal relationship with reactive oxygen intermediates (ROI) provide perfectly tuned and highly localized cellular redox domains and gradients. In this dynamic NO-controlled microenvironment, redox signaling provides a wide-range mechanism that integrates and functionally links different cell's compartments. In fact, the NO “ties the cell's different commitments to its metabolic budget” (116). In other words, the apparently non-specific and widespread chemistry of RNIs and ROIs act as the universal volume signaling or 3D coupling mechanism that integrates the complex cell biochemical machinery and dynamics of cellular energetics. Recent data linking NO functions to the biogenesis of mitochondria reveal a novel systemic level of long-term integration of cellular respiration and cell biochemistry (117-126).
In summary, NO is an extremely dynamic molecule and its behavior in living cells, tissues and organisms must be considered in the context of a specific chemical microenvironment and time (i.e. the biological half-life of NO can be as short as a few milliseconds and as long as days). In some ways NO biology has its own analogy with a well-recognized saying in the real estate business: location, location and location, and ... oxygen – meaning that biological effects of NO are critically depend upon its site-specific synthesis and degradation as well as oxygen concentrations in the same highly localized cellular compartments.
Considering the radical chemistry of NO, it is not surprising that the practial measurement of NO production in living cells is one of the barriers to understanding its functions and mechanisms of action, especially when new models and tissues are to be investigated. Here we will not discuss the methodology of NO detection, which is extensively reviewed elsewhere (76, 127-141). However, we would like to note that as a rule a combination of several complimentary approaches should be employed. For example, it is reasonable to start with preliminary screening for the presence of NOS using NADPH-diaphorase histochemistry (54-57, 83) or spin-trapping to detect gaseous NO (65, 142). Then, for the majority of comparative models, a reasonable strategy would be to clone NOS following analysis of its expression by in situ hybridization, and finally to detect products of NOS activity or NO oxidation using analytical techniques such as capillary electrophoresis (61, 137, 143-147). Considering all the potential non-enzymatic sources of NO production (see below) a combination of physiological studies with application of various NO scavengers is important. In contrast, for a number of species and tissues the use of NOS inhibitors, NO electrodes and fluorescent markers should be used with careful controls and evaluation of the selectivity and specificity of these techniques or probes to avoid interference from a variety of non-specific effects (142, 148, 149).
4. NO AS THE ANCIENT MESSENGER MOLECULE IN THE HISTORY OF LIFE
As a signal molecule, NO has widespread distribution both in prokaryotes and eukaryotes. NO has been found among practically all animal groups investigated so far as well as in plants, diatoms, slime molds and bacteria, where it is involved in countless biological phenomena (see reviews in (67, 68, 82, 150, 151)). Still, the comparative physiology, NO synthesis and associated transduction pathways even within major animal clades are poorly described. For example, among more that 30 extant animal phyla (with about 100 classes, see refs in (152)) functional and biochemical data about NO signaling is mostly limited to the class Mammalia (one of 12 classes comprising the Phylum Chordata) and the class Insecta (one of 13 classes of the Phylum Arthropoda). Consequently, the reconstruction of its evolution both as a toxic intermediate and a modulator of physiological processes is not yet apparent. Interestingly, NO itself was “officially rediscovered” by animal physiologists in prokaryotes (NO formation by bacteria was known since 1956) where it acts not only as a transient intermediate in denitrification pathways, but also operates as a potential endogenous regulator of gene expression (150, 151). Different classes of arginine-depedent NOSs (genetically unrelated to mammalian NOSs) were proposed for plants (153, 154) but mechanisms of NO synthesis by these plant NOSs are obscure. Evolutionary relationships between NO synthetic enzymes in animal and non-animal groups are not clear, although deep phylogenetic roots for parallel origins of numerous NO-dependent signaling pathways are obvious.
Comparative data point out that NO coupled regulatory systems may be as old as cellular organization itself, tracing their origins to the very dawn of biological evolution ~3.8-3.5 billion years ago (Gya). In fact, geological conditions were favorable for NO synthesis and accumulation on the anoxic primitive Earth. During the Hadean and Archean eras (from 4.5 Gya to 2.5 Gya) NO could be produced in the ancient atmosphere and early oceans (155-157) as an important environmental factor contributing to the origin of life itself.
Under this scenario, it is suggested that NO was a crucial intermediate in the utilization of chemically inert molecular nitrogen (N2). Indeed, nitrogen is an essential element for life and, although the Earth atmosphere is the major reservoir of dinitrogen, most organisms are not able to use it directly because of the high energy (948 kJ mol-1) required to dissociate the nitrogen triple bond (N≡N). Thus, to be used by various organisms nitrogen must be “fixed” in reduced (ammonia- NH3/NH+4, hydrogen cyanide - HCN, acetonitrite/methyl cyanide - CH3CN) or oxidized forms (N2O, NO, nitrites – NO-2 or nitrates – NO3-). In other words, NO and its related species were the principal forms of fixed nitrogen in the early Earth. NO also could be produced efficiently in a neutral Hadean and Archean atmosphere (composed primarily of CO2 and N2 (157, 158); under these conditions HCN and related compounds are not synthesized by lightning and other electrical discharges associated with volcanic eruptions, meteorite impacts and thunderstorms.
The estimated accumulated annual production rate of NO in early Earth was on the order of ~1013 g per year (the global production of NO by lighting at the present time is estimated as ~1012 g/year (159)). This NO would have been converted into nitric and nitrous acids and delivered to the Ancient Ocean and early lithosphere as acid rain. NO and related species were then reduced by various minerals (primarily Fe(II), which was present in earlier oceans at much higher concentrations than today) to produce NH+4 (160, 161) providing a prebiotic mechanism for nitrogen fixation on early anoxic Earth and, potentially allowing accumulation of ammonia in localized spots. At the same time, as components of the ancestral nitrogen cycle, NO and nitrites were ideal electron acceptors (oxidizing agents) for the further development and evolution of ancient cellular respiratory machinery and the first biological denitrification pathways in the early Archean biosphere.
These steps are required for further synthesis and buildup of more complex organic nitrogen-containing molecules and should be a factor for the origin of life and establishment of the earliest ecosystems on primitive Earth. Gradually, during the Archean eon (3.8-2.5 Gya), NO production by lighting discharge decreased (due to the decrease of CO2) by two orders of magnitude until about 2.2 Gya. After this time, the rise in oxygen (or methane) concentrations probably initiated the development of mechanisms to utilize other abiotic sources of nitrogen (see also (162-164)).
It was hypothesized (165) that although the temporal Archean reduction in NO production may have lasted for only 100 Myr or less (see also (157),), this was potentially long enough to cause the ecological crisis that triggered the rapid development of biological nitrogen fixation (again with NO as an important electron acceptor in NO respiration and possibly a signal molecule). These new enzymatic mechanisms using NO/nitrite/nitrate reductases had emerged in some prokaryotic cells providing the foundation for the global nitrogen cycle (Figure 1) in the biosphere and representing ~70% of the total quantity of fixed nitrogen today. Thus, as stated by W. Zumft (1993) “NO is not an obscure chemical and certainly no newcomer to the life sciences, as often stated in hyperbole. Early in evolution NO took its role as a central player in bacterial bioenergetics and the global N cycle vital to all organisms.”
Figure 1.
Nitrogen cycle: Enzymatic NOS-independent synthesis. The simplified sequence of the nitrogen cycle reactions is shown. The reactions of nitrification and denitrification are associated with electron transport systems and coupled to cellular energetics and ATP production. NH3 is widely used in the synthesis of various amine-containing biomolecules and, therefore, the NOS catalyzed oxidation of L-arginine can be considered as a heterotrophic nitrification route, in addition to bacterial autotrophic nitrification. Nitrification and NO synthesis by NOS are aerobic pathways, where molecular oxygen is required for the oxidation of L-arginine and NH3. In contrast, denitrification usually occurs under either anaerobic or hypoxic conditions. Additionally, nitrates can be reversibly reduced to ammonia in a nitrate/nitrite type of anaerobic respiration (see (150, 151, 464, 465)) for a review of these and associated pathways).
The rise of oxygen concentrations in Earth atmosphere (~2.4-2.3 Gya) did not eliminate “ancient” functions of NO in the new environment. More likely, these functions still co-exist with newly developed systems and provide important regulatory and coupling mechanisms that are mostly expressed under hypoxic conditions (or during localized acidification) when oxygen-dependent processes are suppressed. It is possible to hypothesize that the development of an L-arginine-NO pathway might be a result of adaptive responses to a novel oxygen rich biosphere where the antioxidant properties of NO could be highly adaptive under oxidative stress. Thus, it is not surprising that multiple pathways leading to NO production and its homeostatic regulation evolved in different eukaryotic lineages following the big oxidation event about 2.4 Gya.
Below, we outline different mechanisms of NO synthesis in biological systems including multiple nonenzymatic and enzymatic pathways suggesting parallel and, possibly, convergent evolution as well as recruitment of NO synthetic pathways and different classes of NO synthases (NOSs).
5. Multiplicity of NO synthetic pathways
At least seven pathways of enzymatic NO synthesis can be found in living systems:
classical multi-domain NOS-type enzymes (166, 167) in animals and slime molds;
prokaryotic “truncated” NOS found in a majority of bacteria and some archaea (151, 168);
plant-type synthetic pathways that possibly involve proteins distantly related to GTPases (153, 169);
prokaryotic and eukaryotic enzymes involved in reduction of nitrites as part of the nitrogen cycle/denitrification pathways (see Figure 1 and (151, 170));
nitrite reductase activity (171-174) of complexes of the mitochondrial electron transport chain under hypoxic conditions (124, 175-182);
Finally, endothelial nitric oxide synthase can also reduce nitrites to NO and recycle bioactive NO under hypoxic/anoxic conditions (212-214).
In addition to these seven pathways, NO can be produced non-enzymatically from nitrites in various cells and tissues under certain chemical conditions (e.g. acidic reduction in the presence of polyphenols (215-217), ascorbate (142, 218) or even some classical NOS inhibitors (142)) using various redox mechanisms. Anoxic and hypoxic states are more favorable for nitrite reduction in biological tissues, but this requirement is not absolute. In the next three sections (parts 6, 7 and 8, we will first discuss non-enzymatic NO formation, and then briefly overview comparative aspects of “more conventional” NOS activity in animals.
6. ABIOTIC REDUCTION OF NITRITES: ALTERNATIVE SOURCES OF NO FORMATION
The requirement of an enzyme for NO synthesis in biological systems is not absolute. The fact that NO can be generated non-enzymatically (without NOS or denitrification pathways - see below) from a nitrite solution is crucial for any comparative and evolutionary analysis of NO synthesis and signaling. In reality, abiotic NO formation is well known from the chemistry of NOx species, but until recently it was not considered to be an endogenous source of NO in animals (67, 147, 219-225).
The chemistry of NO oxidation is very complex, with many transient nitrogen/oxygen species (76, 226-231), where nitrites (NO-2) and nitrates (NO-3) are major sequential products of NO oxidation. However, NO-2 is the only stable product formed by the spontaneous oxidation of NO in oxygenated solutions (102, 103, 226).
In mammalian plasma, ceruloplasmin (a multi-copper oxidase) catalizes nitrite formation from NO (i.e. it acts as endogenous NO oxidase = Nitrite synthase, see (232)). Hemoproteins (e.g. oxyhaemoglobin) in various tissues convert NO and NO-2 to NO-3 (103, 233). As a result, the normal human plasma NO-3 concentrations are within 20-40 mkM while NO-2 varies from 50 to 300 nM; tissue concentrations can be significantly higher (e.g. (137, 143, 147) and discussion below).
Substantial amounts of nitrites and nitrates also come from the diet and, in mammals, they can be accumulated in salivary glands (up to 10 mM for NO-3 (221)) and in saliva. Mammalian cells do not have nitrate reductases, but numerous symbiotic (facultative anaerobic) bacteria living in the oral cavity can efficiently reduce nitrates back to nitrites providing a stable storage pool or reservoir for NO-like bioactivity independently from oxygen dependent NOSs (225).
Indeed, in an acidic environment nitrites are easily converted to NO according to the equation:
Again, the actual mechanisms are more complicated (102, 226, 228, 231, 234, 235), but without describing secondary pathways, the sequence can be presented as follows:
Ascorbate (141, 218, 236-239) and some reducing compounds, such as NADPH, L-cysteine, reduced gluthatione and other thiols (142, 240-244), have been reported to stimulate NO formation from nitrites with less generation of N2O and .NO2 (225). For example, vitamin C is an important co-factor and antioxidant that can accumulate in both intercellular spaces and living cells, where it can enhance reduction of nitrites and generate NO non-enzymatically:
7. BIOLOGICAL FUNCTIONS OF NONENZYMATIC NO SYNTHESIS
Non-enzymatic NO production from dietary nitrites in vivo was originally demonstrated in the gut (245, 246) and in the human oral cavity (247). In both cases NO concentrations were sufficient to be involved in primary antimicrobial, non-immune defense reactions (223, 248-253) and, probably, in the control of digestive functions, such as mucosal blood flow, motility, and possibly secretion and absorption (254). Estimated nitrite concentrations were between 0.1-1000 μM in different parts of the digestive system, approaching a concentration of 1-2 mM in saliva following a high nitrate/nitrite test meal (221, 247, 255).
Non-enzymatic NO formation has also been demonstrated in human skin (256, 257) and urine (218, 258), with a suggested physiological role in the inhibition of infection by pathogenic microscopic fungi (251), as well as in the modulation of cutaneous T-cell function, skin blood flow, and keratinocyte differentiation (259). Evidently, nitrites formed in the skin can act as important bacteriostatic agents and be directly involved in wound healing (260).
Similarly, non-enzymatically generated NO and related species in the digestive tract can be both toxic for potential pathogens and able to perform regulatory functions such as control of circulation, uptake mechanisms and modulation of muscle contractions in the gut. The maintenance of a very acidic stomach pH favors reduction of nitrites from various food sources and supposedly would be essential for ecological adaptations in herbivore species. A combination of low pH and high nitrite levels (~1mM) results in NO levels that sometimes can exceed 4μM (i.e. more than 10,000 times higher than the levels required for vasodilatation) (223).
Non-enzymatic NO formation from nitrites also plays an important role in vascular control (222, 261) and related pathologies – the latter present unique therapeutic opportunities for the use of NO -2 and drug development based upon organic nitro compounds (225). Surprizingly, even low nitrite concentrations (140-300 nM) in human plasma have a significant vasodilation effect and decrease in blood pressure (262, 263). Large quantities of NO (similar to, or even higher than, those produced by NOS) can be formed in ischemic heart tissues by a mechanism that is not enzyme dependent and not blocked by inhibitors of NOS (264-266). The authors conclude that this NO formation is a consequence of acidification (pH ~5.5), which serves to reduce the large pool of nitrites present within tissue. The mean nitrite concentration of the ischemic myocardium (12μM) is sufficient to generate the detected amount of NO. It was suggested that enzyme-independent NO formation not only contributes to the process of postischemic injury, but also eliminates the protective effect of NOS inhibitors (265).
Thus, tissue nitrite can serve as a significant “hypoxic buffer” of NO that can be released during hypoxia when oxygen-dependent NOS activity is suppressed or even eliminated (267). Importantly, the half-life of nitrites in biological tissues is significantly longer than NO. For example, in blood/plasma under physiological conditions t½ for nitrites can be 1-5 minutes while for NO t½ = 1-2 milliseconds. It might be much longer in some specific cells (147). In other words, the recycling pool of nitrites can function as the dynamic but long-term storage of NO in cell and tissues and localized NO release can be triggered by specific signals or redox metabolites at their physiological concentrations. It was also shown than NO generation from nitrites under physiological conditions and induced aortic ring relaxation is further increased by ascorbate (268).
8. CONDITIONS FOR INTRACELLULAR NONENZYMATIC NO FORMATION: NEURONS AS MODELS
Non-enzymatic NO formation can be substantial inside NOS-containing cells and NOS-containing neurons in particular. First, the intracellular nitrite concentrations can be significantly higher than those estimated from plasma or tissue homogenates. Estimations from homogenates of whole neuronal tissues (269) give values of 50-200 μM which are substantially higher than those in the ischemic heart, but only 1-2% of neurons expressed NOS. Our recent direct single cell measurements (147) indicate that actual intracellular nitrite levels can be as high as 1-5mM, more than enough to produce physiological concentrations of NO non-enzymatically even following minor changes of pH or endogenous redox species (e.g. NAD(P)H, ascorbate, cysteine, etc). Furthermore, the distribution of NO --2 is not uniform, but instead appears to be neuron-specific, with higher concentrations being found in neurons expressing NOS. Yet, some NOS-negative neurons might also have relatively high nitrite levels, possibly due to extensive synaptic inputs from NOS-containing presynaptic neurons (55, 147).
Most importantly, the brain has almost the highest ascorbate level in the human body (270, 271) and its distribution is highly compartmentalized (272, 273). Up to 400-600 mkM of L-ascorbate have been detected extracellularly (274, 275). Intracellular concentrations are estimated at 2-5 mM in neurons (276) and ≥7 mM in glial cells (277). Direct single-cell measurements of intracellular ascorbate concentrations using capillary electrophoresis have confirmed millimolar levels of ascorbate in neurons (149, 278). Ascorbate accumulation differs among various neuronal groups and can be modified by electrical activity and neurotransmitters (276). Thus, we hypothesize that either intracellular ascorbate or ascorbate released from neurons during activity can generate NO from a conventional chemical reduction of intracellular and extracellular nitrite ions (137, 142, 147, 278). It has been suggested that extracellular ascorbate-induced NO production could subtly and accurately match oxygen transport to the local metabolic demands of the nerve cells by vasodilatation of cerebral blood vessels under hypoxic conditions (279). Ascorbate can be released from neurons following electrical activity and synaptic transmission providing additional factors than can modulate nonenzymatic NO production following the dynamics of neuronal dischargers in neural circuits.
Are physiological variations in pH sufficient to induce non-enzymatic NO release from nitrites in nervous tissues? pH homeostasis in nervous tissue has been studied in some detail; various transporters such as Na+/H+, Na+-dependent Cl-/HCO-, and Ca2+/H+ exchangers as well as intra- and extracellular carbonic anhydrase are involved in pH regulation (280). Moreover, V-ATPases translocate H+ into vacuoles and synaptic vesicles, providing further avenues for variations in intracellular pH.
Reported physiological pH variations in selected intracellular compartments are very broad and the range of intracellular pH changes can be greater that 2 units (280, 281). The pH values of lysosomes, synaptic vesicles and some other organelles can be as low as 4.5 – 5.7 units (282, 283). It is also known that the release of synaptic vesicles results in acidification (~ 0.6-1 pH unit) of the synaptic cleft (284-287). Furthermore, there is evidence that protons themselves may act as true transmitters (288-291). Based on model chemical measurements (292), these pH values, in combination with the measured nitrite/ascorbate levels, are more than sufficient to generate high micromolar concentrations of NO non-enzymatically in neurons. We suggest that a similar situation can also be found in other cell types (e.g. secretory cells) as well as in many physiological processes such as development, phagocytosis, etc.
It should be noted that high chemical reactivity of nitrites and NO might lead to direct modification of a number of important intercellular messengers such as classical, peptide transmitters and hormones (293, 294) or even second messengers such as cGMP (295) providing the next level of regulation of signaling mechanisms in nervous tissue.
One of the unexpected fundings related to nonenzymatic NO synthesis was the demonstration that “classical” NO inhibitors can produce NO nonenzymatically (142). While NO is generated enzymatically by NO synthase (NOS) from L-arginine, overproduction of NO contributes to cell and tissue damage as sequelae of infection and stroke. Strategies to suppress NO synthesis rely heavily on guanidino-substituted L-arginine analogs (L-NAME, L-NA, L-NMMA, L-NIO) as competitive inhibitors of NOS, which are often used in high doses to compete with millimolar concentrations of intracellular arginine. Surprisingly, these analogs are also a source for non-enzymatically produced NO. Enzyme-independent NO release occurs in the presence of NADPH, glutathione, L-cysteine, dithiothreitol and ascorbate (142). This nonenzymatic synthesis of NO can produce potentially toxic, micromolar concentrations of NO and can oppose the effects of NOS inhibition. NO production driven by NOS inhibitors was also demonstrated ex vivo in the central nervous and peripheral tissues of gastropod molluscs Aplysia and Pleurobranchaea using electron paramagnetic resonance and spin-trapping techniques.
9. NITRITE PHOTOLYSIS AS A SOURCE OF NO IN BIOLOGICAL SYSTEMS
Nitrite/Nitrate photolysis (296-298) is another non-enzymatic, but biologically relevant, chemical process associated with NO signaling. Photochemical generation of NO from nitrites is a notable component of the nitrogen cycle in the Earth’s biosphere. Nitrite absorbs maximally at 356 nm, and the process can be presented as follows (299):
This reaction occurs naturally in the surface layers of the world's oceans, predominantly in the central equatorial areas. NO formed by this mechanism may play an important role in marine ecosystems, and must be considered as an NO source to the atmosphere (299). The measured partial pressure of NO in the air was less than 8×10-12 atm compared with a PNO calculated for surface seawater of 7×10-8 atm (300). The estimated concentrations of NO in the surface film of tropical waters depend strongly on the nitrite distribution; they could be in the subnanomolar or nanomolar range during the day, dropping to a practically undetectable level after sunset.
Nitrites (and, likely, NO) are important endogenous regulators of the biological clocks in planktonic organisms such as the unicellular dinoflagellate Goniaulax polyedra (301). We might therefore speculate that the involvement of NO in the regulation of circadian rhythm, observed in higher animals (302-306), might be traced back to the earlier day-night conditions in the ancient oceanic waters.
One practical aspect of nitrite photolysis is UV-induced generation of NO in human skin (reviewed by (224)). Even a short 3-5 min exposure to sun in the Central European summer leads to significant non-enzymatic NO formation from nitrites (~10 mkM in sweat) or NO-thiols present in normal human skin (307, 308). This UV induced cutaneous NO formation was comparable to or higher than that found in maximally activated human keratinocytes (309). Suschek et al (2006) have calculated that human skin can be “the largest storage organ for NO derivatives such as nitrites and RSNOs” and “non-enzymatic NO generation might represent an initial screen function in human skin”(224). As indicated above, NO can regulate skin pigmentation, growth, and differentiation as well as perform antifungal and antibacterial protective functions. Clearly, this type of mechanism of surface pH/UV-dependent NO formation can be widely distributed across animal and plant kingdoms and be an important player in various ecological adaptations and in development.
In summary, there are two sourses of nitrites in animals: (i) the diet and (ii) the enzymatic synthesis of NO by NOS and subsequent NO oxidation. The last decade of research in the field of NO biology revealed novel physiological functions of abiotic non-enzymatic NO synthesis from nitrites. Furthermore, the biological role of nitrite itself has evolved from an inert by-product of organic nitrate metabolism or NO oxidation into an endogenous signaling molecule and regulator of gene expression (73, 310-312). Indeed, recent findings clearly indicate than certain functions of nitrites are unique and they can not be effected by NO scavengers (i.e. they are not mediated by NO - (311)). Thus, in addition to NO, nitrite can act as an independent player in the family of inter- and intracellular signal molecules and in some ways fits the criteria of a truly endocrine molecule (225). Such regulatory functions of nitrites and nitrates, although relatively novel for animal biology, have many parallels in plants and basal eukaryotes (e.g. (313)).
10. THE DIVERSITY OF ANIMAL (CONVENTIONAL) NITRIC OXIDE SYNTHASES: MODELS, GENES AND PHYLOGENY OF NOS
Four years after the discovery of the signaling role of NO the first constitutive isoform of nitric oxide synthase (NOS) was cloned from the rat brain (314). Then, in 1992, two other isoforms were cloned nearly simultaneously: the inducible type of NOS (iNOS) from macrophages (315-317) and the second constitutive (eNOS) from endothelium (318-322). All of these three NOS types share close sequence similarity with cytochrome P450 reductase in their C-terminal reductase domains and have a common oxygenase domain.
During the next 15 years several dozen NOSs have been identified from different organisms. Tables 1 and 2 summarize the current status of molecular cloning and identification of NOS-related proteins across all major eukaryotic and prokaryotic groups. Figure 2 illustrates phylogenetic relationships between different classes of NOSs. As discussed below these NOSs do not fit into the simple tripartite classification originally established from mammalian studies but rather represent multiple events of parallel evolution in different lineages. Yet, they apparently share a common origin from a truncated NOS with the oxygenase domain, and their ancestry can be traced to earlier bacteria living more than 1 billion years ago.
Table 1.
The comparative characteristics of all known NOSs*
| Taxon/species | Abbreviations | GenBank accession # |
Length | Comments | Exon no. |
Genomic information |
|---|---|---|---|---|---|---|
| Deuterostomes Phylum Chordata. Class Mammalia | ||||||
| Homo sapiens | hNOSn | NP_000611 | 1434 aa | nNOS; constitutive; Ca-dependent, NOS1 | 29 | 12q24.2–12q24.31; GeneID 4842 |
| H. sapiens | hNOSe | NP_000594 | 1203 aa | eNOS; constitutive; Ca-dependent, NOS3 | 27 | 7q36; GeneID 4846 |
| H. sapiens | hNOSi | NP_000616 | 1153 aa | iNOS,Ca-indepen - dent NOS2A | 27 | 17q11.2–17q12; GeneID 4843 |
| Pan troglodytes | PtNOSn | XP_522539 | 1433 aa | nNOS, NOS1 | 42 | Chromosme 12; GeneID 467139 |
| P. troglodytes | PtNOSe | XP_519525 | 1323 aa | eNOS, NOS3 | 22 | Chromosme 7; GeneID 463893 |
| P. troglodytes | PtNOSi | XP_511794 | 1153 aa | iNOS; NOS2A | 27 | Chromosome 17 GeneID: 455026 |
| Macaca mulatta | MnNOSn | XP_001083352 | 1431 aa | nNOS, NOS1 | ||
| M. mulatta | MnNOSe | XR_012468 | 1792 aa | eNOS, NOS3 | ||
| M. mulatta | MnNOSi | XP_001106185 | 1114 aa | iNOS; NOS2A | ||
| Mus musculus | mNOSn | NP_032738 | 1429 aa | nNOS, NOS1 | 28 | Chromosome 5 GeneID: 18125 |
| M. musculus | mNOSe | NP_032739 | 1202 aa | eNOS, NOS3 | 28 | Chromosome 5 GeneID: 18127 |
| M. musculus | mNOSi | NP_035057 | 1144 aa | iNOS; NOS2 | 23 | Chromosome 11 GeneID: 18126 |
| Rattus norvegicus | rNOSn | NP_434686 | 1463 aa | nNOS | 26 | Chromosome 12 GeneID: 24598 |
| R. norvegicus | rNOSe | NP_068610 | 1202 aa | eNOS, NOS3 | 26 | Chromosome 4 GeneID: 24600 |
| R. norvegicus | rNOSi | NP_036743 | 1147 aa | iNOS; NOS2 | 25 | Chromosome 10 GeneID: 24599 |
| Cavia porcellus | CpNOSn | AAD29751 | 397 aa | nNOS, NOS1 Partial sequence | ||
| C. porcellus | CpNOSe | AAD29753 | 1206 aa | eNOS, NOS3 | ||
| C. porcellus | CpNOSi | O54705 | 1149 aa | iNOS; NOS2 | ||
| Canis lupus familiaris | ClfNOSn | XP_534695 | 1431 aa | nNOS, NOS1 | 29 | Chromosome 26GeneID: 477498 |
| C. familiaris | ClfNOSe | NP_001003158 | 1205 aa | eNOS, NOS3 | 26 | Chromosome 16GeneID: 403784 |
| C. familiaris | ClfNOSi | NP_001003186 | 1154 aa | iNOS; NOS2 | 21 | Chromosome 9 GeneID: 403822 |
| Bos taurus | BtNOSn | XP_872723 | 1434 aa | nNOS, NOS1 | 25 | Chromosome 17GeneID: 536132 |
| B. taurus | BtNOSe | NP_851380 | 1205 aa | eNOS, NOS3 | 25 | Chromosome 4 GeneID: 287024 |
| B. taurus | BtNOSi | NP_001070267 | 1156 aa | iNOS; NOS2 | 23 | Chromosome 19 GeneID: 282876 |
| Sus scrofa | SsNOSn | XP_001924891 | 1300 aa | nNOS, NOS1 | ||
| S. scrofa | SsNOSe | AAR27960 | 1205 aa | eNOS, NOS3 | ||
| S. scrofa | SsNOSi | NP_001137162 | 1064 aa | iNOS; NOS2 | ||
| Ornithorhynchus anatinus | OsNOSn | XP_001510768 | 389 aa | nNOS; NOS1 Partial sequence | Annotation not complete | |
| O. anatinus | OsNOSe | XP_001520429 | 612 aa | eNOS; NOS3 Partial sequence | Annotation not complete | |
| O. anatinus | OsNOSi | XP_001506887 | 1152 aa | iNOS; NOS2 | Annotation not complete | |
| Equus caballus | EcNOSn | XP_001915005 | 1434 aa | nNOS, NOS1 | ||
| E. caballus | EcNOSe | XP_001504700 | 1205 aa | eNOS, NOS3 | ||
| E. caballus | EcNOSi | NP_001075238 | 1311 aa | iNOS; NOS2 | ||
| Oryctolagus cuniculus | OcNOSn | NP_001075854 | 1435 aa | nNOS, NOS1 | Annotation not complete | |
| O. cuniculus | OcNOSe | NP_001076202 | 1209 aa | eNOS, NOS3 | Annotation not complete | |
| O. cuniculus | OcNOSi | O19114 | 496 aa | iNOS; NOS2 Partial sequence | Annotation not complete | |
| Monodelphis domestica | MdNOSn | XP_001362705 | 1430 aa | nNOS, NOS1 | ||
| M. domestica | MdNOSe | XP_001375582 | 1212 aa | eNOS, NOS3 | ||
| M. domestica | MdNOSi | XP_001371412.1 | 1203 aa | iNOS; NOS2 | ||
| Class Aves (birds) | 2 NOS genes | |||||
| Gallus gallus | GgNOSn | XP_425296 | 1609 aa | nNOS, NOS1 | 29 | Chromosome 15 GeneID 427721 |
| G. gallus | GgNOSi | NP_990292 | 1136 aa | iNOS, NOS2 | 28 | Chromosome 19GeneID 395807 |
| Taeniopygia guttata | TgNOSn | XP_002196989 | 1321 aa | nNOS, NOS1 | ||
| T. guttata | TgNOSi | XP_002197005 | 1118 aa | iNOS, NOS2 | ||
| Class Reptilia | ||||||
| Aspidoscelis uniparens (lizard) | AuNOSn | AAZ76558 | 872 aa | nNOS-like; Partial missing a part of reductase domain | ||
| Class Amphibia | ||||||
| Xenopus tropicalis | XtNOSn | ENSXETG00000022354 | 1416 aa | nNOS, NOS1 | 27 | |
| X. tropicalis | XtNOSe | ENSXETT00000025059 | 1138 aa | eNOS, NOS3 | 26 | GeneID 373705 |
| X. laevis | XlNOSn | AAD55136 | 1419 aa | nNOS, NOS1 | ||
| X. laevis | XlNOSe | AW765292, AW764664 | ESTs | eNOS-like, NOS3 | ||
| Class Actinopterygii | 1–3 NOS genes, can be two iNOS in genomes | |||||
| Danio rerio | DrNOSn | NP_571735 | 1431 aa | nNOS, NOS1 | ||
| D. rerio | DrNOSai | XP_692454 | 1145 aa | iNOS, NOS2A | 28 | Chromosome 15GeneID 564002 |
| D. rerio | DrNOSbi | XP_692103 | 1130 aa | iNOS, NOS2B | 26 | Chromosome 15GeneID 563654 |
| Tetraodon nigroviridis | TnNOSn | CAG08158 | 1429 aa | nNOS, NOS1 | Chromosome 12 1 NOS gene in genome | |
| Takifugu rubripes | TrNOSn | AAL82736 | 1418 aa | nNOS, NOS1 | ||
| Takifugu poecilonotus | TpNOSn | AAM46138 | 1418 aa | nNOS, NOS1 | ||
| Oryzias latipes | OlNOSn | BAD11808 | 1424 aa | nNOS, NOS1 | ||
| Sciaenops ocellatus | SoNOSn | ACU98970 | 1437 aa | nNOS, NOS1 | ||
| Fundulus heteroclitus | FhNOSn | AAS21300 | 1420 aa | nNOS, NOS1 | ||
| Carassius auratus | CaNOSi | AAX85387 | 1127 aa | iNOS, NOS2a | ||
| C. auratus | CaNOSi | AAX85386 | 1126 aa | iNOS, NOS2b | ||
| Cyprinus carpio | CcNOSi | CAB60197 | 1137 aa | iNOS, NOS2 | ||
| Oncorhynchus mykiss | OmNOSi | NP_001117831 | 1083 aa | iNOS, NOS2 | ||
| Class Chondrichthyes | ||||||
| Scyliorhinus canicula | ScNOSi | AAX85385 | 1125 aa | iNOS, NOS2 | ||
| Order Petromyzontifoes | ||||||
| Petromyzon marinus | PmNOSn | EB720322, EB083238 | ESTs | nNOS-like, NOS1 | ||
| Class Cephalochordata | ||||||
| Branchiostoma floridae | BfNOS1 | XP_002608547 | 1332 aa | nNOS-like PDZ domain | ||
| B. floridae | BfNOS2 | XP_002605826 | 1441 aa | nNOS-like PDZ domain | ||
| Class Urochordata (tunicates) | ||||||
| Ciona savignyi | CsNOS | ENSCSAVG00000009725 | 1128 aa | nNOS-like PDZ domain | 25 | |
| C. intestinalis | CiNOS | ENSCING00000002710 | 1379 aa | nNOS-like PDZ domain | 32 | 10q31 |
| Phylum Hemichordata | ||||||
| Balanoglossus sp. | nNOS-like, partial | |||||
| Phylum Echinodermata | ||||||
| Strongylocentrotus purpuratus | SpNOS1 | XM_001179342 | 1386 aa | iNOS-like | GeneID:587111 | |
| S. purpuratus | SpNOS2 | SPU_002328 | 1590 | iNOS-like | ||
| S. purpuratus | SpNOS | REMOVED | 442 aa | Truncated NOS-like | SPU_013373 | |
| S. purpuratus | SpNOS | REMOVED | 539 aa | Truncated NOS-like | SPU_019970 | |
| Arbacia punctulata | ApNOS | AF191751 AF191750 | ESTs | nNOS-like | ||
| Phylum Arthropoda | ||||||
| Class Crustacea (Decapoda) | ||||||
| Gecarcinus lateralis (crab) | GlNOS | AAT46681 | 1199 aa | nNOS-like | ||
| Marsupenaeus japonicus | MjNOS | BAI67609 | 1187 aa | nNOS-like | ||
| Homarus americanus (lobster) | HaNOS | CN853572 | EST | NOS-like | ||
| Panulirus argus | PaNOS | ACZ60615 | 1200 aa | nNOS-like | ||
| Daphnia pulex | DpNOS1 | jgi|Dappu1|49977| | 1137 aa | nNOS-like | 25 | |
| Daphnia pulex | DpNOS2 | jgi|Dappu1|238200| | 853 aa | Partial sequence | ||
| Daphnia magna | DmNOS1 | ACQ55298 | 1182 aa | nNOS-like | ||
| Daphnia magna | DmNOS2 | ACQ55299 | 1183 aa | |||
| Class Insecta | ||||||
| Luciola lateralis | LlNOS | BAF63160 | 1133 aa | nNOS-like | ||
| Acyrthosiphon pisum | ApNOS | XP_001946209 | 1189 aa | nNOS-like | ||
| Tribolium castaneum | TcNOS | XP_967195 | 1105 aa | nNOS-like | 13 | Chromosome LG9GeneID 655549 |
| Rhodnius prolixus | RpNOS | Q26240 | 1174 aa | nNOS-like | ||
| Bombyx mori | BmNOS | BAB85836 | 1209 aa | nNOS-like | ||
| Manduca sexta | MsNOS | AAC61262 | 1206 aa | nNOS-like | ||
| Apis mellifera | AmNOS | NP_001012980 | 1143 aa | nNOS-like | 25 | Chromosome LG3GeneID 503861 |
| Drosophila melanogaster | DmNOS | NP_523541 | 1349 aa | nNOS-like | 19 | Chromosome 2L; Location 32B1; GeneID 34495 |
| D. pseudoobscura | DpNOS | EAL33128 | 1348 aa | nNOS-like | ||
| Anopheles gambiae | AgNOS | XP_317213 | 1113 aa | nNOS-like | Chromosome 3R;GeneID 1277726 | |
| Anopheles stephensi | AsNOS | O61608 | 1247 aa | nNOS-like | ||
| Aedes aegypti | AaNOS | EAT38354 | 1112 aa | nNOS-like | ||
| Acheta domesticus | AdNOS | AAR88326 | 210 aa | Partial sequence | ||
| Gryllus bimaculatu | GbNOS | BAH14964 | 1163 aa | nNOS-like | ||
| Pediculus humanus corporis | PhcNOS | EEB12398 | 1104 aa | nNOS-like | ||
| Nasonia vitripennis | NvNOS | NP_001161704 | 1145 aa | nNOS-like | ||
| Ixodes scapularis | IsNOS | EEC05792 | 1110 aa | nNOS-like | ||
| Glossina morsitans | GmNOS | AY152725, DV603147 | 429 bp | NOS-like; ESTs, Partial sequence | ||
| Phylum Mollusca | ||||||
| Class Gastropoda | ||||||
| Aplysia californica | AcNOS1 | AAK83069 | 1387 aa | nNOS-like(2 genes) | Genome not completed | |
| A. californica | AcNOS2 | AAK92211 | 1175 aa | nNOS-like | >20 | Genome not completed |
| Lymnaea stagnalis | LsNOS1 | O61309 | 1153 aa | nNOS-like (2 genes) | ||
| L. stagnalis | LsNOS2 | AAW88577 | 1218 aa | nNOS-like | No genome | |
| Lymnaea sp. | AAM21319 | 397 aa | NOS-related protein; partial sequence | |||
| L. stagnalis | LsNOS3 | nNOS-like PDZ domain | ||||
| Lottia gigantean | LgNOS | jgi|Lotgi1|223312 | 1270 aa | nNOS-like | 25 | |
| Limax marginatus | LmNOS | BAC80150 | 209 aa | nNOS-like; partial | ||
| Ilyanassa obsoleta | IoNOS | AAV31753 | 77 aa | nNOS-like; partial sequence | ||
| Lehmannia valentiana | LvNOS | BAF73722 | 1632 | nNOS-like | 33 | |
| Helisoma trivolvis | HtNOS | ACY64755 | 240 aa | nNOS-like; partial | ||
| Class Cephalopoda | ||||||
| Sepia officinalis | SoNOSa | AAS93626 | 1133 aa | nNOSa-like | ||
| S. officinalis | SoNOSb | AAS93627 | 1139 aa | nNOSb-like | ||
| Phylum Nematoda | ||||||
| Caenorhabditis elegans | Not found in genome | NOS gene loss in this lineage | ||||
| C. briggsae | Not found in genome | NOS gene loss in this lineage | ||||
| Phylum Platyhelminthes (Flatworms) No data | Histochemical NADPHd labeling of putative NOS | |||||
| Phylum Cnidaria | ||||||
| Discosoma striata | DsNOS | AAK61379 | 1115 aa | iNOS-like | ||
| Hydra vulgaris | HvNOS | DY449349 | EST | NOS-like | ||
| Hydra magnipapillata | HmNOS | XP_002158785 | 1066 aa | iNOS-like | ||
| Nematostella vectensis | NvNOS | XP_001631503 | 1116 aa | iNOS-like | 29 | |
| Phylum Porifera (sponges) | No data | |||||
| Amphimedon queenslandica | NOS; genome trace archive | |||||
| Phylum Placozoa | ||||||
| Trichoplax adhaerens | TaNOS1 | XP_002108333 | 1028 aa | iNOS-like(3 genes) Missing 5’ | 28 | |
| T. adhaerens | TaNOS2 | XP_002116862 | 1112 aa | iNOS-like | 30 | |
| T. adhaerens | TaNOS3 | XP_002108887 | 1124 aa | iNOS-like | 30 | |
| Other Eukaryotes | ||||||
| Mycetozoa | ||||||
| Physarum polycephalum | PpNOSa | AAK43730 | 1055 aa | iNOSa-like | ||
| P. polycephalum | PpNOSb | AAK43729 | 1046 aa | iNOSb-like | ||
| Giardia lamblia ATCC 50803 | EDO81993 | 604 aa | p450 reductase domain, Not NOS | |||
| Fungi | ||||||
| Aspergillus flavus NRRL3357 | AfNOS | XP_002381643 | 1024 aa | iNOS-like | ||
| Aspergillus oryzae | AoOx | BAE64541 | 178 aa | NOS-like NOS oxygenase domain, 5’-end | ||
| Phylum Chlorophyta (Green plants) Class Prasinophyceae | ||||||
| Ostreococcus tauri | OtaNOS | CAL57731 | 1081 aa | iNOS-like | ||
| Ostreococcus lucimarinus CCE9901 | OluNOS | XP_001421937 | 1059 aa | iNOS-like | ||
| Ostreococcus RCC809 | Os | OstRCC809_1 | 571 aa | p450 reductase Not NOS | ||
The most up-to-date references about individual NOS sequences can be found at http://www.ncbi.nlm.nih.gov using GenBank accession numbers. aa, amino acids. bp, base pairs. EST, expressed sequence tag. bbreviations and GenBank accession numbers were used for the Figures 2 and 3 for all trees.
Table 2.
The comparative characteristics of selected prokaryotic NOSs/NOS-like proteins*
| Prokaryotes: bacteria | ProteinName | Gene Bank Accession # | Length | Similarity to animal NOSs |
|---|---|---|---|---|
| Staphylococcus epidermidis | SeOx | NP_765153 | 355 aa | NOS oxygenase domain |
| Streptomyces sp. Mg1 | SsmOx | ZP_03175002 | 382 aa | NOS oxygenase domain |
| S. saprophyticus | SsOx | YP_300967 | 354 aa | NOS oxygenase domain |
| S. aureus | SaOx | P0A092 | 358 aa | NOS oxygenase domain |
| Oceanobacillus iheyensis | OiOx | NP_693612 | 369 aa | NOS oxygenase domain |
| Deinococcus radiodurans | DrOx | Q9RR97 | 356 aa | NOS oxygenase domain |
| D. geothermalis | DgOx | YP_603740 | 375 aa | NOS oxygenase domain |
| Bacillus subtilis | BsOx | O34453 | 336 aa | NOS oxygenase domain |
| B. halodurans | BhOx | NP_241689 | 366 aa | NOS oxygenase domain |
| B. anthracis | BaOx | ZP_00390385 | 356 aa | NOS oxygenase domain |
| B. thuringiensis | BtOx | ZP_00741647 | 215 aa | NOS oxygenase domain |
| B. thuringiensis | BtOx2 | YP_039435 | 356 aa | NOS oxygenase domain |
| B. clausii | BcOx | YP_174766 | 363 aa | NOS oxygenase domain |
| B. licheniformis | BlOx | YP_090413 | 365 aa | NOS oxygenase domain |
| B. cereus | BceOx | AAU20271 | 440 aa | NOS oxygenase domain |
| Geobacillus kaustophilus | GkOx | YP_147529 | 440 aa | NOS oxygenase domain |
| Exiguobacterium sibiricum | EsOx | ZP_00539087 | 366 aa | NOS oxygenase domain |
| Streptomyces avermitilis | SavOx | NP_822706 | 516 aa | NOS oxygenase domain |
| S. turgidiscabies | StOx | AAW49313 | 400 aa | NOS oxygenase domain |
| S. scabiei | SscOx | AAO53225 | 400 aa | NOS oxygenase domain |
| Archaea (halobacteria) | ||||
| Natronomonas pharaonis | NpOx | CAI49045 | 378 aa | NOS oxygenase domain |
The most up-to-date references about individual sequences can be found at http://www.ncbi.nlm.nih.gov or JGI using GenBank accession numbers. aa, amino acids (see also Figure 1 for phylogenetic relationships among these and eukaryotic NOSs).
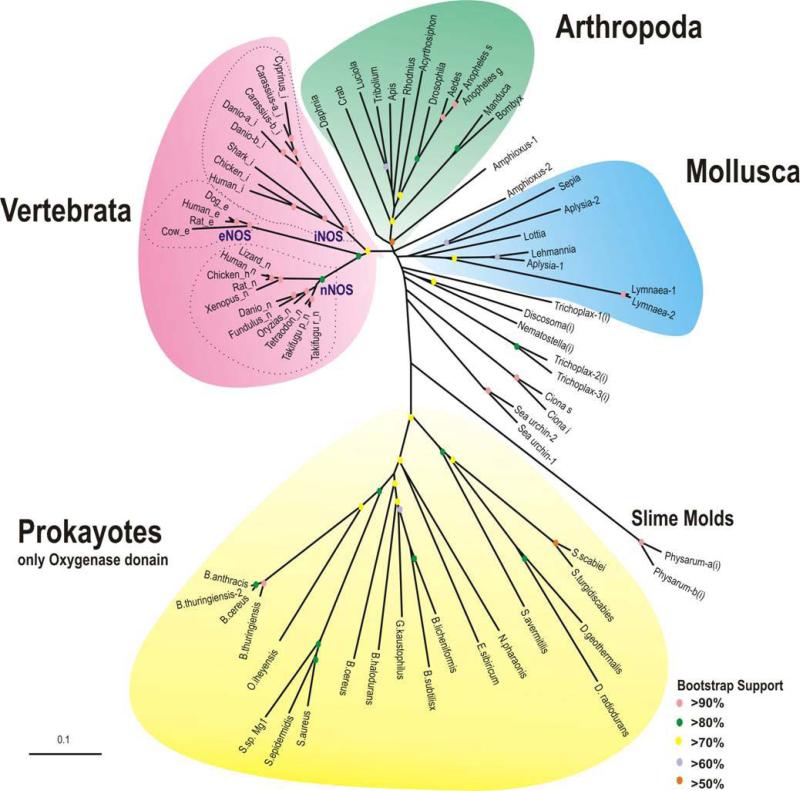
Figure 2.
Phylogeny of all known NOSs (eukaryotes and prokaryotes). Phylogram of NOS oxygenase domains (see Tables 1 and 2). Full-length amino acid sequences were aligned in “ClustalX” version 1.83 using default parameters (466); all gaps were manually removed then trimmed to include only the oxygenase domains, in “GeneDoc”(467).The unrooted phylogenetic tree was generated using default parameters and 10,000 iterations of the maximum likelihood algorithm implemented in the program “TREE-PUZZLE” (http://www.tree-puzzle.de). Numbers at branches represent bootstrap values for 10,000 iterations. The branch-length scale bar represents 0.1 amino acid substitutions per site. The colored dots correspond to the key code for bootstrap support. The graphic output was generated using “Treeview” (468). Bootstrap support is color coded on the figure and located at a node of each branch. See abbreviations for species and gene bank accession numbers for NOSs in Table 1 and 2; iNOS, vertebrate inducible like NOS; eNOS, vertebrate endothelial like NOS; nNOS, vertebrate neuronal-like NOS. The (i) represent invertebrate inducible like NOSs. Note both clustering of NOSs subtypes in major taxons as well as the basal position of iNOS-like proteins.
In all animal tissues NOS catalyzes the synthesis of NO according to the following reaction:
The nitrogen atom in NO comes from L-arginine and O comes from molecular oxygen.
Three NOS isoforms originally described in mammalian tissues are encoded by three distinct genes (Table 1). These three NOSs have been extensively characterized biochemically and their homologs were cloned from many mammalian species with numerous splicing variants (167, 323, 324). Two isoforms, neuronal NOS (nNOS; type I) and endothelial NOS (eNOS; type III), are Ca2+/calmodulin-dependent and are constitutive isoforms (i.e. isoforms permanently expressed in their target tissues (325, 326)). Ca2+ influxes associated with either ligand-gated, voltage activated Ca2+ channels or derived from internal stores up-regulate NOS activity and result in transient activation of NO synthesis. NO is locally released and acts on neighboring cells (e.g. smooth muscles or neuronal terminals).
On the other hand, inducible NOS (iNOS; type II) is a Ca2+-independent enzyme which normally cannot be detected in most tissues, but its expression is dramatically activated after appropriate stimulation (e.g. in the presence of lipopolysaccharides or in response to potentially damaging stimuli), resulting in a high and long-term NO yield. iNOS is primarily involved in defense reactions and cytotoxicity.
Structurally, all NOSs consist of two major domains: the oxygenase or catalytic domain (the N-terminal part of NOS with L-arginine, H4 biopterin and heme (Fe) binding sites) and the reductase domain (with FMN, FAD, and NADPH binding sites). The formation of dimers comprised of identical subunits is essential for NOS activity (324, 327, 328).
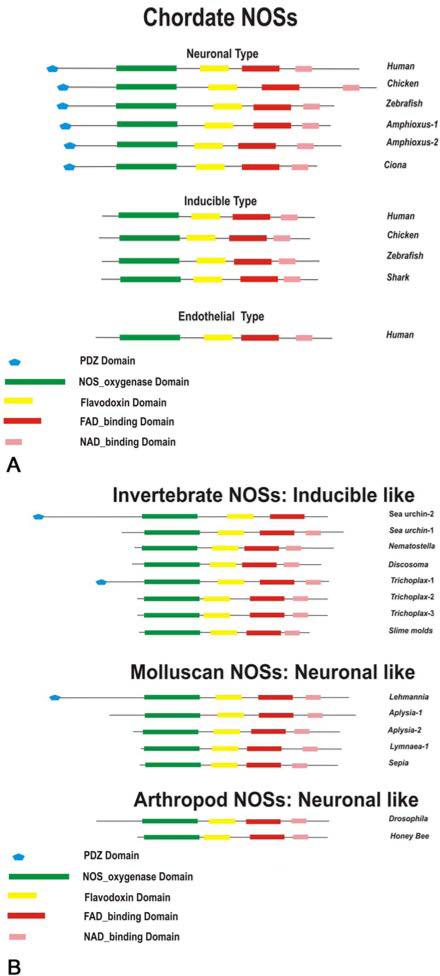
Figure 3 summarizes the domain organization of major animal and slime mold NOSs as multi-domain proteins (166). It is obvious that NOS can not be clearly classified in terms of mammalian/vertebrate subtypes and represents an extensive example of parallel evolution and adaptive radiation in different lineages. It is also clear that many basal metazoans such as Trichoplax have 3 different NOSs while more derived lineages such as insects have only one NOS. Combined with the topology of the presented NOS tree, these data imply multiple events of gene loss and gene gain in various lineages. Interestingly, only selected groups of invertebrate NOSs have PDZ domains. Further, their presence does not correlate with the phylogenetic positions, also suggesting an independent incorporation of this motif in the basic NOS structure. In contrast, all prokaryotic NOSs are “truncated” and consist of the oxygenase domain alone (168, 329, 330).
Figure 3.
Domain organization of Nitric Oxide Synthases (NOSs). See text and Table 1 for details and accession numbers. A. Chordate NOS domain organization. Three distinct types are observed neuronal, inducible and endothelial in chordates. Interestingly, only neuronal like NOSs have PDZ domain in their N-terminal. Note: the length of NOSs in the schematic illustrations correspond to the size of the NOS proteins. All domains were analyzed using SMART (a Simple Modular Architecture Research Tool) database (469) http://smart.embl-heidelberg.de/. B. Invertebrate NOS domain organization (see the next page).
Thus, the most likely scenario for the origin of eukaryotic NOSs is the fusion of two previously independent genes: one gene representing the oxygenase domain such as bacterial NOSs and the second gene encoding the reductase domain such as is found in cytochrome P450 reductase type enzymes. This fusion event might have occurred at the time of the origin of major eukaryotic groups (or at least in the common ancestor of animals and fungi) as a response to the rise of the oxygen concentration in Proterozoic time. It is quite possible that the origin of oxygen-dependent NO synthesis from L-arginine could be an adaptive response against emerging oxygen toxicity more than 2Gya. It is not surprising that this is the major molecular function of the precursor of the reductase domain in NOSs – cytochrome P450 (96). Classical NOS genes have not been found in any of the sequenced genomes from plants, suggesting that NOS was evolved after the split of plant and animal/fungal lineages. The alternative, more likely scenario is that land plants have lost this gene from a commong ancestor of all green plants including green algae. Incoming novel genomic information from basal plants and other eukaryotic lineages will help to solve this puzzle.
11. NOS IN INVERTEBRATES AND THEIR NERVOUS SYSTEMS
The first evidence for the synthesis and functional role of NO in invertebrates was provided in 1991 by Radomski using the basal arthropod, Limulus as a model (58). Independently, in 1992 Moroz and collaborators performed a global survey for the presence of NOS in several major invertebrate phyla confirming the wide-spread distribution NOS across the animal kingdom including molluscs and arthropods (54, 57). Using the freshwater gastropod mollusc, Lymnaea stagnalis, we have shown both the presence of enzymatic L-arginine/L-citrulline conversion and the role of NO as an activator of feeding motor patterns, as well as identifying large NOS-containing neurons (54, 56, 331, 332). In fact, the occurrence of truly gigantic (sometimes more 0.2 -0.5 mm) nitrergic (NOS-containing) neurons in molluscs, plus their reliable functional identification and mapping (55, 62, 83, 333, 334) present unique opportunities to study nearly all aspects of NO biology with subcellular resolution and in real physiological time. Apart from molluscs, only the relatively simpler nervous system of the medicinal leech (59, 60) and the salamander retina (335, 336) has been shown to posses large and individually identified classes of nitrergic cells. Today, the data about the distribution and function of NO in invertebrates is growing and covers a relatively large spectrum of organisms from jellyfish (61) and hydra (337-339) to fly (64) and parasitic worms (340, 341). However, the diversity of proposed functions of NO in invertebrates (reviewed by (63, 82, 342)) contrasts dramatically with the limited information available about the molecular biology and enzymology of NOS from different animal groups (343). The Drosophila NOS is the only enzyme characterized in sufficient detail from more than 30 invertebrate phyla (344, 345).
The first invertebrate NOS enzymes were cloned from insects (346-348) and molluscs (349-353); they all have greater overall sequence similarity to a constitutive neuronal-like NOS than to other isoforms (iNOS or eNOS) in vertebrates. At first glance, it might suggest that this type of NOS could be evolutionarily close to an “ancestral” NOS prototype. However, the recent discoveries of cnidarian and slime mold NOS as well as identification of novel NOS genes from recently sequenced genomes of prebilaterian metazons such as Trichoplax illuminate both the enormous diversity and parallel evolution of different NOS classes (Figure 2).
Cnidarian (Discosoma) and slime mold (Physarum - (354)) NOSs lack a distinct structural element that is present as an insertion in the reductase domains of constitutive NOSs but absent in iNOSs of vertebrates. This insert of ≈ 45 amino acids (residues 835 – 876 in human nNOS) is thought to be an autoinhibitory loop which impedes Ca2+-free CaM binding and enzymatic activation.
Since Discosoma NOS is structurally similar both to the only known non-animal conventional NOS and to vertebrate iNOS isoforms, the inducible type of the enzyme may be evolutionally basal for animal NOSs.
Novel NOSs found in sequenced genomes from basal animal lineages (Table 1) also share sequence features comparable to iNOS-like organization (i.e. without the insertion in the reductase domain). These consist of the only NOS in the Nematostella genome (sea anemone) and one of the three placazoan NOSs in the Trichoplax genome. iNOS-type might also present in Daphnia (355). We also found inducible-like NOS in recently sequenced genomes from the simple eukaryotes Ostreococcus tauri and O. lucimarinus. Ostreococcus belongs to the Prasinophyceae, an early-diverging class within the green plant lineage. Considering the lack of recognizable NOS in land plants (see section 10 below), the presence of a canonical iNOS-like gene in the basal plant lineage is especially interesting. It implies that an iNOS-like enzyme was present in earlier green plants but was lost in at least some higher plant lineages.
All of this information further supports the hypothesis that the inducible like (possibly Ca-independent) NOS is the ancestral prototype of the enzyme (see also Fig 2, 3). Under this scenario, multiple lineages of constitutive-like animal NOSs could be independently derived from iNOS-like proteins, in a process that might include independent events of insertion of the autoinhibitory loop in the NOS reductase domain. This insert provides coupling to Ca2+ regulatory mechanisms and reduces potentially toxic NO yields following the activation of inducible NOS.
It is interesting that, in contrast to vertebrate species which have three NOS genes, only one type of NOS isoform has been found in the genomes from insects and tunicates sequenced to date. Molluscs, sea urchins, and cephalochordates have at least two NOS genes but no NOS genes have been identified in nematodes (C.elegans) (Table 1 and Figure 2). This situation implies the very interesting possibility that more than one NOS co-existed in the common ancestor of all animals (urmetazoan) and then was lost in some animal taxa in the course of evolution. On the other hand, in some lineages such as molluscs (with at least two different types of NOS) and chordates (2-3 NOS genes) duplication events for NOS genes might happen more than once. For example duplication of the constitutive type NOS might occur in both deuterostomes and basal metazoans. Apparently, duplication events might also have happened independently in the evolution of inducible type NOS since some fishes have more than one iNOS-like gene (Table 1) and even basal Placozoa (Trichoplax adherens) has distinct iNOS-like groups.
Phylogram showing relatedness of the predicted oxygenase domains for NOSs is shown in Figure 2. As expected, the diversification of vertebrate NOSs occurs in parallel in multiple lineages and clusters into three distinct roups corresponding to mammalian iNOS, eNOS and nNOS, with the iNOS possibly being most basal. More likely eNOS is the latest innovation associated with the mammalian lineage. However, the diversity of all non-vertebrate iNOS is quite significant and iNOS in fishes might possess enzymatic and functional features distinct from those in mammals.
Interestingly, Molluscs, Arthropods, and Prokaryotes also show appropriate relatedness in their clusters with slime mold NOS as a potential out-group for all animals. NOSs from Placozoa, Cnidaria, echinoderms (the sea urchin) and basal chordates (amphioxus and two ascidians) seem to be the most derived and distinct NOS subtypes. It also appears that several classes of neuronal-type NOS from Deuterostomes (the sea urchin NOS and NOSs in all chordates) and molluscs as well as from Placozoa have developed the PDZ domain independently. This domain probably evolved to support specific subcellular targeting of NOS and formation of highly dynamic protein complexes, as shown in the postsynaptic regions of mammalian neurons.
Unfortunately, lack of detailed biochemical characterization and pharmacological profiles of such a diverse protein family as invertebrate NOSs is one of the major obstacles in the field. Future experiments using purified or expressed NOSs from various phyla is one of the crucial steps in our understanding of the evolution of NO signaling. Such an analysis might also provide valuable information for the biomedical industry as part of the development of more specific and efficient NOS inhibitors for different NOS subtypes. In summary, there is obvious evidence of parallel evolution of NOSs that occurs independently in major animal lineages. In practical terms of experimental design dealing with analysis of NO signaling in representatives of many invertebrate taxa while interpreting the pharmacology in terms of mammalian NOSs can be misleading.
12. DO NON-CONVENTIONAL L-ARGININE/NO SYNTHASES EXIST IN PLANTS AND ANIMALS?
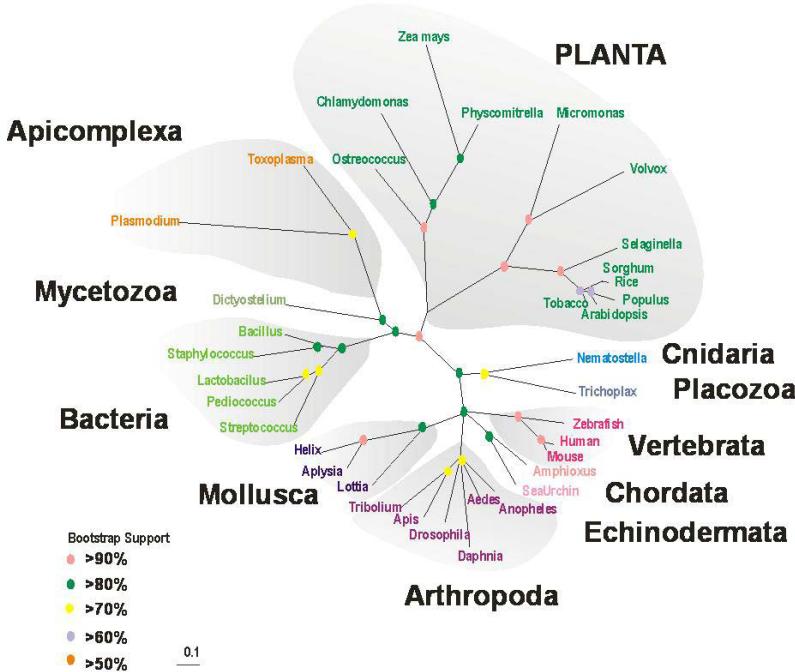
NOS-like activity (with the pharmacological and biochemical properties of mammalian NOS) was described in fungi (356) and has been reported in land plants (357-360). Proposed NO synthesis (via L-arginine to L-citrulline conversion) in some plant tissues is Ca2+-dependent (361-363), whereas in others it is Ca2+-independent (362), resembling the inducible type of NOS of mammals. However, conventional NOS-type enzymes from these species have neither been cloned nor purified. Although some green algae (e.g. Ostreococcus) have canonical NOS-type genes, a search of the sequenced genomes from land plants (e.g. Arabidopsis thaliana or Orysa) showed no recognizable animal-like NOS genes in their genomes.
This paradox has been partially resolved with a suggestion for the presence of non-conventional NOS in plants but with pharmacology similar to mammalian NOSs (reviewed by (78)). This proposal leads to an extensive literature about novel plant-like NOSs. Indeed, an Arabidopsis mutant was identified that had impaired NO production shown to be attributed to a gene which the authors name AtNOS1 (154). Expression of AtNOS1 on a viral promoter in the mutant plants resulted in overproduction of NO (154). This AtNOS1 shares highest identity with hypothetical NOS-related protein of the land pulmonate snail Helix (364), and the bacterial GTPase domain proteins. These authors expressed the cDNA from this hypothetical NOS, and showed a 15 fold increased in NOS-type activity in bacteria – a potentially convincing experiment confirming enzymatic activity of this snail protein.
To further ensure that this novel plant AtNOS1 gene also encoded an enzyme that had NOS activity, the AtNOS1 protein was expressed in bacteria. Again, Guo and collaborators showed elevated levels of NOS activity (154). These authors further characterized the purified protein and showed animal type NOS activity that is dependent on key co-factors (nicotinamide adenine phosphate, calmodulin, and Ca2+) and inhibited by L-NAME. These and additional data from other laboratories (365-368) resulted in two major conclusions: (i) confirmation that NO is an important signal molecule in plant physiology, and (ii) potential discovery of a novel class of NOSs with pharmacology similar to conventional NOSs from mammals but that is completely structurally different from all NOSs known from animals. The second conclusion was somewhat surprising for the field of classical NOS enzymology because of the lack of understanding of how NO synthesis in plants might occur with this protein.
Recently, putative homologs of such “plant-like” NOSs with a conserved GTP-ase domain have been described in virtually all eukaryotes and many bacteria (153); see also Table 3. None of the members of this family of proteins resemble any known conventional isoforms of NOSs, containing neither heme-oxygenase nor flavin reductase domains. Subsequently a mammalian homolog was cloned from mouse, and named mAtNOS1 (369). These authors constructed a fusion gene and expressed it in cell lines which showed the fusion protein localized to the inner-mitochondria compartment. Zemojtel et al. (2006) also suggested that the mAtNOS1 has a role in development of neural, hematopoietic and bone organ system (369, 370).
Table 3.
The comparative characteristics of small GTPases (AtNOS1/AtNOA1 Orthologs formally known as plant nitric oxide synthases, see text for details)
| Eukaryotic and prokaryotic AtNOS1/AtNOA1 orthologs* | |||
|---|---|---|---|
| Common names | Accession N | Length (aa) | |
| Class Mammalia | |||
| Homo sapiens | Human | BAC05262 | 623 aa |
| Pan troglodytes | Chimpanzee | XP_001136575 | 698 aa |
| Mus musculus | Mouse | Q99LH1 | 693 aa |
| Rattus norvegicus | Rat | EDL89873 | 694 aa |
| Monodelphis domestica | Opossum | XP_001369899 | 685 aa |
| Bos taurus | Cow | AAI09655 | 694 aa |
| Canis familiaris | Dog | XP_854207 | 700 aa |
| Class Aves (birds) | |||
| Gallus gallus | Chicken | XP_420579 | 566 aa |
| Class Actinopterygii | |||
| Danio rerio | Zebrafish | Xp_693129 | 702 aa |
| Class Cephalochordata | |||
| Branchiostoma floridae | Amphioxus | EEA52011 | 398 aa |
| Phylum Echinodermata | |||
| Strongylocentrotus purpuratus | Sea Urchin | XP_782008 | 761 aa |
| Phylum Nematoda | |||
| Caenorhabditis elegans | C. elegans | NP_001045614 | 388 aa |
| Phylum Arthropoda | |||
| Class Crustacea | |||
| Daphnia pulex | Daphnia | Dappu1|222051|estExt_fgenesh1_pg.C_70359 | 1162 aa |
| Class Insecta | |||
| Tribolium castaneum | Tribolium | XP_967515 | 709 aa |
| Apis mellifera | Honey bee | XP_396974 | 533 aa |
| Drosophila melanogaster | Fruit fly | NP_611297 | 624 aa |
| Anopheles gambiae | Mosquito | XP_319462 | 536 aa |
| Aedes aegypti | Mosquito | EAT40167 | 689 aa |
| Phylum Mollusca | |||
| Class Gastropoda | |||
| Aplysia californica | Sea slug | EF043280 | 712 aa |
| Helix pomatia | Land snail | CAA65719 | 433 aa |
| Lottia gigantea | Giant limpet | Lotgi1|164844| | 828 aa |
| Phylum Cnidaria | |||
| Nematostella vectensis | Nematostella | XP_001629200.1 | 501 aa (partial) |
| Phylum Placozoa | |||
| Trichoplax adhaerens | Trichoplax | EDV20812 | 659 aa |
| Phylum Mycetozoa | |||
| Dictyostelium discoideum | Dictyostelium | XP_640529 | 650 aa |
| Phylum Apicomplexa | |||
| Toxoplasma gondii ME49 | Toxoplasma | EEA98053 | 1036 aa |
| Plasmodium knowlesi strain H | Plasmodium | CAQ41547 | 824 aa |
| Planta | |||
| Arabidopsis thaliana | Arabidopsis | NP_850666 | 561 aa |
| Oryza sativa | Rice | NP_001045614 | 547 aa |
| Nicotiana benthamiana | Tobaco | BAF93184.1 | 568 aa |
| Sorghum bicolor | Sorghum | jgi|Sorbi1|3987737|gw1.4.296.1 | 508 aa (partial) |
| Selaginella moellendorffii | Selaginella | jgi|Selmo1|164227|estExt_Genewise1Plus.C_01842 | 600 aa |
| Volvox carteri | Volvox | jgi|Volca1|120437|estExt_fgenesh5_synt.C_50054 | 927 aa |
| Populus trichocarpa | Populus | jgi gw1.I.6621.1 | 577 aa (partial) |
| Physcomitrella patens | Physcomitrella | XP_001758508 | 456 aa (partial) |
| Zea mays | Zea mays | ACG28725.1 | 668 aa |
| Micromonas strain RCC299 | Micromonas | jgi|MicpuN2|84071|e_gw2.08.98 | 669 aa |
| Chlamydomonas reinhardtii | Chlamydomonas | XP_001700794.1 | 665 aa |
| Ostreococcus lucimarinus CCE9901 | Ostreococcus | XP_001418282.1 | 604 aa |
| Ostreococcus tauri | Ostreococcus | CAL53631 | 519 aa |
| Prokaryotes: bacteria | |||
| Lactobacillus plantarum WCFS1 | Lactobacillus | NP_785132l | 378 aa |
| Pediococcus pentosaceus ATCC 25745 | Pediococcus | ZP_00323186 | 370 aa |
| Bacillus anthracis str. A2012 | Bacillus | ZP_00394666 | 368 aa |
| Staphylococcus aureus subsp. aureus NCTC | Staphylococcus | YP_500210 | 366 aa |
| Streptococcus pneumoniae R6 | Streptococcus | NP_359186 | 368 aa |
The most up-to-date references about individual sequences can be found at http://www.ncbi.nlm.nih.gov or JGI using GenBank accession numbers. aa, amino acids
We also cloned one of the AtNOS1 homologs from the marine opisthobranch mollusc, Aplysia californica. AcAtNOS1 (GB# EF043280) has a deduced amino acid sequence of 712 residues. The putative AcAtNOS1 has a longer N terminus as do all the eukaryotic orthologs compared to their prokaryotic orthologs (153). We analyzed the structure of this putative AcAtNOS1 to predict the subcellular location of eukaryotic proteins using Target-P. This program uses N-terminal sequence information only to discriminate between proteins destined for the mitochondrion, the chloroplast, the secretory pathway, and “other” localizations with a success rate of 85% (plant) or 90% (non-plant) on redundancy-reduced test sets (371). Similarly to mammalian mAtNOS1, molluscan AcAtNOS1 contains a mitochondrial targeting peptide as predicted by Target-P with a score of 0.93 (a score of 1 indicates the strongest prediction) (371).
The summary phylogram showing relatedness of AtNOS-like proteins with the GTPase domains is shown in Figure 4. The clades of vertebrates, gastropods, insects, plants, and bacteria form four distinct clusters. As predicted, the molluscan AtNOSs are most closely related to each other. However, the molecular mechanisms of NO synthesis by these hypothetical plant-type NOSs are unresolved, suggesting that these proteins are not NO generating enzymes themselves.
Figure 4.
Phylogeny of Small GTPases (AtNOS1/AtNOA1 Orthologs formally known as plant nitric oxide synthases). This dendrogram shows relatedness of prokaryotic and eukaryotic GTPases (see text and Table 3 for detail). Full length amino acid sequences were aligned in ClustalX ver. 1.83 using default parameters (466). All gaps were manually removed to include only the GTPase-binding domain in GeneDoc. The unrooted phylogenetic tree was generated using default parameters and 10,000 iterations of the maximum likelihood algorithm implemented in the program “TREE-PUZZLE” (http://www.tree-puzzle.de). Numbers at branches represent bootstrap values for 10,000 iterations. The branch-length scale bar represents 0.1 amino acid substitutions per site. The colored dots correspond to the key code for bootstrap support.The graphic output was generated using Treeview (468). Bootstrap support is color coded on the figure and located at a node of each branch. See abbreviations for species and gene bank accession numbers for GTPases in Table 3.
It is likely that this protein can be an important component of a larger complex or chaperone indirectly contributing to cellular or systemic NO production both in plants and other eukaryotes. The most recent biochemical and structural data indicate that AtNOS itself does not bind L-arginine and synthesize NO (370, 372, 373). It was proposed to rename this class as NO associated protein or AtNOA1. Since the eukaryotic AtNOA1 can be an RNA binding protein, it might be involved in regulation and assembly of large molecular complexes associated with NO synthesis via alternative molecular mechanisms. One of the explanations of the earlier biochemical data about NO synthesis may be the fact that AtNOA1 is specifically targeted to mitochondria, and therefore might be a part of the process associated with mitochondrial ribosome biogenesis. Thus, down regulation or deletion of AtNOA1 might lead to the observed decrease in NO formation by mitochondria under hypoxic conditions (370). Moreau and collaborators suggested that the disregulation/disfunction of AtNOA1 might also result in the increase of reactive oxygen species (ROS) that can reduce NO accumulation, since NO can rapidly interact with .O -2 or related free radicals generated from the respiratory chain (372). Novel experimental data confirms that AtNOA1 is a slow Mg2+-/K+-dependent GTPase because it binds GDP more tightly than GTP. Thus AtNOA1 might require additional guanine nucleotide exchange factor(s) or GTPase-activating protein(s) to reach physiological relevant activity (372).
Bacterial homologs of AtNOA1 are associated with ribosome particles, and are involved in their biogenesis and local protein synthesis. Thus, they can act as G-proteins that modulate nucleic acid recognition; specifically, they serve as a molecular switch that couples GTP hydrolysis to nucleic acid and/or protein binding (373). The molecular function(s) of AtNOA1 in plants and animals still needs to be determined. Today, it can be broadly described as a long-term modulation of a ribosomal function based up on its assembly and subcellular localization in mitochondria. As a result, the observed changes in NO formation can have downstream consequences modulating cellular bioenergetics. Thus, earlier data from plants and Helix with identification of novel non-conventional NOS classes with with the GTPase domain need to be critically reevaluated.
13. THE ROLE OF NO IN DEVELOPMENT AND PLASTICITY: INSIGHTS FROM INSECTS AND SNAILS
13.1. Overview
In this last chapter we will briefly summarize the existing knowledge and challenges related to the analysis of NO in memory mechanisms and plasticity as it reflects the ultimate level of complexity of NO signaling. The role of NO in long-term plasticity is one of the emerging frontiers in biomedicine. It appears that virtually all mechanisms of NO action discovered in non-neuronal tissues and across species can be found to co-exist in a given neural circuit but the mechanisms of integration of different signaling pathways resulting in novel memory traces are obscure. Here, we will focus upon use of the relatively simpler nervous systems of invertebrates as promising experimental models in the field. A recent summary of ideas and controversies about the role of NO in memory mechanisms of vertebrates can be found elsewhere (374).
Before the current literature is reviewed, we would like to make a note about two levels of complexity that we face in the analysis of NO signaling mechanisms in nervous systems. As indicated in the previous sections, the emerging diversity of more than a dozen parallel NO synthetic and degradative pathways create the first level of complexity in analysis of NO signal transduction within highly heterogeneous cell populations, such as a human brain with more than 100 billion neurons and 100 trillion synaptic connections.
With a few exceptions (e.g. circulatory, digestive and immune systems in mammals), alternative sources of NO are rarely considered in present experimental designs to characterize neural functions or neurodevelopment. Indeed, dynamic reorganization of cell-cell interactions following differentiation, organogenesis, learning and memory requires precise timing in integration of internal and environmental stimuli, following even more critical timing in co-activation of underlying signal transduction cascades within the same or functionally coupled cells. NO is known to be critically involved in these integrative processes across all the animal phyla so far investigated, but the accurate dynamics of NO action in defined neural circuits is largely unknown. It is proposed that NO can act as a “true” volume transmitter acting in 3D space and providing precise spacial and temporal coupling within a localized group of nerve and effector cells or their processes. For example, as a volume transmitter localized NO release might modulate electrical activity or synaptic inputs within its diffusion range, and induce its own downstream chain of molecular events on multiple targets simultaneously, although with a different time-course (depending upon local concentration, redox state, pH, etc). Therefore, NO can functionally couple parallel or converging signal transduction pathways within specific neurons of a memory-forming circuit following associative and non-associative forms of learning.
The second level of complexity becomes apparent from observations that mechanisms of NO action are cell-/neuron-specific (i.e. non-identical in various neuronal types), and the way such integration (or coordinated activity of multiple neurons) is achieved is not clearly understood. Traditionally, in comparative neuroscience a validation of the presence of canonical NOS activity and involvement of NO/cGMP signaling are the two most familiar steps to test for the role of NO in a given process or circuit. Hitherto, using such biased approaches, the diversity of NO synthetic pathways in different components of neural circuits is frequently ignored. The widespread presence of other components involved in enzymatic and non-enzymatic nitrite reduction supports the existence of alternative (NOS-independent) sources of NO formation (Section 8) in a majority of biological tissues in general and nervous systems in particular. This knowledge is crucial to understanding the numerous contradictions in the field of NO-dependent mechanisms of synaptic plasticity. Nevertheless, we know very little about regional microenvironments in specific neurons and synapses.
As a result, the development of simpler experimental models is a vital step toward mechanistic understanding of NO signaling and its role in learning and memory. Although there is compelling evidence that NO is one of the key players in long-term plasticity mechanisms in mammals, there is no agreement about the contributions and mechanisms of NO action in memory-forming circuits. The enormous heterogeneity of neuronal populations, their complexity, and an insufficient knowledge about wiring diagrams in a brain are just a few of many gaps in our knowledge which have led to multiple controversies (see (374)). One of the major challenges in the use of vertebrate preparations is lack of identified and accessible neurons amenable for direct molecular and physiological analysis. In fact, nitrergic neurons are one of the least understood cell populations in our brain. In contrast, numerically simpler nervous systems of invertebrate animals offer unique prospects for reliable identification of nitrergic neurons and for characterization of NO-dependent signaling in memory mechanisms.
Historically, after the first discovery that NO is widely distributed in nervous systems of invertebrates (54, 56, 57, 331, 375), two lines of research were naturally established, focusing on the biology of NO in insects and gastropod molluscs respectively. More likely such a biased (from a comparative point of view) target selection is a reflection of the fact that both instects and gastropods are two of the largest animal groups and offer well established models. Plus, representatives of these two invertebrate classes became famous in the analysis of learning and memory mechanisms. Not surprisingly, for both groups it was demonstrated that NO controlled long-term plasticity; similar mechanisms are expected to be found for many other animal taxa.
Today, the neurobiology of NO is better studied in insects then in molluscs. Here the analysis of NO signaling is facilitated by the availability of a dozen sequenced genomes as well as by the unique opportunities offered by that unsurpassed genetically traceable model – Drosophila melanogaster. As in Drosophila, all studied insects have a single NOS gene with multiple splice forms; some of these might act as dominant negative regulators of functional NOS transcripts (376). In the insect brain, NOS is expressed in distinct neuronal populations including those associated with chemosensory structures (taste, olfaction) and the visual system (377-379). NO/cGMP pathways are shown to be involved in control of development (380-384), synaptic functions (385-387), and sensory processing (346, 388, 389). The relationship between NO signaling and long-term plasticity has been extensively studied in the honey bee, Apis melifera – a wonderful experimental paradigm for analysis of complex learned behaviors including social relationships in colonies.
13.2. NO signaling in the honey bee
The inhibition of NO synthesis impairs a distinct type of long-term memory and affects olfactory learning in the honey bee (390, 391). This type of associative learning properly occurs when an odor representing food stimulation (conditioned stimuli, CS) precedes an actual sucrose reward (unconditioned stimuli, US). In the honey bee, only three repetitive precisely paired CS/US trials induce a stable memory lasting more than 7 days (392). Even a single pairing, that lasts only a few seconds, induces an associative memory lasting more than 1-2 days. In Apis, as in many other models, prolonged activation of cAMP-dependent protein kinase A (PKA) is required for long-term memory formation (>3 days). Interestingly, a single CS/US pairing test leads a transient increase in the PKA activity lasting approximately one minute, while three repetitions of CS/US trials prolong PKA activation to more than 3 minutes. It is amazing that such relatively small 2 minute time differences in the same molecular event have such dramatic consequences, resulting in many days difference in the preservation of a long-term memory about a short olfactory stimulus. Clearly, it allows bees to quickly adjust their foraging strategy and social communications within a colony.
The possible molecular mechanisms of this type of NO-dependent associative learning have been investigated by Muller and collaborators (375, 377, 378, 391, 393, 394) and can be summarized as follows. One of the transmitters that mediates US and is involved in insect appetite learning is octopamine. It can induce a short term activation of PKA lasting several seconds via the cAMP system. If a single US is paired with an additional increase of cAMP from a caged compound, the prolonged activation of PKA is induced and a long-term memory (>3 days) is formed. The second messenger, apparently associated with a CS in the antennal lobes of the brain, can be NO. NO released from local AL interneurons (and possibly from sensory neurons) can also induce a prolonged activation of PKA but via the cGMP system, because the inhibition of both NOS and the soluble guanylyl cyclase (sGC) impair the prolonged activation of PKA. Thus, NO operates as a signal integrator and a volume/space transmitter in this system, temporally coupling at least two different processes and leading to long-term associated learning. Nevertheless, the precise molecular mechanisms of NO/cGMP mediated prolonged activation of PKA are still unknown. Muller and collaborators suggest that such an effect can be direct, although other mechanisms are not excluded (e.g. activation of cGMP dependent protein kinases, cGMP regulated phosphodiesterases and cyclic nucleotide gated channels (392).
Interestingly, habituation (a type of non-associative learning) of the same reflex can also be mediated by the NO/cGMP/PKA pathway but it follows different temporal dynamics and a different time-course of PKA activation in the same brain regions. For example, in habituation only 10 repetitive US result in detectable activation of PKA via the NO/cGMP cascade; whereas 3 paired CS/US trials are sufficient for a prolonged PKA activation. The release sites of NO might be very different during associative learning (e.g. interneurons) versus nonassociative learning or habituation (unknown), leading to different temporal dynamics of both NO synthesis and the scale of PKA activation. Considering the high-density packing of nitrergic terminals in the antennal lobe glomeruli with numerous NOS-negative postsynaptic cells, it would be reasonable to assume that NO can diffuse into these neurons then be oxidized to nitrites. Accumulated nitrites can serve as an additional dynamic NO storage or a mobile secondary pool for NO release by non-enzymatic mechanisms (section 8 of this manuscript). It would be a testable hypothesis that stronger and more repetitive stimulation following habituation or other type of training protocols might require alternative NOS-independent sources of NO.
In the honey bee NO can mediate additional functions (including cGMP independent signaling) in the same brain regions and even circuits. Surprisingly, global release of NO from a caged NO-donor in the antennal lobe impairs memory formation (i.e. it has an opposite effect compared to that induced by cGMP). It is likely that a very narrow time window exists for the NO/cGMP system to induce different types of long-term plasticity by recruiting parallel signaling cascades. Obviously, the interaction of concentration-dependence, spatial and temporal parameters of NO gradients, and cGMP dynamics are all crucial and can be coupled with alternative sources of NO formation and its inactivation. Other gaseous molecules (CO and H2S) might also be a part of cGMP/PKA signaling system.
The molecular processes triggered by prolonged activation of PKA are also unknown in the honey bee; they might involve transport of PKA to the nucleus (as found in Aplysia and other systems – (374, 395)) as well as interactions with other signal transduction cascades such as glutamatergic systems, Ca2+, PKC, unknown molecular events involved in satiation, etc. The emerging theme here, as in many other species, is the realization that memory formation is based upon multiple parallel (sometime redundant) molecular transduction pathways with hundreds of components organized in complex signal and gene-regulatory circuits with distinct temporal and spatial dynamics in various neurons. The complete sequencing of the Apis genome, correlated with well-characterized stereotyped and learned behaviors of this species, will further facilitate the analysis of gaseous signaling in learning and memory (379). Nevertheless, Apis has a very complex brain with about 1 million relatively small neurons and largely uncharacterized neuronal circuits. All of these are serious obstacles in the cellular analysis of behavior comparable with the challenges in the biology of vertebrate brain circuits.
13.3. NOS and NO signaling in molluscs
13.3.1. Overview
In contrast to insects and many other invertebrate lineages, representatives of two groups of gastropod molluscs (Opisthobranchia – sea slugs; and Pulmonata -freshwater and land air breathing snails) have truly gigantic neurons in their central nervous system (CNS). These cells can reach 0.5-1 mm in diameter with remarkable growth cones up to 0.6 mm (396, 397) - these are the largest neurons and neurites ever found in the animal kingdom. Gastropod neurons are located at the surface of ganglia and can be individually identified based upon their position, size, color, electrophysiological properties and synaptic connections. As a result, neural circuits controlling different stereotyped and learned behaviors have been identified in several model species, such as Aplysia, Lymnaea, Pleurobranchaea, Tritonia, Helix, Hermissenda, etc.. Naturally, these models have already provided the first insights in molecular deciphering of memory mechanisms at the level of individual neurons (395, 398-400). Furthermore, single identified molluscan neurons can be directly analyzed both biochemically (137, 146, 147, 401) and now at the genome scale (351, 397, 402, 403) leading to complete molecular characterization of all signal and gene regulatory pathways in a given characterized neuron within a circuit as the animal learns and remembers.
It is not surprising that the quest to find large identified NO releasing neurons in the CNS has been initiated in dozens of molluscan species (55, 56, 62, 83, 334), in parallel with characterization of the role of NO in behavior and memory mechanisms (404-406). The cephalopod molluscs were also investigated for the presence and function of NO signaling (83, 352, 407-417). Yet the progress in this field was quite modest, mostly due to limited NOS cloning efforts (NOSs were cloned only from three gastropod species and the cuttlefish: Aplysia (351, 418), Lymnaea (349) and Lehmannia valentiana (353), Sepia officinalis (419)); no sufficient mapping studies were performed where histochemical localization of NOS was accompanied by electrophysiological analysis to identify the position of nitrergic neurons in a given neuronal circuit. Thus, we will use only two illustrative examples (Lymnaea and Aplysia) that provide initial insights into the role and mechanisms of NO-dependent plasticity in molluscs.
13.3.2. NO signaling in Lymnaea
The fresh-water snail, Lymnaea stagnalis was the first invertebrate species where both histochemical and functional effects of NO were tested. It was shown that a significant fraction of nitrergic neurons is located in the buccal ganglia and that NO activates buccal motor patterns (331). A pair of symmetrical giant nitrergic neurons has been identified as esophageal B2 motoneurons with homologs in related species Helisoma and Biomphalaria (55, 56, 147, 332). In addition, it was found that numerous nitrergic (56) neurons are located at the periphery, including gut and chemosensory structures (420). In general, a similar pattern is observed for other molluscs, with detectable trends that the localization of NOS containing neurons is linked to feeding and chemosensory structures (62, 68, 82, 333, 334). Functional tests further confirmed that NO can be an important endogenous modulator of feeding in all molluscs tested so far (e.g. (62, 331, 421).
Considering the fact that feeding behavior can be highly adaptive and modified by various stimuli, it was reasonable to assume that NO might be involved in various forms of short and long-term plasticity within these circuits. Indeed, it was shown that inhibitors of NOS in Lymnaea disturb single trail training in Lymnaea and that apparently NO/cGMP signaling and plasticity can be coupled with PKA (422), similar to the events described earlier in the honey bee (392). In the Lymnaea feeding system, NO can also act within a defined time window (10 min to 6 hrs postraining) to promote long-term memory formation (406, 422). The requirement for NO/soluble guanylyl cyclase (sGC) is wider than the time window for local protein synthesis and PKA (up to 2 hrs posttraining, see (406, 422)). Importantly, protein kinase G (PKG), a potential downstream target of NO, is not involved in earlier stages of memory consolidation (< 2hrs). Instead it is required for later memory consolidation processes (between 2 and 8 hrs) and, therefore, overlaps with the critical window of NO/sGC action in the system. Specific linkages between NO, PKA, MAPK and PKG signaling or other memory related pathways have not been determined.
Unfortunately, the precise localization of specific sources of NO in the feeding circuit which are associated with long-term memory in Lymnaea is unknown. Apart from the buccal B2 motoneurons that we identified as nitrergic cells (56, 147, 331, 332), other NO-releasing neurons are not characterized. Recent data provided by Korneev et. al. (349, 423, 424) suggest that one of the key serotonergic modulatoty neurons in the circuit CGC can be a “silent” nitrergic cell where NOS translation and subsequent enzymatic NOS activity may not be detected in control preparations but may be activated during training or memory consolidation. One of the mechanisms can be transcriptional regulation of NOS by antisense transcripts (423, 424). However, neither biochemical nor histochemical evidence confirms the presence of NOS-dependent NO synthesis in these neurons (147). Thus, although NO is apparently involved in long-term memory in Lymnaea further investigations are needed to determine the contribution of enzymatic or non-enzymatic sources of NO formation. It will be crucial to identify NO-dependent signal transduction pathways as well as the role, diversity, relative contribution and regulation of NOS antisense transcripts. It will also be important to identify novel NOS isoforms (e.g. see (425)), in addition to previously reported truncated NOSs, in the CNS of Lymnaea generally and in the feeding circuit in particular. Interestingly, Lymnaea has NMDA like receptors (426, 427) and it would be important to estimate their contribution in NO-dependent plasticity. This was demonstrated in mammals using long-term potentiation and long-term depression paradigms.
13.3.3. NO signaling in Aplysia
The second molluscan experimental model developed for analysis of learning and memory mechanisms (395, 398) is the sea slug Aplysia californica. Although Aplysia has two NOS genes, only one isoform (NOS-1; GB# AAK83069) is expressed in the CNS in about a hundred NOS-1 containing neurons that are primarily located in the buccal, cerebral and pedal ganglia. NOS-1 is not expressed in any known sensory or motor neuron, and it appears that the majority of NOS-containing cells are interneurons. The abdominal ganglia have only 3 NOS-containing cells (L29 interneurons) making the gill-withdrawal reflex circuit one of the simplest in terms of composition and number of identified NOS containing neurons. Moreover, because NOS-1 is localized in the CNS, it was possible to characterize this enzyme biochemically, confirming that as with mammalian neuronal NOS, it is a NADPH-, Ca2+-calmodulin-dependent enzyme and that NOS activity can be suppressed by a variety of mammalian NOS inhibitors (428).
In this species, as in Lymnaea, the various components of the feeding system are also richly innervated by putative nitrergic neurons including well-recognized glomeruli (333, 429). It was also shown that NO is a powerful modulator of feeding patterns. In Aplysia, NO elicited normal appetitive and consummatory behaviors leading to the deposition of cordons containing egg capsules without eggs (430). NO is also involved in conditioning feeding behavior and mediates the process determining if food is inedible (404, 405). More than a dozen neurons are located in the buccal ganglia but their precise identification and their contribution to the feeding program and plasticity are unknown. Apparently feeding can be quite a complex behavior for cellular analysis of NO signaling, with a widely distributed neural circuit and the presence of numerous nitrergic neurons and mechanisms in both buccal and cerebral ganglia (293, 333, 431-435).
In contrast, the Aplysia gill withdrawal reflex is a relatively simple behavior and the underlying neural circuit is widely used for cellular analysis of two types of memory paradigms – sensitization and plasticity (395, 398, 436). The Hawkins and Moroz groups had therefore investigated the possible role of the extracellular signaling molecule NO in facilitation during behavioral conditioning in the siphon-withdrawal preparation (418). The facilitation is produced in part by L29 interneurons (437, 438), but the facilitatory transmitter of the L29 neurons had not previously been identified. We first used in situ hybridization to show that the L29 neurons express NO synthase (NOS) and that exogenous NO produces facilitation of sensory-motor neuron EPSPs. Next, we found that an inhibitor of NO synthase or an NO scavenger blocked behavioral conditioning. Furthermore, application of the scavenger to the ganglion or injection into a sensory neuron blocked facilitation of the EPSP and changes in the sensory neuron membrane properties during conditioning. Injection of the scavenger into the motor neuron reduced facilitation without affecting sensory neuron membrane properties, and injection of an inhibitor of NO synthase had no effect (418). These results indicate that NO makes an important contribution to classical conditioning and facilitation, and acts directly in both the sensory and motor neurons to affect different processes of facilitation at the synapses between them. Those results are also consistent with the idea that NO acts as a local “volume messenger” in the circuit for the reflex. In addition, our data indicate that NO does not come from either the sensory or motor neurons but rather comes from another source, perhaps the L29 interneurons (see Figure 5).
Figure 5.
A simplified diagram of the neural circuit controlling the gill-withdrawal reflex in Aplysia with the position of NOS containing cells as a subpopulation of facilitatory interneurons (cells L29). Transmitters of the second subgroup of L29 interneurons are currently unknown; one of them may be the molluscan small cardioactive peptide (SCP, a neuropeptide). VC and LE are mechanosensory sensory neurons with glutamate and sensorin as co-transmitters (see details in (470)). LFs are motor neurons located in the abdominal ganglion. CB1 are serononin containing interneurons located in the cerebral ganglion. The dotted lines represent polysynaptic connections. US - unconditioned stimulus and CS - conditioned stimulus used for the classical conditioning in Aplysia (see (418, 442) and the text for additional details).
Previous experiments in Aplysia (439-443) suggested that facilitation of sensory-motor neuron EPSPs during conditioning of the gill-withdrawal reflex involves at least two distinguishable processes – one that is associated with changes in the membrane properties of the sensory (LE) neurons, including spike broadening, and one or more processes that are independent of those changes and may include presynaptic vesicle mobilization (444) or insertion of postsynaptic receptors (445, 446). All of these effects can be produced by serotonin (5-HT - the best studied transmitter known to induce long-term plasticity in this model) acting on pre- and postsynaptic receptors. Antonov et al. (2007) found (418) that NO may be involved in all of these effects as well: (1) both the facilitation and changes in LE membrane properties during conditioning are blocked by injection of an NO scavenger into the LE neuron, (2) the facilitation but not the changes in LE membrane properties is also reduced by injection of an NO scavenger into the motor (LFS) neuron, (3) application of an NO donor produces facilitation but not changes in LE membrane properties, and (4) facilitation by an NO donor is blocked by injection of an NO scavenger into the sensory LE neuron but not the motor LFS neuron. These results suggest that NO is both necessary and sufficient for a membrane property independent mechanism of facilitation, such as vesicle mobilization, in the LE neurons. However, whereas 5-HT acting through presynaptic PKA is thought to be both necessary and sufficient for changes in the LE membrane properties (439, 442), NO is necessary but not sufficient – that is, an NO donor alone does not produce those changes. Likewise, Hawkins and collaborators showed that NO is necessary but not sufficient for another mechanism of facilitation, such as receptor insertion, in the motor LFS neurons.
13.3.4. Molecular targets of NO in Aplysia
The evolutionarily conservative NO-cGMP signaling is present and widely distributed in the CNS of Aplysia (447-449). We have found biochemically (428) that NO increases cGMP production by activating soluble guanylyl cyclase (sGC); ODQ, a specific inhibitor of sGC, eliminated this effect while inhibitors of phosphodiesterases enhanced it. Surprisingly, when we mapped the location of sGC subunits in Aplysia, we did not find any labeling in mechanosensory neurons (Moroz and Bodnarova, unpublished observations). In contrast, sGC is abundant in motor neurons including those involved in the gill-/siphon withdrawal reflex. Recent results (418) also indicate that classical conditioning is blocked by bathing the ganglion with either an NOS inhibitor, an NO scavenger, or an inhibitor of PKG suggesting that, like in associative learning in Lymnaea, PKG acts as one of the downstream targets of NO in the classical conditioning of gill-withdrawal in Aplysia.
Large-scale unbiased searches for potential molecular targets of NO in the CNS of Aplysia and related species can be achieved using transcriptome profiling with microarrays (351, 397) or by direct sequencing (351). As a result, we have identified and cloned several genes related to the NO transduction cascade (Kohn, Bodnarova, Moroz, unpublished data) such as PIN (protein inhibitor of NOS), CAPON (a NOS-associated chaperon in cGMP-independent signaling), and Eip75B (a nuclear hormone type of receptor for NO). Eip75B is the Aplysia homolog of a heme-containing nuclear receptor found in Drosophila shown to be activated by NO and CO (450); this is an interesting NO target and possibly a direct link to transcriptional cGMP-independent regulation. As for downstream cGMP-dependent NO targets in the CNS of Aplysia, we found cyclic nucleotide phosphodiesterases, subunits of sGC (only in motor neurons so far) and at least one type of PKG (GB# AY362340.1) (in both sensory and motor neurons). One of the most interesting components of NO-signaling is a class of hyperpolarization-activated cyclic nucleotide gated channels (HCN, GB# AAX98669). In general, HCN expression in the CNS overlaps with NO sensitive sGC (Moroz and Kuzyk, unpublished observations). We did not detect their expression in the sensory neurons but they are quite abundant in motor neurons, suggesting that HCNs are involved in the NO/cGMP cascade in motor neurons and are associated with their electrical activity. Surprisingly, we did not detect heme-oxygenase 2 type activity (known to produce the second gaseous messenger - CO) in central ganglia, and apparently CO-mediated signaling is not involved in neuronal signaling in Aplysia (Moroz, Kohn, unpublished observations). Finally, we have identified and cloned from the CNS several related components of signal transduction, including 2 isoforms of CaMK II and 8 iGluR receptors including NMDA receptors (426). Some of these were also independently cloned by the Glanzman/Martin laboratories, bringing the total number of iGluRs involved in sensory-motor synapses to 15 (Kohn, Moroz, unpublished observations). It is known that NMDA receptors have a critical role in learning mechanisms (as coincident detectors) and can also be direct targets of NO in mammals (451-453) suggesting that similar mechanisms of modulation of long-term plasticity might exist in Aplysia as well.
Collectively, the outlined findings from a variety of invertebrate preparations point out that NO acts as an important messenger in learning and memory mechanisms. The developed model systems and the ongoing genome projects open unique opportunities to characterize molecular mechanisms of interactions of NO with other known transmitter systems as well as to study genomic bases of associative and non-associative forms of learning and memory with resolution that is difficult to achieve elsewhere. For example, since giant Aplysia neurons are well-suited for direct genomic and epigenomic profiling (351, 397, 454, 455) one can monitor transcriptional and methylome output of the entire genome of individually identified neurons as they sense NO or learn and remember. It was recently discovered that NO controls chromatin reorganization (456-463) - a fact that provides the mechanistical bases for molecular analysis of chromatin remodeling and the deciphering of the logic of gene regulation during long-term plasticity.
14. SUMMARY AND PERSPECTIVE
It appears that life itself could not have evolved on our planet without NO as a critical intermediate of nitrogen fixation in biological systems. Indeed, it is a long way from the diversity of pathways for nitrogen fixation at the dawn of biological evolution to the present day diversity of NO functions in memory mechanisms; from the abiotic synthesis of NO by lighting to its production now by multiple NOS isoforms and other components of NO synthesis. Many gaps still exist in our understanding of the role of this ubiquitous but enigmatic signal molecule. The growth of genomic and molecular information about comparative NO biology revealed an unexpected complexity of synthetic enzymatic and non-enzymatic pathways in virtually all studied systems, from plants to animals, with dozens of novel proteins and low molecular weight redox compounds. Many of these pathways co-exist within the same cells and tissues, providing molecular bases for homeostatic regulation of NO microenvironments and signaling under normoxic and hypoxic conditions. Although comparative data are still incomplete, it is obvious that classification of NOS based upon three mammalian NOSs types needs to be reevaluated. Apparently, there are multiple examples of parallel evolution of NOS in different lineages with the inducible type NOS as probably the most basal ancestral prototype.
The obvious next step to advance the field is cloning of NOS from all major invertebrate groups and phyla, especially from non-metazoan taxa, followed by enzymatic and functional characterization of the expressed proteins. Significant discoveries can also be expected in the analysis of alternative NOS independent pathways, the role of NO in ecological adaptations and evolution, and the contribution of NO to symbiotic interactions. Although the canonical NO/sGC/cGMP/cGK pathway seems to be present in many invertebrate lineages, novel molecular targets of NO are emerging. The molecular analysis of memory and learning mechanisms in a variety of invertebrate preparations (most notably in insects and gastropod molluscs) opens conceptually different perspectives to understand the logic of recruiting evolutionarily conserved pathways for novel functions. Giant uniquely identified cells and recently sequenced genomes from Aplysia and related species open unprecedented opportunities for integrative analysis of NO signaling at the single cell level and in defined neural circuits or behaviors.
ACKNOWLEDGEMENTS
This research was supported by NIH (P50HG002806 and RO1NS06076, R21RR025699, R21DA030118), NSF (0744649) and McKnight Brain Research Fooundation grants to LLM. We thank Jim Netherton for his comments and technical support of the work and R. Hawkins for discussion of NO related plasticity mechanisms in Aplysia and insights into the neuronal organization of the gill-withdrawal reflex.
Abbreviations
- NO
nitric oxide
- NOS
nitric oxide synthase
- sGC
soluble guanylyl cyclase
- CNS
central nervous system
- PKA
cAMP-dependent protein kinase A
REFERENCES
- 1.Somerville C. The twentieth century trajectory of plant biology. Cell. 2000;100:13–25. doi: 10.1016/s0092-8674(00)81680-3. [DOI] [PubMed] [Google Scholar]
- 2.Thimann KV. Fifty Years of Plant Hormone Research. Plant Physiol. 1974;54:450–453. doi: 10.1104/pp.54.4.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang C, Shockey JA. The ethylene-response pathway: signal perception to gene regulation. Curr Opin Plant Biol. 1999;2:352–8. doi: 10.1016/s1369-5266(99)00004-7. [DOI] [PubMed] [Google Scholar]
- 4.Chow B, McCourt P. Plant hormone receptors: perception is everything. Genes Dev. 2006;20:1998–2008. doi: 10.1101/gad.1432806. [DOI] [PubMed] [Google Scholar]
- 5.Benavente LM, Alonso JM. Molecular mechanisms of ethylene signaling in Arabidopsis. Mol Biosyst. 2006;2:165–73. doi: 10.1039/b513874d. [DOI] [PubMed] [Google Scholar]
- 6.van Loon LC, Geraats BP, Linthorst HJ. Ethylene as a modulator of disease resistance in plants. Trends Plant Sci. 2006;11:184–91. doi: 10.1016/j.tplants.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Hedrick MS, Chen AK, Jessop KL. Nitric oxide changes its role as a modulator of respiratory motor activity during development in the bullfrog (Rana catesbeiana) Comp Biochem Physiol A Mol Integr Physiol. 2005;142:231–40. doi: 10.1016/j.cbpb.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Chang C, Bleecker AB. Ethylene biology. More than a gas. Plant Physiol. 2004;136:2895–9. doi: 10.1104/pp.104.900122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kendrick MD, Chang C. Ethylene signaling: new levels of complexity and regulation. Curr Opin Plant Biol. 2008;11:479–85. doi: 10.1016/j.pbi.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu Z, Guo H. Genetic basis of ethylene perception and signal transduction in Arabidopsis. J Integr Plant Biol. 2008;50:808–15. doi: 10.1111/j.1744-7909.2008.00710.x. [DOI] [PubMed] [Google Scholar]
- 11.Krasko A, Schroder HC, Perovic S, Steffen R, Kruse M, Reichert W, Muller IM, Muller WE. Ethylene modulates gene expression in cells of the marine sponge Suberites domuncula and reduces the degree of apoptosis. J Biol Chem. 1999;274:31524–30. doi: 10.1074/jbc.274.44.31524. [DOI] [PubMed] [Google Scholar]
- 12.Perovic S, Seack J, Gamulin V, Muller WE, Schroder HC. Modulation of intracellular calcium and proliferative activity of invertebrate and vertebrate cells by ethylene. BMC Cell Biol. 2001;2:7. doi: 10.1186/1471-2121-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seack J, Perovic S, Gamulin V, Schroder HC, Beutelmann P, Muller IM, Muller WE. Identification of highly conserved genes: SNZ and SNO in the marine sponge Suberites domuncula: their gene structure and promoter activity in mammalian cells (1) Biochim Biophys Acta. 2001;1520:21–34. doi: 10.1016/s0167-4781(01)00246-9. [DOI] [PubMed] [Google Scholar]
- 14.Muller WE, Ushijima H, Batel R, Krasko A, Borejko A, Muller IM, Schroder HC. Novel mechanism for the radiation-induced bystander effect: nitric oxide and ethylene determine the response in sponge cells. Mutat Res. 2006;597:62–72. doi: 10.1016/j.mrfmmm.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 15.Marks GS, Brien JF, Nakatsu K, McLaughlin BE. Does carbon monoxide have a physiological function? Trends Pharmacol Sci. 1991;12:185–8. doi: 10.1016/0165-6147(91)90544-3. [DOI] [PubMed] [Google Scholar]
- 16.Verma A, Hirsch DJ, Glatt CE, Ronnett GV, Snyder SH. Carbon monoxide: a putative neural messenger. Science. 1993;259:381–4. doi: 10.1126/science.7678352. [DOI] [PubMed] [Google Scholar]
- 17.Dawson TM, Snyder SH. Gases as biological messengers: nitric oxide and carbon monoxide in the brain. J Neurosci. 1994;14:5147–59. doi: 10.1523/JNEUROSCI.14-09-05147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boehning D, Sedaghat L, Sedlak TW, Snyder SH. Heme oxygenase-2 is activated by calcium-calmodulin. J Biol Chem. 2004;279:30927–30. doi: 10.1074/jbc.C400222200. [DOI] [PubMed] [Google Scholar]
- 19.Boehning D, Snyder SH. Novel neural modulators. Annu Rev Neurosci. 2003;26:105–31. doi: 10.1146/annurev.neuro.26.041002.131047. [DOI] [PubMed] [Google Scholar]
- 20.Gelperin A, Flores J, Raccuia-Behling F, Cooke IR. Nitric oxide and carbon monoxide modulate oscillations of olfactory interneurons in a terrestrial mollusk. J Neurophysiol. 2000;83:116–27. doi: 10.1152/jn.2000.83.1.116. [DOI] [PubMed] [Google Scholar]
- 21.Kim HP, Ryter SW, Choi AM. CO as a cellular signaling molecule. Annu Rev Pharmacol Toxicol. 2006;46:411–49. doi: 10.1146/annurev.pharmtox.46.120604.141053. [DOI] [PubMed] [Google Scholar]
- 22.Leffler CW, Parfenova H, Jaggar JH, Wang R. Carbon monoxide and hydrogen sulfide: gaseous messengers in cerebrovascular circulation. J Appl Physiol. 2006;100:1065–76. doi: 10.1152/japplphysiol.00793.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryter SW, Alam J, Choi AM. Heme oxygenase-1/carbon monoxide: from basic science to therapeutic applications. Physiol Rev. 2006;86:583–650. doi: 10.1152/physrev.00011.2005. [DOI] [PubMed] [Google Scholar]
- 24.Ryter SW, Morse D, Choi AM. Carbon monoxide: to boldly go where NO has gone before. Sci STKE. 2004;2004:RE6. doi: 10.1126/stke.2302004re6. [DOI] [PubMed] [Google Scholar]
- 25.Ryter SW, Otterbein LE. Carbon monoxide in biology and medicine. Bioessays. 2004;26:270–80. doi: 10.1002/bies.20005. [DOI] [PubMed] [Google Scholar]
- 26.Piantadosi CA. Carbon monoxide, reactive oxygen signaling, and oxidative stress. Free Radic Biol Med. 2008;45:562–9. doi: 10.1016/j.freeradbiomed.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bilban M, Haschemi A, Wegiel B, Chin BY, Wagner O, Otterbein LE. Heme oxygenase and carbon monoxide initiate homeostatic signaling. J Mol Med. 2008;86:267–79. doi: 10.1007/s00109-007-0276-0. [DOI] [PubMed] [Google Scholar]
- 28.Syapin PJ. Regulation of haeme oxygenase-1 for treatment of neuroinflammation and brain disorders. Br J Pharmacol. 2008;155:623–40. doi: 10.1038/bjp.2008.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watkins CC, Boehning D, Kaplin AI, Rao M, Ferris CD, Snyder SH. Carbon monoxide mediates vasoactive intestinal polypeptide-associated nonadrenergic/noncholinergic neurotransmission. Proc Natl Acad Sci U S A. 2004;101:2631–5. doi: 10.1073/pnas.0308695100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boehning D, Snyder SH. Circadian rhythms. Carbon monoxide and clocks. Science. 2002;298:2339–40. doi: 10.1126/science.1080339. [DOI] [PubMed] [Google Scholar]
- 31.Xue L, Farrugia G, Miller SM, Ferris CD, Snyder SH, Szurszewski JH. Carbon monoxide and nitric oxide as coneurotransmitters in the enteric nervous system: evidence from genomic deletion of biosynthetic enzymes. Proc Natl Acad Sci U S A. 2000;97:1851–5. doi: 10.1073/pnas.97.4.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burnett AL, Johns DG, Kriegsfeld LJ, Klein SL, Calvin DC, Demas GE, Schramm LP, Tonegawa S, Nelson RJ, Snyder SH, Poss KD. Ejaculatory abnormalities in mice with targeted disruption of the gene for heme oxygenase-2. Nat Med. 1998;4:84–7. doi: 10.1038/nm0198-084. [DOI] [PubMed] [Google Scholar]
- 33.Abe K, Kimura H. The possible role of hydrogen sulfide as an endogenous neuromodulator. J Neurosci. 1996;16:1066–71. doi: 10.1523/JNEUROSCI.16-03-01066.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stipanuk MH, Dominy JE, Jr., Lee JI, Coloso RM. Mammalian cysteine metabolism: new insights into regulation of cysteine metabolism. J Nutr. 2006;136:1652S–1659S. doi: 10.1093/jn/136.6.1652S. [DOI] [PubMed] [Google Scholar]
- 35.Kimura H. Hydrogen sulfide as a neuromodulator. Mol Neurobiol. 2002;26:13–9. doi: 10.1385/MN:26:1:013. [DOI] [PubMed] [Google Scholar]
- 36.Lowicka E, Beltowski J. Hydrogen sulfide (H2S) - the third gas of interest for pharmacologists. Pharmacol Rep. 2007;59:4–24. [PubMed] [Google Scholar]
- 37.Papenbrock J, Riemenschneider A, Kamp A, Schulz-Vogt HN, Schmidt A. Characterization of cysteine-degrading and H2S-releasing enzymes of higher plants - from the field to the test tube and back. Plant Biol (Stuttg) 2007;9:582–8. doi: 10.1055/s-2007-965424. [DOI] [PubMed] [Google Scholar]
- 38.Qu K, Lee SW, Bian JS, Low CM, Wong PT. Hydrogen sulfide: neurochemistry and neurobiology. Neurochem Int. 2008;52:155–65. doi: 10.1016/j.neuint.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 39.Szabo C. Hydrogen sulphide and its therapeutic potential. Nat Rev Drug Discov. 2007;6:917–35. doi: 10.1038/nrd2425. [DOI] [PubMed] [Google Scholar]
- 40.Eto K, Ogasawara M, Umemura K, Nagai Y, Kimura H. Hydrogen sulfide is produced in response to neuronal excitation. J Neurosci. 2002;22:3386–91. doi: 10.1523/JNEUROSCI.22-09-03386.2002. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Hosoki R, Matsuki N, Kimura H. The possible role of hydrogen sulfide as an endogenous smooth muscle relaxant in synergy with nitric oxide. Biochem Biophys Res Commun. 1997;237:527–31. doi: 10.1006/bbrc.1997.6878. [DOI] [PubMed] [Google Scholar]
- 42.Kimura H, Nagai Y, Umemura K, Kimura Y. Physiological roles of hydrogen sulfide: synaptic modulation, neuroprotection, and smooth muscle relaxation. Antioxid Redox Signal. 2005;7:795–803. doi: 10.1089/ars.2005.7.795. [DOI] [PubMed] [Google Scholar]
- 43.Nagai Y, Tsugane M, Oka J, Kimura H. Hydrogen sulfide induces calcium waves in astrocytes. Faseb J. 2004;18:557–9. doi: 10.1096/fj.03-1052fje. [DOI] [PubMed] [Google Scholar]
- 44.Yang G, Wu L, Jiang B, Yang W, Qi J, Cao K, Meng Q, Mustafa AK, Mu W, Zhang S, Snyder SH, Wang R. H2S as a physiologic vasorelaxant: hypertension in mice with deletion of cystathionine gamma-lyase. Science. 2008;322:587–90. doi: 10.1126/science.1162667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhatia M. Hydrogen sulfide as a vasodilator. IUBMB Life. 2005;57:603–6. doi: 10.1080/15216540500217875. [DOI] [PubMed] [Google Scholar]
- 46.Chahl LA. Hydrogen sulphide: an endogenous stimulant of capsaicin-sensitive primary afferent neurons? Br J Pharmacol. 2004;142:1–2. doi: 10.1038/sj.bjp.0705765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fiorucci S, Distrutti E, Cirino G, Wallace JL. The emerging roles of hydrogen sulfide in the gastrointestinal tract and liver. Gastroenterology. 2006;131:259–71. doi: 10.1053/j.gastro.2006.02.033. [DOI] [PubMed] [Google Scholar]
- 48.Li L, Bhatia M, Moore PK. Hydrogen sulphide--a novel mediator of inflammation? Curr Opin Pharmacol. 2006;6:125–9. doi: 10.1016/j.coph.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 49.Tang C, Li X, Du J. Hydrogen sulfide as a new endogenous gaseous transmitter in the cardiovascular system. Curr Vasc Pharmacol. 2006;4:17–22. doi: 10.2174/157016106775203144. [DOI] [PubMed] [Google Scholar]
- 50.Ebrahimkhani MR, Mani AR, Moore K. Hydrogen sulphide and the hyperdynamic circulation in cirrhosis: a hypothesis. Gut. 2005;54:1668–71. doi: 10.1136/gut.2004.056556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elsey DJ, Fowkes RC, Baxter GF. Regulation of cardiovascular cell function by hydrogen sulfide (H (2)S) Cell Biochem Funct. 2010;28:95–105. doi: 10.1002/cbf.1618. [DOI] [PubMed] [Google Scholar]
- 52.Smith HS. Hydrogen sulfide's involvement in modulating nociception. Pain Physician. 2009;12:901–10. [PubMed] [Google Scholar]
- 53.Wang R. Hydrogen sulfide: a new EDRF. Kidney Int. 2009;76:700–4. doi: 10.1038/ki.2009.221. [DOI] [PubMed] [Google Scholar]
- 54.Elofsson R, Carlberg M, Moroz L, Nezlin L, Sakharov D. Is nitric oxide (NO) produced by invertebrate neurones? Neuroreport. 1993;4:279–82. doi: 10.1097/00001756-199303000-00013. [DOI] [PubMed] [Google Scholar]
- 55.Moroz LL. Giant identified NO-releasing neurons and comparative histochemistry of putative nitrergic systems in gastropod molluscs. Microsc Res Tech. 2000;49:557–69. doi: 10.1002/1097-0029(20000615)49:6<557::AID-JEMT6>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 56.Moroz LL, Bulloch AGM, Lukowiak K, Syed NI. Putative NO-synthesizing neurons of Lymnaea in vivo and in vitro. Netherlands Journal of Zoology. 1994;44:535–549. [Google Scholar]
- 57.Moroz LL, Nezlin LP, Carlberg M, Elofsson R, Sakharov DA. (Do nitric oxide producing neurons occur in invertebrate animals). Biological Membranes. 1992;9:1119–1121. in Russian. [Google Scholar]
- 58.Radomski MW, Martin JF, Moncada S. Synthesis of nitric oxide by the haemocytes of the American horseshoe crab (Limulus polyphemus) Phil. Trans. R. Soc. Lond. 1991;334B:129–133. [Google Scholar]
- 59.Leake LD, Davis MP, Chen D, Moroz LL. An unique model for the analysis of neuronal nitric oxide signaling: the leech CNS. Acta Biol Hung. 1995;46:135–43. [PubMed] [Google Scholar]
- 60.Leake LD, Moroz LL. Putative nitric oxide synthase (NOS)-containing cells in the central nervous system of the leech, Hirudo medicinalis: NADPH-diaphorase histochemistry. Brain Res. 1996;723:115–24. doi: 10.1016/0006-8993(96)00220-x. [DOI] [PubMed] [Google Scholar]
- 61.Moroz LL, Meech RW, Sweedler JV, Mackie GO. Nitric oxide regulates swimming in the jellyfish Aglantha digitale. J Comp Neurol. 2004;471:26–36. doi: 10.1002/cne.20023. [DOI] [PubMed] [Google Scholar]
- 62.Moroz LL, Norekian TP, Pirtle TJ, Robertson KJ, Satterlie RA. Distribution of NADPH-diaphorase reactivity and effects of nitric oxide on feeding and locomotory circuitry in the pteropod mollusc, Clione limacina. J Comp Neurol. 2000;427:274–84. [PubMed] [Google Scholar]
- 63.Palumbo A. Nitric oxide in marine invertebrates: a comparative perspective. Comp Biochem Physiol A Mol Integr Physiol. 2005;142:241–8. doi: 10.1016/j.cbpb.2005.05.043. [DOI] [PubMed] [Google Scholar]
- 64.Trimmer BA, Aprille J, Modica-Napolitano J. Nitric oxide signalling: insect brains and photocytes. Biochem Soc Symp. 2004:65–83. doi: 10.1042/bss0710065. [DOI] [PubMed] [Google Scholar]
- 65.Bishop CD, Pires A, Norby SW, Boudko D, Moroz LL, Hadfield MG. Analysis of nitric oxide-cyclic guanosine monophosphate signaling during metamorphosis of the nudibranch. Phestilla sibogae Bergh (Gastropoda: Opisthobranchia) Evol Dev. 2008;10:288–99. doi: 10.1111/j.1525-142X.2008.00238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moncada S, Martin JF. Evolution of nitric oxide. Lancet. 1993;341:1511. doi: 10.1016/0140-6736(93)90641-s. [DOI] [PubMed] [Google Scholar]
- 67.Moroz LL. On the origin and early evolution of neuronal NO signaling: A comparative analysis. In: Kalsner S, editor. Nitric oxide and free radicals in peripheral neurotransmission. Springer-Verlag; New York: 2000. [Google Scholar]
- 68.Moroz LL, Kohn AB. On the comparative biology of Nitric Oxide (NO) synthetic pathways: Parallel evolution of NO-mediated signaling. In: Toda B, Trimmer B, editors. Nitric oxide. Elselvier; Amsterdam: 2007. [Google Scholar]
- 69.Feelisch M, Martin JE. The early role of nitric oxide in evolution. Trends in Ecology and Evolution. 1995;10:496–499. doi: 10.1016/s0169-5347(00)89206-x. [DOI] [PubMed] [Google Scholar]
- 70.Ignarro LJ, Buga GM, Wood KS, Byrns RE, Chaudhuri G. Endothelium-derived relaxing factor produced and released from artery and vein is nitric oxide. Proc Natl Acad Sci U S A. 1987;84:9265–9. doi: 10.1073/pnas.84.24.9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moncada S, Palmer RM, Higgs EA. Nitric oxide: physiology, pathophysiology, and pharmacology. Pharmacol Rev. 1991;43:109–42. [PubMed] [Google Scholar]
- 72.Palmer RM, Ferrige AG, Moncada S. Nitric oxide release accounts for the biological activity of endothelium-derived relaxing factor. Nature. 1987;327:524–6. doi: 10.1038/327524a0. [DOI] [PubMed] [Google Scholar]
- 73.Gladwin MT, Schechter AN, Kim-Shapiro DB, Patel RP, Hogg N, Shiva S, Cannon RO, 3rd, Kelm M, Wink DA, Espey MG, Oldfield EH, Pluta RM, Freeman BA, Lancaster JR, Jr., Feelisch M, Lundberg JO. The emerging biology of the nitrite anion. Nat Chem Biol. 2005;1:308–14. doi: 10.1038/nchembio1105-308. [DOI] [PubMed] [Google Scholar]
- 74.Ignarro LJ. Nitric Oxide: Biology and Pathobiology. Vol. 1003. Academic Press; 2000. [Google Scholar]
- 75.Stamler JS, Singel DJ, Loscalzo J. Biochemistry of nitric oxide and its redox-activated forms. Science. 1992;258:1898–902. doi: 10.1126/science.1281928. [DOI] [PubMed] [Google Scholar]
- 76.Flores-Santana W, Switzer C, Ridnour LA, Basudhar D, Mancardi D, Donzelli S, Thomas DD, Miranda KM, Fukuto JM, Wink DA. Comparing the chemical biology of NO and HNO. Arch Pharm Res. 2009;32:1139–53. doi: 10.1007/s12272-009-1805-x. [DOI] [PubMed] [Google Scholar]
- 77.Lowenstein CJ, Snyder SH. Nitric oxide, a novel biologic messenger. Cell. 1992;70:705–7. doi: 10.1016/0092-8674(92)90301-r. [DOI] [PubMed] [Google Scholar]
- 78.Crawford NM. Mechanisms for nitric oxide synthesis in plants. J Exp Bot. 2006;57:471–8. doi: 10.1093/jxb/erj050. [DOI] [PubMed] [Google Scholar]
- 79.Garthwaite J. Dynamics of cellular NO-cGMP signaling. Front Biosci. 2005;10:1868–80. doi: 10.2741/1666. [DOI] [PubMed] [Google Scholar]
- 80.Mur LA, Carver TL, Prats E. NO way to live; the various roles of nitric oxide in plant-pathogen interactions. J Exp Bot. 2006;57:489–505. doi: 10.1093/jxb/erj052. [DOI] [PubMed] [Google Scholar]
- 81.Nathan C. Nitric oxide as a secretory product of mammalian cells. Faseb J. 1992;6:3051–64. [PubMed] [Google Scholar]
- 82.Moroz LL. Gaseous transmission across time and species. American Zoologist. 2001;41:304–320. [Google Scholar]
- 83.Moroz LL, Gillette R. From Polyplacophora to Cephalopoda: comparative analysis of nitric oxide signalling in mollusca. Acta Biol Hung. 1995;46:169–82. [PubMed] [Google Scholar]
- 84.Thomas DD, Liu X, Kantrow SP, Lancaster JR., Jr. The biological lifetime of nitric oxide: implications for the perivascular dynamics of NO and O2. Proc Natl Acad Sci U S A. 2001;98:355–60. doi: 10.1073/pnas.011379598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lancaster JR., Jr. A tutorial on the diffusibility and reactivity of free nitric oxide. Nitric Oxide. 1997;1:18–30. doi: 10.1006/niox.1996.0112. [DOI] [PubMed] [Google Scholar]
- 86.Lancaster JR., Jr. Diffusion of free nitric oxide. Methods Enzymol. 1996;268:31–50. doi: 10.1016/s0076-6879(96)68007-0. [DOI] [PubMed] [Google Scholar]
- 87.Lancaster JR., Jr. Simulation of the diffusion and reaction of endogenously produced nitric oxide. Proc Natl Acad Sci U S A. 1994;91:8137–41. doi: 10.1073/pnas.91.17.8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wood J, Garthwaite J. Models of the diffusional spread of nitric oxide: implications for neural nitric oxide signalling and its pharmacological properties. Neuropharmacology. 1994;33:1235–44. doi: 10.1016/0028-3908(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 89.Donzelli S, Switzer CH, Thomas DD, Ridnour LA, Espey MG, Isenberg JS, Tocchetti CG, King SB, Lazzarino G, Miranda KM, Roberts DD, Feelisch M, Wink DA. The activation of metabolites of nitric oxide synthase by metals is both redox and oxygen dependent: a new feature of nitrogen oxide signaling. Antioxid Redox Signal. 2006;8:1363–71. doi: 10.1089/ars.2006.8.1363. [DOI] [PubMed] [Google Scholar]
- 90.Miranda KM, Katori T, Torres de Holding CL, Thomas L, Ridnour LA, McLendon WJ, Cologna SM, Dutton AS, Champion HC, Mancardi D, Tocchetti CG, Saavedra JE, Keefer LK, Houk KN, Fukuto JM, Kass DA, Paolocci N, Wink DA. Comparison of the NO and HNO donating properties of diazeniumdiolates: primary amine adducts release HNO in vivo. J Med Chem. 2005;48:8220–8. doi: 10.1021/jm050151i. [DOI] [PubMed] [Google Scholar]
- 91.Mancardi D, Ridnour LA, Thomas DD, Katori T, Tocchetti CG, Espey MG, Miranda KM, Paolocci N, Wink DA. The chemical dynamics of NO and reactive nitrogen oxides: a practical guide. Curr Mol Med. 2004;4:723–40. doi: 10.2174/1566524043359854. [DOI] [PubMed] [Google Scholar]
- 92.Thomas DD, Miranda KM, Espey MG, Citrin D, Jourd'heuil D, Paolocci N, Hewett SJ, Colton CA, Grisham MB, Feelisch M, Wink DA. Guide for the use of nitric oxide (NO) donors as probes of the chemistry of NO and related redox species in biological systems. Methods Enzymol. 2002;359:84–105. doi: 10.1016/s0076-6879(02)59174-6. [DOI] [PubMed] [Google Scholar]
- 93.Bartberger MD, Liu W, Ford E, Miranda KM, Switzer C, Fukuto JM, Farmer PJ, Wink DA, Houk KN. The reduction potential of nitric oxide (NO) and its importance to NO biochemistry. Proc Natl Acad Sci U S A. 2002;99:10958–63. doi: 10.1073/pnas.162095599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wink DA, Hanbauer I, Grisham MB, Laval F, Nims RW, Laval J, Cook J, Pacelli R, Liebmann J, Krishna M, Ford PC, Mitchell JB. Chemical biology of nitric oxide: regulation and protective and toxic mechanisms. Curr Top Cell Regul. 1996;34:159–87. doi: 10.1016/s0070-2137(96)80006-9. [DOI] [PubMed] [Google Scholar]
- 95.Roy B, Garthwaite J. Nitric oxide activation of guanylyl cyclase in cells revisited. Proc Natl Acad Sci U S A. 2006;103:12185–90. doi: 10.1073/pnas.0602544103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gorren AC, Mayer B. Nitric-oxide synthase: A cytochrome P450 family foster child. Biochim Biophys Acta. 2007;1770:432–45. doi: 10.1016/j.bbagen.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 97.Singel DJ, Stamler JS. Chemical physiology of blood flow regulation by red blood cells: the role of nitric oxide and S-nitrosohemoglobin. Annu Rev Physiol. 2005;67:99–145. doi: 10.1146/annurev.physiol.67.060603.090918. [DOI] [PubMed] [Google Scholar]
- 98.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–66. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 99.Stamler JS, Lamas S, Fang FC. Nitrosylation: the prototypic redox-based signaling mechanism. Cell. 2001;106:675–83. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- 100.Miranda KM, Espey MG, Jour'heuil D, Grisham MB, Fukuto JM, Feelisch M, Wink DA. In: The chemical biology of nitric oxide. In: Nitric Oxide: Biology and Pathobiology. Ignarro LJ, editor. Academic Press; San Diego: 2000. [Google Scholar]
- 101.Fukuto JM, Cho JY, Switzer CH. The chemical properties of nitric oxide and related nitrogen oxides. In: Ignarro LJ, editor. Nitric Oxide: Biology and Pathobiology. Academic Press; San Diego: 2000. [Google Scholar]
- 102.Lewis RS, Deen WM. Kinetics of the reaction of nitric oxide with oxygen in aqueous solutions. Chem Res Toxicol. 1994;7:568–74. doi: 10.1021/tx00040a013. [DOI] [PubMed] [Google Scholar]
- 103.Ignarro LJ, Fukuto JM, Griscarage JM, Rogers NE, Byrns RE. Oxidation of nitric oxide in aqueous solution to nitrite but not nitrate: comparison with enzymatically formed nitric oxide from L-arginine. Proc. Natl. Acad. Sci. USA. 1993;90:8103–8107. doi: 10.1073/pnas.90.17.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ford PC, Wink DA, Stanbury DM. Autoxidation kinetics of aqueous nitric oxide. FEBS Lett. 1993;326:1–3. doi: 10.1016/0014-5793(93)81748-o. [DOI] [PubMed] [Google Scholar]
- 105.Shaw AW, Vosper AJ. Solubility of nitric oxide in aqueous and nonaqueous solvents. J. Chem. Soc. Faraday Trans. I. 1977;8:1239–1244. [Google Scholar]
- 106.Lancaster JR., Jr. The physical properties of nitric oxide. In: Ignarro LJ, editor. Nitric Oxide: Biology and Pathobiology. Academic Press; San Diego: 2000. [Google Scholar]
- 107.Moller MN, Li Q, Lancaster JR, Jr., Denicola A. Acceleration of nitric oxide autoxidation and nitrosation by membranes. IUBMB Life. 2007;59:243–8. doi: 10.1080/15216540701311147. [DOI] [PubMed] [Google Scholar]
- 108.Lima ES, Bonini MG, Augusto O, Barbeiro HV, Souza HP, Abdalla DS. Nitrated lipids decompose to nitric oxide and lipid radicals and cause vasorelaxation. Free Radic Biol Med. 2005;39:532–9. doi: 10.1016/j.freeradbiomed.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 109.Botti H, Trostchansky A, Batthyany C, Rubbo H. Reactivity of peroxynitrite and nitric oxide with LDL. IUBMB Life. 2005;57:407–12. doi: 10.1080/15216540500137701. [DOI] [PubMed] [Google Scholar]
- 110.Baker PR, Schopfer FJ, Sweeney S, Freeman BA. Red cell membrane and plasma linoleic acid nitration products: synthesis, clinical identification, and quantitation. Proc Natl Acad Sci U S A. 2004;101:11577–82. doi: 10.1073/pnas.0402587101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Baker PR, Lin Y, Schopfer FJ, Woodcock SR, Groeger AL, Batthyany C, Sweeney S, Long MH, Iles KE, Baker LM, Branchaud BP, Chen YE, Freeman BA. Fatty acid transduction of nitric oxide signaling: multiple nitrated unsaturated fatty acid derivatives exist in human blood and urine and serve as endogenous peroxisome proliferator-activated receptor ligands. J Biol Chem. 2005;280:42464–75. doi: 10.1074/jbc.M504212200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kalyanaraman B. Nitrated lipids: a class of cell-signaling molecules. Proc Natl Acad Sci U S A. 2004;101:11527–8. doi: 10.1073/pnas.0404309101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wright MM, Schopfer FJ, Baker PR, Vidyasagar V, Powell P, Chumley P, Iles KE, Freeman BA, Agarwal A. Fatty acid transduction of nitric oxide signaling: nitrolinoleic acid potently activates endothelial heme oxygenase 1 expression. Proc Natl Acad Sci U S A. 2006;103:4299–304. doi: 10.1073/pnas.0506541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schopfer FJ, Baker PR, Giles G, Chumley P, Batthyany C, Crawford J, Patel RP, Hogg N, Branchaud BP, Lancaster JR, Jr., Freeman BA. Fatty acid transduction of nitric oxide signaling. Nitrolinoleic acid is a hydrophobically stabilized nitric oxide donor. J Biol Chem. 2005;280:19289–97. doi: 10.1074/jbc.M414689200. [DOI] [PubMed] [Google Scholar]
- 115.Bryan NS, Rassaf T, Maloney RE, Rodriguez CM, Saijo F, Rodriguez JR, Feelisch M. Cellular targets and mechanisms of nitros (yl)ation: an insight into their nature and kinetics in vivo. Proc Natl Acad Sci U S A. 2004;101:4308–13. doi: 10.1073/pnas.0306706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nathan C. The moving frontier in nitric oxide-dependent signaling. Sci STKE. 2004;2004:pe52. doi: 10.1126/stke.2572004pe52. [DOI] [PubMed] [Google Scholar]
- 117.Quintero M, Colombo SL, Godfrey A, Moncada S. Mitochondria as signaling organelles in the vascular endothelium. Proc Natl Acad Sci U S A. 2006;103:5379–84. doi: 10.1073/pnas.0601026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, Falcone S, Valerio A, Cantoni O, Clementi E, Moncada S, Carruba MO. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–7. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- 119.Nisoli E, Falcone S, Tonello C, Cozzi V, Palomba L, Fiorani M, Pisconti A, Brunelli S, Cardile A, Francolini M, Cantoni O, Carruba MO, Moncada S, Clementi E. Mitochondrial biogenesis by NO yields functionally active mitochondria in mammals. Proc Natl Acad Sci U S A. 2004;101:16507–12. doi: 10.1073/pnas.0405432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Moncada S, Bolanos JP. Nitric oxide, cell bioenergetics and neurodegeneration. J Neurochem. 2006;97:1676–89. doi: 10.1111/j.1471-4159.2006.03988.x. [DOI] [PubMed] [Google Scholar]
- 121.Navarro A. Mitochondrial nitric oxide synthase and the regulation of heart rate. Am J Hypertens. 2008;21:485. doi: 10.1038/ajh.2008.22. [DOI] [PubMed] [Google Scholar]
- 122.Lopez-Lluch G, Irusta PM, Navas P, de Cabo R. Mitochondrial biogenesis and healthy aging. Exp Gerontol. 2008;43:813–9. doi: 10.1016/j.exger.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hendgen-Cotta UB, Merx MW, Shiva S, Schmitz J, Becher S, Klare JP, Steinhoff HJ, Goedecke A, Schrader J, Gladwin MT, Kelm M, Rassaf T. Nitrite reductase activity of myoglobin regulates respiration and cellular viability in myocardial ischemiareperfusion injury. Proc Natl Acad Sci U S A. 2008;105:10256–61. doi: 10.1073/pnas.0801336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Benamar A, Rolletschek H, Borisjuk L, Avelange-Macherel MH, Curien G, Mostefai HA, Andriantsitohaina R, Macherel D. Nitrite-nitric oxide control of mitochondrial respiration at the frontier of anoxia. Biochim Biophys Acta. 2008;1777:1268–75. doi: 10.1016/j.bbabio.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 125.Valdez LB, Boveris A. Mitochondrial nitric oxide synthase, a voltage-dependent enzyme, is responsible for nitric oxide diffusion to cytosol. Front Biosci. 2007;12:1210–9. doi: 10.2741/2139. [DOI] [PubMed] [Google Scholar]
- 126.Hill BG, Darley-Usmar VM. S-nitrosation and thiol switching in the mitochondrion: a new paradigm for cardioprotection in ischaemic preconditioning. Biochem J. 2008;412:e11–3. doi: 10.1042/BJ20080716. [DOI] [PubMed] [Google Scholar]
- 127.Vanin A, Poltorakov A. NO spin trapping in biological systems. Front Biosci. 2009;14:4427–35. doi: 10.2741/3538. [DOI] [PubMed] [Google Scholar]
- 128.Nagano T. Bioimaging probes for reactive oxygen species and reactive nitrogen species. J Clin Biochem Nutr. 2009;45:111–24. doi: 10.3164/jcbn.R09-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lopez-Sanchez LM, Muntane J, de la Mata M, Rodriguez-Ariza A. Unraveling the S-nitrosoproteome: tools and strategies. Proteomics. 2009;9:808–18. doi: 10.1002/pmic.200800546. [DOI] [PubMed] [Google Scholar]
- 130.Likhtenshtein GI. Novel fluorescent methods for biotechnological and biomedical sensoring: assessing antioxidants, reactive radicals, NO dynamics, immunoassay, and biomembranes fluidity. Appl Biochem Biotechnol. 2009;152:135–55. doi: 10.1007/s12010-008-8219-y. [DOI] [PubMed] [Google Scholar]
- 131.Ye X, Rubakhin SS, Sweedler JV. Detection of nitric oxide in single cells. Analyst. 2008;133:423–33. doi: 10.1039/b716174c. [DOI] [PubMed] [Google Scholar]
- 132.Teng X, Scott Isbell T, Crawford JH, Bosworth CA, Giles GI, Koenitzer JR, Lancaster JR, Doeller JE, Kraus DW, Patel RP. Novel method for measuring S-nitrosothiols using hydrogen sulfide. Methods Enzymol. 2008;441:161–72. doi: 10.1016/S0076-6879(08)01209-3. [DOI] [PubMed] [Google Scholar]
- 133.Ghafourifar P, Parihar MS, Nazarewicz R, Zenebe WJ, Parihar A. Detection assays for determination of mitochondrial nitric oxide synthase activity; advantages and limitations. Methods Enzymol. 2008;440:317–34. doi: 10.1016/S0076-6879(07)00821-X. [DOI] [PubMed] [Google Scholar]
- 134.Davies IR, Zhang X. Nitric oxide selective electrodes. Methods Enzymol. 2008;436:63–95. doi: 10.1016/S0076-6879(08)36005-4. [DOI] [PubMed] [Google Scholar]
- 135.Barbosa RM, Lourenco CF, Santos RM, Pomerleau F, Huettl P, Gerhardt GA, Laranjinha J. In vivo real-time measurement of nitric oxide in anesthetized rat brain. Methods Enzymol. 2008;441:351–67. doi: 10.1016/S0076-6879(08)01220-2. [DOI] [PubMed] [Google Scholar]
- 136.Nagano T. Practical methods for detection of nitric oxide. Luminescence. 1999;14:283–90. doi: 10.1002/(SICI)1522-7243(199911/12)14:6<283::AID-BIO572>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 137.Moroz LL, Gillette R, Sweedler JV. Single-cell analyses of nitrergic neurons in simple nervous systems. J Exp Biol. 1999;202:333–41. doi: 10.1242/jeb.202.4.333. [DOI] [PubMed] [Google Scholar]
- 138.Kiechle FL, Malinski T. Indirect detection of nitric oxide effects: a review. Ann Clin Lab Sci. 1996;26:501–11. [PubMed] [Google Scholar]
- 139.Kiechle FL, Malinski T. Nitric oxide. Biochemistry, pathophysiology, and detection. Am J Clin Pathol. 1993;100:567–75. doi: 10.1093/ajcp/100.5.567. [DOI] [PubMed] [Google Scholar]
- 140.Henry Y, Lepoivre M, Drapier JC, Ducrocq C, Boucher JL, Guissani A. EPR characterization of molecular targets for NO in mammalian cells and organelles. Faseb J. 1993;7:1124–34. doi: 10.1096/fasebj.7.12.8397130. [DOI] [PubMed] [Google Scholar]
- 141.Archer S. Measurement of nitric oxide in biological models. FASEB J. 1993;7:349–360. doi: 10.1096/fasebj.7.2.8440411. [DOI] [PubMed] [Google Scholar]
- 142.Moroz LL, Norby SW, Cruz L, Sweedler JV, Gillette R, Clarkson RB. Non-enzymatic production of nitric oxide (NO) from NO synthase inhibitors. Biochem Biophys Res Commun. 1998;253:571–6. doi: 10.1006/bbrc.1998.9810. [DOI] [PubMed] [Google Scholar]
- 143.Boudko DY, Cooper BY, Harvey WR, Moroz LL. High-resolution microanalysis of nitrite and nitrate in neuronal tissues by capillary electrophoresis with conductivity detection. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;774:97–104. doi: 10.1016/s1570-0232(02)00219-2. [DOI] [PubMed] [Google Scholar]
- 144.Cruz L, Moroz LL, Gillette R, Sweedler JV. Nitrite and nitrate levels in individual molluscan neurons: single-cell capillary electrophoresis analysis. J Neurochem. 1997;69:110–5. doi: 10.1046/j.1471-4159.1997.69010110.x. [DOI] [PubMed] [Google Scholar]
- 145.Floyd PD, Moroz LL, Gillette R, Sweedler JV. Capillary electrophoresis analysis of nitric oxide synthase related metabolites in single identified neurons. Anal Chem. 1998;70:2243–7. doi: 10.1021/ac9713013. [DOI] [PubMed] [Google Scholar]
- 146.Fuller RR, Moroz LL, Gillette R, Sweedler JV. Single neuron analysis by capillary electrophoresis with fluorescence spectroscopy. Neuron. 1998;20:173–81. doi: 10.1016/s0896-6273(00)80446-8. [DOI] [PubMed] [Google Scholar]
- 147.Moroz LL, Dahlgren RL, Boudko D, Sweedler JV, Lovell P. Direct single cell determination of nitric oxide synthase related metabolites in identified nitrergic neurons. J Inorg Biochem. 2005;99:929–39. doi: 10.1016/j.jinorgbio.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 148.Moroz LL, Radbourne S, Winlow W. The use of NO-sensitive microelectrodes for direct detection of nitric oxide (NO) production in molluscs. Acta Biol Hung. 1995;46:155–67. [PubMed] [Google Scholar]
- 149.Zhang X, Kim WS, Hatcher N, Potgieter K, Moroz LL, Gillette R, Sweedler JV. Interfering with nitric oxide measurements. 4,5-diaminofluorescein reacts with dehydroascorbic acid and ascorbic acid. J Biol Chem. 2002;277:48472–8. doi: 10.1074/jbc.M209130200. [DOI] [PubMed] [Google Scholar]
- 150.Zumft WG. The biological role of nitric oxide in bacteria. Arch Microbiol. 1993;160:253–64. doi: 10.1007/BF00292074. [DOI] [PubMed] [Google Scholar]
- 151.Zumft WG. Nitric oxide reductases of prokaryotes with emphasis on the respiratory, heme-copper oxidase type. J Inorg Biochem. 2005;99:194–215. doi: 10.1016/j.jinorgbio.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 152.Brusca RC, Brusca GJ. Invertebrates. Sinauer Associates, Inc.; Sunderland, Massachusetts: 2003. [Google Scholar]
- 153.Zemojtel T, Penzkofer T, Dandekar T, Schultz J. A novel conserved family of nitric oxide synthase? Trends Biochem Sci. 2004;29:224–6. doi: 10.1016/j.tibs.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 154.Guo FQ, Okamoto M, Crawford NM. Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science. 2003;302:100–3. doi: 10.1126/science.1086770. [DOI] [PubMed] [Google Scholar]
- 155.Nna Mvondo D, Navarro-Gonzalez R, McKay CP, Coll P, Raulin F. Production of nitrogen oxides by lightning and coronae discharges in simulated early Earth, Venus and Mars environments. Adv Space Res. 2001;27:217–23. doi: 10.1016/s0273-1177(01)00050-3. [DOI] [PubMed] [Google Scholar]
- 156.Nna-Mvondo D, Navarro-Gonzalez R, Raulin F, Coll P. Nitrogen fixation by corona discharge on the early precambrian Earth. Orig Life Evol Biosph. 2005;35:401–9. doi: 10.1007/s11084-005-1972-9. [DOI] [PubMed] [Google Scholar]
- 157.Martin RS, Mather TA, Pyle DM. Volcanic emissions and the early Earth atmosphere. Geochimica et Cosmochimica Acta. 2007;71:3673–3685. [Google Scholar]
- 158.Kasting JF. Bolide impacts and the oxidation state of carbon in the Earth's early atmosphere. Orig Life Evol Biosph. 1992;20:199–231. doi: 10.1007/BF01808105. [DOI] [PubMed] [Google Scholar]
- 159.Wang Y, DeSilva AW, Goldenbaum GC, Dickerson RR. Nitrogen oxide production by simulated lighting: Dependence on current, energy, and pressure. Journal of Geophysical Research. 1998;103:19149–19160. [Google Scholar]
- 160.Summers DP, Chang S. Prebiotic ammonia from reduction of nitrite by iron (II) on the early Earth. Nature. 1993;365:630–3. doi: 10.1038/365630a0. [DOI] [PubMed] [Google Scholar]
- 161.Brandes JA, Boctor NZ, Cody GD, Cooper BA, Hazen RM, Yoder HS., Jr. Abiotic nitrogen reduction on the early Earth. Nature. 1998;395:365–7. doi: 10.1038/26450. [DOI] [PubMed] [Google Scholar]
- 162.Kasting JF, Howard MT. Atmospheric composition and climate on the early Earth. Philos Trans R Soc Lond B Biol Sci. 2006;361:1733–42. doi: 10.1098/rstb.2006.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kasting JF, Ono S. Palaeoclimates: the first two billion years. Philos Trans R Soc Lond B Biol Sci. 2006;361:917–29. doi: 10.1098/rstb.2006.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kasting JF, Siefert JL. Biogeochemistry. The nitrogen fix. Nature. 2001;412:26–7. doi: 10.1038/35083660. [DOI] [PubMed] [Google Scholar]
- 165.Navarro-Gonzalez R, McKay CP, Mvondo DN. A possible nitrogen crisis for Archaean life due to reduced nitrogen fixation by lightning. Nature. 2001;412:61–4. doi: 10.1038/35083537. [DOI] [PubMed] [Google Scholar]
- 166.Ghosh DK, Salerno JC. Nitric oxide synthases: domain structure and alignment in enzyme function and control. Front Biosci. 2003;8:d193–209. doi: 10.2741/959. [DOI] [PubMed] [Google Scholar]
- 167.Griffith OW, Stuehr DJ. Nitric oxide synthases: properties and catalytic mechanism. Annu Rev Physiol. 1995;57:707–36. doi: 10.1146/annurev.ph.57.030195.003423. [DOI] [PubMed] [Google Scholar]
- 168.Zemojtel T, Wade RC, Dandekar T. In search of the prototype of nitric oxide synthase. FEBS Lett. 2003;554:1–5. doi: 10.1016/s0014-5793(03)01081-0. [DOI] [PubMed] [Google Scholar]
- 169.Neill S, Bright J, Desikan R, Hancock J, Harrison J, Wilson I. Nitric oxide evolution and perception. J Exp Bot. 2008;59:25–35. doi: 10.1093/jxb/erm218. [DOI] [PubMed] [Google Scholar]
- 170.Philippot L. Denitrifying genes in bacterial and Archaeal genomes. Biochim Biophys Acta. 2002;1577:355–76. doi: 10.1016/s0167-4781(02)00420-7. [DOI] [PubMed] [Google Scholar]
- 171.Reutov VP. Nitric oxide cycle in mammals and the cyclicity principle. Biochemistry (Mosc) 2002;67:293–311. doi: 10.1023/a:1014832416073. [DOI] [PubMed] [Google Scholar]
- 172.Reutov VP, Sorokina EG. NO-synthase and nitritereductase components of nitric oxide cycle. Biochemistry (Mosc) 1998;63:874–84. [PubMed] [Google Scholar]
- 173.Meyer DJ. Interaction of cytochrome oxidases aa3 and d with nitrite. Nature New Biology. 1973;245:276–277. doi: 10.1038/newbio245276a0. [DOI] [PubMed] [Google Scholar]
- 174.Walters CL, Taylor AM. The Reduction of Nitrite by Skeletal-Muscle Mitochondria. Biochim Biophys Acta. 1965;96:522–4. doi: 10.1016/0005-2787(65)90570-8. [DOI] [PubMed] [Google Scholar]
- 175.Castello PR, David PS, McClure T, Crook Z, Poyton RO. Mitochondrial cytochrome oxidase produces nitric oxide under hypoxic conditions: implications for oxygen sensing and hypoxic signaling in eukaryotes. Cell Metab. 2006;3:277–87. doi: 10.1016/j.cmet.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 176.Castello PR, Woo DK, Ball K, Wojcik J, Liu L, Poyton RO. Oxygen-regulated isoforms of cytochrome c oxidase have differential effects on its nitric oxide production and on hypoxic signaling. Proc Natl Acad Sci U S A. 2008;105:8203–8. doi: 10.1073/pnas.0709461105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kozlov AV, Staniek K, Nohl H. Nitrite reductase activity is a novel function of mammalian mitochondria. FEBS Lett. 1999;454:127–30. doi: 10.1016/s0014-5793(99)00788-7. [DOI] [PubMed] [Google Scholar]
- 178.Nohl H, Staniek K, Kozlov AV. The existence and significance of a mitochondrial nitrite reductase. Redox Rep. 2005;10:281–6. doi: 10.1179/135100005X83707. [DOI] [PubMed] [Google Scholar]
- 179.Nohl H, Staniek K, Sobhian B, Bahrami S, Redl H, Kozlov AV. Mitochondria recycle nitrite back to the bioregulator nitric monoxide. Acta Biochim Pol. 2000;47:913–21. [PubMed] [Google Scholar]
- 180.Igamberdiev AU, Hill RD. Plant Mitochondrial Function During Anaerobiosis. Ann Bot (Lond) 2009;103:259–68. doi: 10.1093/aob/mcn100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kollau A, Beretta M, Russwurm M, Koesling D, Keung WM, Schmidt K, Mayer B. Mitochondrial nitrite reduction coupled to soluble guanylate cyclase activation: lack of evidence for a role in the bioactivation of nitroglycerin. Nitric Oxide. 2009;20:53–60. doi: 10.1016/j.niox.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 182.Basu S, Azarova NA, Font MD, King SB, Hogg N, Gladwin MT, Shiva S, Kim-Shapiro DB. Nitrite reductase activity of cytochrome C. J Biol Chem. 2008;283:32590–7. doi: 10.1074/jbc.M806934200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Cosby K, Partovi KS, Crawford JH, Patel RP, Reiter CD, Martyr S, Yang BK, Waclawiw MA, Zalos G, Xu X, Huang KT, Shields H, Kim-Shapiro DB, Schechter AN, Cannon RO, 3rd, Gladwin MT. Nitrite reduction to nitric oxide by deoxyhemoglobin vasodilates the human circulation. Nat Med. 2003;9:1498–505. doi: 10.1038/nm954. [DOI] [PubMed] [Google Scholar]
- 184.Nagababu E, Ramasamy S, Abernethy DR, Rifkind JM. Active nitric oxide produced in the red cell under hypoxic conditions by deoxyhemoglobin-mediated nitrite reduction. J Biol Chem. 2003;278:46349–56. doi: 10.1074/jbc.M307572200. [DOI] [PubMed] [Google Scholar]
- 185.Nagababu E, Ramasamy S, Rifkind JM. S-nitrosohemoglobin: a mechanism for its formation in conjunction with nitrite reduction by deoxyhemoglobin. Nitric Oxide. 2006;15:20–9. doi: 10.1016/j.niox.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 186.Angelo M, Hausladen A, Singel DJ, Stamler JS. Interactions of NO with hemoglobin: from microbes to man. Methods Enzymol. 2008;436:131–68. doi: 10.1016/S0076-6879(08)36008-X. [DOI] [PubMed] [Google Scholar]
- 187.Minneci PC, Deans KJ, Shiva S, Zhi H, Banks SM, Kern S, Natanson C, Solomon SB, Gladwin MT. Nitrite reductase activity of hemoglobin as a systemic nitric oxide generator mechanism to detoxify plasma hemoglobin produced during hemolysis. Am J Physiol Heart Circ Physiol. 2008;295:H743–54. doi: 10.1152/ajpheart.00151.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Perissinotti LL, Marti MA, Doctorovich F, Luque FJ, Estrin DA. A microscopic study of the deoxyhemoglobin-catalyzed generation of nitric oxide from nitrite anion. Biochemistry. 2008;47:9793–802. doi: 10.1021/bi801104c. [DOI] [PubMed] [Google Scholar]
- 189.Mittermayr R, Osipov A, Piskernik C, Haindl S, Dungel P, Weber C, Vladimirov YA, Redl H, Kozlov AV. Blue laser light increases perfusion of a skin flap via release of nitric oxide from hemoglobin. Mol Med. 2007;13:22–9. doi: 10.2119/2006-00035.Mittermayr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Grubina R, Huang Z, Shiva S, Joshi MS, Azarov I, Basu S, Ringwood LA, Jiang A, Hogg N, Kim-Shapiro DB, Gladwin MT. Concerted nitric oxide formation and release from the simultaneous reactions of nitrite with deoxy- and oxyhemoglobin. J Biol Chem. 2007;282:12916–27. doi: 10.1074/jbc.M700546200. [DOI] [PubMed] [Google Scholar]
- 191.Huang Z, Shiva S, Kim-Shapiro DB, Patel RP, Ringwood LA, Irby CE, Huang KT, Ho C, Hogg N, Schechter AN, Gladwin MT. Enzymatic function of hemoglobin as a nitrite reductase that produces NO under allosteric control. J Clin Invest. 2005;115:2099–107. doi: 10.1172/JCI24650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Doyle MP, LePoire DM, Pickering RA. Oxidation of hemoglobin and myoglobin by alkyl nitrites inhibition by oxygen. J Biol Chem. 1981;256:12399–404. [PubMed] [Google Scholar]
- 193.Doyle MP, Pickering RA, DeWeert TM, Hoekstra JW, Pater D. Kinetics and mechanism of the oxidation of human deoxyhemoglobin by nitrites. J Biol Chem. 1981;256:12393–8. [PubMed] [Google Scholar]
- 194.Brooks J. The action of nitrite on haemoglobin in the absence of oxygen. Proc. R. Soc. Med. 1937;137:368–382. [Google Scholar]
- 195.Shiva S, Huang Z, Grubina R, Sun J, Ringwood LA, MacArthur PH, Xu X, Murphy E, Darley-Usmar VM, Gladwin MT. Deoxymyoglobin is a nitrite reductase that generates nitric oxide and regulates mitochondrial respiration. Circ Res. 2007;100:654–61. doi: 10.1161/01.RES.0000260171.52224.6b. [DOI] [PubMed] [Google Scholar]
- 196.Rassaf T, Flogel U, Drexhage C, Hendgen-Cotta U, Kelm M, Schrader J. Nitrite reductase function of deoxymyoglobin: oxygen sensor and regulator of cardiac energetics and function. Circ Res. 2007;100:1749–54. doi: 10.1161/CIRCRESAHA.107.152488. [DOI] [PubMed] [Google Scholar]
- 197.Kozlov AV, Dietrich B, Nohl H. Various intracellular compartments cooperate in the release of nitric oxide from glycerol trinitrate in liver. Br J Pharmacol. 2003;139:989–97. doi: 10.1038/sj.bjp.0705323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Li H, Cui H, Liu X, Zweier JL. Xanthine oxidase catalyzes anaerobic transformation of organic nitrates to nitric oxide and nitrosothiols: characterization of this mechanism and the link between organic nitrate and guanylyl cyclase activation. J Biol Chem. 2005;280:16594–600. doi: 10.1074/jbc.M411905200. [DOI] [PubMed] [Google Scholar]
- 199.Doel JJ, Godber BL, Eisenthal R, Harrison R. Reduction of organic nitrates catalysed by xanthine oxidoreductase under anaerobic conditions. Biochim Biophys Acta. 2001;1527:81–7. doi: 10.1016/s0304-4165(01)00148-9. [DOI] [PubMed] [Google Scholar]
- 200.Li H, Samouilov A, Liu X, Zweier JL. Characterization of the magnitude and kinetics of xanthine oxidase-catalyzed nitrate reduction: evaluation of its role in nitrite and nitric oxide generation in anoxic tissues. Biochemistry. 2003;42:1150–9. doi: 10.1021/bi026385a. [DOI] [PubMed] [Google Scholar]
- 201.Millar TM, Stevens CR, Benjamin N, Eisenthal R, Harrison R, Blake DR. Xanthine oxidoreductase catalyses the reduction of nitrates and nitrite to nitric oxide under hypoxic conditions. FEBS Lett. 1998;427:225–8. doi: 10.1016/s0014-5793(98)00430-x. [DOI] [PubMed] [Google Scholar]
- 202.Zhang Z, Naughton D, Winyard PG, Benjamin N, Blake DR, Symons MC. Generation of nitric oxide by a nitrite reductase activity of xanthine oxidase: a potential pathway for nitric oxide formation in the absence of nitric oxide synthase activity. Biochem Biophys Res Commun. 1998;249:767–72. doi: 10.1006/bbrc.1998.9226. [DOI] [PubMed] [Google Scholar]
- 203.Webb AJ, Milsom AB, Rathod KS, Chu WL, Qureshi S, Lovell MJ, Lecomte FM, Perrett D, Raimondo C, Khoshbin E, Ahmed Z, Uppal R, Benjamin N, Hobbs AJ, Ahluwalia A. Mechanisms underlying erythrocyte and endothelial nitrite reduction to nitric oxide in hypoxia: role for xanthine oxidoreductase and endothelial nitric oxide synthase. Circ Res. 2008;103:957–64. doi: 10.1161/CIRCRESAHA.108.175810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Webb A, Bond R, McLean P, Uppal R, Benjamin N, Ahluwalia A. Reduction of nitrite to nitric oxide during ischemia protects against myocardial ischemia-reperfusion damage. Proc Natl Acad Sci U S A. 2004;101:13683–8. doi: 10.1073/pnas.0402927101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Sinha SS, Shiva S, Gladwin MT. Myocardial protection by nitrite: evidence that this reperfusion therapeutic will not be lost in translation. Trends Cardiovasc Med. 2008;18:163–72. doi: 10.1016/j.tcm.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 206.Jansson EA, Huang L, Malkey R, Govoni M, Nihlen C, Olsson A, Stensdotter M, Petersson J, Holm L, Weitzberg E, Lundberg JO. A mammalian functional nitrate reductase that regulates nitrite and nitric oxide homeostasis. Nat Chem Biol. 2008;4:411–7. doi: 10.1038/nchembio.92. [DOI] [PubMed] [Google Scholar]
- 207.Li H, Cui H, Kundu TK, Alzawahra W, Zweier JL. Nitric oxide production from nitrite occurs primarily in tissues not in the blood: critical role of xanthine oxidase and aldehyde oxidase. J Biol Chem. 2008;283:17855–63. doi: 10.1074/jbc.M801785200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Baker JE, Su J, Fu X, Hsu A, Gross GJ, Tweddell JS, Hogg N. Nitrite confers protection against myocardial infarction: role of xanthine oxidoreductase, NADPH oxidase and K (ATP) channels. J Mol Cell Cardiol. 2007;43:437–44. doi: 10.1016/j.yjmcc.2007.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Zhang Z, Naughton DP, Blake DR, Benjamin N, Stevens CR, Winyard PG, Symons MC, Harrison R. Human xanthine oxidase converts nitrite ions into nitric oxide (NO) Biochem Soc Trans. 1997;25:524S. doi: 10.1042/bst025524s. [DOI] [PubMed] [Google Scholar]
- 210.Godber BL, Doel JJ, Goult TA, Eisenthal R, Harrison R. Suicide inactivation of xanthine oxidoreductase during reduction of inorganic nitrite to nitric oxide. Biochem J. 2001;358:325–33. doi: 10.1042/0264-6021:3580325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Godber BL, Doel JJ, Sapkota GP, Blake DR, Stevens CR, Eisenthal R, Harrison R. Reduction of nitrite to nitric oxide catalyzed by xanthine oxidoreductase. J Biol Chem. 2000;275:7757–63. doi: 10.1074/jbc.275.11.7757. [DOI] [PubMed] [Google Scholar]
- 212.Gautier C, van Faassen E, Mikula I, Martasek P, Slama-Schwok A. Endothelial nitric oxide synthase reduces nitrite anions to NO under anoxia. Biochem Biophys Res Commun. 2006;341:816–21. doi: 10.1016/j.bbrc.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 213.Vanin AF, Bevers LM, Slama-Schwok A, van Faassen EE. Nitric oxide synthase reduces nitrite to NO under anoxia. Cell Mol Life Sci. 2007;64:96–103. doi: 10.1007/s00018-006-6374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Mikula I, Durocher S, Martasek P, Mutus B, Slama-Schwok A. Isoform-specific differences in the nitrite reductase activity of nitric oxide synthases under hypoxia. Biochem J. 2008 doi: 10.1042/BJ20080987. [DOI] [PubMed] [Google Scholar]
- 215.Peri L, Pietraforte D, Scorza G, Napolitano A, Fogliano V, Minetti M. Apples increase nitric oxide production by human saliva at the acidic pH of the stomach: a new biological function for polyphenols with a catechol group? Free Radic Biol Med. 2005;39:668–81. doi: 10.1016/j.freeradbiomed.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 216.Gago B, Lundberg JO, Barbosa RM, Laranjinha J. Red wine-dependent reduction of nitrite to nitric oxide in the stomach. Free Radic Biol Med. 2007;43:1233–42. doi: 10.1016/j.freeradbiomed.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 217.Gago B, Nystrom T, Cavaleiro C, Rocha BS, Barbosa RM, Laranjinha J, Lundberg JO. The potent vasodilator ethyl nitrite is formed upon reaction of nitrite and ethanol under gastric conditions. Free Radic Biol Med. 2008;45:404–12. doi: 10.1016/j.freeradbiomed.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 218.Carlsson S, Wiklund NP, Engstrand L, Weitzberg E, Lundberg JO. Effects of pH, nitrite, and ascorbic acid on nonenzymatic nitric oxide generation and bacterial growth in urine. Nitric Oxide. 2001;5:580–6. doi: 10.1006/niox.2001.0371. [DOI] [PubMed] [Google Scholar]
- 219.Weitzberg E, Lundberg JO. Nonenzymatic nitric oxide production in humans. Nitric Oxide. 1998;2:1–7. doi: 10.1006/niox.1997.0162. [DOI] [PubMed] [Google Scholar]
- 220.Zweier JL, Samouilov A, Kuppusamy P. Non-enzymatic nitric oxide synthesis in biological systems. Biochim Biophys Acta. 1999;1411:250–62. doi: 10.1016/s0005-2728(99)00018-3. [DOI] [PubMed] [Google Scholar]
- 221.Lundberg JO, Govoni M. Inorganic nitrate is a possible source for systemic generation of nitric oxide. Free Radic Biol Med. 2004;37:395–400. doi: 10.1016/j.freeradbiomed.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 222.Lundberg JO, Weitzberg E. NO generation from nitrite and its role in vascular control. Arterioscler Thromb Vasc Biol. 2005;25:915–22. doi: 10.1161/01.ATV.0000161048.72004.c2. [DOI] [PubMed] [Google Scholar]
- 223.Lundberg JO, Weitzberg E, Cole JA, Benjamin N. Nitrate, bacteria and human health. Nat Rev Microbiol. 2004;2:593–602. doi: 10.1038/nrmicro929. [DOI] [PubMed] [Google Scholar]
- 224.Suschek CV, Schewe T, Sies H, Kroncke KD. Nitrite, a naturally occurring precursor of nitric oxide that acts like a ‘prodrug’. Biol Chem. 2006;387:499–506. doi: 10.1515/BC.2006.065. [DOI] [PubMed] [Google Scholar]
- 225.Lundberg JO, Weitzberg E, Gladwin MT. The nitrate-nitrite-nitric oxide pathway in physiology and therapeutics. Nat Rev Drug Discov. 2008;7:156–67. doi: 10.1038/nrd2466. [DOI] [PubMed] [Google Scholar]
- 226.Kharitonov VG, Sundquist AR, Sharma VS. Kinetics of nitric oxide autoxidation in aqueous solution. J Biol Chem. 1994;269:5881–3. [PubMed] [Google Scholar]
- 227.Kharitonov VG, Sundquist AR, Sharma VS. Kinetics of nitrosation of thiols by nitric oxide in the presence of oxygen. J Biol Chem. 1995;270:28158–28164. doi: 10.1074/jbc.270.47.28158. [DOI] [PubMed] [Google Scholar]
- 228.Saran M, Bors W. Pulse radiolysis for investigation of nitric oxide related reactions. Methods in Enzymology. 1994;233:20–34. [Google Scholar]
- 229.Wink DA, Grisham MB, Miles AM, Nims RW, Krishna MC, Pacelli R, Teague D, Poore CM, Cook JA, Ford PC. Determination of selectivity of reactive nitrogen oxide species for various substrates. Methods Enzymol. 1996;268:120–30. doi: 10.1016/s0076-6879(96)68014-8. [DOI] [PubMed] [Google Scholar]
- 230.Wink DA, Grisham MB, Mitchell JB, Ford PC. Direct and indirect effects of nitric oxide in chemical reactions relevant to biology. Methods Enzymol. 1996;268:12–31. doi: 10.1016/s0076-6879(96)68006-9. [DOI] [PubMed] [Google Scholar]
- 231.Wink DA, Mitchell JB. Chemical biology of nitric oxide: Insights into regulatory, cytotoxic, and cytoprotective mechanisms of nitric oxide. Free Radic Biol Med. 1998;25:434–56. doi: 10.1016/s0891-5849(98)00092-6. [DOI] [PubMed] [Google Scholar]
- 232.Shiva S, Wang X, Ringwood LA, Xu X, Yuditskaya S, Annavajjhala V, Miyajima H, Hogg N, Harris ZL, Gladwin MT. Ceruloplasmin is a NO oxidase and nitrite synthase that determines endocrine NO homeostasis. Nat Chem Biol. 2006;2:486–93. doi: 10.1038/nchembio813. [DOI] [PubMed] [Google Scholar]
- 233.Grisham MB, Johnson GG, Lancaster JR., Jr. Quantitation of nitrate and nitrite in extracellular fluids. Methods Enzymol. 1996;268:237–46. doi: 10.1016/s0076-6879(96)68026-4. [DOI] [PubMed] [Google Scholar]
- 234.Butler AR, Flitney FW, Williams DL. NO, nitrosonium ions, nitroxide ions, nitrosothiols and iron-nitrosyls in biology: a chemist's perspective. Trends Pharmacol Sci. 1995;16:18–22. doi: 10.1016/s0165-6147(00)88968-3. [DOI] [PubMed] [Google Scholar]
- 235.Feelisch M, Stamler JS. Donors of nitrogen oxides. In: Feelisch M, Stamler JS, editors. Methods in nitric oxide research. John Wiley & Sons; Chichester, New York: 1996. [Google Scholar]
- 236.Mirvish SS, Wallcave L, Eagen M, Shubik P. Ascorbate-nitrite reaction: possible means of blocking the formation of carcinogenic N-nitroso compounds. Science. 1972;177:65–8. doi: 10.1126/science.177.4043.65. [DOI] [PubMed] [Google Scholar]
- 237.Dahn H, Loewe L, Luscher E, Menasse R. Uber die Oxydation von Ascorbinsaure durch salpetrige Saule. TeilI: Stochiometrie und Kinetische Messtechnik. Helv. Chim. Acta. 1960;43:287–293. [Google Scholar]
- 238.Yamasaki H. Nitrite-dependent nitric oxide production pathway: implications for involvement of active nitrogen species in photoinhibition in vivo. Philos Trans R Soc Lond B Biol Sci. 2000;355:1477–88. doi: 10.1098/rstb.2000.0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Kytzia A, Korth HG, Sustmann R, de Groot H, Kirsch M. On the mechanism of the ascorbic acid-induced release of nitric oxide from N-nitrosated tryptophan derivatives: scavenging of NO by ascorbyl radicals. Chemistry. 2006;12:8786–97. doi: 10.1002/chem.200600405. [DOI] [PubMed] [Google Scholar]
- 240.Feelisch M. Biotransformation to nitric oxide of organic nitrates in comparison to other nitrovasodilators. Eur Heart J. 1993;14(Suppl I):123–32. [PubMed] [Google Scholar]
- 241.Feelisch M, Kelm M. Biotransformation of organic nitrates to nitric oxide by vascular smooth muscle and endothelial cells. Biochem Biophys Res Commun. 1991;180:286–93. doi: 10.1016/s0006-291x(05)81290-2. [DOI] [PubMed] [Google Scholar]
- 242.Feelisch M, Noack EA. Correlation between nitric oxide formation during degradation of organic nitrates and activation of guanylate cyclase. Eur J Pharmacol. 1987;139:19–30. doi: 10.1016/0014-2999(87)90493-6. [DOI] [PubMed] [Google Scholar]
- 243.Scorza G, Minetti M. One-electron oxidation pathway of thiols by peroxynitrite in biological fluids: bicarbonate and ascorbate promote the formation of albumin disulphide dimers in human blood plasma. Biochem J. 1998;329(Pt 2):405–13. doi: 10.1042/bj3290405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Scorza G, Pietraforte D, Minetti M. Role of ascorbate and protein thiols in the release of nitric oxide from S-nitroso-albumin and S-nitroso-glutathione in human plasma. Free Radic Biol Med. 1997;22:633–42. doi: 10.1016/s0891-5849(96)00378-4. [DOI] [PubMed] [Google Scholar]
- 245.Benjamin N, O'Driscoll F, Dougall H, Duncan C, Smith L, Golden M, McKenzie H. Stomach NO synthesis. Nature. 1994;368:502. doi: 10.1038/368502a0. [DOI] [PubMed] [Google Scholar]
- 246.Lundberg JO, Weitzberg E, Lundberg JM, Alving K. Intragastric nitric oxide production in humans: measurements in expelled air. Gut. 1994;35:1543–6. doi: 10.1136/gut.35.11.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Duncan C, Dougall H, Johnston P, Green S, Brogan R, Leifert C, Smith L, Golden M, Benjamin N. Chemical generation of nitric oxide in the mouth from the enterosalivary circulation of dietary nitrate. Nat Med. 1995;1:546–51. doi: 10.1038/nm0695-546. [DOI] [PubMed] [Google Scholar]
- 248.Dykhuizen RS, Fraser A, McKenzie H, Golden M, Leifert C, Benjamin N. Helicobacter pylori is killed by nitrite under acidic conditions. Gut. 1998;42:334–7. doi: 10.1136/gut.42.3.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Dykhuizen RS, Frazer R, Duncan C, Smith CC, Golden M, Benjamin N, Leifert C. Antimicrobial effect of acidified nitrite on gut pathogens: importance of dietary nitrate in host defense. Antimicrob Agents Chemother. 1996;40:1422–5. doi: 10.1128/aac.40.6.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Allaker RP, Silva Mendez LS, Hardie JM, Benjamin N. Antimicrobial effect of acidified nitrite on periodontal bacteria. Oral Microbiol Immunol. 2001;16:253–6. doi: 10.1034/j.1399-302x.2001.160410.x. [DOI] [PubMed] [Google Scholar]
- 251.Anyim M, Benjamin N, Wilks M. Acidified nitrite as a potential antifungal agent. Int J Antimicrob Agents. 2005;26:85–7. doi: 10.1016/j.ijantimicag.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 252.Silva Mendez LS, Allaker RP, Hardie JM, Benjamin N. Antimicrobial effect of acidified nitrite on cariogenic bacteria. Oral Microbiol Immunol. 1999;14:391–2. doi: 10.1034/j.1399-302x.1999.140612.x. [DOI] [PubMed] [Google Scholar]
- 253.Phillips R, Kuijper S, Benjamin N, Wansbrough-Jones M, Wilks M, Kolk AH. In vitro killing of Mycobacterium ulcerans by acidified nitrite. Antimicrob Agents Chemother. 2004;48:3130–2. doi: 10.1128/AAC.48.8.3130-3132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Duncan C, Li H, Dykhuizen R, Frazer R, Johnston P, MacKnight G, Smith L, Lamza K, McKenzie H, Batt L, Kelly D, Golden M, Benjamin N, Leifert C. Protection against oral and gastrointestinal diseases: importance of dietary nitrate uptake, oral nitrate reduction and enterosalivary nitrate circulation. Comparative Biochemistry Physiology. 1997;118A:939–948. doi: 10.1016/s0300-9629(97)00023-6. [DOI] [PubMed] [Google Scholar]
- 255.McKnight GM, Smith LM, Drummond RS, Duncan CW, Golden M, Benjamin N. Chemical synthesis of nitric oxide in the stomach from dietary nitrate in humans. Gut. 1997;40:211–4. doi: 10.1136/gut.40.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Weller R, Pattullo S, Smith L, Golden M, Ormerod A, Benjamin N. Nitric oxide is generated on the skin surface by reduction of sweat nitrate. J Invest Dermatol. 1996;107:327–31. doi: 10.1111/1523-1747.ep12363167. [DOI] [PubMed] [Google Scholar]
- 257.Ma SX, Li XY, Sakurai T, Pandjaitan M. Evidence of enhanced non-enzymatic generation of nitric oxide on the skin surface of acupuncture points: An innovative approach in humans. Nitric Oxide. 2007;17:60–8. doi: 10.1016/j.niox.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 258.Lundberg JO, Carlsson S, Engstrand L, Morcos E, Wiklund NP, Weitzberg E. Urinary nitrite: more than a marker of infection. Urology. 1997;50:189–91. doi: 10.1016/S0090-4295(97)00257-4. [DOI] [PubMed] [Google Scholar]
- 259.Vallette G, Tenaud I, Branka JE, Jarry A, Sainte-Marie I, Dreno B, Laboisse CL. Control of growth and differentiation of normal human epithelial cells through the manipulation of reactive nitrogen species. Biochem J. 1998;331(Pt 3):713–7. doi: 10.1042/bj3310713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Benjamin N, Pattullo S, Weller R, Smith L, Ormerod A. Wound licking and nitric oxide. Lancet. 1997;349:1776. doi: 10.1016/S0140-6736(05)63002-4. [DOI] [PubMed] [Google Scholar]
- 261.Gribbe O, Gustafsson LE, Wiklund NP. Transdermally administered nitric oxide by application of acidified nitrite increases blood flow in rat epigastric island skin flaps. Eur J Pharmacol. 2008;578:51–6. doi: 10.1016/j.ejphar.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 262.Dejam A, Hunter CJ, Gladwin MT. Effects of dietary nitrate on blood pressure. N Engl J Med. 2007;356:1590. doi: 10.1056/NEJMc070163. author reply 1590. [DOI] [PubMed] [Google Scholar]
- 263.Larsen FJ, Ekblom B, Sahlin K, Lundberg JO, Weitzberg E. Effects of dietary nitrate on blood pressure in healthy volunteers. N Engl J Med. 2006;355:2792–3. doi: 10.1056/NEJMc062800. [DOI] [PubMed] [Google Scholar]
- 264.Zweier JL, Wang P, Samouilov A, Kuppusamy P. Enzyme-independent formation of nitric oxide in biological tissues. Nat Med. 1995;1:804–9. doi: 10.1038/nm0895-804. [DOI] [PubMed] [Google Scholar]
- 265.Zweier JL, Wang P, Samouilov A, Kuppusamy P. Enzymatic/non-enzymatic formation of nitric oxide. Nature Medicine. 1995;1:1103–1104. doi: 10.1038/nm0895-804. [DOI] [PubMed] [Google Scholar]
- 266.Zweier JL, Li H, Samouilov A, Liu X. Mechanisms of nitrite reduction to nitric oxide in the heart and vessel wall. Nitric Oxide. 2010;22:83–90. doi: 10.1016/j.niox.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Gladwin MT, Raat NJ, Shiva S, Dezfulian C, Hogg N, Kim-Shapiro DB, Patel RP. Nitrite as a vascular endocrine nitric oxide reservoir that contributes to hypoxic signaling, cytoprotection, and vasodilation. Am J Physiol Heart Circ Physiol. 2006;291:H2026–35. doi: 10.1152/ajpheart.00407.2006. [DOI] [PubMed] [Google Scholar]
- 268.Modin A, Bjorne H, Herulf M, Alving K, Weitzberg E, Lundberg JO. Nitrite-derived nitric oxide: a possible mediator of ‘acidic-metabolic’ vasodilation. Acta Physiol Scand. 2001;171:9–16. doi: 10.1046/j.1365-201X.2001.00771.x. [DOI] [PubMed] [Google Scholar]
- 269.Salter M, Duffy C, Garthwaite J, Strigbos PJLM. Ex vivo measurement of brain tissue nitrite and nitrate accurately reflects nitric oxide synthase activity in vivo. J. Neurochem. 1996;66:1683–1690. doi: 10.1046/j.1471-4159.1996.66041683.x. [DOI] [PubMed] [Google Scholar]
- 270.Oke AF, May L, Adams RN. Ascorbic acid distribution patterns in human brain. A comparison with nonhuman mammalian species. Ann N. Y. Acad Sci. 1987;498:1–12. doi: 10.1111/j.1749-6632.1987.tb23747.x. [DOI] [PubMed] [Google Scholar]
- 271.Hornig D. Distribution of ascorbic acid, metabolites and analogues in man and animals. Annu. N.Y.Acad. Sci. 1975;258:103–115. doi: 10.1111/j.1749-6632.1975.tb29271.x. [DOI] [PubMed] [Google Scholar]
- 272.Rice ME. Ascorbate compartmentalization in the CNS. Neurotox Res. 1999;1:81–90. doi: 10.1007/BF03033272. [DOI] [PubMed] [Google Scholar]
- 273.Grunewald RA. Ascorbic acid in the brain. Brain Res Brain Res Rev. 1993;18:123–33. doi: 10.1016/0165-0173(93)90010-w. [DOI] [PubMed] [Google Scholar]
- 274.Miele M, Fillenz M. In vivo determination of extracellular brain ascorbate. J. Neurosci. Meth. 1996;70:15–19. doi: 10.1016/S0165-0270(96)00094-5. [DOI] [PubMed] [Google Scholar]
- 275.Rice ME, Nicholson C. Interstitial ascorbate in turtle brain is modulated by release and extracellular volume changes. J. Neurochem. 1987;49:1096–1104. doi: 10.1111/j.1471-4159.1987.tb09999.x. [DOI] [PubMed] [Google Scholar]
- 276.Grunewald RA. Ascorbic acid in the brain. Brain Res. Rev. 1993;18:123–133. doi: 10.1016/0165-0173(93)90010-w. [DOI] [PubMed] [Google Scholar]
- 277.Siushansian R, Wilson JX. Ascorbate transport and intracellular concentrations in cerebral astrocytes. J. Neurochem. 1993;65:41–49. doi: 10.1046/j.1471-4159.1995.65010041.x. [DOI] [PubMed] [Google Scholar]
- 278.Kim WS, Dahlgren RL, Moroz LL, Sweedler JV. Ascorbic acid assays of individual neurons and neuronal tissues using capillary electrophoresis with laser-induced fluorescence detection. Anal Chem. 2002;74:5614–20. doi: 10.1021/ac025917q. [DOI] [PubMed] [Google Scholar]
- 279.Millar J. The nitric oxide/ascorbate cycle: how neurones may control their own oxygen supply. Med Hypotheses. 1995;45:21–6. doi: 10.1016/0306-9877(95)90194-9. [DOI] [PubMed] [Google Scholar]
- 280.Casey JR, Grinstein S, Orlowski J. Sensors and regulators of intracellular pH. Nature Reviews in Molecular and Cell Biology. 2010;11:50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- 281.Mellman I. The importance of being acid: the role of acidification in intracellular membrane traffic. J. Exp. Biol. 1992;172:39–45. doi: 10.1242/jeb.172.1.39. [DOI] [PubMed] [Google Scholar]
- 282.Kornfeld S, Mellaman I. The biogenesis of lysosomes. Annu. Rev. Cell Biol. 1989;5:483–525. doi: 10.1146/annurev.cb.05.110189.002411. [DOI] [PubMed] [Google Scholar]
- 283.Miesenbock G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394:192–5. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 284.Palmer MJ, Hull C, Vigh J, von Gersdorff H. Synaptic cleft acidification and modulation of short-term depression by exocytosed protons in retinal bipolar cells. J Neurosci. 2003;23:11332–41. doi: 10.1523/JNEUROSCI.23-36-11332.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Traynelis SF, Chesler M. Proton release as a modulator of presynaptic function. Neuron. 2001;32:960–2. doi: 10.1016/s0896-6273(01)00549-9. [DOI] [PubMed] [Google Scholar]
- 286.DeVries SH. Exocytosed protons feedback to suppress the Ca2+ current in mammalian cone photoreceptors. Neuron. 2001;32:1107–17. doi: 10.1016/s0896-6273(01)00535-9. [DOI] [PubMed] [Google Scholar]
- 287.Yuste R, Miller RB, Holthoff K, Zhang S, Miesenbock G. Synapto-pHluorins: chimeras between pH-sensitive mutants of green fluorescent protein and synaptic vesicle membrane proteins as reporters of neurotransmitter release. Methods Enzymol. 2000;327:522–46. doi: 10.1016/s0076-6879(00)27300-x. [DOI] [PubMed] [Google Scholar]
- 288.Beg AA, Ernstrom GG, Nix P, Davis MW, Jorgensen EM. Protons act as a transmitter for muscle contraction in C. elegans. Cell. 2008;132:149–60. doi: 10.1016/j.cell.2007.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Kim JS, Zhen M. Protons as intercellular messengers. Cell. 2008;132:21–2. doi: 10.1016/j.cell.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 290.Pfeiffer J, Johnson D, Nehrke K. Oscillatory transepithelial H (+) flux regulates a rhythmic behavior in C. elegans. Curr Biol. 2008;18:297–302. doi: 10.1016/j.cub.2008.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Prolo LM, Goodman MB. Physiology: Keeping it regular with protons. Nature. 2008;452:35–6. doi: 10.1038/452035a. [DOI] [PubMed] [Google Scholar]
- 292.Nehrke K. Intracellular pH measurements in vivo using green fluorescent protein variants. Methods Mol Biol. 2006;351:223–39. doi: 10.1385/1-59745-151-7:223. [DOI] [PubMed] [Google Scholar]
- 293.Fossier P, Blanchard B, Ducrocq C, Leprince C, Tauc L, Baux G. Nitric oxide transforms serotonin into an inactive form and this affects neuromodulation. Neuroscience. 1999;93:597–603. doi: 10.1016/s0306-4522(99)00165-7. [DOI] [PubMed] [Google Scholar]
- 294.Kirsch M, de Groot H. N-nitrosomelatonin: synthesis, chemical properties, potential prodrug. J Pineal Res. 2009;46:121–7. doi: 10.1111/j.1600-079X.2008.00655.x. [DOI] [PubMed] [Google Scholar]
- 295.Feelisch M. Nitrated cyclic GMP as a new cellular signal. Nat Chem Biol. 2007;3:687–8. doi: 10.1038/nchembio1107-687. [DOI] [PubMed] [Google Scholar]
- 296.Goldstein S, Rabani J. Mechanism of nitrite formation by nitrate photolysis in aqueous solutions: the role of peroxynitrite, nitrogen dioxide, and hydroxyl radical. J Am Chem Soc. 2007;129:10597–601. doi: 10.1021/ja073609+. [DOI] [PubMed] [Google Scholar]
- 297.Roca M, Zahardis J, Bone J, El-Maazawi M, Grassian VH. 310 nm Irradiation of Atmospherically Relevant Concentrated Aqueous Nitrate Solutions: Nitrite Production and Quantum Yields. J Phys Chem A. 2008;112:13275–81. doi: 10.1021/jp809017b. [DOI] [PubMed] [Google Scholar]
- 298.Dejam A, Kleinbongard P, Rassaf T, Hamada S, Gharini P, Rodriguez J, Feelisch M, Kelm M. Thiols enhance NO formation from nitrate photolysis. Free Radic Biol Med. 2003;35:1551–9. doi: 10.1016/j.freeradbiomed.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 299.Zafiriou OC, McFarland M, Bromund RH. Nitric Oxide in Seawater. Science. 1980;207:637–639. doi: 10.1126/science.207.4431.637. [DOI] [PubMed] [Google Scholar]
- 300.McFarland M, K. D., Drummond JW, Schmeltekopf AL, Winkler RM. Nitric oxide measurements in the equatorial Pacific region. Geophys Res Lett. 1979;6:605–609. [Google Scholar]
- 301.Roenneberg T, Rehman J. Nitrate, a nonphotic signal for the circadian system. Faseb J. 1996;10:1443–7. doi: 10.1096/fasebj.10.12.8903515. [DOI] [PubMed] [Google Scholar]
- 302.Artinian LR, Ding JM, Gillette MU. Carbon monoxide and nitric oxide: interacting messengers in muscarinic signaling to the brain's circadian clock. Exp Neurol. 2001;171:293–300. doi: 10.1006/exnr.2001.7781. [DOI] [PubMed] [Google Scholar]
- 303.Ding JM, Chen D, Weber ET, Faiman LE, Rea MA, Gillette MU. Resetting the biological clock: mediation of nocturnal circadian shifts by glutamate and NO. Science. 1994;266:1713–7. doi: 10.1126/science.7527589. [DOI] [PubMed] [Google Scholar]
- 304.Golombek DA, Agostino PV, Plano SA, Ferreyra GA. Signaling in the mammalian circadian clock: the NO/cGMP pathway. Neurochem Int. 2004;45:929–36. doi: 10.1016/j.neuint.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 305.Golombek DA, Ferreyra GA, Agostino PV, Murad AD, Rubio MF, Pizzio GA, Katz ME, Marpegan L, Bekinschtein TA. From light to genes: moving the hands of the circadian clock. Front Biosci. 2003;8:s285–93. doi: 10.2741/1038. [DOI] [PubMed] [Google Scholar]
- 306.Zhang DQ, Zhou T, Ruan GX, McMahon DG. Circadian rhythm of Period1 clock gene expression in NOS amacrine cells of the mouse retina. Brain Res. 2005;1050:101–9. doi: 10.1016/j.brainres.2005.05.042. [DOI] [PubMed] [Google Scholar]
- 307.Paunel AN, Dejam A, Thelen S, Kirsch M, Horstjann M, Gharini P, Murtz M, Kelm M, de Groot H, Kolb-Bachofen V, Suschek CV. Enzyme-independent nitric oxide formation during UVA challenge of human skin: characterization, molecular sources, and mechanisms. Free Radic Biol Med. 2005;38:606–15. doi: 10.1016/j.freeradbiomed.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 308.Suschek CV, Paunel A, Kolb-Bachofen V. Nonenzymatic nitric oxide formation during UVA irradiation of human skin: experimental setups and ways to measure. Methods Enzymol. 2005;396:568–78. doi: 10.1016/S0076-6879(05)96048-5. [DOI] [PubMed] [Google Scholar]
- 309.Bruch-Gerharz D, Schnorr O, Suschek C, Beck KF, Pfeilschifter J, Ruzicka T, Kolb-Bachofen V. Arginase 1 overexpression in psoriasis: limitation of inducible nitric oxide synthase activity as a molecular mechanism for keratinocyte hyperproliferation. Am J Pathol. 2003;162:203–11. doi: 10.1016/S0002-9440(10)63811-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Bryan NS. Nitrite in nitric oxide biology: cause or consequence? A systems-based review. Free Radic Biol Med. 2006;41:691–701. doi: 10.1016/j.freeradbiomed.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 311.Bryan NS, Fernandez BO, Bauer SM, Garcia-Saura MF, Milsom AB, Rassaf T, Maloney RE, Bharti A, Rodriguez J, Feelisch M. Nitrite is a signaling molecule and regulator of gene expression in mammalian tissues. Nat Chem Biol. 2005;1:290–7. doi: 10.1038/nchembio734. [DOI] [PubMed] [Google Scholar]
- 312.Gladwin MT. Nitrite as an intrinsic signaling molecule. Nat Chem Biol. 2005;1:245–6. doi: 10.1038/nchembio1005-245. [DOI] [PubMed] [Google Scholar]
- 313.Camargo A, Llamas A, Schnell RA, Higuera JJ, Gonzalez-Ballester D, Lefebvre PA, Fernandez E, Galvan A. Nitrate signaling by the regulatory gene NIT2 in Chlamydomonas. Plant Cell. 2007;19:3491–503. doi: 10.1105/tpc.106.045922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Bredt DS, Hwang PM, Glatt CE, Lowenstein C, Reed RR, Snyder SH. Cloned and expressed nitric oxide synthase structurally resembles cytochrome P-450 reductase. Nature. 1991;351:714–8. doi: 10.1038/351714a0. [DOI] [PubMed] [Google Scholar]
- 315.Lowenstein CJ, Glatt CS, Bredt DS, Snyder SH. Cloned and expressed macrophage nitric oxide synthase contrasts with the brain enzyme. Proc Natl Acad Sci U S A. 1992;89:6711–5. doi: 10.1073/pnas.89.15.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Lyons CR, Orloff GJ, Cunningham JM. Molecular cloning and functional expression of an inducible nitric oxide synthase from a murine macrophage cell line. J Biol Chem. 1992;267:6370–4. [PubMed] [Google Scholar]
- 317.Xie QW, Cho HJ, Calaycay J, Mumford RA, Swiderek KM, Lee TD, Ding A, Troso T, Nathan C. Cloning and characterization of inducible nitric oxide synthase from mouse macrophages. Science. 1992;256:225–8. doi: 10.1126/science.1373522. [DOI] [PubMed] [Google Scholar]
- 318.Janssens SP, Shimouchi A, Quertermous T, Bloch DB, Bloch KD. Cloning and expression of a cDNA encoding human endothelium-derived relaxing factor/nitric oxide synthase. J Biol Chem. 1992;267:14519–22. [PubMed] [Google Scholar]
- 319.Marsden PA, Schappert KT, Chen HS, Flowers M, Sundell CL, Wilcox JN, Lamas S, Michel T. Molecular cloning and characterization of human endothelial nitric oxide synthase. FEBS Lett. 1992;307:287–93. doi: 10.1016/0014-5793(92)80697-f. [DOI] [PubMed] [Google Scholar]
- 320.Lamas S, Marsden PA, Li GK, Tempst P, Michel T. Endothelial nitric oxide synthase: molecular cloning and characterization of a distinct constitutive enzyme isoform. Proc Natl Acad Sci U S A. 1992;89:6348–52. doi: 10.1073/pnas.89.14.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Nishida K, Harrison DG, Navas JP, Fisher AA, Dockery SP, Uematsu M, Nerem RM, Alexander RW, Murphy TJ. Molecular cloning and characterization of the constitutive bovine aortic endothelial cell nitric oxide synthase. J Clin Invest. 1992;90:2092–6. doi: 10.1172/JCI116092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Sessa WC, Harrison JK, Barber CM, Zeng D, Durieux ME, D'Angelo DD, Lynch KR, Peach MJ. Molecular cloning and expression of a cDNA encoding endothelial cell nitric oxide synthase. J Biol Chem. 1992;267:15274–6. [PubMed] [Google Scholar]
- 323.Roman LJ, Martasek P, Masters BS. Intrinsic and extrinsic modulation of nitric oxide synthase activity. Chem Rev. 2002;102:1179–90. doi: 10.1021/cr000661e. [DOI] [PubMed] [Google Scholar]
- 324.Stuehr DJ. Mammalian nitric oxide synthases. Biochim Biophys Acta. 1999;1411:217–30. doi: 10.1016/s0005-2728(99)00016-x. [DOI] [PubMed] [Google Scholar]
- 325.Knowles RG, Moncada S. Nitric oxide synthases in mammals. Biochem J. 1994;298:249–258. doi: 10.1042/bj2980249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Zhou L, Zhu DY. Neuronal nitric oxide synthase: structure, subcellular localization, regulation, and clinical implications. Nitric Oxide. 2009;20:223–30. doi: 10.1016/j.niox.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 327.Garcin ED, Bruns CM, Lloyd SJ, Hosfield DJ, Tiso M, Gachhui R, Stuehr DJ, Tainer JA, Getzoff ED. Structural basis for isozyme-specific regulation of electron transfer in nitric-oxide synthase. J Biol Chem. 2004;279:37918–27. doi: 10.1074/jbc.M406204200. [DOI] [PubMed] [Google Scholar]
- 328.Stuehr DJ, Santolini J, Wang ZQ, Wei CC, Adak S. Update on mechanism and catalytic regulation in the NO synthases. J Biol Chem. 2004;279:36167–70. doi: 10.1074/jbc.R400017200. [DOI] [PubMed] [Google Scholar]
- 329.Adak S, Aulak KS, Stuehr DJ. Direct evidence for nitric oxide production by a nitric-oxide synthase-like protein from Bacillus subtilis. J Biol Chem. 2002;277:16167–71. doi: 10.1074/jbc.M201136200. [DOI] [PubMed] [Google Scholar]
- 330.Adak S, Bilwes AM, Panda K, Hosfield D, Aulak KS, McDonald JF, Tainer JA, Getzoff ED, Crane BR, Stuehr DJ. Cloning, expression, and characterization of a nitric oxide synthase protein from Deinococcus radiodurans. Proc Natl Acad Sci U S A. 2002;99:107–12. doi: 10.1073/pnas.012470099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Moroz LL, Park JH, Winlow W. Nitric oxide activates buccal motor patterns in Lymnaea stagnalis. Neuroreport. 1993;4:643–6. doi: 10.1097/00001756-199306000-00010. [DOI] [PubMed] [Google Scholar]
- 332.Moroz LL, Winlow W, Turner RW, Bulloch AG, Lukowiak K, Syed NI. Nitric oxide synthaseimmunoreactive cells in the CNS and periphery of Lymnaea. Neuroreport. 1994;5:1277–80. doi: 10.1097/00001756-199406020-00031. [DOI] [PubMed] [Google Scholar]
- 333.Moroz LL. Localization of putative nitrergic neurons in peripheral chemosensory areas and the central nervous system of Aplysia californica. J Comp Neurol. 2006;495:10–20. doi: 10.1002/cne.20842. [DOI] [PubMed] [Google Scholar]
- 334.Moroz LL, Gillette R. NADPH-diaphorase localization in the CNS and peripheral tissues of the predatory sea-slug Pleurobranchaea californica. J Comp Neurol. 1996;367:607–22. doi: 10.1002/(SICI)1096-9861(19960415)367:4<607::AID-CNE10>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 335.Kurenni DE, Thurlow GA, Turner RW, Moroz LL, Sharkey KA, Barnes S. Nitric oxide synthase in tiger salamander retina. J Comp Neurol. 1995;361:525–36. doi: 10.1002/cne.903610314. [DOI] [PubMed] [Google Scholar]
- 336.Kurenny DE, Moroz LL, Turner RW, Sharkey KA, Barnes S. Modulation of ion channels in rod photoreceptors by nitric oxide. Neuron. 1994;13:315–24. doi: 10.1016/0896-6273(94)90349-2. [DOI] [PubMed] [Google Scholar]
- 337.Colasanti M, Venturini G, Merante A, Musci G, Lauro GM. Nitric oxide involvement in Hydra vulgaris very primitive olfactory-like system. J Neurosci. 1997;17:493–9. doi: 10.1523/JNEUROSCI.17-01-00493.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Cristino L, Guglielmotti V, Cotugno A, Musio C, Santillo S. Nitric oxide signaling pathways at neural level in invertebrates: functional implications in cnidarians. Brain Res. 2008;1225:17–25. doi: 10.1016/j.brainres.2008.04.056. [DOI] [PubMed] [Google Scholar]
- 339.Colasanti M, Mazzone V, Mancinelli L, Leone S, Venturini G. Involvement of nitric oxide in the head regeneration of Hydra vulgaris. Nitric Oxide. 2009;21:164–70. doi: 10.1016/j.niox.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 340.Rivero A. Nitric oxide: an antiparasitic molecule of invertebrates. Trends Parasitol. 2006;22:219–25. doi: 10.1016/j.pt.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 341.Kohn AB, Lea JM, Moroz LL, Greenberg RM. Schistosoma mansoni: use of a fluorescent indicator to detect nitric oxide and related species in living parasites. Exp Parasitol. 2006;113:130–3. doi: 10.1016/j.exppara.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 342.Morton DB. Invertebrates yield a plethora of atypical guanylyl cyclases. Mol Neurobiol. 2004;29:97–116. doi: 10.1385/MN:29:2:097. [DOI] [PubMed] [Google Scholar]
- 343.Toda N, Ayajiki K. Phylogenesis of constitutively formed nitric oxide in non-mammals. Rev Physiol Biochem Pharmacol. 2006;157:31–80. doi: 10.1007/112_0601. [DOI] [PubMed] [Google Scholar]
- 344.Ray SS, Sengupta R, Tiso M, Haque MM, Sahoo R, Konas DW, Aulak K, Regulski M, Tully T, Stuehr DJ, Ghosh S. Reductase domain of Drosophila melanogaster nitric-oxide synthase: redox transformations, regulation, and similarity to mammalian homologues. Biochemistry. 2007;46:11865–73. doi: 10.1021/bi700805x. [DOI] [PubMed] [Google Scholar]
- 345.Wang XW, Tan NS, Ho B, Ding JL. Evidence for the ancient origin of the NF-kappaB/IkappaB cascade: its archaic role in pathogen infection and immunity. Proc Natl Acad Sci U S A. 2006;103:4204–9. doi: 10.1073/pnas.0507044103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Nighorn A, Gibson NJ, Rivers DM, Hildebrand JG, Morton DB. The nitric oxide-cGMP pathway may mediate communication between sensory afferents and projection neurons in the antennal lobe of Manduca sexta. J Neurosci. 1998;18:7244–55. doi: 10.1523/JNEUROSCI.18-18-07244.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Regulski M, Tully T. Molecular and biochemical characterization of dNOS: a Drosophila Ca2+/calmodulin-dependent nitric oxide synthase. Proc Natl Acad Sci U S A. 1995;92:9072–6. doi: 10.1073/pnas.92.20.9072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Yuda M, Hirai M, Miura K, Matsumura H, Ando K, Chinzei Y. cDna cloning, expression and characterization of nitric-oxide synthase from the salivary glands of the blood-sucking insect Rhodnius prolixus. Eur J Biochem. 1996;242:807–12. doi: 10.1111/j.1432-1033.1996.0807r.x. [DOI] [PubMed] [Google Scholar]
- 349.Korneev SA, Piper MR, Picot J, Phillips R, Korneeva EI, O'Shea M. Molecular characterization of NOS in a mollusc: expression in a giant modulatory neuron. J Neurobiol. 1998;35:65–76. doi: 10.1002/(sici)1097-4695(199804)35:1<65::aid-neu6>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 350.Sadreyev RI, Panchin Y, Uvarov P, Belyavski A, Matz M, Moroz LL. Cloning of nitric oxide synthase (NOS) from Aplysia californica. Soc Neurosci Abstracts. 2000;26 [Google Scholar]
- 351.Moroz LL, Edwards JR, Puthanveettil SV, Kohn AB, Ha T, Heyland A, Knudsen B, Sahni A, Yu F, Liu L, Jezzini S, Lovell P, Iannucculli W, Chen M, Nguyen T, Sheng H, Shaw R, Kalachikov S, Panchin YV, Farmerie W, Russo JJ, Ju J, Kandel ER. Neuronal transcriptome of Aplysia: neuronal compartments and circuitry. Cell. 2006;127:1453–67. doi: 10.1016/j.cell.2006.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Di Cristo C, Fiore G, Scheinker V, Enikolopov G, d'Ischia M, Palumbo A, Di Cosmo A. Nitric oxide synthase expression in the central nervous system of Sepia officinalis: an in situ hybridization study. Eur J Neurosci. 2007;26:1599–610. doi: 10.1111/j.1460-9568.2007.05765.x. [DOI] [PubMed] [Google Scholar]
- 353.Matsuo R, Misawa K, Ito E. Genomic structure of nitric oxide synthase in the terrestrial slug is highly conserved. Gene. 2008;415:74–81. doi: 10.1016/j.gene.2008.02.021. [DOI] [PubMed] [Google Scholar]
- 354.Golderer G, Werner ER, Leitner S, Grobner P, Werner-Felmayer G. Nitric oxide synthase is induced in sporulation of Physarum polycephalum. Genes Dev. 2001;15:1299–309. doi: 10.1101/gad.890501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Labbe P, McTaggart SJ, Little TJ. An ancient immunity gene duplication in Daphnia magna: RNA expression and sequence analysis of two nitric oxide synthase genes. Dev Comp Immunol. 2009;33:1000–10. doi: 10.1016/j.dci.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Ninnemann H, Maier J. Indications for the occurrence of nitric oxide synthases in fungi and plants and the involvement in photoconidiation of Neurospora crassa. Photochem Photobiol. 1996;64:393–8. doi: 10.1111/j.1751-1097.1996.tb02477.x. [DOI] [PubMed] [Google Scholar]
- 357.Durner J, Klessig DF. Nitric oxide as a signal in plants. Curr Opin Plant Biol. 1999;2:369–74. doi: 10.1016/s1369-5266(99)00007-2. [DOI] [PubMed] [Google Scholar]
- 358.Durner J, Wendehenne D, Klessig DF. Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc Natl Acad Sci U S A. 1998;95:10328–33. doi: 10.1073/pnas.95.17.10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Caro A, Puntarulo S. Free Radic Res. Vol. 31. Suppl: 1999. Nitric oxide generation by soybean embryonic axes. Possible effect on mitochondrial function. pp. S205–12. [DOI] [PubMed] [Google Scholar]
- 360.Ribeiro EA, Jr., Cunha FQ, Tamashiro WM, Martins IS. Growth phase-dependent subcellular localization of nitric oxide synthase in maize cells. FEBS Lett. 1999;445:283–6. doi: 10.1016/s0014-5793(99)00138-6. [DOI] [PubMed] [Google Scholar]
- 361.Delledonne M, Xia Y, Dixon RA, Lamb C. Nitric oxide functions as a signal in plant disease resistance. Nature. 1998;394:585–8. doi: 10.1038/29087. [DOI] [PubMed] [Google Scholar]
- 362.Cueto M, Hernandez-Perera O, Martin R, Bentura ML, Rodrigo J, Lamas S, Golvano MP. Presence of nitric oxide synthase activity in roots and nodules of Lupinus albus. FEBS Lett. 1996;398:159–64. doi: 10.1016/s0014-5793(96)01232-x. [DOI] [PubMed] [Google Scholar]
- 363.Barroso JB, Corpas FJ, Carreras A, Sandalio LM, Valderrama R, Palma JM, Lupianez JA, del Rio LA. Localization of nitric-oxide synthase in plant peroxisomes. J Biol Chem. 1999;274:36729–33. doi: 10.1074/jbc.274.51.36729. [DOI] [PubMed] [Google Scholar]
- 364.Huang S, Kerschbaum HH, Engel E, Hermann A. Biochemical characterization and histochemical localization of nitric oxide synthase in the nervous system of the snail, Helix pomatia. J Neurochem. 1997;69:2516–28. doi: 10.1046/j.1471-4159.1997.69062516.x. [DOI] [PubMed] [Google Scholar]
- 365.Ahlfors R, Brosche M, Kollist H, Kangasjarvi J. Nitric Oxide Modulates Ozone-Induced Cell Death, Hormone Biosynthesis and Gene Expression in Arabidopsis thaliana. Plant J. 2009;58:1–12. doi: 10.1111/j.1365-313X.2008.03756.x. [DOI] [PubMed] [Google Scholar]
- 366.Kolbert Z, Bartha B, Erdei L. Exogenous auxin-induced NO synthesis is nitrate reductase-associated in Arabidopsis thaliana root primordia. J Plant Physiol. 2008;165:967–75. doi: 10.1016/j.jplph.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 367.Tun NN, Livaja M, Kieber JJ, Scherer GF. Zeatin-induced nitric oxide (NO) biosynthesis in Arabidopsis thaliana mutants of NO biosynthesis and of two-component signaling genes. New Phytol. 2008;178:515–31. doi: 10.1111/j.1469-8137.2008.02383.x. [DOI] [PubMed] [Google Scholar]
- 368.Zhao MG, Tian QY, Zhang WH. Nitric oxide synthase-dependent nitric oxide production is associated with salt tolerance in Arabidopsis. Plant Physiol. 2007;144:206–17. doi: 10.1104/pp.107.096842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Zemojtel T, Kolanczyk M, Kossler N, Stricker S, Lurz R, Mikula I, Duchniewicz M, Schuelke M, Ghafourifar P, Martasek P, Vingron M, Mundlos S. Mammalian mitochondrial nitric oxide synthase: characterization of a novel candidate. FEBS Lett. 2006;580:455–62. doi: 10.1016/j.febslet.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 370.Zemojtel T, Frohlich A, Palmieri MC, Kolanczyk M, Mikula I, Wyrwicz LS, Wanker EE, Mundlos S, Vingron M, Martasek P, Durner J. Plant nitric oxide synthase: a never-ending story? Trends Plant Sci. 2006;11:524–5. doi: 10.1016/j.tplants.2006.09.008. author reply 526-8. [DOI] [PubMed] [Google Scholar]
- 371.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–16. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 372.Moreau M, Lee GI, Wang Y, Crane BR, Klessig DF. AtNOS/AtNOA1 Is a Functional Arabidopsis thaliana cGTPase and Not a Nitric-oxide Synthase. J Biol Chem. 2008;283:32957–67. doi: 10.1074/jbc.M804838200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Sudhamsu J, Lee GI, Klessig DF, Crane BR. The Structure of YqeH: AN AtNOS1/AtNOA1 Ortholog that couples GTP hydrolysis to molecular recognition. J Biol Chem. 2008;283:32968–76. doi: 10.1074/jbc.M804837200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Hawkins RD. Trans-synaptic signaling by NO during learning-related synaptic plasticity. In: Byrne JH, editor. Learning and memory -- A comprehensive reference. Elsevier; Oxford, U.K: 2008. [Google Scholar]
- 375.Muller U, Buchner E. Histochemical localization of NADPH-diaphorase in the adult Drosophila brain. Is nitric oxide a neuronal messenger also in insects? Naturwissenschaften. 1993;80:524–6. doi: 10.1007/BF01140811. [DOI] [PubMed] [Google Scholar]
- 376.Stasiv Y, Regulski M, Kuzin B, Tully T, Enikolopov G. The Drosophila nitric-oxide synthase gene (dNOS) encodes a family of proteins that can modulate NOS activity by acting as dominant negative regulators. J Biol Chem. 2001;276:42241–51. doi: 10.1074/jbc.M105066200. [DOI] [PubMed] [Google Scholar]
- 377.Muller U. Ca2+/calmodulin-dependent nitric oxide synthase in Apis mellifera and Drosophila melanogaster. Eur J Neurosci. 1994;6:1362–70. doi: 10.1111/j.1460-9568.1994.tb00327.x. [DOI] [PubMed] [Google Scholar]
- 378.Muller U, Hildebrandt H. The nitric oxide/cGMP system in the antennal lobe of Apis mellifera is implicated in integrative processing of chemosensory stimuli. Eur J Neurosci. 1995;7:2240–8. doi: 10.1111/j.1460-9568.1995.tb00645.x. [DOI] [PubMed] [Google Scholar]
- 379.Watanabe T, Kikuchi M, Hatakeyama D, Shiga T, Yamamoto T, Aonuma H, Takahata M, Suzuki N, Ito E. Gaseous neuromodulator-related genes expressed in the brain of honeybee Apis mellifera. Dev Neurobiol. 2007;67:456–73. doi: 10.1002/dneu.20359. [DOI] [PubMed] [Google Scholar]
- 380.Truman JW, De Vente J, Ball EE. Nitric oxide-sensitive guanylate cyclase activity is associated with the maturational phase of neuronal development in insects. Development. 1996;122:3949–58. doi: 10.1242/dev.122.12.3949. [DOI] [PubMed] [Google Scholar]
- 381.Enikolopov G, Banerji J, Kuzin B. Nitric oxide and Drosophila development. Cell Death Differ. 1999;6:956–63. doi: 10.1038/sj.cdd.4400577. [DOI] [PubMed] [Google Scholar]
- 382.Wildemann B, Bicker G. Developmental expression of nitric oxide/cyclic GMP synthesizing cells in the nervous system of Drosophila melanogaster. J Neurobiol. 1999;38:1–15. doi: 10.1002/(sici)1097-4695(199901)38:1<1::aid-neu1>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 383.Gibbs SM, Truman JW. Nitric oxide and cyclic GMP regulate retinal patterning in the optic lobe of Drosophila. Neuron. 1998;20:83–93. doi: 10.1016/s0896-6273(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 384.Kuzin B, Roberts I, Peunova N, Enikolopov G. Nitric oxide regulates cell proliferation during Drosophila development. Cell. 1996;87:639–49. doi: 10.1016/s0092-8674(00)81384-7. [DOI] [PubMed] [Google Scholar]
- 385.Shakiryanova D, Levitan ES. Prolonged presynaptic posttetanic cyclic GMP signaling in Drosophila motoneurons. Proc Natl Acad Sci U S A. 2008;105:13610–3. doi: 10.1073/pnas.0802131105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Wildemann B, Bicker G. Nitric oxide and cyclic GMP induce vesicle release at Drosophila neuromuscular junction. J Neurobiol. 1999;39:337–46. doi: 10.1002/(sici)1097-4695(19990605)39:3<337::aid-neu1>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 387.Hata Y, Nakanishi H, Takai Y. Synaptic PDZ domain-containing proteins. Neurosci Res. 1998;32:1–7. doi: 10.1016/s0168-0102(98)00069-8. [DOI] [PubMed] [Google Scholar]
- 388.Wilson CH, Christensen TA, Nighorn AJ. Inhibition of nitric oxide and soluble guanylyl cyclase signaling affects olfactory neuron activity in the moth, Manduca sexta. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2007;193:715–28. doi: 10.1007/s00359-007-0227-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Bicker G. Pharmacological approaches to nitric oxide signalling during neural development of locusts and other model insects. Arch Insect Biochem Physiol. 2007;64:43–58. doi: 10.1002/arch.20161. [DOI] [PubMed] [Google Scholar]
- 390.Dacher M, Gauthier M. Involvement of NO-synthase and nicotinic receptors in learning in the honey bee. Physiol Behav. 2008;95:200–7. doi: 10.1016/j.physbeh.2008.05.019. [DOI] [PubMed] [Google Scholar]
- 391.Muller U. Inhibition of nitric oxide synthase impairs a distinct form of long-term memory in the honeybee, Apis mellifera. Neuron. 1996;16:541–9. doi: 10.1016/s0896-6273(00)80073-2. [DOI] [PubMed] [Google Scholar]
- 392.Muller U. Molecular mechanisms of associative learning in the bee. In: Byrne JH, editor. Learning and memory -- A comprehensive reference. Elsevier; Oxford, U.K: 2008. [Google Scholar]
- 393.Muller U. Prolonged activation of cAMP-dependent protein kinase during conditioning induces long-term memory in honeybees. Neuron. 2000;27:159–68. doi: 10.1016/s0896-6273(00)00017-9. [DOI] [PubMed] [Google Scholar]
- 394.Muller U, Hildebrandt H. Nitric oxide/cGMP-mediated protein kinase A activation in the antennal lobes plays an important role in appetitive reflex habituation in the honeybee. J Neurosci. 2002;22:8739–47. doi: 10.1523/JNEUROSCI.22-19-08739.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–8. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- 396.Lovell P, Moroz LL. The largest growth cones in the animal kingdom and dynamics of neuronal growth in cell culture of Aplysia. Integrative and Comparative Biology. 2006;46:847–870. doi: 10.1093/icb/icl042. [DOI] [PubMed] [Google Scholar]
- 397.Moroz LL, Kohn AB. Do different neurons age differently? Direct genome-wide analysis of aging in single identified cholinergic neurons. Frontiers in Aging Neuroscience. 2010;2:1–18. doi: 10.3389/neuro.24.006.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Kandel ER. Cellular Basis of Behavior. W.H. Freeman and Company; San Francisco: 1976. [Google Scholar]
- 399.Kandel ER. Behavioral Biology of Aplysia. W.H. Freeman and Company; San Francisco: 1979. [Google Scholar]
- 400.Chase RB. Behavior & its neural control in gastropod molluscs. Oxford University Press; Oxford: 2002. [Google Scholar]
- 401.Li L, Garden RW, Sweedler JV. Single-cell MALDI: a new tool for direct peptide profiling. Trends Biotechnol. 2000;18:151–60. doi: 10.1016/s0167-7799(00)01427-x. [DOI] [PubMed] [Google Scholar]
- 402.Cummins SF, Erpenbeck D, Zou Z, Claudianos C, Moroz LL, Nagle GT, Degnan BM. Candidate chemoreceptor subfamilies differentially expressed in the chemosensory organs of the mollusc Aplysia. BMC Biol. 2009;7:28. doi: 10.1186/1741-7007-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Lee YS, Choi SL, Kim TH, Lee JA, Kim HK, Kim H, Jang DJ, Lee JJ, Lee S, Sin GS, Kim CB, Suzuki Y, Sugano S, Kubo T, Moroz LL, Kandel ER, Bhak J, Kaang BK. Transcriptome analysis and identification of regulators for long-term plasticity in Aplysia kurodai. Proc Natl Acad Sci U S A. 2008;105:18602–7. doi: 10.1073/pnas.0808893105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Katzoff A, Ben-Gedalya T, Hurwitz I, Miller N, Susswein YZ, Susswein AJ. Nitric oxide signals that aplysia have attempted to eat, a necessary component of memory formation after learning that food is inedible. J Neurophysiol. 2006;96:1247–57. doi: 10.1152/jn.00056.2006. [DOI] [PubMed] [Google Scholar]
- 405.Katzoff A, Ben-Gedalya T, Susswein AJ. Nitric oxide is necessary for multiple memory processes after learning that a food is inedible in aplysia. J Neurosci. 2002;22:9581–94. doi: 10.1523/JNEUROSCI.22-21-09581.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Kemenes I, Kemenes G, Andrew RJ, Benjamin PR, O'Shea M. Critical time-window for NO-cGMP-dependent long-term memory formation after one-trial appetitive conditioning. J Neurosci. 2002;22:1414–25. doi: 10.1523/JNEUROSCI.22-04-01414.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Chichery R, Chichery MP. NADPH-diaphorase in a cephalopod brain (Sepia): presence in an analogue of the cerebellum. Neuroreport. 1994;5:1273–6. doi: 10.1097/00001756-199406020-00030. [DOI] [PubMed] [Google Scholar]
- 408.Davidson SK, Koropatnick TA, Kossmehl R, Sycuro L, McFall-Ngai MJ. NO means ‘yes’ in the squid-vibrio symbiosis: nitric oxide (NO) during the initial stages of a beneficial association. Cell Microbiol. 2004;6:1139–51. doi: 10.1111/j.1462-5822.2004.00429.x. [DOI] [PubMed] [Google Scholar]
- 409.Di Cosmo A, Di Cristo C, Palumbo A, d'Ischia M, Messenger JB. Nitric oxide synthase (NOS) in the brain of the cephalopod Sepia officinalis. J Comp Neurol. 2000;428:411–27. doi: 10.1002/1096-9861(20001218)428:3<411::aid-cne3>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 410.Kimura T, Shouno O, Matsumoto G. NADPH-diaphorase containing cells and fibers in the central nervous system of squid, Loligo bleekeri keferstein. Life Sci. 1997;61:2375–81. doi: 10.1016/s0024-3205(97)00954-5. [DOI] [PubMed] [Google Scholar]
- 411.Palumbo A, Di Cosmo A, Gesualdo I, d'Ischia M. A calcium-dependent nitric oxide synthase and NMDA R1 glutamate receptor in the ink gland of Sepia officinalis: a hint to a regulatory role of nitric oxide in melanogenesis? Biochem Biophys Res Commun. 1997;235:429–32. doi: 10.1006/bbrc.1997.6734. [DOI] [PubMed] [Google Scholar]
- 412.Robertson JD, Bonaventura J, Kohm A, Hiscat M. Nitric oxide is necessary for visual learning in Octopus vulgaris. Proc Biol Sci. 1996;263:1739–43. doi: 10.1098/rspb.1996.0254. [DOI] [PubMed] [Google Scholar]
- 413.Robertson JD, Bonaventura J, Kohm AP. Nitric oxide is required for tactile learning in Octopus vulgaris. Proc Biol Sci. 1994;256:269–73. doi: 10.1098/rspb.1994.0080. [DOI] [PubMed] [Google Scholar]
- 414.Schipp R, Gebauer M. Nitric oxide: a vasodilatatory mediator in the cephalic aorta of Sepia officinalis (L.) (Cephalopoda) Invert Neurosci. 1999;4:9–15. doi: 10.1007/pl00022364. [DOI] [PubMed] [Google Scholar]
- 415.Tu Y, Budelmann BU. Effects of L-arginine on the afferent resting activity in the cephalopod statocyst. Brain Res. 1999;845:35–49. doi: 10.1016/s0006-8993(99)01929-0. [DOI] [PubMed] [Google Scholar]
- 416.Tu Y, Budelmann BU. Inhibitory effect of cyclic guanosine 3′,5′-monophosphate (cGMP) on the afferent resting activity in the cephalopod statocyst. Brain Res. 2000;880:65–9. doi: 10.1016/s0006-8993(00)02777-3. [DOI] [PubMed] [Google Scholar]
- 417.Tu Y, Budelmann BU. Effects of nitric oxide donors on the afferent resting activity in the cephalopod statocyst. Brain Res. 2000;865:211–20. doi: 10.1016/s0006-8993(00)02222-8. [DOI] [PubMed] [Google Scholar]
- 418.Antonov I, Ha T, Antonova I, Moroz LL, Hawkins RD. Role of nitric oxide in classical conditioning of siphon withdrawal in Aplysia. J Neurosci. 2007;27:10993–1002. doi: 10.1523/JNEUROSCI.2357-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Scheinker V, Fiore G, Di Cristo C, Di Cosmo A, d'Ischia M, Enikolopov G, Palumbo A. Nitric oxide synthase in the nervous system and ink gland of the cuttlefish Sepia officinalis: molecular cloning and expression. Biochem Biophys Res Commun. 2005;338:1204–15. doi: 10.1016/j.bbrc.2005.10.069. [DOI] [PubMed] [Google Scholar]
- 420.Moroz LL, Roger I, Syed NIAGM, Bulloch AGM, Lukowiak K. Abundance of putative NO-synthesizing cells in the oesophagus of gastropod molluscs. J. Physiol. 1993;473 [Google Scholar]
- 421.Hatcher NG, Sudlow LC, Moroz LL, Gillette R. Nitric oxide potentiates cAMP-gated cation current in feeding neurons of Pleurobranchaea californica independent of cAMP and cGMP signaling pathways. J Neurophysiol. 2006;95:3219–27. doi: 10.1152/jn.00815.2005. [DOI] [PubMed] [Google Scholar]
- 422.Kemenes G. Molecular mechanisms of associative learning in Lymnaea. In: Byrne JH, editor. Learning and memory -- A comprehensive reference. Elsevier; Oxford, U.K: 2008. [Google Scholar]
- 423.Korneev SA, Park JH, O'Shea M. Neuronal expression of neural nitric oxide synthase (nNOS) protein is suppressed by an antisense RNA transcribed from an NOS pseudogene. J Neurosci. 1999;19:7711–20. doi: 10.1523/JNEUROSCI.19-18-07711.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Korneev SA, Straub V, Kemenes I, Korneeva EI, Ott SR, Benjamin PR, O'Shea M. Timed and targeted differential regulation of nitric oxide synthase (NOS) and anti-NOS genes by reward conditioning leading to long-term memory formation. J Neurosci. 2005;25:1188–92. doi: 10.1523/JNEUROSCI.4671-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Sadamoto H, Ito E. Neuronal nitric oxide synthase in the pond snail, Lymnaea stagnalis. Society for Neuroscience Meeting, Abstracts. 2009;889:6/GG31. [Google Scholar]
- 426.Ha TJ, Kohn AB, Bobkova YV, Moroz LL. Molecular characterization of NMDA-like receptors in Aplysia and Lymnaea: relevance to memory mechanisms. Biol Bull. 2006;210:255–70. doi: 10.2307/4134562. [DOI] [PubMed] [Google Scholar]
- 427.Moroz LL, Gyori J, Salanki J. NMDA-like receptors in the CNS of molluscs. Neuroreport. 1993;4:201–4. doi: 10.1097/00001756-199302000-00022. [DOI] [PubMed] [Google Scholar]
- 428.Bodnarova M, Martasek P, Moroz LL. Calcium/calmodulin-dependent nitric oxide synthase activity in the CNS of Aplysia californica: biochemical characterization and link to cGMP pathways. J Inorg Biochem. 2005;99:922–8. doi: 10.1016/j.jinorgbio.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 429.Jacklet JW, Gruhn M. Co-localization of NADPH-diaphorase and myomodulin in synaptic glomeruli of Aplysia. Neuroreport. 1994;5:1841–4. doi: 10.1097/00001756-199409080-00040. [DOI] [PubMed] [Google Scholar]
- 430.Miller N, Katzoff A, Susswein AJ. Nitric oxide induces aspects of egg-laying behavior in Aplysia. J Exp Biol. 2008;211:2388–96. doi: 10.1242/jeb.015040. [DOI] [PubMed] [Google Scholar]
- 431.Jacklet J, Grizzaffi J, Tieman D. Serotonin, nitric oxide and histamine enhance the excitability of neuron MCC by diverse mechanisms. Acta Biol Hung. 2004;55:201–10. doi: 10.1556/ABiol.55.2004.1-4.25. [DOI] [PubMed] [Google Scholar]
- 432.Jacklet JW. Nitric oxide is used as an orthograde cotransmitter at identified histaminergic synapses. J Neurophysiol. 1995;74:891–5. doi: 10.1152/jn.1995.74.2.891. [DOI] [PubMed] [Google Scholar]
- 433.Jacklet JW, Tieman DG. Nitric oxide and histamine induce neuronal excitability by blocking background currents in neuron MCC of Aplysia. J Neurophysiol. 2004;91:656–65. doi: 10.1152/jn.00409.2003. [DOI] [PubMed] [Google Scholar]
- 434.Mothet JP, Fossier P, Tauc L, Baux G. Opposite actions of nitric oxide on cholinergic synapses: which pathways? Proc Natl Acad Sci U S A. 1996;93:8721–6. doi: 10.1073/pnas.93.16.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Mothet JP, Fossier P, Tauc L, Baux G. NO decreases evoked quantal ACh release at a synapse of Aplysia by a mechanism independent of Ca2+ influx and protein kinase G. J Physiol. 1996;493(Pt 3):769–84. doi: 10.1113/jphysiol.1996.sp021421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Bailey CH, Barco A, Hawkins RD, Kandel ER. Molecular studies of learning and memory in Aplysia and the hyppocampus: A comparative anaysis of implicit and explicit memory storage. In: Byrne JH, editor. Learning and memory -- A comprehensive reference. Elsevier; Oxford, U.K: 2008. [Google Scholar]
- 437.Hawkins RD, Castellucci VF, Kandel ER. Interneurons involved in mediation and modulation of gill-withdrawal reflex in Aplysia. II. Identified neurons produce heterosynaptic facilitation contributing to behavioral sensitization. J Neurophysiol. 1981;45:315–28. doi: 10.1152/jn.1981.45.2.315. [DOI] [PubMed] [Google Scholar]
- 438.Hawkins RD, Castellucci VF, Kandel ER. Interneurons involved in mediation and modulation of gill-withdrawal reflex in Aplysia. I. Identification and characterization. J Neurophysiol. 1981;45:304–14. doi: 10.1152/jn.1981.45.2.304. [DOI] [PubMed] [Google Scholar]
- 439.Byrne JH, Kandel ER. Presynaptic facilitation revisited: state and time dependence. J Neurosci. 1996;16:425–35. doi: 10.1523/JNEUROSCI.16-02-00425.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Clark GA, Hawkins RD, Kandel ER. Activity-dependent enhancement of presynaptic facilitation provides a cellular mechanism for the temporal specificity of classical conditioning in Aplysia. Learn Mem. 1994;1:243–57. [PubMed] [Google Scholar]
- 441.Bao JX, Kandel ER, Hawkins RD. Involvement of presynaptic and postsynaptic mechanisms in a cellular analog of classical conditioning at Aplysia sensory-motor neuron synapses in isolated cell culture. J Neurosci. 1998;18:458–66. doi: 10.1523/JNEUROSCI.18-01-00458.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Antonov I, Antonova I, Kandel ER, Hawkins RD. Activity-dependent presynaptic facilitation and hebbian LTP are both required and interact during classical conditioning in Aplysia. Neuron. 2003;37:135–47. doi: 10.1016/s0896-6273(02)01129-7. [DOI] [PubMed] [Google Scholar]
- 443.Roberts AC, Glanzman DL. Learning in Aplysia: looking at synaptic plasticity from both sides. Trends Neurosci. 2003;26:662–70. doi: 10.1016/j.tins.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 444.Gingrich KJ, Byrne JH. Simulation of synaptic depression, posttetanic potentiation, and presynaptic facilitation of synaptic potentials from sensory neurons mediating gill-withdrawal reflex in Aplysia. J Neurophysiol. 1985;53:652–69. doi: 10.1152/jn.1985.53.3.652. [DOI] [PubMed] [Google Scholar]
- 445.Chitwood RA, Li Q, Glanzman DL. Serotonin facilitates AMPA-type responses in isolated siphon motor neurons of Aplysia in culture. J Physiol. 2001;534:501–10. doi: 10.1111/j.1469-7793.2001.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Li Q, Roberts AC, Glanzman DL. Synaptic facilitation and behavioral dishabituation in Aplysia: dependence on release of Ca2+ from postsynaptic intracellular stores, postsynaptic exocytosis, and modulation of postsynaptic AMPA receptor efficacy. J Neurosci. 2005;25:5623–37. doi: 10.1523/JNEUROSCI.5305-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Koh HY, Jacklet JW. Nitric oxide stimulates cGMP production and mimics synaptic responses in metacerebral neurons of Aplysia. J Neurosci. 1999;19:3818–26. doi: 10.1523/JNEUROSCI.19-10-03818.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Koh HY, Jacklet JW. Nitric oxide induces cGMP immunoreactivity and modulates membrane conductance in identified central neurons of Aplysia. Eur J Neurosci. 2001;13:553–60. doi: 10.1046/j.0953-816x.2000.01416.x. [DOI] [PubMed] [Google Scholar]
- 449.Lewin MR, Walters ET. Cyclic GMP pathway is critical for inducing long-term sensitization of nociceptive sensory neurons. Nat Neurosci. 1999;2:18–23. doi: 10.1038/4520. [DOI] [PubMed] [Google Scholar]
- 450.Reinking J, Lam MM, Pardee K, Sampson HM, Liu S, Yang P, Williams S, White W, Lajoie G, Edwards A, Krause HM. The Drosophila nuclear receptor e75 contains heme and is gas responsive. Cell. 2005;122:195–207. doi: 10.1016/j.cell.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 451.Lei SZ, Pan ZH, Aggarwal SK, Chen HS, Hartman J, Sucher NJ, Lipton SA. Effect of nitric oxide production on the redox modulatory site of the NMDA receptor-channel complex. Neuron. 1992;8:1087–99. doi: 10.1016/0896-6273(92)90130-6. [DOI] [PubMed] [Google Scholar]
- 452.Lipton SA, Choi YB, Takahashi H, Zhang D, Li W, Godzik A, Bankston LA. Cysteine regulation of protein function--as exemplified by NMDA-receptor modulation. Trends Neurosci. 2002;25:474–80. doi: 10.1016/s0166-2236(02)02245-2. [DOI] [PubMed] [Google Scholar]
- 453.Takahashi H, Shin Y, Cho SJ, Zago WM, Nakamura T, Gu Z, Ma Y, Furukawa H, Liddington R, Zhang D, Tong G, Chen HS, Lipton SA. Hypoxia enhances S-nitrosylation-mediated NMDA receptor inhibition via a thiol oxygen sensor motif. Neuron. 2007;53:53–64. doi: 10.1016/j.neuron.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Kohn AB, Jezzini S, Ha T, Bentley-Sloan J, C. M., Bobkova E, Lee C, Yu F, Shaw R, Li J, Colman B, Clouser C, Barbaciorus M, Shan A, Farmerie W, Moroz LL. Nearly complete genomic profiling of individual identified neurons: SOLiD approach. Society for Neuroscience Meeting. 2008;880.1 [Google Scholar]
- 455.Moroz LL, Jezzini S, Ha T, Yu F, Shaw R, Farmerie W, Panchin Y, Kohn A. The genomic portrait of a characterized neuron: single-cell high-throughput expression profiling of identified neurons in Aplysia. Society for Neuroscience Meeting. 2007;208.23 [Google Scholar]
- 456.Wachsman JT. DNA methylation and the association between genetic and epigenetic changes: relation to carcinogenesis. Mutat Res. 1997;375:1–8. doi: 10.1016/s0027-5107(97)00003-1. [DOI] [PubMed] [Google Scholar]
- 457.Tanaka T, Kurose A, Halicka HD, Huang X, Traganos F, Darzynkiewicz Z. Nitrogen oxide-releasing aspirin induces histone H2AX phosphorylation, ATM activation and apoptosis preferentially in S-phase cells: involvement of reactive oxygen species. Cell Cycle. 2006;5:1669–74. doi: 10.4161/cc.5.15.3100. [DOI] [PubMed] [Google Scholar]
- 458.Nott A, Watson PM, Robinson JD, Crepaldi L, Riccio A. S-Nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455:411–5. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]
- 459.Matouk CC, Marsden PA. Epigenetic regulation of vascular endothelial gene expression. Circ Res. 2008;102:873–87. doi: 10.1161/CIRCRESAHA.107.171025. [DOI] [PubMed] [Google Scholar]
- 460.Huang X, Orucevic A, Li M, Gorelik E. Nitric oxide (NO), methylation and TIMP-1 expression in BL6 melanoma cells transfected with MHC class I genes. Clin Exp Metastasis. 2000;18:329–35. doi: 10.1023/a:1010867618014. [DOI] [PubMed] [Google Scholar]
- 461.Gan Y, Shen YH, Wang J, Wang X, Utama B, Wang J, Wang XL. Role of histone deacetylation in cell-specific expression of endothelial nitric-oxide synthase. J Biol Chem. 2005;280:16467–75. doi: 10.1074/jbc.M412960200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Clemons NJ, McColl KE, Fitzgerald RC. Nitric oxide and acid induce double-strand DNA breaks in Barrett's esophagus carcinogenesis via distinct mechanisms. Gastroenterology. 2007;133:1198–209. doi: 10.1053/j.gastro.2007.06.061. [DOI] [PubMed] [Google Scholar]
- 463.Nott A, Riccio A. Nitric oxide-mediated epigenetic mechanisms in developing neurons. Cell Cycle. 2009;8:725–30. doi: 10.4161/cc.8.5.7805. [DOI] [PubMed] [Google Scholar]
- 464.Zumft WG. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Hollocher TC, Hibbs JBJ. Enzymes of bacteria, plants and fungi that process free nitrogen oxides. In: Feelisch M, Stamler JS, editors. Methods in nitric oxide research. John Wiley & Sons; Chichester, New York: 1996. [Google Scholar]
- 466.Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–5. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 467.Nicholas KB, Nicholas HBJ, Deerfield DWI. GeneDoc: Analysis and Visualization of Genetic Variation. EMBNEW.NEWS. 1997 [Google Scholar]
- 468.Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–8. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 469.Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–60. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Walters ET, Bodnarova M, Billy AJ, Dulin MF, Diaz-Rios M, Miller MW, Moroz LL. Somatotopic organization and functional properties of mechanosensory neurons expressing sensorin-A mRNA in Aplysia californica. J Comp Neurol. 2004;471:219–40. doi: 10.1002/cne.20042. [DOI] [PubMed] [Google Scholar]