Abstract
Individual bacteria and shifts in the composition of the microbiome have been associated with human diseases including cancer. To investigate changes in the microbiome associated with oral cancers, we profiled cancers and anatomically matched contralateral normal tissue from the same patient by sequencing 16S rDNA hypervariable region amplicons. In cancer samples from both a discovery and a subsequent confirmation cohort, abundance of Firmicutes (especially Streptococcus) and Actinobacteria (especially Rothia) was significantly decreased relative to contralateral normal samples from the same patient. Significant decreases in abundance of these phyla were observed for pre-cancers, but not when comparing samples from contralateral sites (tongue and floor of mouth) from healthy individuals. Weighted UniFrac principal coordinates analysis based on 12 taxa separated most cancers from other samples with greatest separation of node positive cases. These studies begin to develop a framework for exploiting the oral microbiome for monitoring oral cancer development, progression and recurrence.
Introduction
Annually, ∼22,000 Americans are diagnosed with oral cancer of which 90% are squamous cell carcinoma (SCC). The five-year survival, at 40% has not improved in the past 40 years, and it is one of the lowest of the major cancer sites, resulting in more people dying from oral cancer than melanoma, cervical or ovarian cancer in the USA. Worldwide there are 350,000–400,000 new cases diagnosed each year. Unlike most other anatomic sites, which have decreased in cancer incidence, the incidence of oral cancer is increasing, particularly among young people and women [1], [2]. The major risk factors, tobacco and alcohol use, cannot explain the changes in incidence, because oral cancer also commonly occurs in patients without a history of tobacco or alcohol exposure [3]. Recently, human papillomavirus (HPV) has been identified as an etiologic agent for oropharyngeal cancer, but HPV infection is not a significant contributor to oral cancer, as the virus is rarely found in these cancers (2–4% of cases) [4]. Thus, contributions from other, possibly environmental factors remain to be found.
A role for bacterial infection in causing or promoting cancer is well known with respect to the association of Helicobacter pylori with gastric cancer [5], and other cancers, including gallbladder, colon, lung and prostate, have been associated with particular bacterial infections [6], [7], [8]. It is reasonable to ask, therefore, if shifts in the composition of the normal oral cavity microbiome, comprised of more than 600 different bacterial species [9] and/or chronic bacterial infection could be promoters or causes of oral cancer. Indeed, changes in the microbial community are commonly associated with dental diseases such as periodontal disease, which is most likely a polymicrobial disease characterized by outgrowth of certain pathologic organisms [10], and chronic periodontitis has been reported to be a risk factor for oral premalignant lesions and cancers [11]. Elevated levels and changes in the composition of bacterial and fungal microbiota of the oral cavity have been reported in association with oral pre-cancers and cancers [12]. There is, however, no consensus amongst reports regarding cancer-associated changes in the oral microbiome. This confusion may have arisen because early studies were limited to analysis of the relatively small numbers of known and cultivable oral bacterial species [13], [14], and later studies using molecular methods focused on particular phyla [15] or cloned and sequenced small numbers of clones per sample [16], [17].
Culture independent methods, particularly those employing next generation sequencing of the hypervariable region of the 16S ribosomal subunit, provide a means to more comprehensively and accurately profile the microbiome in health and disease [18]. Such studies of the oral microbiome [16], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28] reveal, on the one hand, that the healthy oral microbiome is characterized by a relatively small number of bacterial phyla (9–13), the most commonly reported abundant phyla being Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria, and Fusobacteria [16], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28]. On the other hand, the majority of inter-individual variation has been attributed to diversity at the species or strain level [24]. Streptococcus is most often observed to be the dominant genus in the healthy oral microbiome, and less frequently Prevotella, Veillonella, Neisseria, and Haemophilus dominate an individual's oral microbiome [19], [24]. Variation is also observed in the microbial community composition of biofilms at each intraoral habitat (e.g., tooth surface, lateral and dorsal tongue, etc.), most likely reflecting the different surface properties and microenvironments [21], [24].
To properly investigate possible shifts in the composition of the oral microbiome in oral cancer, therefore, it is necessary to control for differences between oral subsites and inter-individual variation. In addition, high recurrence rates and prevalence of second primary oral cancers support the proposal that these cancers develop out of a field of genetically altered cells, the concept of “field cancerization” [29]. Such fields have been reported to extend as much as 7 cm from a tumor and to appear clinically normal [30]. For these reasons, we investigated the oral cancer associated microbiome by non-invasively sampling the cancer lesion and an anatomically matched contralateral region of normal tissue from each individual. We subjected DNA isolated from these samples to 16S ribosomal subunit amplification and sequencing. The aim of these studies was to begin to lay a foundation that would allow exploitation of the oral microbiome for treatment and monitoring of oral cancer initiation, progression and recurrence.
Results
To investigate changes in the oral microbiome associated with oral cancer, we prospectively collected cancer and anatomically matched patient clinically normal samples from five patients (Table 1, Study 1 Discovery Cohort, Table S1). To confirm and extend our initial observations, we performed a second study (Study 2, Tables S2 – S6, total number of samples = 83) in which we prospectively collected an independent set of lesional and anatomically matched clinically normal samples from oral cancer (Table 2, Study 2 Confirmation Cohort, Table S2), carcinoma in situ (CIS, Table S3) and pre-cancer patients (Table S4), as well as from the left and right sides of the lateral tongue and floor of mouth of healthy normal individuals (Table S5). In Study 2, we also included an independent analysis of the initial five cancer patients from Study 1 and six pairs of replicate samples (three cancer and three pre-cancer patients, Table S6). The latter were included to assess reproducibility of sample collection and processing and were not included in any of the analyses (see further discussion in Methods).
Table 1. Discovery Cohort Cancer Patient Characteristics.
| Patient | Age | Sex | Site | Tumor size (cm) | pTNM1 | Immunocompromised | Smoker | Alcohol |
| 113 | 62 | M | Right buccal mucosa | 2.5×3.0 | pT2N2bMx | Yes | Previous | Current |
| 116 | 84 | F | Right retromolar trigone | 6.0 | pT4aN0Mx | No | Previous | Previous |
| 117 | 68 | M | Left posterior lateral tongue | 1.5×1.0 | pT2N0Mx | No | Previous | Current |
| 136 | 68 | M | Left lateral tongue | 5.0×2.0 | pT2N2bMx | No | Never | Current |
| 142 | 64 | M | Right dorsum of tongue | 0.3×1.5 | pT3N0M0 | Yes | Current | Current |
Pathologic tumor stage according to the American Joint Committee on Cancer guidelines (T = tumor size, N = regional lymph node metastasis, M = distant metastasis)
Table 2. Conformation Cohort Cancer Patient Characteristics.
| Patient | Age | Sex | Site | Tumor size (cm) | pTNM1 | Immunocompromised | Smoker | Alcohol |
| 104 | 55 | F | Right lateral tongue | 2.5×2.5 | pT4N2M0 | No | Current | Current |
| 108 | 39 | F | Right lateral tongue corner | 3.0 | pT4N2bM0 | No | Previous | Non-drinker |
| 114 | 48 | M | Left post. mandibular gingiva | 1.5 | pT1N0M0 | Yes | Non-smoker | Current |
| 114 | 48 | M | Left post. maxillary gingiva | 2.5 | pT2N0M0 | Yes | Non-smoker | Current |
| 128 | 71 | M | Left alveolar ridge | 4.0×3.0 | pT4bN0M0 | No | Previous | Previous |
| 143 | 61 | M | Right floor of mouth | 1.6×1.0 | pT1N0M0 | No | Current | Current |
| 144 | 70 | M | Left floor of mouth | 3.5×1.5 | pT1N0M0 | No | Previous | Current |
| 146 | 78 | M | Right retromolar trigone | 1.5×1.5 | pT1N2bMx | No | Previous | Current |
| 153 | 51 | F | Left ventral tongue | 8.0×8.0 | pT2N1Mx | No | Non-smoker | Current |
| 156 | 69 | F | Right mandibular gingiva | 1.0×1.0 | pT1N0M0 | No | Non-smoker | Current |
Pathologic tumor stage according to the American Joint Committee on Cancer guidelines (T = tumor size, N = regional lymph node metastasis, M = distant metastasis
Study 1. Discovery cohort
We swabbed the oral cancer lesion and a corresponding anatomically matched clinically normal tissue area from the Discovery Cohort of five patients (Table 1). Using the Roche GS Junior instrument to perform pyrosequencing, we obtained, in a single run, a total of 104,380 sequences from amplicons that spanned the V4 hypervariable region of the bacterial 16S small ribosomal subunit (Table S7). The number of raw sequence reads varied by >10-fold across samples, ranging from 1,231 to a maximum of 17,682 raw reads. Quality filtered sequences were searched against the Greengenes reference database of 16S sequences, clustered at 97%, and Operational Taxonomic Units (OTUs) were assigned taxonomic classification using mothur's Bayesian classifier. Of the 92,987 sequences that passed quality filtering, 81,308 were similar to known bacteria and most could be classified to the genus level (65,037) with fewer classified at the species level (17,115). Sequence coverage was variable across samples; the number of reads per sample assigned to OTUs (excluding those filtered due to poor quality or lack of a related sequence in the Greengenes reference database) ranged from 1,038 to a maximum of 14,359, and comprised 76–85% of raw sequences (Table S7). A total of 276 OTUs were identified (per sample range, 37–161, (Table S8). Rarefaction analysis performed at the family level demonstrated a fairly wide range of α diversity with the number of detected families ranging from ∼15–28 (Figure S1a). Three patient samples, normal and cancer from patient 117 and the normal sample from patient 142, plateaued at fewer families than the other samples, indicating somewhat reduced diversity in these samples. All samples plateaued to some extent, though not entirely; further sequencing would likely reveal additional families. On the other hand, pyrosequencing noise and PCR errors could erroneously increase OTU numbers [31]. A similar effect was seen at the genus level (Figure S1b).
Diversity of microbiomes associated with anatomically matched oral cancer and normal samples
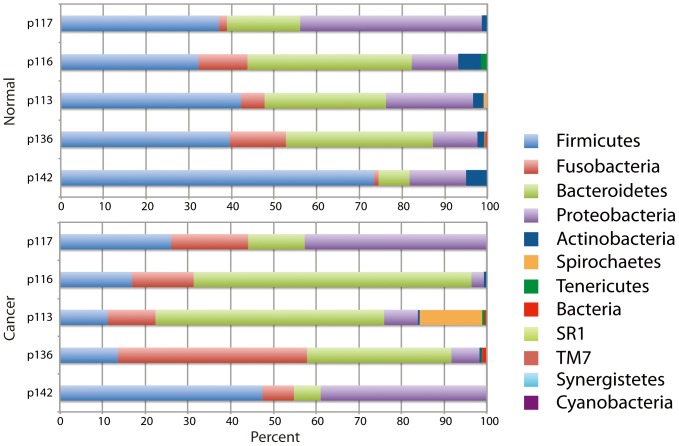
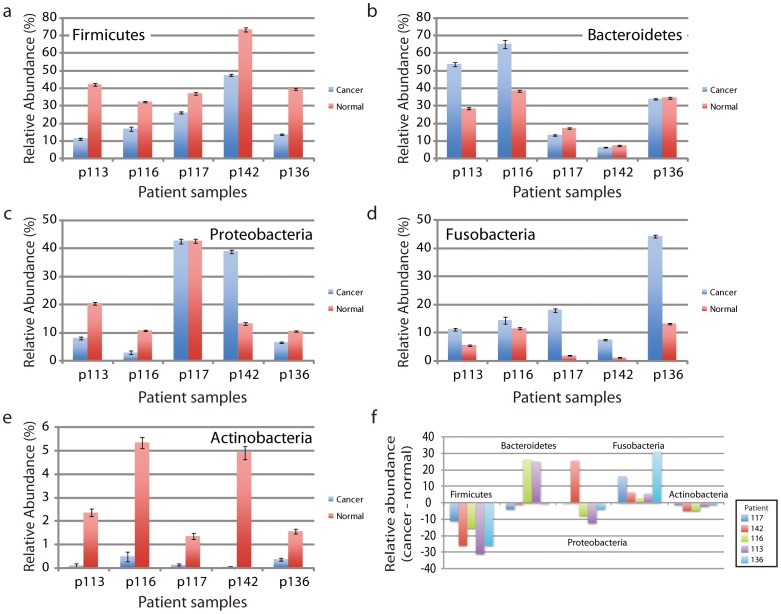
The OTUs of the cancer and clinically normal samples in the Discovery cohort were classified into 12 phyla (Table S7). The majority belonged to one of five phyla (99.2%, normal, 98.0% cancers) with the more abundant phyla being Firmicutes, Bacteroidetes, Proteobacteria, Fusobacteria, and Actinobacteria (Table 7, Figure 1). Although the distribution in cancer and clinically normal samples of these five common phyla varied amongst individuals (Figure 1), in all patients, we observed a significant reduction in the abundance of Firmicutes and Actinobacteria in cancer compared to the anatomically matched contralateral clinically normal patient sample, (p = 0.004, FDR adjusted p = 0.02 and p = 0.028, FDR adjusted p = 0.07, respectively). We also observed that the proportion of Fusobacteria was increased in all patients, but the change in abundance did not reach statistical significance (p = 0.074, Figure 2). We note, however, that to observe consistent changes in abundance of the three phyla in this small cohort is highly significant. Ignoring interactions among the relative abundances of different phyla, a binomial test yields p = 0.0022 as the probability (under the null hypothesis) of having three or more phyla where (all five patients share an increase) or (all five share a decrease).
Figure 1. Distribution of phyla in normal and cancer samples in Study 1.
The relative distribution of phyla (percent of sequences) is shown for each patient sample with clinically normal samples shown together on top and cancer samples on the bottom.
Figure 2. Change in relative abundance of phyla associated with cancer compared to anatomically matched contralateral clinically normal samples in Study 1.
(a – e) Relative abundance of each of the five more abundant phyla in cancers compared to clinically normal samples from each of five patients. Note, that data are shown on different scales, reflecting the abundance of the phyla. The magnitudes of the changes in abundance are clearly greater than the statistical counting noise, as indicated by the error bar estimates, which are based on the square root of the actual number of reads. (f) Change in relative abundance shown as the difference in abundance of phyla associated with cancers compared to anatomically matched contralateral clinically normal samples. In cancers, decreases in the relative abundance of Firmicutes and Actinobacteria were seen in all patients, while the relative abundance of Fusobacteria was elevated in cancers from all patients.
Study 2. Confirmation cohort
To confirm these initial observations, we performed Study 2. As before, we swabbed both the lesion (cancer or pre-cancer) and a contralateral anatomically clinically normal matched site. We swabbed the left and right sides of the lateral tongue and floor of mouth of the healthy individuals. Using the Illumina MiSeq instrument, we sequenced amplicons spanning the 16S rDNA V4 hypervariable region. We obtained 4,486,196 raw sequence reads with a range of 31,109 to 125,847 reads per sample after excluding two samples that failed in sequencing (Tables S9 – S13). We assigned taxonomic classification to the OTUs as before. Of the sequences that passed quality filtering, 4,444,432 were similar to known bacteria and most could be classified to the genus level (4,148,785) with fewer classified at the species level (1,650,037). We identified a total of 2,107 OTUs (per sample range, 90–482, Tables S9 – S13). Rarefaction analysis demonstrated a fairly wide range of α diversity with almost all samples plateauing to some extent (Figure S2). As discussed above, sequencing noise and PCR errors may increase OTU numbers [32].
Diversity of microbiomes associated with anatomically matched samples from oral cancer, pre-cancer and healthy normal individuals in Study 2
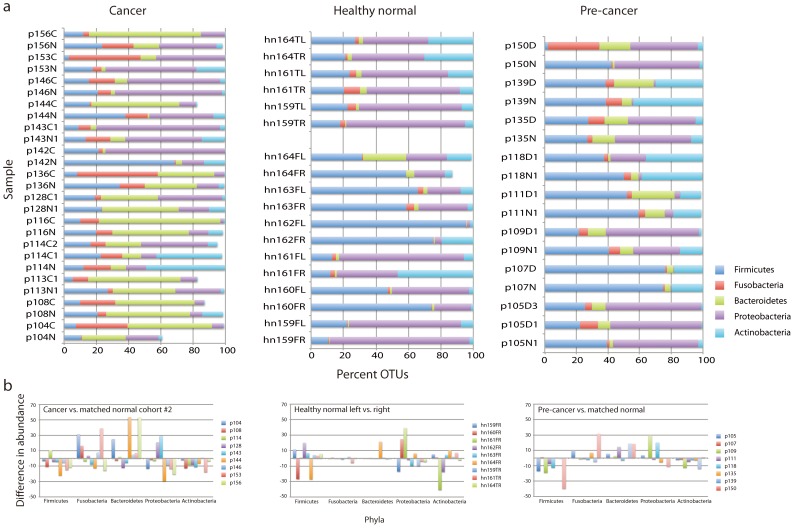
We first determined that data collected on nine of the 10 Discovery Cohort samples that were successfully profiled in Study 2 (Table S9) were highly correlated with the original data obtained by 454 pyrosequencing (Figure S3). We then considered the Confirmation Cohort comprised only of samples from cancer patients not included in the Discovery Cohort, i.e., we excluded the nine Discovery cohort samples (Table 2, Table S9). We again found that the majority of OTUs (61–100%) belonged to one of the five more abundant phyla (Firmicutes, Bacteroidetes, Proteobacteria, Fusobacteria, and Actinobacteria) (Figure 3a, Table S14). We then asked if there were significant reductions in the abundance of Firmicutes and Actinobacteria in cancer compared to the anatomically matched contralateral clinically normal patient sample. Indeed, as we had observed in the Discovery cohort (Study 1), there was a significant reduction in abundance of these two phyla in cancers compared to the anatomically matched contralateral clinically normal patient samples (p = 0.042 and p = 0.004, respectively), confirming our initial observations (Figure 3b, Figure S4).
Figure 3. Distribution of phyla in cancer, pre-cancer and healthy normal samples in Study 2.
(a) Shown is the relative distribution of phyla (percent of sequences). For cancers, we included only patients for which both the cancer and contralateral clinically normal samples were available. (b) Change in relative abundance of phyla shown as the difference in abundance of phyla associated with cancers or pre-cancers compared to anatomically matched contralateral clinically normal samples. For healthy normal samples, we compared left and right sides of the lateral tongue or floor of mouth.
In Study 2, we also observed that the majority of OTUs in samples from pre-cancer patients and healthy normals (Figure 3a) belonged to one of the five phyla Firmicutes, Bacteroidetes, Proteobacteria, Fusobacteria, and Actinobacteria (pre-cancers = 99–100%, healthy normal = 87–100%). We again asked if significant reductions in abundance of Firmicutes and Actinobacteria might also be present when comparing pre-cancers and anatomically matched contralateral clinically normal patient samples. We did find that abundance of Firmicutes and Actinobacteria was reduced (p = 0.048 and p = 0.037, respectively), suggesting that changes in abundance of these two phyla may occur early (Figure 3b). By contrast, we did not find significant differences in abundance of these two phyla or any of the five more abundant phyla when comparing the left and right sides of the lateral tongue and floor of mouth of healthy normal individuals (Figure 3b).
To investigate changes in abundance at the genus level for the five more abundant phyla, we considered patient matched samples from the Confirmation Cohort and also included four of the five Discovery Cohort cases that were successfully analyzed in Study 2 (14 cancers from 13 patients, Table S9). We normalized the number of OTUs to one million counts, (Table S15) and for each phylum, we determined changes in abundance of the genera that represented >10% of OTUs in more than 20% of samples (Figure S5 and S6). Significant reduction in abundance was observed for Streptococcus (p = 0.003) and Rothia (p = 0.021) in cancers relative to anatomically matched clinically normal samples (Table S16). By contrast, we observed increased abundance of Fusobacterium (p = 0.044) relative to matched clinically normal samples from the cancer patients. In pre-cancers, we observed significantly reduced abundance of Streptococcus (p = 0.042) (Table S16). We found no significant differences in abundance of these more common genera when comparing abundance in samples taken from the left and right sides of the lateral tongue and floor of mouth of healthy individuals (Table S16).
We also noted that although we found no consistent changes in abundance of Bacteroidetes when comparing within individuals (Figure 3b), samples from cancer and pre-cancer patients (both lesion and anatomically matched contralateral clinically normal tissue) were associated with increased abundance of Bacteroidetes compared to samples from healthy normal individuals (Figure 3a). Prevotella species, in particular, differed and included, for example, OTUs corresponding to P. intermedia, P. melaninogenica, P. nanceiensis, P. oris, P. tannerae and unclassified species (Figure S5, Table S15). Elevated levels of P. melaninogenica have been reported previously as a potential salivary biomarker of oral cancer [33] and P. intermedia is a periodontal pathogen [34]. Further studies will be required to understand the contributions of general (Bacteroidetes) and lesion-specific changes (Actinobacteria, Firmicutes) in the microbiome of oral cancer and pre-cancer patients.
OTUs distinguishing cancer from anatomically matched contralateral normal patient samples
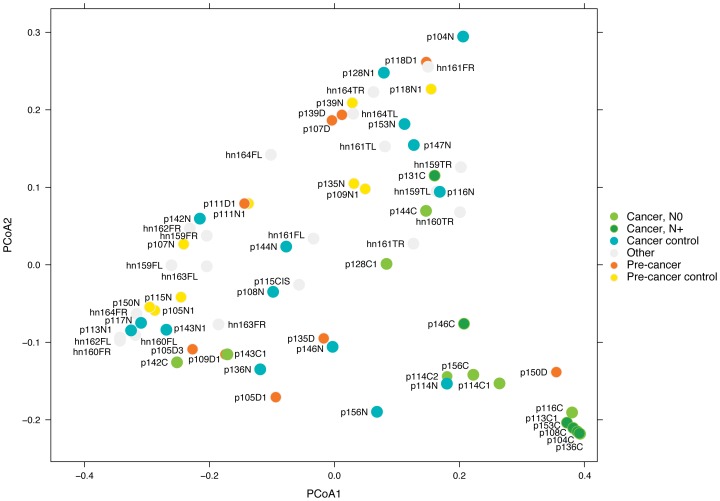
To address the need for biomarkers to predict behavior of oral cancers that could be assayed by non-invasive tests, we asked whether cancer samples could be distinguished by bacterial composition. Considering cancer/normal paired samples, we identified 11 OTUs from the phyla Actinobacteria (Actinomyces and Rothia, 2 OTUs from each genus) and Firmicutes (Streptococcus, 7 OTUs) that were significantly decreased in cancers and one OTU from the phylum Fusobacteria (Fusobacterium) that was increased in cancers compared to anatomically matched contralateral normal patient samples (Table S17). Weighted UniFrac principal coordinates analysis (PCoA) based on this set of OTUs separated most cancers from normal and pre-cancer samples with five of the seven lymph node positive cases forming a tight cluster in the lower right corner of the plot (Figure 4). Metastasis to the cervical (neck) lymph nodes is a major determinant of oral cancer patient survival [35]. The current lack of reliable methods to assess risk of metastasis results in patients being routinely subjected to additional surgery to remove the lymph nodes, even though the majority will not benefit from the procedure. The tight clustering of samples from node positive patients in Figure 4 suggests that shifts in the composition of the oral cancer microbiome may also hold promise as a tumor associated biomarker of risk of metastasis.
Figure 4. Distinguishing cancer and normal samples.
PCoA based on Weighted UniFrac distance between samples given abundance of 12 OTUs. Axis 1 (PCoA1): 54% of variation explained. Axis 2 (PCoA2): 24% of variation explained. N0 and N+ indicate the nodal status of the cancer patient, N0 = node negative, N+ = node positive. Cancer control and pre-cancer control are contralateral clinically normal patient samples. Other identifies samples from healthy normal individuals.
Whole micrbiome – β diversity
To investigate sample-to-sample dissimilarity, we subsampled the data by randomly selecting 31,109 sequences from each community to adjust for variation in sequencing depth (Table S18). When considering the three sample groups (Cancers, Healthy Normals, and Pre-cancers), we observed significant microbiome differences for patient identity based on both weighted UniFrac (abundance) and Unweighted UniFrac (presence/absence) metrics (Table S19, Figures S7 and S8). No other comparisons, such as left vs. right, number of sequences or lesion (cancer or pre-cancer) vs. control normal sites revealed significant differences. These observations are consistent with other studies that have highlighted the inter-individual differences in the oral microbiome [24]. Further, they support our study design, which measures changes in the microbiome within individuals, i.e., using each patient as his or her own control.
Discussion
To study oral malignancy-associated microbiome changes, we performed a Discovery screen (Study 1), in which we non-invasively sampled cancers and contralateral clinically normal tissue samples from each individual. Comparison of the composition of the microbial communities within patients identified changes in abundance of Actinobacteria and Firmicutes. We confirmed these observations in a second Confirmation Cohort (Study 2) and further found significant changes in the abundance of the Actinobacteria genus Rothia and the Firmicutes genus Streptococcus when considering all cancers in Study 2 (Table S16). Although we did not see a significant change in abundance of the phylum Fusobacteria in either the Discovery or Confirmation Cohorts, we did find a significant increase in abundance of the Fusobacteria genus, Fusobacterium when considering all cancer patients in Study 2. We note that while the cohorts of patients studied here are small and heterogeneous, our findings regarding abundance of phyla are similar to published studies, which focused on comprehensive analysis of the oral microbiome. Moreover, the use of different sequencing technologies to measure the abundance of 16S rDNA amplicons in Studies 1 and 2 supports the robustness of our observations. Nevertheless, further larger studies should help to better define the oral cancer associated changes in abundance of these phyla and genera. Moreover, we observed changes in abundance of Firmicutes (Streptococcus) in association with oral pre-cancers, suggesting that oral lesion associated shifts in the composition of the microbial community may occur early in oral cancer development and/or herald cancer progression.
Smoking is a risk factor for oral diseases, including cancer and periodontitis. Studies have established that smoking impacts the composition of the bacterial communities in the oral cavity, including, for example, the salivary microbiome of healthy smokers and non-smokers [36] and the subgingival microbiomes of patients with periodontal disease [37], as well as the formation of plaque biofilms [38]. In our study, we found no evidence of overall differences in the microbiomes that could be attributed to smoking, but with only three current smokers and four non-smokers in our cancer patient cohorts for example, we cannot draw any conclusions at this time. On the one hand, because we used each patient as his/her own control, we would not expect to see smoking associated differences in abundance of microbiota associated with cancer, since smoking would affect both the control site and the cancer. On the other hand, in smokers, cells at the clinically normal sites and the cancers my respond differently to smoking induced changes in biofilm formation, for example, raising the possibility that while cancer associated changes in the abundance of microbiota in smokers and non-smokers appear similar, the formation and functional consequences of the altered microbiomes may differ in these patient groups. Similar considerations could apply to immunosuppressed individuals.
The presence of bacteria in oral cancers and/or differences in the bacterial communities associated with oral cancer have been reported previously using either culture dependent [13], [14] or molecular methods [15], [16], [17], yet no consistent observations have been reported across these studies. It is, however, difficult to make comparisons even amongst two recent studies reporting abundance of bacteria [16], [17] and this study, because of differences in (a) sample type (swab, this study vs. tissue sample [16], [17]), (b) oral cavity site, (c) source of patient matched normal control samples (anatomically matched contralateral clinically normal, this study, upper aerodigestive tract mucosae [16], adjacent normal [17]), (d) amplified region of the 16S ribosomal gene (V4, this study, V4-V5 [16] or V1-V4 [17]), (e) methodology (Sanger [16], [17] vs. pyrosequencing or MiSeq, this study), and (f) number of clones or sequence reads assigned to OTUs per sample (average 8000 and 55,000 reads per sample, Study 1 and 2, respectively, compared to ∼90 or ∼250 clones per sample [16], [17]). For example, Pushalkar and colleagues [16] reporting on 10 patients found that 75 and 80% of clones (normal and cancer, respectively) were assigned to the phylum Firmicutes. This proportion is not only higher than the 40–0% reported in other studies of the oral cavity of healthy or cancer patients, but since only ∼90 clones were sequenced per sample, there are too few clones to reliably determine relative abundance of other phyla for comparisons. A phylum level analysis of only the 16 tongue, floor of mouth and oral cavity cancers reported by Bebek and colleagues [17], however, revealed cancer associated increase in abundance of the phylum Fusobacteria, consistent with our observations, but decreased abundance of Streptococcus could not be seen, as few clones were assigned to this genus.
We cannot distinguish whether the observed shifts in the microbial community reflect the fact that certain bacteria are more suited to adhere and grow in the cancer microenvironment or whether they are cancer promoting. Further, it is unclear how to weigh the potential contributions from changes in abundant genera such as Streptococcus compared to the less abundant Actinobacteria genera. Potential roles for bacteria and fungi in cancer promotion include generation of carcinogenic substances, such as nitrosamine or other pro-carcinogenic chemicals, chronic inflammation and direct effects on signaling in epithelial cells resulting in enhanced proliferation or suppression of apoptosis [6], [7], [12], [39]. Only a minority of the oral microbial community can adhere to hard and soft oral tissues, and assembly of the complex oral biofilm is accomplished by subsequent adherence of secondary colonizers. Streptococcus is an early colonizer and Fusobacterium (e.g., F. nucleatum) has a propensity for co-aggregation with many genera, forming a bridge between early and late colonizers in the oral biofilm [40]. Thus, on the one hand, the observed decrease in prevalence of Streptococcus and increased abundance of Fusobacterium genera in pre-cancers could reflect the altered surface properties of the cancer cells and stroma, which might no longer support adhesion of streptococci. On the other hand, we can hypothesize that shifts in abundance of these two genera could result in an enhanced pro-inflammatory environment, since Streptococcus species have been reported to attenuate Fusobacterium nucleatum induced pro-inflammatory responses of oral epithelial cells [41], [42]. We also note that Fusobacterium nucleatum grown as a biofilm is capable of invading organotypic cultures [43], and secondly, that the organism has recently been reported in colon cancers [44], [45], further supporting a potential role in oral cancer.
The oral cavity offers a relatively unique opportunity to screen at risk individuals for (oral) cancer, because the lesions can be seen, and as we report here, the shift in the microbiome of the cancer and pre-cancer lesions compared to anatomically matched clinically normal tissue from the same individual can be detected in non-invasively collected swab samples. Saliva is another non-invasively collected oral sample composed largely of bacterial cells, but also shed epithelial and immune cells. A variety of “omics” biomarkers in saliva have been proposed for use in diagnosis of oral cancer, including metabolites, proteins, transcribed genes, miRNAs, genome alterations and epigenomic changes, as well as the microbiome [16], [46], [47], [48], [49]. For the microbiome, however, saliva may not be optimal. Saliva bathes the entire oral cavity, resulting in a loss of information on the subsite specific composition of bacterial communities. Moreover, with saliva, there is no possibility to use each individual as his or her own control, and so account for the substantial variation in the oral microbiome amongst individuals.
Non-invasively sampling the microbiome of oral lesions and corresponding normal tissue opens the possibility to not only detect cancer-associated changes at one time point, but the relative stability of the adult oral microbiome [25], [28], [50] also offers the opportunity to monitor shifts in bacterial communities over time. Here we observed changes in the microbiome, which, in future larger studies, may be confirmed as a potential biomarker of oral cancers or pre-cancers, and may even have utility to discriminate patients with lymph node metastases (Figure 4). In addition, there are other challenges in clinical management of oral cancers and pre-cancers that would benefit from better diagnostic tools. Most oral cancers are preceded by oral epithelial dysplasia (pre-cancer), a lesion, which unpredictably transforms to cancer. Oral cancer patients are also at risk of second primary cancers and recurrences. The microbiome may provide signatures that can be used as a biomarker for (a) progression of pre-cancers to cancer, (b) distinguishing oral cancer subtypes, (c) monitoring field changes associated with the high rate of second primary oral cancers and recurrences, and (d) predicting clinical behavior such as metastasis (Figure 4). We also highlight the possibility of medically modulating the oral microbiome for treatment of oral pre-cancers and damaged fields (field cancerization).
Methods
Ethics statement
The study was approved by the Institutional Review Board of New York University College of Dentistry and all patients provided written informed consent.
Study population and biospecimen collection
In Study 1, samples from cancer and anatomically matched contralateral clinically normal regions of the oral cavity were obtained from five patients with oral cancer who were referred to the Bluestone Center for Clinical Research, New York University College of Dentistry in the period July 2011 to March 2012. Individuals enrolled in Study 2 included cancer and pre-cancer patients who were referred to the Bluestone Center for Clinical Research during the period April 2011 to August 2012 and individuals with no history of oral cancer (healthy normal). For this study, we used a well-defined clinical protocol for swabbing the oral lesion and a contralateral normal site. In addition to sampling the microbiome, the procedure provides a tumor genomic DNA sample with a DNA copy number profile that, when tested, reflects the profile obtained with the genomic DNA isolated from an incisional biopsy of the lesion; however, the procedure is not designed to optimally sample the entire oral microbiome.
Specifically, to collect samples from the cancer or pre-cancer, the lesion was dried by blotting with gauze and then the lesion was stroked with an Isohelix SK-2 swab (Cell Projects Ltd., Harrietsham, UK). The swab was held at an angle of approximately 20° relative to the surface of the lesion and one side of the swab was stroked across the lesion 10 times applying gentle downward pressure. The swab was then rotated 180° and the other side of the swab was stroked 10 times across the lesion in the same manner. Anatomically matched contralateral normal tissue and tissues from healthy normal individuals were sampled using an Isohelix swab, the brush from the OralCDx Brush Test (CDx Diagnostics, Suffern, NY) or a Rovers Orcellex brush (Rovers Medical Devices B.V., Oss, The Netherlands). The swabs and the brushes provided with the OralCDX Brush Test were placed into the tube provided as part of the Isohelix SK-2 swab or a microfuge tube, respectively and kept on ice for no more than 30 minutes prior to adding the Isohelix cell lysis and DNA stabilization solution (LS solution, 500 µL) and proteinase K solution (20 µL) both provided in the Isohelix DSK-2 kit. Samples were stored at room temperature and subsequently shipped to the University of California San Francisco for nucleic acid extraction.
To assess the reproducibility of the swab procedure, we included six pairs of replicate swabs taken at the same visit as the original swab as part of Study 2 (Tables S6 and S13). Five pairs of samples showed good concordance (Pearson correlation, minimum = 0.628, first quartile = 0.801, median = 0.868, mean = 0.843, third quartile = 0.912, maximum = 0.962), indicating that in most cases the procedure reproducibly samples the microbial communities of oral lesions and normal sites. Sample p113N2, however, showed little concordance with p113N1 (R2 = 0.116), but good concordance with p113C2 (R2 = 0.702). In other analyses, p113N2 behaved similarly to the cancer samples from patient 113 (data not shown), suggesting an error had occurred at some point during collection and processing of this sample.
DNA extraction
Swabs in Isohelix tubes with 500 µL solution were vortexed, then centrifuged briefly and the liquid removed to a fresh 1.5 mL microfuge tube. This process was repeated 2–3 more times before transferring the swab to a fresh 1.5 mL microfuge tube and centrifuging at 14,000 rpm for 1 minute to extract the remaining cell lysate from the swab. The DNeasy blood and tissue kit (Qiagen Corp.) was used to extract nucleic acid from the solution recovered from the swab, following the manufacturer's protocol and including an initial incubation with Proteinase K (addition of 20 µL solution provided as part of the DNeasy blood and tissue kit) at 56°C for 10 minutes. The DNA concentration was determined using the Qubit v2.0 fluorometer.
16S rRNA amplicon preparation and 454 pyrosequencing
The V4 region of the small subunit ribosomal RNA gene (16S rRNA) was selected for study, because it is suited to analysis on multiple high throughput platforms yielding short reads. It has been reported to give low error rates when assigning taxonomy [51], [52] and to be suitable for community clustering [53]. For Study 1, the region was amplified using the primer set 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACVSGGGTATCTAAT-3′) [32], [54]. The complete forward primer construct (5′-3′) consisted of the Roche 454 Life Sciences Sequencing FLX Adaptor A (Roche Applied Science, Branford, CT, USA), a 12 bp Golay nucleotide barcode, and a GT linker followed by the 515F primer sequence. The 806R was similarly constructed but incorporated the Roche 454 Life Sciences Sequencing FLX Adaptor B, and a GC linker followed by the 806R primer sequence.
For each patient sample in Study 1, three separate amplifications were carried out in 25 µL reaction volumes. Each reaction contained 1 7L each of forward and reverse primers at a 10 µM concentration, 10 µL of 5 Prime HotMasterMix (5 Prime Inc., Gaithersburg, MD, USA), and 1 µL of extracted genomic DNA from each patient sample (concentration range = 2.3–64.2 ng/ µL). After denaturation at 94°C for 3 minutes, 35 cycles of incubation at 94°C for 45 seconds, 50°C for 30 seconds, and 72°C for 1.5 minutes were carried out, followed by a 10 minute final elongation step at 72°C. The replicate reactions from each patient sample were pooled together by sample and quantified using the Quant-IT PicoGreen dsDNA Assay (Invitrogen, Carlsbad, CA, USA). Barcoded samples were then normalized to equimolar amounts to ensure equal sequencing depth for each sample, and finally pooled into one combined sample, which was further purified using the UltraClean PCR Clean-Up Kit (Mo Bio Laboratories, Inc., Carlsbad, CA). A final quantification was performed using the Quant-IT PicoGreen dsDNA Assay. Pooled amplicon libraries were sequenced unidirectionally in a single run on the Roche GS Junior instrument (100,000 reads per run) using the A beads for emulsion PCR.
For Study 2, the 16S V4 region was amplified using 515F and 806R fusion primers tailed with sequences to incorporate Illumina flow cell adapters with indexing barcodes [32]. Each sample was PCR amplified in a 10 µL reaction. The reaction contained 1 µL of a mixture of forward and reverse primers at a 5 µM concentration, 5 µL of Qiagen HotStar Master Mix (Qiagen Inc., Gaithersburg, MD, USA), 0.5 µL genomic DNA from each patient sample (20–50 ng) and nuclease free water. After incubation at 94°C for 15 minutes, 35 cycles of incubation at 94°C for 30 seconds, 50°C for 30 seconds and 72°C for 30 seconds were carried out, followed by incubation at 72°C for 10 minutes and storage at 4°C. Primer dimers and low molecular weight products were removed using Agencourt Ampure Beads (Beckman Coulter, Inc., Indianapolis, IN) and samples were quantified and quality checked for amplicon size using the Agilent Bioanalyzer. Amplicons (1×1010 molecules) were pooled. The pooled sample was diluted to 3.5 pM and 8 pM phiX DNA was spiked in to a final concentration of 2 pM. Amplicons were sequenced from both ends for 250 cycles using primers designed for paired-end sequencing avoiding the PCR amplification primers. The indexing barcode was sequenced using a third sequencing primer and an additional 13 cycles. Sequence data are available at the European Bioinformatics Institute, accession number PRJEB4953.
Data analysis
We used QIIME [55] and custom scripts to process the sequencing data. Sequences were quality filtered and de-multiplexed using exact matches to the supplied DNA barcodes. Paired end sequencing performed on the MiSeq resulted in forward and reverse reads with some overlap in the V4 hypervariable region of the 16S. Each forward and reverse pair were stitched together using pandaseq with parameters "-F -l 220 -L 300 -t 0.5". Read pairs without sufficient overlap to allow stitching were discarded. For both MiSeq and 454 sequences, the resulting sequences were then searched against the Greengenes reference database of 16S sequences from 4 February 2011 [56], clustered at 97% by uclust. Sequences not matching any Greengenes reference sequence at >97% sequence identity were discarded (closed-reference OTU picking). The longest sequence from each Operational Taxonomic Unit (OTU) thus formed was then used as the OTU representative sequence, and assigned taxonomic classification using mothur's bayesian classifier, trained against the Greengenes reference database of 16S sequences from 4 February 2011 [56], clustered at 98% (bootstrap confidence level = 80%). We note that ambiguous sequence identifications between species, for example, were addressed by not assigning any species identification to such reads, rather they were identified at broader levels (e.g., phylum, genus) where confident assignments could be made.
After pooling sequences into OTUs as described above, we generated a table listing the abundance of each OTU in each microbial community in this study (the OTU table). To obtain a summary statistic representing the overall dissimilarity between any two microbial communities in this study, we used the phylogenetically aware dissimilarity measures Unweighted UniFrac [57] and Weighted UniFrac [58]. Because some OTUs are closely related, while others are more distantly related, these two community-wide dissimilarity measures provide a more informative assessment of community resemblance than a more naive approach of e.g., counting shared OTUs between communities. Unweighted UniFrac considers only presence/absence of taxa and counts the fraction of the branch length unique to either community. In weighted UniFrac, the branch distance is weighted by difference in abundance. In both cases, the result is a scalar measure of dissimilarity between each pair of communities in this study, and is based on the information contained in the OTU table as well as the phylogeny relating the OTUs. We used the supplied Greengenes phylogeny (which is a curated version of a FastTree constructed phylogeny built from full length Greengenes 16S/SSU sequences). Because these measures are sensitive to differences in sequencing depth amongst samples, we randomly selected 1,038 (Study 1) or 31,109 (Study 2) sequences from each sample (subsampled dataset), and generated the distance matrix by a pair-wise inter-comparison of the profiles in the subsampled dataset. We used PCoA to compare samples based on the Unweighted and Weighted UniFrac distances. We clustered samples by applying the average neighbor (HC-AN) method as implemented in hclust (R package http://www.R-project.org) to the distance matrix.
Statistical analysis
Statistical tests with p-values less than 0.05 were considered significant. We used a binomial test to evaluate the significance of finding consistent changes in abundance of three phyla in the five Discovery cohort patients (Study 1). In the absence of any cancer effects, we expect that in a single patient, the chance that a microbial phylum would increase in the cancer sample is 0.5, as is the chance that the phylum would decrease. Although our counts are discrete, the probability of "no change in abundance at all" is so improbable that we can neglect it. Therefore we can calculate the probability that (all five patients show an increase or all five patients show a decrease) – a two-sided binomial with p = 2*(1/2)∧5 = .0625. In our data, three of the five major phyla show such a unanimous shift (all five patients show an increase or all five patients show a decrease). The probability that (three or more phyla have such an unanimous shift) absent any cancer effects is thus the CDF of a binomial with B(n = 5,p = 0.0625) to x = 3. p = 0.0022 in this case.
The adonis function implemented in the vegan package [59] was used to test for whole microbiome differences between sample groups.
For comparisons of specific sample groups, we used the relative abundance of OTUs after normalizing samples to one million counts. The normalization was done only for ease of display – no imputation of missing sequences was performed. A paired t-test was performed to identify OTUs significantly increased or decreased across cancer samples relative to the normal samples from the same patient, and Welch's t test (a two tailed, unequal variance test) was used to identify OTUs significantly increased or decreased across samples from two groups of patients.
Supporting Information
Rarefaction curves displaying average number of families and genera detected vs. sequencing depth in Study 1. For each point, sequences were subsampled without replacement 10 times and displayed is the average number of families (a) or genera (b) found. There is a fairly wide range of α diversity. For example, at 5,000 sequences per sample, the number of families detected ranges from ∼15–28.
(TIF)
Rarefaction curves displaying average number of OTUs detected vs. sequencing depth in Study 2. For each point, sequences were subsampled without replacement 10 times and displayed is the average number of OTUs found. Sample hn164FL has relatively high OTU diversity. Note, also that here we show diversity at the OTU level, whereas in Figure S1, the rarefaction curves are shown at the Family and Genus levels.
(TIF)
Data from studies 1 and 2 are highly correlated. (a) Scatterplots comparing sequence counts for OTUs determined in Study 1 (454, X-axis) with Study 2 (MiSeq, Y-axis). Shown is the Pearson correlation for each pair of samples. (b) Summary of correlations. The sequence counts were square root transformed to bring in the outliers prior to computing the Pearson and concordance correlation coefficients.
(TIF)
Relative abundance of phyla in paired samples in Study 2. Shown are the percent of OTUs corresponding to the five more abundant phyla in cancer and pre-cancer samples and their anatomically matched contralateral normal samples and left and right samples from healthy normal subjects.
(TIF)
Diversity of Firmicutes, Fusobacteria, Bacteroidetes, Proteobacteria and Actinobacteria genera in cancer, pre-cancers and healthy normal samples. Read counts normalized to one million counts are shown for the genera accounting for >10% of OTUs in more than 20% of samples for each phylum.
(TIF)
Change in relative abundance of genera in Study 2. Change in relative abundance of genera representing 10% of OTUs in more than 20% of samples. Shown is the difference in abundance of genera associated with cancers and pre-cancers compared to anatomically matched contralateral normal samples. For healthy normal samples, we compared left and right sides of the lateral tongue or floor of mouth.
(TIF)
Hierarchical clustering based on Weighted (left) and Unweighted (right) UniFrac for three sample types (Cancer and contralateral normal, healthy normal left/right and Pre-cancer and contralateral normal).
(TIF)
Whole microbiome PCoA based on Weighted (left) and Unweighted (right) UniFrac for three sample types (Cancer and contralateral normal, healthy normal left/right and Pre-cancer and contralateral normal). Significant microbiome differences were observed for patient identity.
(TIF)
Discovery cohort clinical characteristics.
(XLSX)
Study 2 cancer patients.
(XLSX)
Study 2 Carcinoma in situ patient.
(XLSX)
Study 2 pre-cancer patients.
(XLSX)
Study 2 healthy normal individuals.
(XLSX)
Replicate samples from cancer and pre-cancer patients.
(XLSX)
Sequence reads and OTUs in five patient matched cancer and normal samples (Discovery cohort).
(XLSX)
Study 1 OUT table.
(XLSX)
Study 2 cancer patients sequence reads and OTUs.
(XLSX)
Study 2 pre-cancer patients sequence reads and OTUs.
(XLSX)
Study 2 healthy normal individuals sequence reads and OTUs.
(XLSX)
Study 2 carcinoma in situ patient sequence reads and OTUs.
(XLSX)
Study 2 sequence reads and OTUs for replicate samples.
(XLSX)
Study 2 OTU table.
(XLSX)
Study 2 OTU table normalized to one million counts.
(XLSX)
Changes in abundance of genera from the five more abundant phyla.
(XLSX)
OTUs discriminating cancer and matched normal samples.
(XLSX)
Study 2 OTU table adjusted for variation in sequencing depth. Data were subsampled by randomly selecting 31,109 sequences from each community.
(XLSX)
Whole microbiome analysis - β diversity.
(XLSX)
Acknowledgments
We thank Matthew James Gebert and Rob Knight for preparation of the 16S rDNA amplicons for Study 1.
Funding Statement
This work was supported in part by an award of a GS Junior 454 Sequencing run from Roche, the National Center for Research Resources, the National Center for Advancing Translational Sciences, and the Office of the Director, National Institutes of Health, through University of San Francisco (UCSF)--CTSI grant number UL1 RR024129, and individual investigator awards from the National Cancer Institute grant (R01 CA131286, R21 CA 941186215) and the National Institute of Dental and Craniofacial Research (R01 DE019796). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Parkin DM, Pisani P, Ferlay J (1999) Global cancer statistics. CA Cancer J Clin 49: 33–64, 31. [DOI] [PubMed]
- 2. Shiboski CH, Schmidt BL, Jordan RC (2005) Tongue and tonsil carcinoma: increasing trends in the U.S. population ages 20–44 years. Cancer 103: 1843–1849. [DOI] [PubMed] [Google Scholar]
- 3. Schmidt BL, Dierks EJ, Homer L, Potter B (2004) Tobacco smoking history and presentation of oral squamous cell carcinoma. J Oral Maxillofac Surg 62: 1055–1058. [DOI] [PubMed] [Google Scholar]
- 4. Gillison ML (2004) Human papillomavirus-associated head and neck cancer is a distinct epidemiologic, clinical, and molecular entity. Semin Oncol 31: 744–754. [DOI] [PubMed] [Google Scholar]
- 5. Correa P, Haenszel W, Cuello C, Tannenbaum S, Archer M (1975) A model for gastric cancer epidemiology. Lancet 2: 58–60. [DOI] [PubMed] [Google Scholar]
- 6. Lax AJ, Thomas W (2002) How bacteria could cause cancer: one step at a time. Trends in microbiology 10: 293–299. [DOI] [PubMed] [Google Scholar]
- 7. Mager DL (2006) Bacteria and cancer: cause, coincidence or cure? A review. Journal of translational medicine 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hooper SJ, Wilson MJ, Crean SJ (2009) Exploring the link between microorganisms and oral cancer: a systematic review of the literature. Head & neck 31: 1228–1239. [DOI] [PubMed] [Google Scholar]
- 9. Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, et al. (2010) The human oral microbiome. Journal of bacteriology 192: 5002–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jenkinson HF, Lamont RJ (2005) Oral microbial communities in sickness and in health. Trends Microbiol 13: 589–595. [DOI] [PubMed] [Google Scholar]
- 11. Tezal M, Sullivan MA, Hyland A, Marshall JR, Stoler D, et al. (2009) Chronic periodontitis and the incidence of head and neck squamous cell carcinoma. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 18: 2406–2412. [DOI] [PubMed] [Google Scholar]
- 12. Hooper SJ, Wilson MJ, Crean SJ (2009) Exploring the link between microorganisms and oral cancer: a systematic review of the literature. Head Neck 31: 1228–1239. [DOI] [PubMed] [Google Scholar]
- 13. Nagy KN, Sonkodi I, Szoke I, Nagy E, Newman HN (1998) The microflora associated with human oral carcinomas. Oral oncology 34: 304–308. [PubMed] [Google Scholar]
- 14. Hooper SJ, Crean SJ, Lewis MA, Spratt DA, Wade WG, et al. (2006) Viable bacteria present within oral squamous cell carcinoma tissue. J Clin Microbiol 44: 1719–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hooper SJ, Crean SJ, Fardy MJ, Lewis MA, Spratt DA, et al. (2007) A molecular analysis of the bacteria present within oral squamous cell carcinoma. Journal of medical microbiology 56: 1651–1659. [DOI] [PubMed] [Google Scholar]
- 16. Pushalkar S, Ji X, Li Y, Estilo C, Yegnanarayana R, et al. (2012) Comparison of oral microbiota in tumor and non-tumor tissues of patients with oral squamous cell carcinoma. BMC microbiology 12: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bebek G, Bennett KL, Funchain P, Campbell R, Seth R, et al. (2012) Microbiomic subprofiles and MDR1 promoter methylation in head and neck squamous cell carcinoma. Human molecular genetics 21: 1557–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, et al. (2012) Experimental and analytical tools for studying the human microbiome. Nature reviews Genetics 13: 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE (2005) Defining the normal bacterial flora of the oral cavity. Journal of clinical microbiology 43: 5721–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Keijser BJ, Zaura E, Huse SM, van der Vossen JM, Schuren FH, et al. (2008) Pyrosequencing analysis of the oral microflora of healthy adults. Journal of dental research 87: 1016–1020. [DOI] [PubMed] [Google Scholar]
- 21. Zaura E, Keijser BJ, Huse SM, Crielaard W (2009) Defining the healthy "core microbiome" of oral microbial communities. BMC Microbiol 9: 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nasidze I, Li J, Quinque D, Tang K, Stoneking M (2009) Global diversity in the human salivary microbiome. Genome research 19: 636–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nasidze I, Quinque D, Li J, Li M, Tang K, et al. (2009) Comparative analysis of human saliva microbiome diversity by barcoded pyrosequencing and cloning approaches. Analytical biochemistry 391: 64–68. [DOI] [PubMed] [Google Scholar]
- 24. Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, et al. (2010) Bacterial diversity in the oral cavity of 10 healthy individuals. The ISME journal 4: 962–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lazarevic V, Whiteson K, Hernandez D, Francois P, Schrenzel J (2010) Study of inter- and intra-individual variations in the salivary microbiota. BMC genomics 11: 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Contreras M, Costello EK, Hidalgo G, Magris M, Knight R, et al. (2010) The bacterial microbiota in the oral mucosa of rural Amerindians. Microbiology 156: 3282–3287. [DOI] [PubMed] [Google Scholar]
- 27. Pushalkar S, Mane SP, Ji X, Li Y, Evans C, et al. (2011) Microbial diversity in saliva of oral squamous cell carcinoma. FEMS immunology and medical microbiology 61: 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stahringer SS, Clemente JC, Corley RP, Hewitt J, Knights D, et al. (2012) Nurture trumps nature in a longitudinal survey of salivary bacterial communities in twins from early adolescence to early adulthood. Genome research. [DOI] [PMC free article] [PubMed]
- 29. Braakhuis BJ, Tabor MP, Kummer JA, Leemans CR, Brakenhoff RH (2003) A genetic explanation of Slaughter's concept of field cancerization: evidence and clinical implications. Cancer research 63: 1727–1730. [PubMed] [Google Scholar]
- 30. Tabor MP, Brakenhoff RH, van Houten VM, Kummer JA, Snel MH, et al. (2001) Persistence of genetically altered fields in head and neck cancer patients: biological and clinical implications. Clin Cancer Res 7: 1523–1532. [PubMed] [Google Scholar]
- 31. Quince C, Lanzen A, Curtis TP, Davenport RJ, Hall N, et al. (2009) Accurate determination of microbial diversity from 454 pyrosequencing data. Nature methods 6: 639–641. [DOI] [PubMed] [Google Scholar]
- 32. Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, et al. (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the United States of America 108 Suppl 14516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mager DL, Haffajee AD, Devlin PM, Norris CM, Posner MR, et al. (2005) The salivary microbiota as a diagnostic indicator of oral cancer: a descriptive, non-randomized study of cancer-free and oral squamous cell carcinoma subjects. Journal of translational medicine 3: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guan SM, He JJ, Zhang M, Shu L (2011) Prevotella intermedia stimulates tissue-type plasminogen activator and plasminogen activator inhibitor-2 expression via multiple signaling pathways in human periodontal ligament cells. FEMS immunology and medical microbiology 62: 91–100. [DOI] [PubMed] [Google Scholar]
- 35. Cheng A, Schmidt BL (2008) Management of the N0 neck in oral squamous cell carcinoma. Oral Maxillofac Surg Clin North Am 20: 477–497. [DOI] [PubMed] [Google Scholar]
- 36. Chen J, Bittinger K, Charlson ES, Hoffmann C, Lewis J, et al. (2012) Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics 28: 2106–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bizzarro S, Loos BG, Laine ML, Crielaard W, Zaura E (2013) Subgingival microbiome in smokers and non-smokers in periodontitis: an exploratory study using traditional targeted techniques and a next-generation sequencing. Journal of clinical periodontology 40: 483–492. [DOI] [PubMed] [Google Scholar]
- 38. Kumar PS, Matthews CR, Joshi V, de Jager M, Aspiras M (2011) Tobacco smoking affects bacterial acquisition and colonization in oral biofilms. Infection and immunity 79: 4730–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chang AH, Parsonnet J (2010) Role of bacteria in oncogenesis. Clinical microbiology reviews 23: 837–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nobbs AH, Jenkinson HF, Jakubovics NS (2011) Stick to your gums: mechanisms of oral microbial adherence. Journal of dental research 90: 1271–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang G, Chen R, Rudney JD (2011) Streptococcus cristatus modulates the Fusobacterium nucleatum-induced epithelial interleukin-8 response through the nuclear factor-kappa B pathway. Journal of periodontal research 46: 558–567. [DOI] [PubMed] [Google Scholar]
- 42. Zhang G, Rudney JD (2011) Streptococcus cristatus attenuates Fusobacterium nucleatum-induced cytokine expression by influencing pathways converging on nuclear factor-kappaB. Molecular oral microbiology 26: 150–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gursoy UK, Pollanen M, Kononen E, Uitto VJ (2010) Biofilm formation enhances the oxygen tolerance and invasiveness of Fusobacterium nucleatum in an oral mucosa culture model. Journal of periodontology 81: 1084–1091. [DOI] [PubMed] [Google Scholar]
- 44. Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, et al. (2012) Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome research 22: 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, et al. (2012) Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome research 22: 292–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Markopoulos AK, Michailidou EZ, Tzimagiorgis G (2010) Salivary markers for oral cancer detection. The open dentistry journal 4: 172–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matthews AM, Kaur H, Dodd M, D'Souza J, Liloglou T, et al. (2012) Saliva collection methods for DNA biomarker analysis in oral cancer patients. The British journal of oral & maxillofacial surgery. [DOI] [PubMed]
- 48. Wei J, Xie G, Zhou Z, Shi P, Qiu Y, et al. (2011) Salivary metabolite signatures of oral cancer and leukoplakia. International journal of cancer Journal international du cancer 129: 2207–2217. [DOI] [PubMed] [Google Scholar]
- 49. Mager DL, Haffajee AD, Devlin PM, Norris CM, Posner MR, et al. (2005) The salivary microbiota as a diagnostic indicator of oral cancer: a descriptive, non-randomized study of cancer-free and oral squamous cell carcinoma subjects. J Transl Med 3: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Charlson ES, Chen J, Custers-Allen R, Bittinger K, Li H, et al. (2010) Disordered microbial communities in the upper respiratory tract of cigarette smokers. PLoS One 5: e15216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and environmental microbiology 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu Z, DeSantis TZ, Andersen GL, Knight R (2008) Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic acids research 36: e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R (2007) Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic acids research 35: e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, et al. (2011) Examining the global distribution of dominant archaeal populations in soil. The ISME journal 5: 908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nature methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, et al. (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and environmental microbiology 72: 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Applied and environmental microbiology 71: 8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lozupone CA, Hamady M, Kelley ST, Knight R (2007) Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Applied and environmental microbiology 73: 1576–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oksanen j, Blanchet FG, Kindt R, Legendre P, Minchin PR, et al. (2012) vegan: Community Ecology Package. R package version 2.0–5.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rarefaction curves displaying average number of families and genera detected vs. sequencing depth in Study 1. For each point, sequences were subsampled without replacement 10 times and displayed is the average number of families (a) or genera (b) found. There is a fairly wide range of α diversity. For example, at 5,000 sequences per sample, the number of families detected ranges from ∼15–28.
(TIF)
Rarefaction curves displaying average number of OTUs detected vs. sequencing depth in Study 2. For each point, sequences were subsampled without replacement 10 times and displayed is the average number of OTUs found. Sample hn164FL has relatively high OTU diversity. Note, also that here we show diversity at the OTU level, whereas in Figure S1, the rarefaction curves are shown at the Family and Genus levels.
(TIF)
Data from studies 1 and 2 are highly correlated. (a) Scatterplots comparing sequence counts for OTUs determined in Study 1 (454, X-axis) with Study 2 (MiSeq, Y-axis). Shown is the Pearson correlation for each pair of samples. (b) Summary of correlations. The sequence counts were square root transformed to bring in the outliers prior to computing the Pearson and concordance correlation coefficients.
(TIF)
Relative abundance of phyla in paired samples in Study 2. Shown are the percent of OTUs corresponding to the five more abundant phyla in cancer and pre-cancer samples and their anatomically matched contralateral normal samples and left and right samples from healthy normal subjects.
(TIF)
Diversity of Firmicutes, Fusobacteria, Bacteroidetes, Proteobacteria and Actinobacteria genera in cancer, pre-cancers and healthy normal samples. Read counts normalized to one million counts are shown for the genera accounting for >10% of OTUs in more than 20% of samples for each phylum.
(TIF)
Change in relative abundance of genera in Study 2. Change in relative abundance of genera representing 10% of OTUs in more than 20% of samples. Shown is the difference in abundance of genera associated with cancers and pre-cancers compared to anatomically matched contralateral normal samples. For healthy normal samples, we compared left and right sides of the lateral tongue or floor of mouth.
(TIF)
Hierarchical clustering based on Weighted (left) and Unweighted (right) UniFrac for three sample types (Cancer and contralateral normal, healthy normal left/right and Pre-cancer and contralateral normal).
(TIF)
Whole microbiome PCoA based on Weighted (left) and Unweighted (right) UniFrac for three sample types (Cancer and contralateral normal, healthy normal left/right and Pre-cancer and contralateral normal). Significant microbiome differences were observed for patient identity.
(TIF)
Discovery cohort clinical characteristics.
(XLSX)
Study 2 cancer patients.
(XLSX)
Study 2 Carcinoma in situ patient.
(XLSX)
Study 2 pre-cancer patients.
(XLSX)
Study 2 healthy normal individuals.
(XLSX)
Replicate samples from cancer and pre-cancer patients.
(XLSX)
Sequence reads and OTUs in five patient matched cancer and normal samples (Discovery cohort).
(XLSX)
Study 1 OUT table.
(XLSX)
Study 2 cancer patients sequence reads and OTUs.
(XLSX)
Study 2 pre-cancer patients sequence reads and OTUs.
(XLSX)
Study 2 healthy normal individuals sequence reads and OTUs.
(XLSX)
Study 2 carcinoma in situ patient sequence reads and OTUs.
(XLSX)
Study 2 sequence reads and OTUs for replicate samples.
(XLSX)
Study 2 OTU table.
(XLSX)
Study 2 OTU table normalized to one million counts.
(XLSX)
Changes in abundance of genera from the five more abundant phyla.
(XLSX)
OTUs discriminating cancer and matched normal samples.
(XLSX)
Study 2 OTU table adjusted for variation in sequencing depth. Data were subsampled by randomly selecting 31,109 sequences from each community.
(XLSX)
Whole microbiome analysis - β diversity.
(XLSX)