Abstract
Background
Originating from Primordial Germ Cells/gonocytes and developing via a precursor lesion called Carcinoma In Situ (CIS), Germ Cell Cancers (GCC) are the most common cancer in young men, subdivided in seminoma (SE) and non-seminoma (NS). During physiological germ cell formation/maturation, epigenetic processes guard homeostasis by regulating the accessibility of the DNA to facilitate transcription. Epigenetic deregulation through genetic and environmental parameters (i.e. genvironment) could disrupt embryonic germ cell development, resulting in delayed or blocked maturation. This potentially facilitates the formation of CIS and progression to invasive GCC. Therefore, determining the epigenetic and functional genomic landscape in GCC cell lines could provide insight into the pathophysiology and etiology of GCC and provide guidance for targeted functional experiments.
Results
This study aims at identifying epigenetic footprints in SE and EC cell lines in genome-wide profiles by studying the interaction between gene expression, DNA CpG methylation and histone modifications, and their function in the pathophysiology and etiology of GCC. Two well characterized GCC-derived cell lines were compared, one representative for SE (TCam-2) and the other for EC (NCCIT). Data were acquired using the Illumina HumanHT-12-v4 (gene expression) and HumanMethylation450 BeadChip (methylation) microarrays as well as ChIP-sequencing (activating histone modifications (H3K4me3, H3K27ac)). Results indicate known germ cell markers not only to be differentiating between SE and NS at the expression level, but also in the epigenetic landscape.
Conclusion
The overall similarity between TCam-2/NCCIT support an erased embryonic germ cell arrested in early gonadal development as common cell of origin although the exact developmental stage from which the tumor cells are derived might differ. Indeed, subtle difference in the (integrated) epigenetic and expression profiles indicate TCam-2 to exhibit a more germ cell-like profile, whereas NCCIT shows a more pluripotent phenotype. The results provide insight into the functional genome in GCC cell lines.
Introduction
Type II (testicular) germ cell tumors, here referred to as Germ Cell Cancers (GCC), are the most common malignancy in Caucasian adolescents and young adults, and their incidence is still rising [1]–[3]. GCC originate from primordial germ cells or gonocytes, and are subdivided into seminomas (SE) and non-seminomas (NS), with carcinoma in situ (CIS) of the testis as their common precursor lesion [1], also known as Intratubular Germ Cell Neoplasia Unclassified (IGCNU) [3]. In contrast to CIS and SE, the stem cell component of NS (i.e., embryonal carcinoma, EC) is characterized by pluripotent potential [4]. EC can differentiate into somatic lineages and extra–embryonic tissues (teratoma vs yolk sac tumor and choriocarcinoma, respectively), including the germ cell lineage [4]. Various clinical, environmental and genetic risk factors for GCC have been identified, although the exact role of these factors is not completely clear. Clinical risk factors constitute urological/andrological/gonadal aberrations [5]–[8], while environmental factors focus on endocrine disruptors and androgen - estrogen balance [9]–[11]. Genetic risk factors include a number of susceptibility Single Nucleotide Polymorphisms, likely related to early gonadal development [12]–[15] and an association with familial predisposition [16]. Somatic mutations are rarely found in GCC [17]. There are strong indications that the micro-environment of the developing testis is of significant importance in the pathogenesis of GCC. Patients with Testicular Dysgenesis Syndrome (TDS) and specific forms of Disorders of Sex Development (DSD) are known to have an increased risk of developing GCC due to abnormal gonadal development, i.e. hypovirilization [18].
Epigenetic processes have a clear role in both the initiation and protection of pluripotency [19]. Deregulation of these tightly controlled processes is known to be involved in the formation and progression of various cancer types [20]–[24], including GCC [25]. During physiological germ cell formation and maturation, epigenetic processes (e.g. DNA methylation, histone modifications) guard homeostasis by regulating the accessibility of the DNA to facilitate transcription [25], [26]. The epigenome is highly dynamic, and changes occur depending on cell type and developmental stage, influenced by/reflecting the (micro-) environment. In spite of this knowledge, little is known about the role of histone modifications and DNA methylation regarding gene expression in GCC in general, and the possible similarities and differences between SE and EC [25], [27]. Epigenetic deregulation through genetic and environmental parameters (referred to as genvironment) could disrupt physiological embryonic germ cell development, resulting in delayed or blocked maturation, thereby facilitating the formation of CIS, and potentially progression to an invasive GCC [25], [28]–[30]. Therefore, determining the epigenetic and functional genomic landscape in GCC cell lines could provide insight into the pathophysiology and etiology of GCC. The results could provide guidance for targeted functional experiments.
In this study, epigenetic footprints of SE and EC cell lines were identified by studying the interaction between gene expression, DNA methylation and histone modifications. Two well characterized GCC-derived cell lines were used, one representative for SE (TCam-2) [31], [32] and the other for EC (NCCIT) [33]. Two types of epigenetic modifications were investigated and related to genome wide expression analysis: CpG DNA methylation status, and enrichment of activating histone marks (H3K4me3, H3K27ac).
Methods
Cell culture
TCam-2 [31], [32], [34] and NCCIT [33] cells were cultured in DMEM medium (#31966-021, Thermo Fisher Scientific/Life Technologies, Carlsbad, CA, USA) containing 10% fetal calf serum (FCS, GE Healthcare Life Sciences, HyClone Laboratories, Utah, USA) in T75 cm2 flasks to 75–90% confluence. For RNA preparation, fresh medium was added 24 hours before harvest. Cells were washed once with Hanks balanced Salt Solution (HBSS, #14175-053, Thermo Fisher Scientific/Life Technologies, Carlsbad, CA, USA), and lysed with 7 ml of ice-cold RNA-Bee (#Cs-105B, TEL-TEST Inc, Friendswood, Texas, USA). For methylation, gene expression (biological duplicates) and ChiP-seq analyses, different cultures of cells from a single source were used (LEPO lab, Department of Pathology, Erasmus MC Rotterdam). Biological replicates were started as independent cultures at different days and processed similarly.
Methylation profiling
DNA was isolated using the DNeasy kit according to manufacturer's instructions (#69504, QIAGEN, Hilden, Germany). Bisulfite conversion (EZ DNA Methylation Gold Kit, Zymo Research, Irvine, CA, USA) and methylation detection was performed at ServiceXS (ServiceXS B.V., Leiden, The Netherlands). Illumina's HumanMethylation450 BeadChip was used (Illumina, Inc., San Diego, CA, U.S.A, processing and hybridization according to the manufacturer's instructions). Image processing took place on the iScan system and the data was extracted using GenomeStudio, using default analysis settings (including background correction and normalization based on internal controls) and v 1.2 of the annotation manifest (http://support.illumina.com/downloads/humanmethylation450_15017482_v12.ilmn). Further processing was carried out in R using the LUMI (http://www.bioconductor.org/) package [35] following the optimized “lumi: QN+BMIQ” pipeline [36] This includes exclusion of poorly performing probes (p<0.01), color adjustment, quantile normalization and correction for probe type bias (Infinium I vs II) using the BMIQ algorithm [37]. All raw and processed data files are submitted as a GEO SuperSeries and accessible via GSE56454 (http://www.ncbi.nlm.nih.gov/geo/). Differentially methylated regions were identified using the DMRforPairs algorithm using the default settings [38]. DMRforPairs is available via Bioconductor: (http://www.bioconductor.org/packages/release/bioc/html/DMRforPairs.html). Briefly, DMRforPairs defines genomic regions using local probe density and optionally functional homogeneity (e.g. all probes in a region should be gene associated). It quantifies, tests and visualizes (differential) methylation patterns between unique samples. Differences were calculated as NCCIT versus TCam-2.
Gene expression profiling
Approximately 20 µg of RNA was treated with RNase-free DNaseI (#2238, Ambion, Ambion Inc., Austin, TX, U.S.A.) for 30 minutes at 37°C and subsequently purified using the RNeasy mini kit (#74104, Qiagen, Hilden, Germany) according to the manufacturer's instructions. Pure RNA was eluted in 50 µl of water, and quantified using a Nanodrop (Thermo Scientific). Quality control, RNA labeling, hybridization and data extraction were performed at ServiceXS B.V. (Leiden, The Netherlands) according to their in-house protocol. Biotinylated cRNA was prepared using the Illumina TotalPrep RNA Amplification Kit (#AMIL1791, Ambion Inc., Austin, TX, USA) according to the manufacturer's specifications with an input of 200 ng total RNA. Per sample, 750 ng of the biotinylated cRNA was hybridized onto the Illumina HumanHT-12 v4 (Illumina, Inc., San Diego, CA, U.S.A.) according to the Illumina Manual “Direct Hybridization Assay Guide”. Image processing took place on the iScan system and the data was extracted using GenomeStudio (default settings). Further processing was carried out in R using the LUMI package (http://www.bioconductor.org/) [35]. Following the guidelines presented in [39], robust spline normalization was applied to the log2 transformed intensity values. Probes with pdetection>0.05 in >50% of the samples were excluded from the analysis (n = 27,964 out of 47,323). Average log2 intensities of biological replicates and per gene were used to assess expression levels (GEO accession number GSE56454). Log2 ratios (R) of the average intensities in the two cell lines (NCCIT/TCam-2) were used to identify significantly differentially expressed genes. Genes with expression levels outside the 99% confidence interval (CI) of this log ratio were identified as differentially expressed between NCCIT and TCam-2.
Histone modification profiling (H3K27ac, H3K4me3)
The ChIP assay was performed according to the low cell number ChIP protocol from Diagenode (Liege, Belgium), with minor modifications. In brief, 1×106 cells were cross-linked for eight minutes by addition of formaldehyde to a final concentration of 1%, followed by neutralization with 1.25 M glycine. The cells were then lysed, and chromatin was sheared to ∼500 bp fragments using the Covaris sonicator under the following conditions; duty cycle 20%, peak incident power 200 watts, cycles/burst 200, time 5 min, temperature 4°C. Protein A-coated Dynabeads (#10002D, Invitrogen, Thermo Fisher Scientific/Life Technologies, Carlsbad, CA, USA) were incubated with 7 µg of the following antibodies: H3K4me3 (Diagenode pAb-003-050) or H3K27ac (Ab4729, Abcam, Cambridge, UK). The beads were combined with chromatin from 1×106 cells overnight on a rotating wheel. The immunobeads were washed, and DNA was purified using the iPure DNA purification kit (AL-100-0100, Diagenode, Liège, Belgium) according to manufacturer's instructions. DNA fragments were sheared a second time using a Covaris sonicator (duty cycle 10%, peak incidence power 175 watts, cycles/burst 200, time 5 minutes, temperature 4°C). Massively parallel sequencing of ChIP DNA (ChIP-Seq) was performed using the 5500xl SOLiD™ sequencing platform (Applied Biosystems, Foster City, CA, USA) at the Monash Health Translation Precinct Medical Genomics Facility. The sequencing experiments were single-end with 50 nt read length (300 nt average fragment size). Sequencing reads were aligned to the complete hg19 human genome (UCSC version, February 2009) using LifeScope™ Genomic Analysis Software v2.5 (http://www.lifetechnologies.com/lifescope). ChIP-Seq experimental samples were normalized to a total of 107 uniquely mapped sequencing tags. Data was processed in HOMER ([40], http://homer.salk.edu/homer/chipseq/) to detect peaks and motif enrichments using the default settings (except “fold enrichment over input”; used 2; threshold for p-value 0.01) (GEO accession number GSE56454). Peak heights from HOMER were corrected for background (lowest peak height detectable). Heights were then summed per gene (ΣP) as annotated by HOMER and genes without any detectable peak were set to 0. The difference in summed peak heights (ΔΣP = ΣPTCam-2-ΣPNCCIT) was used to quantify differences between the cell lines. Genes with significantly differential histone modification patterns were identified for both marks separately (outside 99% CI of ΔΣP). Association of a peak with TSS was used as annotated by HOMER (1 kb upstream of the TSS - 100 bp downstream).
MLPA-DNaseI analysis
MLPA probes were designed following previously described criteria [41]. Based on differential modification patterns in NCCIT and TCam-2 probes were designed for the following loci: NCCIT: chr3:181425532-181425720, chr5:146699813-146699953, chr3:181577755-181577830, chr6:15240050-15240160, chr15:93191596-93191696, chr3:178908801-178908966, chr5:101550991-101551125, TCam-2: chr11:10613134-10613220, chr1:201278163-201278251, chr12:3091402-3091483, chr19:13985162-13985322, chr2:38323813-38323931, chr9:843018-843110, chr5:140762378-140762475. MLPA-DNaseI was performed as previously described [42], with minor modifications. In brief, nuclei from 1×106 cells were isolated, and treated with a range of DNaseI concentrations (0; 2 and 5 Units). Digested genomic DNA was purified, and 50–100 ng was used in an MLPA reaction. Following PCR amplification of ligated probes, products were separated on an ABI3700 DNA sequencer. Data was analyzed as previously described, with a reduction to 75% of peak height in undigested DNA used as a threshold for defining DNaseI-hypersensitivity [43].
Software
Analyses were performed in R 3.0.1 (Windows 7×64) and 2.15.2 (Redhat Linux ×64). The networks/enrichment analyses were performed in IPA (Ingenuity® Systems, www.ingenuity.com). Genomic positions reported in this manuscript are based on the GRch37/hg19 assembly.
Results
To investigate epigenetic characteristics of SE and EC and their relationship to gene expression, genome-wide histone modification and DNA methylation patterns were investigated in the cell lines TCam-2 (SE) and NCCIT (EC), and matched to gene expression profiles. The differences between the two cell lines with regard to histone modification and DNA methylation status were first investigated separately. Subsequently, the resulting datasets were integrated to identify (target) genes with a strong relationship between primed DNA configuration and higher expression levels.
Histone modification
Histone modification patterns were assessed using chromatin immunoprecipitation combined with high throughput sequencing (ChIP-seq). Data analysis was performed as described in the materials and methods section. Alterations in H3K4me3 and H3K27ac were investigated, which are markers associated with promoter activation (transcription start site (TSS), H3K4me3 and H3K27ac) and enhancer activation (primarily H3K27ac) [44], [45]. In addition to the analysis of (differential) modification patterns, motif enrichment of the modified regions was investigated and compared between the cell lines.
H3K4me3 and H3K27ac do not show differential enrichment near transcription start sites and their peak heights correlated within genes
Depending on the cell line, 10.2/11.3% of the H3K27ac enriched loci were located within 1 kb of a TSS against a comparable 14.7/16.8% of the H3K4me3 loci (TCam-2/NCCIT). This is in line with observations in other cell types showing that, even though H3K4me3 is directly related to promoter activation, a large majority of the H3K4me3 loci are located distally of the TSS [46]. H3K27ac has no reported preferential localization to TSS [47]. The level of summed peaks per gene (ΣP, see Methods) was used to compare histone modification patterns between the two histone marks. There was significant correlation in both cell lines between the peak levels at genes where both were present (ρTCam-2 = 0.62, pTCam-2<0.001, nTCam-2 = 1837; ρNCCIT = 0.37, pNCCIT<0.001, nNCCIT = 746; Spearman's ρ). This is in line with their overlapping function: open chromatin configuration is associated with both marks [19], [22] and allowed us to combine the histone modification results in the subsequent analysis.
H3K4me3 and H3K27ac enrichment patterns in TCam-2 and NCCIT are in accordance with known SE/EC markers specificity
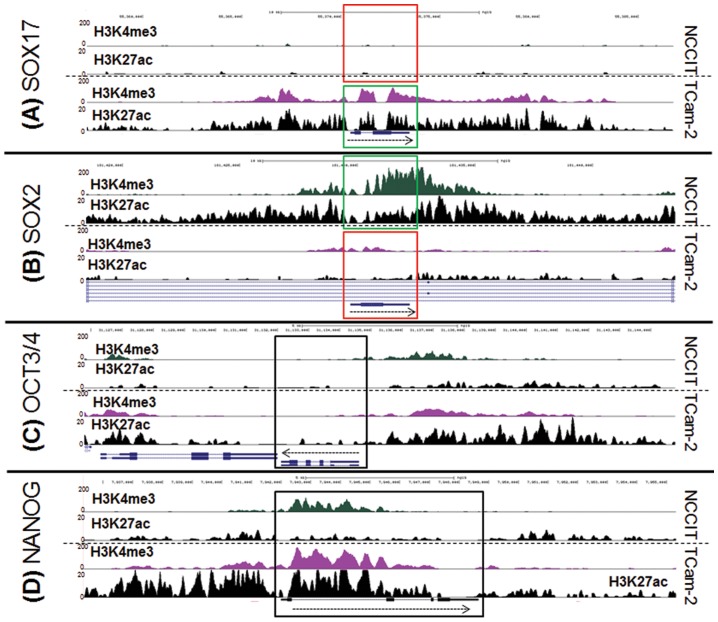
We previously showed that active chromatin modification patterns for SOX17 and SOX2 in the cell lines TCam-2 and NCCIT match the expected pattern, based on gene and protein expression and histological constitution (SOX17 active in TCam-2, SOX2 active in NCCIT) [25]. In line with this, SOX17 and SOX2 were differentially enriched for both H3K4me3 and H3K27ac in TCam-2 and NCCIT respectively (Figure 1). OCT3/4 showed no enrichment within the coding sequence, however there was enrichment of both markers close to the TSS in NCCIT and TCam-2. This is consistent with known OCT3/4 mRNA and protein expression in both cell lines [31], [48]. NANOG was more enriched for both markers in TCam-2, in line with differences in expression level (see below).
Figure 1. Display of H3K4me3 and H3K27ac tracks for both NCCIT and TCam-2. (A) SOX17, (B) SOX2, (C) OCT3/4 (POU5F1), (D) NANOG.
Arrows indicate direction of transcription. Green boxes indicate markers specific for the histological subtype represented by the cell line. Black boxes = no difference between the cell lines; red boxes = not a marker for that cell type. Note the different ranges on the y-axis for H3K4me3 and H3K27ac.
On a genome-wide scale, 29,428 H3K4me3 enriched loci were identified in TCam-2 and 19,015 in NCCIT. 25–41% more enriched loci were identified for H3K27ac than for H3K4me3 (nTCam-2 = 41,569, nNCCIT = 23,763). Genes with significant differences in summed peak height per gene (ΔΣP, see Methods) were selected for further analysis. For TCam-2, genes showing differential histone modifications were higher in number for H3K27ac (nTCam-2 = 433, NNCCIT = 28). For NCCIT, there were substantially more genes selected for H3K4me3 compared to H3K27ac (nTCam-2 = 215, NNCCIT = 325). For both marks, 86/11 genes overlapped between top differentiating lists in TCam-2 and NCCIT respectively. These included the SE marker SOX17 in TCam-2 and the EC marker SOX2 in NCCIT (Figure S1, Table S1). Functionally, the gene lists of both cell lines showed significant enrichment for (embryonic) stem cell maintenance/pluripotency (Table S2). Enrichment of biological functions in TCam-2 indicated similarity to more mature germ cells, which was lacking in the list of NCCIT (GO categories TCam-2 included development of normal testis morphology and germ cell maintenance). Moreover, two germ cell-specific canonical pathways IGF1 signaling (logp = 3.42) and germ cell-Sertoli cell Junction Signaling (logp = 2.11)) showed enrichment. In TCam-2, two functional networks were identified incorporating the AR pathway and lipid metabolism (Table S2).
Germ cell markers AP-2α and AP-2γ are top enriched motifs in TCam-2, while embryonic stem cell specific motifs SOX2/OCT4/TCF/NANOG are enriched in both cell lines
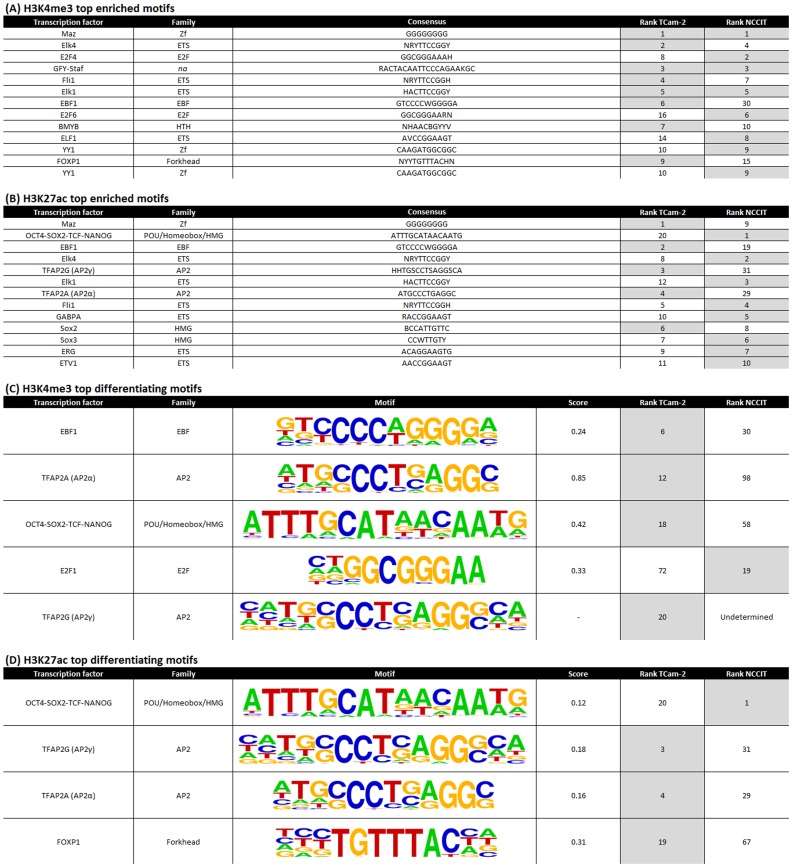
Significantly enriched motifs were identified for each cell line and histone mark (HOMER tool, see Methods). There was strong overlap between the top-ranked enriched motifs in NCCIT and TCam-2. This was true for both activating markers. For example, for H3K4me3 the top enriched motif was MAZ for both TCam-2 and NCCIT, a transcription factor associated with MYC (binds to two sites in its promoter) and known to be involved in transcription initiation as well as termination [49] (Figure 2A,B; Table S3).
Figure 2. Motif enrichment in histone modification data.
All motifs were significantly enriched in target over background sequences (p<0.01). Fold enrichment is indicated relative to background. (A,B) Top ranking motifs in both cell lines showed strong overlap (top 10). (C,D) Motifs that differed strongly between the cell lines with regard to their enrichments were selected. A motif was assessed favorably if its ranking was high (≤20) for one cell line and low for the other cell line (or was absent in the other list of enriched motifs). Score: The difference in ranking was assessed based on the difference in relative position in the list (|1-(rTCam-2/nTCam-2)–1-(rNCCIT/nNCCIT)|≥15%, n = nr of enriched motifs, r is the rank of a specific motif in the list of enriched motifs for either cell line).
A limited number of markers showed differences in enrichment between the cell lines (Figure 2C,D, Table S3). For H3K4me3, five motifs were identified which showed sufficient difference. Four were higher ranked in TCam-2 and one higher in NCCIT. The top ranked TCam-2 motifs presented in this differentiating list were EBF1 (role in developmental processes), AP-2α and AP-2γ (known germ cell markers) and OCT4/SOX2/TCF/NANOG (pluripotency motif). For NCCIT, E2F1 (cell cycle control, action of tumor suppressor proteins, cell proliferation) was ranked higher. For H3K27ac, there were four differentiating motifs, of which three were ranked higher in TCam-2 and one higher in NCCIT. The OCT4/SOX2/TCF/NANOG motif was the most significantly enriched motif for H3K27ac in NCCIT (ranked 20 in TCam-2). This motif is known to be predominantly enriched in embryonic stem (ES) cells [50] as well as embryonic germ cells, which is in line with the stem cell-like origin (EC) of NCCIT and the germ cell-like origin of TCam-2 respectively. Moreover, for H3K27ac, AP-2α and AP-2γ were ranked as 3rd and 4th most enriched motif in TCam-2 (compared to 29th and 31st in NCCIT) reflecting their (embryonic) germ cell origin (Figure 2C,D). For ES cells the enrichment rankings for these two motifs were 87th and 105th [50]. These observations fit with the proposed more differentiated (germ cell lineage) cell of origin of SE as compared to EC [28], and are in line with the findings of related histone peaks (see Discussion section).
Verification of the H3K4me3 & H3K27ac enrichment-based open chromatin configuration was independently confirmed using DNaseI-hypersensitivity
As the investigated chromatin marks (H3K4me3 & H3K27ac) are considered to be associated with active chromatin, we explored whether their presence was associated with another characteristic of active chromatin: DNaseI-hypersensitivity [51]. Using a DNaseI-MLPA approach [43] we targeted 14 regions which showed the greatest differences in either H3K4me3 and/or H3K27ac enrichment between the same two cell lines. Six of seven enriched regions in NCCIT (Figure S2A: N1,N2,N3,N4,N6,N7), and five of seven enriched regions in TCam-2 (Figure S2B: S1,S2,S5,S6,S7), showed significant DNaseI-hypersensitivity in the respective cell lines. In contrast, only one H3K27ac-negative locus in NCCIT (Figure S2A, S1), and no H3K27ac-negative loci in TCam-2 (Figure S2B), showed DNaseI-hypersensitivity.
CpG Methylation
The DMRforPairs algorithm was used to identify differentially methylated regions (DMR) [38]. The algorithm was set to detect strong differences, i.e. using stringent settings. Regions containing a minimum of four probes within 200 bp distance of each other were considered for further analysis (n = 30,306). Regions in which median methylation levels (M-values) between the samples differed at least |1.4| (n = 5,139) were tested for statistical significance (significant: p<0.05; Bonferroni adjusted, Wilcoxon-rank-sum test, n = 143) (Output DMRforPairs: File S1).
Methylation patterns at DMRs are in line with marker positivity in SE and EC
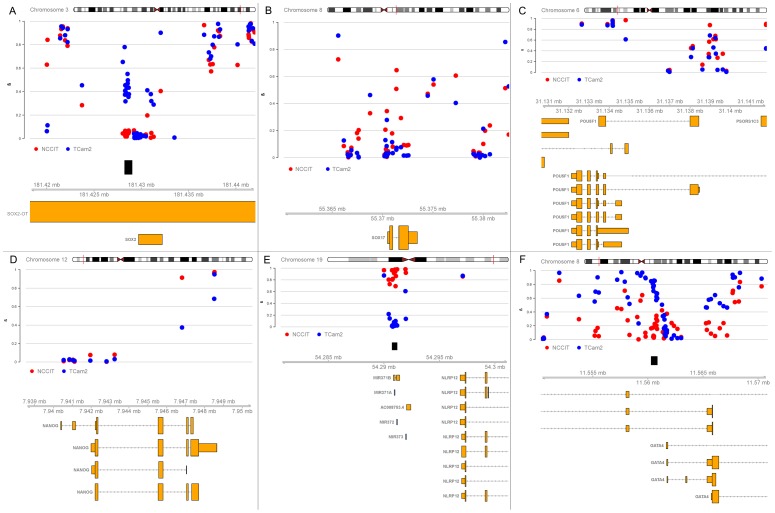
Because of the activating histone modifications investigated, we focused on hypo- or absence of DNA methylation. Global methylation levels were in line with the hyper- and hypomethylated global status of NS and SE respectively, and the previously shown intermediate status of TCam-2 (Figure S3) [52], [53]. After DMR identification using DMRforPairs, a total of 99 DMRs (annotating to 170 unique gene symbols) were hypomethylated in TCam-2, compared to 44 in NCCIT (annotating to 64 unique gene symbols) (Table S1). In line with the histone modifications (see above), the SOX2 promoter region was found to be strongly hypomethylated in NCCIT (Figure 3A). SOX2 was partly methylated in TCam-2, in line with findings illustrating that TCam-2 can differentiate and become SOX2 positive after extra-gonadal injection in mice [54]. A 220 bp region directly upstream of the TSS of SOX2 (chr3:181429712) has previously been shown to be completely hypomethylated in TCam-2 [55]. This is in line with our findings as a consistently hypomethylated region (chr3:181429233-181430485) is shown directly upstream of the TSS while a 652 bp long DMR (chr3:181428046-181428697) between NCCIT and TCam-2 is detected by DMRforPairs in a region ca. 800 bp upstream of the region sequenced by Nettersheim et al. SOX17 did not show a significant differential methylation pattern, indicating that it is, in principle, accessible for transcription in both cell types (Figure 3B). Indeed, SOX17 expression can be induced in NCCIT (unpublished observation). In line with known gene expression in both cell lines, OCT3/4 showed an inconsistent, but non-differential methylation pattern (Figure 3C). The TSS of NANOG was hypomethylated in both cell lines (Figure 3D), in line with the expression data and previous reports [56]. In the list of top DMRs, the miR-371/2/3 cluster stood out by significant differential hypermethylation in NCCIT (Figure 3E). The promoter region of GATA4 was significantly hypermethylated in TCam-2 (Figure 3F). In general, 33% (47/143) DMRs were annotated to TSSs (File S1, according to Illumina's manifest) which is similar to the fraction of TSS associated regions identified by DMRforPairs (12,652/30,306). Functionally, the DMR list of both cell lines showed enrichment for (embryonic) stem cell maintenance/pluripotency. Biological functions indicating similarity to more mature germ cells were enriched in TCam-2 (Table S2).
Figure 3. Methylation patterns of known germ cell markers (A–D) and significant DMRs for both cell lines (E, F).
Dots depict individual CpGs and black boxes denote DMRs identified by DMRforPairs. Percentages below indicate average CG density in the plotted regions (calculated using the Repitools R package, gcContentCalc function (http://www.bioconductor.org/)). (A) SOX2 [44%], (B) SOX17 [46%], (C) OCT3/4 (POU5F1), [52%] (D) NANOG, [44%] (E) miR-371/2/3 cluster, [49%] (F) GATA4 [53%].
DMRs were significantly enriched for imprinted genes, and 59% (51/86) of all imprinted genes showed loss of methylation around their TSS in one or both cell lines
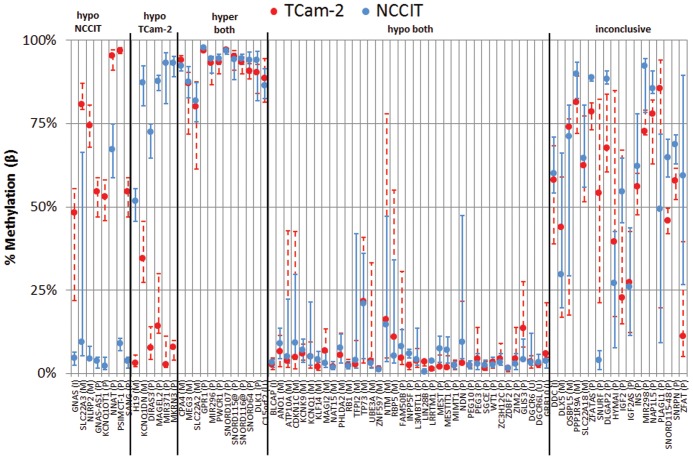
From the list of verified imprinted human genes (n = 88 retrieved from geneimprint.com), 82 were also annotated in Illumina's manifest (total of 21,243 unique gene symbols annotated) and 10 were present in the top DMRs between TCam-2 and NCCIT. This overrepresentation of imprinted genes in the list of DMRs was significant (p<0.0001, χ2 test). When investigating the region surrounding the TSS of the 86 imprinted genes with known genomic localization, 14 showed a differential status between the cell lines (8/6 hypomethylated in NCCIT and TCam-2 respectively). In total, 37 imprinted genes displayed a hypomethylated status in both cell lines, compared with 12 hypermethylated genes (Figure 4). In summary, these results indicate an overall erased status of the imprinted regions in both cell lines. Regarding genes with a differential methylation status, there is no clear difference in number of hypermethylated genes that would indicate a difference in maturation status or environmental disruption.
Figure 4. Methylation patterns in promoter regions of imprinted genes.
Localization of TSS was retrieved from Ensembl and manually corrected for genes with multiple transcripts to select a region with representative coverage on the Illumina BeadChip (nprobes varied between the genes: median = 10, inter-quartile range 6–21). Promoter region was defined as TSS-1000–TSS+100 (or opposite on reverse strand). Genes with >0.25 difference in median β and a consistent (stable) methylation pattern were identified as differentially methylated at the TSS between the two cell lines. Median methylation <25% for both cell lines was interpreted as a hypomethylated state in both cell line. Median methylation >75% was interpreted as hypermethylation. No probes were annotated around the TSS of TCEB3C (M), RNU5D (P), SNORD108 (P), SNORD109A (P) and SNORD109B (P). SANG and GNAS-AS1 are known aliases but separate entities on geneimprint.com and are depicted separately to preserve consistency. P = paternally imprinted/expressed, M = maternally imprinted/expressed, I = isoform-dependent imprinting.
Expression
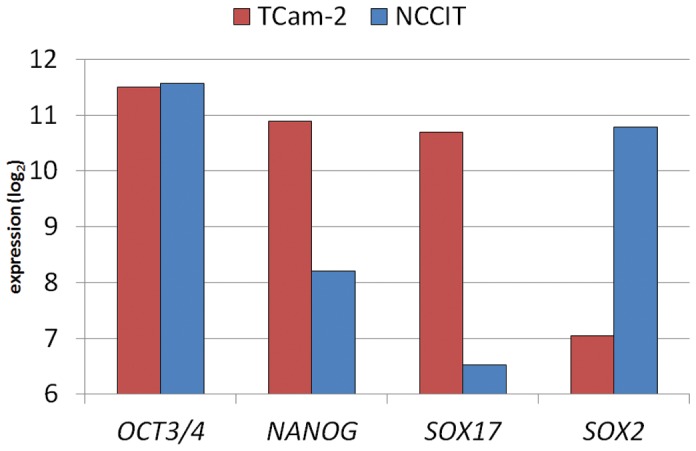
Expression levels of markers matched histological origin of both cell lines
In total, 257 genes were expressed higher in TCam-2, compared to 149 in NCCIT (Table S1). Greater than 3.65 fold difference in expression level (99% confidence interval (CI) of log2-ratio of intensities) was considered significant. The expression levels were in agreement with the classification of the cell lines: SOX17 was higher in TCam-2 compared with NCCIT, with the opposite observed for SOX2 [57] (Figure 5). OCT3/4, a general marker for the stem cell components (SE/EC) of GCC, was expressed at equal levels in both cell lines. NANOG was expressed higher in TCam-2, which is in line with the open histone configuration (Figure 1D, transcription possible in both cell lines). Functionally, the gene lists of both cell lines showed enrichment for (embryonic) stem cell maintenance/pluripotency, and Wnt/β-catenin signaling. Enrichment of biological functions consistent with more mature germ cells was present in TCam-2, and absent in NCCIT. In addition, network analysis revealed the androgen pathway in TCam-2, represented by both the AR and testosterone, thus showing major overlap between the networks found by genes that had differential histone modification patterns (Table S2).
Figure 5. Expression levels of known GCC markers match the histological subtype from which the cell lines originate.
Integration of epigenetic and expression data
Differential gene lists from histone modification, methylation and expression data showed limited overlap
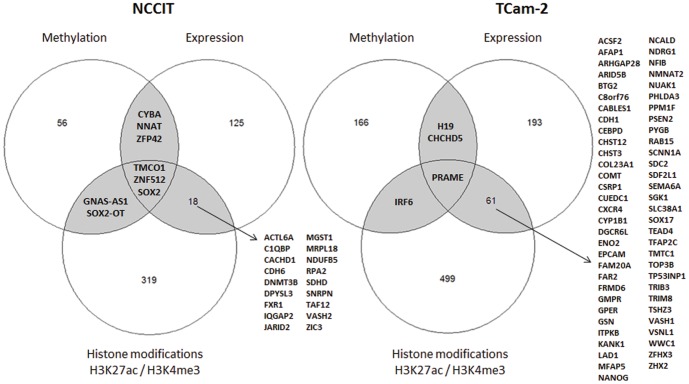
Differentiating gene lists from the separate analyses discussed above were matched based on Gene Symbol to assess the relationship between active histone modifications, the absence of CpG methylation and gene expression. Overlap between gene expression and one of the epigenetic regulatory mechanisms is of interest as expression of a specific gene does not need to be regulated by both mechanisms. Figure 6 shows the overlap of the different variables for the differentiating gene lists between TCam-2 and NCCIT. In general, little overlap between relative hypomethylation/histone marker enrichment and relatively high expression is observed, but this overlap was significant (Figure S4). In TCam-2, one gene, PRAME, was present in all three differential lists. H19 and CHCHD5 were differentially hypomethylated in TCam-2 and showed high expression compared to NCCIT, but no differential enrichment for H3K27ac or H3K4me3. There were 62 genes with overlapping active chromatin marks and expression, including SOX17 and NANOG. In NCCIT, three genes showed overlap between the histone marks, hypomethylation and expression. Significantly, one of these genes was SOX2. An additional 18 genes showed overlap between active chromatin marks and expression.
Figure 6. Overlap between top differentiating genes (methylation/histone modification/gene expression).
(Hypo) methylation, (high) gene expression and histone marker (enrichment) should be interpreted relative to the other cell line. Criteria for selection are described in the main text. Briefly, significant differential methylation of regions with sufficient probe density was identified by DMRforPairs (frequently, but not necessarily close to, the TSS). The difference in histone modification enrichment was assessed by significant differences in summed peak heights between the cell lines. Finally, a fold difference of 3.65 (boundary of 99% CI) was used as cutoff for differential gene expression. Gene lists are presented in Table S1, and overlap was determined based on matching gene symbol.
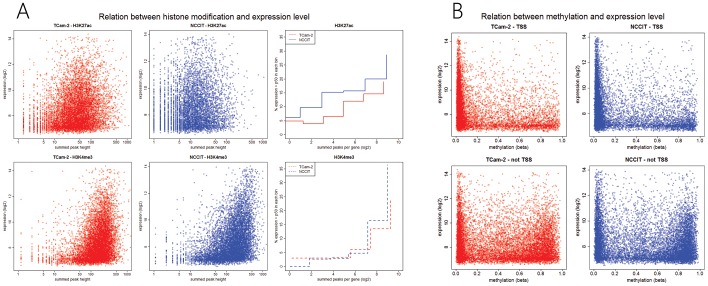
Enrichment of both histone marks in general and absence of DNA methylation around the TSS is related to higher expression levels
Higher expression levels are present when genes are more enriched for either histone mark. Quantification of the fraction of genes with higher than median expression at various intervals of summed peak heights confirmed the general trend towards higher expression at higher levels of enrichment (Figure 7A). DNA methylation levels were only correlated to expression around the TSS: a fully methylated TSS is predominantly associated with low gene expression levels while low methylation status is not predictive of expression level (Figure 7B).
Figure 7. Relation between histone modification level (summed peak heights per gene) and expression level.
Top and bottom right images depict the percentage of highly (>p50) expressed genes calculated for an interval of summed peaks. For example, 5% of genes with a log2(summed peak height) of ≈5.5–7.5 were highly expressed. (B) Relation between CpG methylation (TSS/no TSS) and gene expression.
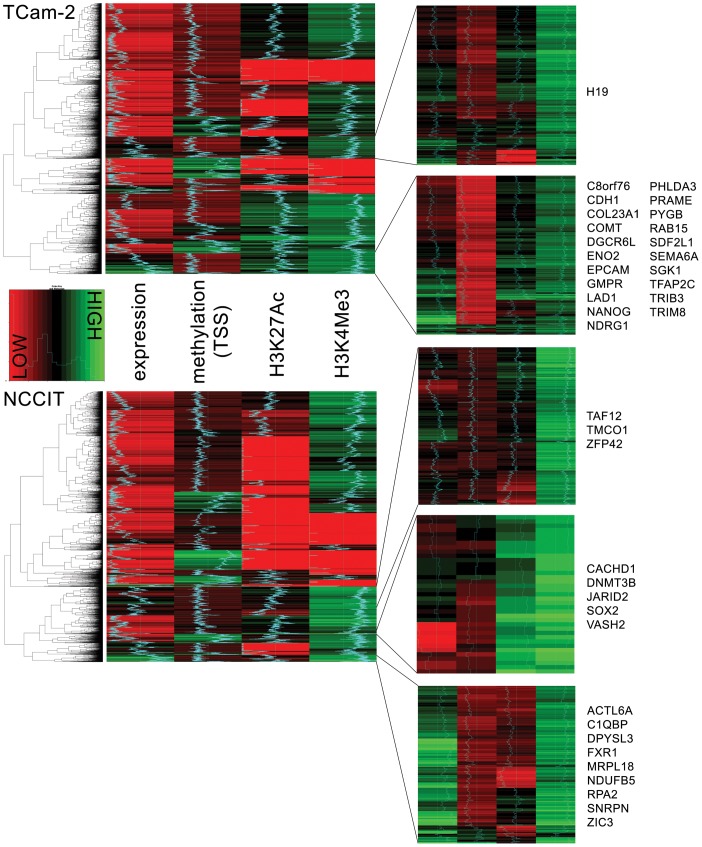
NCCIT and TCam-2 show a largely overlapping epigenetically open network with specific elements that are differentially regulated based on cell type
Histone modification, methylation and gene expression data were analyzed together using hierarchical clustering. This unsupervised clustering procedure revealed specific groups of genes with a profile poised for transcription. These gene clusters showed active histone marks, combined with an activating methylation landscape and are hypothesized to contain genes accessible for transcription (e.g. epigenetically ON = transcription possible, Figure 8). Functionally, the ON network for both cell lines showed a large degree of overlap (related to the androgen pathway, lipid metabolism and pluripotency) (Table S2). There was considerable overlap between the genes found in this separate analysis and the differential gene lists (Figure 6, overlap indicated by gene symbols in Figure 8).
Figure 8. Heat map of epigenetic markers and gene expression profiles.
Genes with quantified methylation status around their TSS (based on Illumina annotation) and valid (see Materials & Methods) measurement of their expression level were included (n = 11,620). Log-2 summed peak heights per gene were used as an estimate of histone marker enrichment. Variables scaled between 0 and 1. Hierarchical clustering was performed using complete linkage. Clusters of interest were identified based on a consistent enriched state for one or more of the active histone markers and a hypomethylated state around the TSS. Number of genes in the displayed right panel (zoomed in heat maps, top→bottom): 899/892 (TCam-2) and 1,224/37/308 (NCCIT). Gene expression was allowed to vary within clusters, but clusters with almost absent expression levels (completely red) were not selected. Gene symbols indicate genes that overlap with the analysis of top differentiating genes between the cell lines (Figure 6). Gene symbols are listed alphabetically. An indication of the level for each gene in each column is presented by the color/shade and a blue line (for each column: left = 0, right = 1).
Discussion
Histone and DNA methylation signatures were studied to explore the epigenetic differences between representative cell lines for the GCC histological subtypes and their relation to expression. TCam-2 and NCCIT cells were used as representatives of SE and EC respectively. Our study includes the generation, integration and interpretation of genome-wide profiles for histone marks (H3K4me3 and H3K27ac), DNA methylation and gene expression. H3K4me3 and H3K27ac are well-characterized markers for active promoter and enhancer sites, respectively. Analysis of the histone marks matched the classification of the cell-lines: SOX17 was strongly enriched for H3K4me3 and H3K27ac in TCam-2 compared to NCCIT cells, whereas the opposite pattern was observed for SOX2. Motifs for germ cell-specific transcription factors AP-2α/γ were enriched in TCam-2, but not in NCCIT. Methylation profiling showed SOX2 to be in the top DMRs, being more methylated in TCam-2 as expected. In addition, SOX2 and SOX17 expression levels confirmed and matched the previously described patterns in SE and EC [2]. General SE/EC markers (NANOG and OCT3/4) showed expression patterns compatible with their epigenetic configurations [58]. Moreover, imprinting patterns confirm the suggested overall erased status of genomic imprinting in both cell lines, in line with the situation in early germ cells [59].
The cell lines were studied for their differences with regard to epigenetic marks and expression. In line with the early germ cell origin of both cell lines (NCCIT more stem cell-like than TCam-2) canonical pathways related to stem cell maintenance and regulation were significantly overrepresented for all three variables studied (NCCIT more pronounced than TCam-2). In TCam-2 (differential histone modification) there was strong overrepresentation of genes involved in IGF1 signaling, a pathway that is implicated in maintenance of spermatogonia [60]. Moreover, genes involved in germ cell – Sertoli cell junction signaling were significantly overrepresented in this cell line, fitting with a more mature type of germ cell depending on the Sertoli cell niche [2]. In the list of top DMRs, the miR-371/2/3 cluster stood out by significant differential hypermethylation in NCCIT. These miRs have shown to be specific biomarkers for GCC in serum and tumor tissue [61]–[65]. Even though GCCs are reported to express these embryonic miRs [63], [66], [67], NCCIT has been shown to exhibit low expression levels due to the absence of a functional TP53 pathway (i.e. lacking the need to inactivate this pathway by miR-372/3 expression via LATS2 inhibition) [67].
Pathway analysis using IPA revealed a network including the androgen receptor (AR) and testosterone targets enriched for open chromatin configuration marks in TCam-2. Such enrichment was also identified based on the expression data of TCam-2, and to a lesser extent in NCCIT. Despite of this observation, no differential AR expression was present between the cell lines, and no enrichment of DMRs in AR targets was identified. Future experiments are needed to validate the differential role of the AR pathway between NCCIT and TCam-2 and their in vivo counterparts. A second network was closely related, focused on lipid metabolism (LEP central, late germ cell differentiation and survival [68]). Androgens are well-known environmental and physiological factors that influence epigenetic marks and phenotypes. They are related to GCC risk and are considered to be crucial for the progression of germ cell development [28] Ammerpohl et al. found significant enrichment of hypermethylated AR target genes in androgen insensitivity syndrome (AIS) patients versus controls [69]. This is consistent with earlier reports that diminished gene activation (due to an AR mutation) results in subsequent increased DNA methylation of target genes [70], [71], linking DSD and GCC at the epigenetic level.
On the other hand, functional analysis for NCCIT predominantly revealed genes involved in embryonic stem cell maintenance (less pronounced in TCam-2). More specifically, an interaction between SOX2 and DMRT1 was the most important network identified. Both are involved in stem cell maintenance in embryonic (mouse) germ cells [28]. It has been reported that Dmrt1 can bind the mouse Sox2 promoter, and Dmrt1 controls expression of Sox2 and other pluripotency genes (such as Nanog and Oct3/4) in the embryonic testis, in part via transcriptional repression [72]. In addition, DMRT1 has a role in sex determination, as it prevents female reprogramming in the postnatal mammalian testis [73]. A study by Murphy et al confirmed the influence of DMRT1 on SOX2 expression while no change in expression of SOX17 was observed [74]. However, these observations in mice are not necessarily representative for the human situation. No consistent networks related to progression of germ cell differentiation (as with AR in TCam-2) were identified in NCCIT.
As stated above, motif enrichment based on the histone modification data showed that the AP-2α and AP-2γ motifs were enriched in TCam-2 only. The ETS family was enriched in both TCam-2 and NCCIT. AP-2γ is a known germ cell marker, abundantly expressed in CIS and SE, and heterogeneously expressed in NS and somatic tumors [75], [76]. AP-2y and KIT are co-expressed in gonocytes [76], which could point to a direct regulation loop which supports proliferation, and agrees with the observation that TCam-2 is more germ cell-like than NCCIT. AP-2y would then serve as a molecule that keeps fetal germ cells in a pluripotent state by suppressing differentiation and supporting proliferation [76], [77]. AP-2y expression is induced by estrogens [78] and AP-2α and AP-2y are able to induce changes in the chromatin structure known to be associated with ERα (ESR1) transcription [79]. The importance of the androgen-estrogen balance is also indicated by the strongly androgen/estrogen-centered gene networks identified in this study. Additionally, the ETS family was present in the top motif enrichments for both TCam-2 and NCCIT. Recently it was shown that overexpression of ETS, combined with loss of PTEN, increases AR binding and restores AR transcriptional activity in prostate [80]. Indeed, disruption of the PTEN pathway has been suggested to be part of the pathogenesis of GCC [81].
Conclusions
In conclusion, this study provides an integrated analysis of the functional genome in GCC cell lines. Our data show that known germ cell markers are not only present and differentiating between SE and NS at the expression level, but also in the epigenetic landscape. The overall similarity between TCam-2/NCCIT support an erased embryonic germ cell arrested in early gonadal development as common cell of origin although the exact developmental stage from which the tumor cells are derived might differ. Indeed, subtle difference in the (integrated) epigenetic and expression profiles indicate TCam-2 to exhibit a more germ cell-like profile, whereas NCCIT shows a more pluripotent phenotype. Future research has already been initiated to investigate primary cancer samples from patients to confirm and further expand the integrated epigenetic EC and SE footprints identified in this study.
Supporting Information
Number of top differentially modified regions between TCam-2 and NCCIT, and their overlap between H3K27ac and H3K4me3 based on associated genes. Genes with significant differences (outside 99% confidence interval) in summed peak height per gene (ΔΣP) were identified as top-differentially modified.
(TIF)
Normalized ratios for each of the 14 loci tested in the MLPA-DNaseI assay. A threshold of <0.75 was defined for DNaseI-hypersensitivity. N = enriched in ChIP-seq analysis in NCCIT (non-seminoma cell model), S = enriched in ChIP-seq analysis in TCam-2 (seminoma model). (A) Analysis of NCCIT cells. (B) Analysis of TCam-2 cells. (C) Overview of loci and interpretation of results. DNaseI hypersensitivity is indicated if present in the cell line in which marker enrichment was also found in the ChIP-seq analysis.
(TIF)
Visualization of global methylation patterns in both cell lines. Depicted is a violin plot of the distribution of methylation values (β) for both cell lines. In general, NS are considered globally hypermethylated in comparison to SE but TCam-2 is known to show an intermediate phenotype with regard to global methylation status (see Wermann et al 2010 and Netto et al 2008 in reference list). Indeed, significantly lower methylation levels were detected in TCam-2 but the quantitative difference in methylation distribution was very moderate (p<0.01, Mann Whitney U test, medianβ (1st–3rd quantileβ): 51%TCam-2 (46%–84%) versus 63%NCCIT (58%–87%)).
(TIFF)
Venn diagrams analogous to Figure 6 corrected for gene symbols that are not represented by valid measurements in the expression or methylation data (histone modification = genome wide assessment). NCCIT: 33 genes differentially methylated were not annotated in the expression data. 17 overexpressed genes were not annotated in the methylation data. 97 genes showing differential histone modifications were not present in the expression or methylation data. For TCam-2 these numbers were 101/28/198. Based on an empirical probability distribution we assessed random overlap using 10,000 draws from simulated genelists with nexpression (EXPR) = 14,525, nhistone-modification (HM) = 22,000 and nmethylation (MEHTY) = 21,243 genes. These numbers correspond with the number of genes with valid measurements on the arrays (histone modification: genome wide proxy). Significant overlap indicates more overlapping genes identified in these venn diagrams than would we expected based on random subsets of genes. Significant overlap is indicated with a * (p<0.05). p-values TCam-2: pEXPR_HM = <0.0001, pEXPR_METHY = 0.0370–0.1604, pHM_METHY = 0.3229–0.6860, pall3 = 0.0003–0.0167. p-values NCCIT: pEXPR_HM = 0–0, pEXPR_METHY = 0.0001–0.0007, pHM_METHY = 0.3110–1.0000, pall3 = <0.0001. (P-values are ranges if in the repeated random draws used to construct the empirical cumulative distribution function a specific count of overlapping genes occurred more than once).
(TIF)
Top differentiating genes in histone modification, CpG methylation and gene expression analyses.
(XLSX)
Results of IPA/functional analysis of (differentiating) gene lists. IPA was performed using the default settings including “testis” as specific tissue of interest, and only incorporating experimentally observed evidence. Green fill indicates overlap between TCam-2 and NCCIT. Reported log(p) values are the result of IPA's internal enrichment tests.
(XLSX)
Detailed results of motif enrichment analysis (HOMER) per cell line and per histone mark.
(PDF)
ZIP file containing DMRforPairs output for significant regions. Please start from the html files.
(ZIP)
Acknowledgments
The authors thank the Department of Bioinformatics, Erasmus MC, Rotterdam, for their support. They especially thank Ms Sylvia de Does and Mr Ivo Palli.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. Array and ChIP sequencing data is available at the Gene Expression Omnibus under accession number GSE56454.
Funding Statement
This work is supported by funding from the European Society for Pediatric Endocrinology Research Fellowship (YZ). MR is supported by a Translational Grant, Erasmus MC. SB is supported by APA and IPRS scholarships from Monash University. This work was supported by funding from Monash University (SW). MIMR receives support from the Victorian Government's Operational Infrastructure Support Program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Looijenga LH (2009) Human testicular (non)seminomatous germ cell tumours: the clinical implications of recent pathobiological insights. J Pathol 218: 146–162. [DOI] [PubMed] [Google Scholar]
- 2. Oosterhuis JW, Looijenga LH (2005) Testicular germ-cell tumours in a broader perspective. Nat Rev Cancer 5: 210–222. [DOI] [PubMed] [Google Scholar]
- 3.Woodward PJ, Heidenreich A, Looijenga LHJ, Oosterhuis JW, McLeod DG, et al.. (2004) Testicular germ cell tumors. In: Eble JN, Sauter G, Epstein JI, Sesterhann IA, editors. World Health Organization Classification of Tumours Pathology and Genetics of the Urinary System and Male Genital Organs. Lyon: IARC Press. pp. 217–278.
- 4. Looijenga LH, Gillis AJ, Stoop H, Biermann K, Oosterhuis JW (2011) Dissecting the molecular pathways of (testicular) germ cell tumour pathogenesis; from initiation to treatment-resistance. Int J Androl 34: e234–251. [DOI] [PubMed] [Google Scholar]
- 5. Banks K, Tuazon E, Berhane K, Koh CJ, De Filippo RE, et al. (2012) Cryptorchidism and testicular germ cell tumors: comprehensive meta-analysis reveals that association between these conditions diminished over time and is modified by clinical characteristics. Front Endocrinol (Lausanne) 3: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dieckmann KP, Pichlmeier U (2004) Clinical epidemiology of testicular germ cell tumors. World J Urol 22: 2–14. [DOI] [PubMed] [Google Scholar]
- 7. Elzinga-Tinke JE, Sirre ME, Looijenga LH, van Casteren N, Wildhagen MF, et al. (2010) The predictive value of testicular ultrasound abnormalities for carcinoma in situ of the testis in men at risk for testicular cancer. Int J Androl 33: 597–603. [DOI] [PubMed] [Google Scholar]
- 8. Horwich A, Shipley J, Huddart R (2006) Testicular germ-cell cancer. Lancet 367: 754–765. [DOI] [PubMed] [Google Scholar]
- 9. Czene K, Lichtenstein P, Hemminki K (2002) Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish Family-Cancer Database. Int J Cancer 99: 260–266. [DOI] [PubMed] [Google Scholar]
- 10. Del-Mazo J, Brieno-Enriquez MA, Garcia-Lopez J, Lopez-Fernandez LA, De-Felici M (2013) Endocrine disruptors, gene deregulation and male germ cell tumors. Int J Dev Biol 57: 225–239. [DOI] [PubMed] [Google Scholar]
- 11. Godmann M, Lambrot R, Kimmins S (2009) The dynamic epigenetic program in male germ cells: Its role in spermatogenesis, testis cancer, and its response to the environment. Microsc Res Tech 72: 603–619. [DOI] [PubMed] [Google Scholar]
- 12. Kanetsky PA, Mitra N, Vardhanabhuti S, Li M, Vaughn DJ, et al. (2009) Common variation in KITLG and at 5q31.3 predisposes to testicular germ cell cancer. Nat Genet 41: 811–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kanetsky PA, Mitra N, Vardhanabhuti S, Vaughn DJ, Li M, et al. (2011) A second independent locus within DMRT1 is associated with testicular germ cell tumor susceptibility. Hum Mol Genet 20: 3109–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rapley EA, Turnbull C, Al Olama AA, Dermitzakis ET, Linger R, et al. (2009) A genome-wide association study of testicular germ cell tumor. Nat Genet 41: 807–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Turnbull C, Rapley EA, Seal S, Pernet D, Renwick A, et al. (2010) Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat Genet 42: 604–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kratz CP, Mai PL, Greene MH (2010) Familial testicular germ cell tumours. Best Pract Res Clin Endocrinol Metab 24: 503–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bignell G, Smith R, Hunter C, Stephens P, Davies H, et al. (2006) Sequence analysis of the protein kinase gene family in human testicular germ-cell tumors of adolescents and adults. Genes Chromosomes Cancer 45: 42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cools M, Drop SL, Wolffenbuttel KP, Oosterhuis JW, Looijenga LH (2006) Germ cell tumors in the intersex gonad: old paths, new directions, moving frontiers. Endocr Rev 27: 468–484. [DOI] [PubMed] [Google Scholar]
- 19. Hawkins RD, Hon GC, Yang C, Antosiewicz-Bourget JE, Lee LK, et al. (2011) Dynamic chromatin states in human ES cells reveal potential regulatory sequences and genes involved in pluripotency. Cell Res 21: 1393–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. De Carvalho DD, Sharma S, You JS, Su SF, Taberlay PC, et al. (2012) DNA methylation screening identifies driver epigenetic events of cancer cell survival. Cancer Cell 21: 655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Esteller M (2008) Epigenetics in cancer. N Engl J Med 358: 1148–1159. [DOI] [PubMed] [Google Scholar]
- 22. Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, et al. (2010) Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell 6: 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jones PA, Baylin SB (2007) The epigenomics of cancer. Cell 128: 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Suva ML, Riggi N, Bernstein BE (2013) Epigenetic reprogramming in cancer. Science 339: 1567–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Van Der Zwan YG, Stoop H, Rossello F, White SJ, Looijenga LH (2013) Role of epigenetics in the etiology of germ cell cancer. Int J Dev Biol 57: 299–308. [DOI] [PubMed] [Google Scholar]
- 26. Seisenberger S, Peat JR, Reik W (2013) Conceptual links between DNA methylation reprogramming in the early embryo and primordial germ cells. Curr Opin Cell Biol 25: 281–288. [DOI] [PubMed] [Google Scholar]
- 27. Amatruda JF, Ross JA, Christensen B, Fustino NJ, Chen KS, et al. (2013) DNA methylation analysis reveals distinct methylation signatures in pediatric germ cell tumors. BMC Cancer 13: 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Looijenga LH, Van Agthoven T, Biermann K (2013) Development of malignant germ cells - the genvironmental hypothesis. Int J Dev Biol 57: 241–253. [DOI] [PubMed] [Google Scholar]
- 29. Mirbahai L, Chipman JK (2014) Epigenetic memory of environmental organisms: A reflection of lifetime stressor exposures. Mutat Res 764–765: 10–17. [DOI] [PubMed] [Google Scholar]
- 30. Skinner MK, Haque CG, Nilsson E, Bhandari R, McCarrey JR (2013) Environmentally induced transgenerational epigenetic reprogramming of primordial germ cells and the subsequent germ line. PLoS One 8: e66318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. de Jong J, Stoop H, Gillis AJ, Hersmus R, van Gurp RJ, et al. (2008) Further characterization of the first seminoma cell line TCam-2. Genes Chromosomes Cancer 47: 185–196. [DOI] [PubMed] [Google Scholar]
- 32. Mizuno Y, Gotoh A, Kamidono S, Kitazawa S (1993) [Establishment and characterization of a new human testicular germ cell tumor cell line (TCam-2)]. Nihon Hinyokika Gakkai Zasshi 84: 1211–1218. [DOI] [PubMed] [Google Scholar]
- 33. Teshima S, Shimosato Y, Hirohashi S, Tome Y, Hayashi I, et al. (1988) Four new human germ cell tumor cell lines. Lab Invest 59: 328–336. [PubMed] [Google Scholar]
- 34. Eckert D, Nettersheim D, Heukamp LC, Kitazawa S, Biermann K, et al. (2008) TCam-2 but not JKT-1 cells resemble seminoma in cell culture. Cell Tissue Res 331: 529–538. [DOI] [PubMed] [Google Scholar]
- 35. Du P, Kibbe WA, Lin SM (2008) lumi: a pipeline for processing Illumina microarray. Bioinformatics 24: 1547–1548. [DOI] [PubMed] [Google Scholar]
- 36. Marabita F, Almgren M, Lindholm ME, Ruhrmann S, Fagerstrom-Billai F, et al. (2013) An evaluation of analysis pipelines for DNA methylation profiling using the Illumina HumanMethylation450 BeadChip platform. Epigenetics 8: 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, et al. (2013) A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 29: 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rijlaarsdam MA, Zwan YG vd, Dorssers LC, Looijenga LH (2014) DMRforPairs: identifying Differentially Methylated Regions between unique samples using array based methylation profiles. BMC Bioinformatics: in press. [DOI] [PMC free article] [PubMed]
- 39. Schmid R, Baum P, Ittrich C, Fundel-Clemens K, Huber W, et al. (2010) Comparison of normalization methods for Illumina BeadChip HumanHT-12 v3. BMC Genomics 11: 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, et al. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. White SJ, Breuning MH, den Dunnen JT (2004) Detecting copy number changes in genomic DNA: MAPH and MLPA. Methods Cell Biol 75: 751–768. [DOI] [PubMed] [Google Scholar]
- 42. Ohnesorg T, Eggers S, Leonhard W, Sinclair A, White S (2009) Rapid high-throughput analysis of DNaseI hypersensitive sites using a modified Multiplex Ligation-dependent Probe Amplification approach. BMC Genomics 10: 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ohnesorg T, Eggers S, White SJ (2012) Detecting DNaseI-hypersensitivity sites with MLPA. Methods Mol Biol 786: 201–210. [DOI] [PubMed] [Google Scholar]
- 44. Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, et al. (2011) A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Clouaire T, Webb S, Skene P, Illingworth R, Kerr A, et al. (2012) Cfp1 integrates both CpG content and gene activity for accurate H3K4me3 deposition in embryonic stem cells. Genes Dev 26: 1714–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA (2007) A Chromatin Landmark and Transcription Initiation at Most Promoters in Human Cells. Cell 130: 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rada-Iglesias A, Wysocka J (2011) Epigenomics of human embryonic stem cells and induced pluripotent stem cells: insights into pluripotency and implications for disease. Genome Med 3: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. de Jong J, Stoop H, Dohle GR, Bangma CH, Kliffen M, et al. (2005) Diagnostic value of OCT3/4 for pre-invasive and invasive testicular germ cell tumours. J Pathol 206: 242–249. [DOI] [PubMed] [Google Scholar]
- 49. Bossone SA, Asselin C, Patel AJ, Marcu KB (1992) MAZ, a zinc finger protein, binds to c-MYC and C2 gene sequences regulating transcriptional initiation and termination. Proc Natl Acad Sci U S A 89: 7452–7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, et al. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shu W, Chen H, Bo X, Wang S (2011) Genome-wide analysis of the relationships between DNaseI HS, histone modifications and gene expression reveals distinct modes of chromatin domains. Nucleic Acids Res 39: 7428–7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wermann H, Stoop H, Gillis AJ, Honecker F, van Gurp RJ, et al. (2010) Global DNA methylation in fetal human germ cells and germ cell tumours: association with differentiation and cisplatin resistance. J Pathol 221: 433–442. [DOI] [PubMed] [Google Scholar]
- 53. Netto GJ, Nakai Y, Nakayama M, Jadallah S, Toubaji A, et al. (2008) Global DNA hypomethylation in intratubular germ cell neoplasia and seminoma, but not in nonseminomatous male germ cell tumors. Mod Pathol 21: 1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nettersheim D, Westernstroer B, Haas N, Leinhaas A, Brustle O, et al. (2012) Establishment of a versatile seminoma model indicates cellular plasticity of germ cell tumor cells. Genes Chromosomes Cancer 51: 717–726. [DOI] [PubMed] [Google Scholar]
- 55. Nettersheim D, Gillis A, Biermann K, Looijenga LH, Schorle H (2011) The seminoma cell line TCam-2 is sensitive to HDAC inhibitor depsipeptide but tolerates various other chemotherapeutic drugs and loss of NANOG expression. Genes Chromosomes Cancer 50: 1033–1042. [DOI] [PubMed] [Google Scholar]
- 56. Nettersheim D, Biermann K, Gillis AJ, Steger K, Looijenga LH, et al. (2011) NANOG promoter methylation and expression correlation during normal and malignant human germ cell development. Epigenetics 6: 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. de Jong J, Stoop H, Gillis AJ, van Gurp RJ, van de Geijn GJ, et al. (2008) Differential expression of SOX17 and SOX2 in germ cells and stem cells has biological and clinical implications. J Pathol 215: 21–30. [DOI] [PubMed] [Google Scholar]
- 58. Rijlaarsdam MA, van Herk HA, Gillis AJ, Stoop H, Jenster G, et al. (2011) Specific detection of OCT3/4 isoform A/B/B1 expression in solid (germ cell) tumours and cell lines: confirmation of OCT3/4 specificity for germ cell tumours. Br J Cancer 6: 854–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. MacDonald WA, Mann MR (2014) Epigenetic regulation of genomic imprinting from germ line to preimplantation. Mol Reprod Dev 81: 126–140. [DOI] [PubMed] [Google Scholar]
- 60. Huang YH, Chin CC, Ho HN, Chou CK, Shen CN, et al. (2009) Pluripotency of mouse spermatogonial stem cells maintained by IGF-1- dependent pathway. FASEB J 23: 2076–2087. [DOI] [PubMed] [Google Scholar]
- 61. Belge G, Dieckmann KP, Spiekermann M, Balks T, Bullerdiek J (2012) Serum levels of microRNAs miR-371-3: a novel class of serum biomarkers for testicular germ cell tumors? Eur Urol 61: 1068–1069. [DOI] [PubMed] [Google Scholar]
- 62. Dieckmann KP, Spiekermann M, Balks T, Flor I, Loning T, et al. (2012) MicroRNAs miR-371-3 in serum as diagnostic tools in the management of testicular germ cell tumours. Br J Cancer 107: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gillis AJ, Rijlaarsdam MA, Eini R, Dorssers LC, Biermann K, et al. (2013) Targeted serum miRNA (TSmiR) test for diagnosis and follow-up of (testicular) germ cell cancer patients: a proof of principle. Mol Oncol 7: 1083–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Murray MJ, Coleman N (2012) Testicular cancer: a new generation of biomarkers for malignant germ cell tumours. Nat Rev Urol 9: 298–300. [DOI] [PubMed] [Google Scholar]
- 65. Murray MJ, Halsall DJ, Hook CE, Williams DM, Nicholson JC, et al. (2011) Identification of microRNAs From the miR-371∼373 and miR-302 clusters as potential serum biomarkers of malignant germ cell tumors. Am J Clin Pathol 135: 119–125. [DOI] [PubMed] [Google Scholar]
- 66. Gillis AJ, Stoop HJ, Hersmus R, Oosterhuis JW, Sun Y, et al. (2007) High-throughput microRNAome analysis in human germ cell tumours. J Pathol 213: 319–328. [DOI] [PubMed] [Google Scholar]
- 67. Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, et al. (2006) A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 124: 1169–1181. [DOI] [PubMed] [Google Scholar]
- 68. Bhat GK, Sea TL, Olatinwo MO, Simorangkir D, Ford GD, et al. (2006) Influence of a leptin deficiency on testicular morphology, germ cell apoptosis, and expression levels of apoptosis-related genes in the mouse. J Androl 27: 302–310. [DOI] [PubMed] [Google Scholar]
- 69. Ammerpohl O, Bens S, Appari M, Werner R, Korn B, et al. (2013) Androgen receptor function links human sexual dimorphism to DNA methylation. PLoS One 8: e73288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, et al. (2011) DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480: 490–495. [DOI] [PubMed] [Google Scholar]
- 71. Clark SJ, Melki J (2002) DNA methylation and gene silencing in cancer: which is the guilty party? Oncogene 21: 5380–5387. [DOI] [PubMed] [Google Scholar]
- 72. Krentz AD, Murphy MW, Kim S, Cook MS, Capel B, et al. (2009) The DM domain protein DMRT1 is a dose-sensitive regulator of fetal germ cell proliferation and pluripotency. Proc Natl Acad Sci U S A 106: 22323–22328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Matson CK, Murphy MW, Sarver AL, Griswold MD, Bardwell VJ, et al. (2011) DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature 476: 101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Murphy MW, Sarver AL, Rice D, Hatzi K, Ye K, et al. (2010) Genome-wide analysis of DNA binding and transcriptional regulation by the mammalian Doublesex homolog DMRT1 in the juvenile testis. Proceedings of the National Academy of Sciences 107: 13360–13365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hoei-Hansen CE, Nielsen JE, Almstrup K, Sonne SB, Graem N, et al. (2004) Transcription factor AP-2gamma is a developmentally regulated marker of testicular carcinoma in situ and germ cell tumors. Clin Cancer Res 10: 8521–8530. [DOI] [PubMed] [Google Scholar]
- 76. Weber S, Eckert D, Nettersheim D, Gillis AJ, Schafer S, et al. (2010) Critical function of AP-2 gamma/TCFAP2C in mouse embryonic germ cell maintenance. Biol Reprod 82: 214–223. [DOI] [PubMed] [Google Scholar]
- 77. Schemmer J, Arauzo-Bravo MJ, Haas N, Schafer S, Weber SN, et al. (2013) Transcription factor TFAP2C regulates major programs required for murine fetal germ cell maintenance and haploinsufficiency predisposes to teratomas in male mice. PLoS One 8: e71113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Orso F, Cottone E, Hasleton MD, Ibbitt JC, Sismondi P, et al. (2004) Activator protein-2gamma (AP-2gamma) expression is specifically induced by oestrogens through binding of the oestrogen receptor to a canonical element within the 5′-untranslated region. Biochem J 377: 429–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Schuur ER, McPherson LA, Yang GP, Weigel RJ (2001) Genomic structure of the promoters of the human estrogen receptor-alpha gene demonstrate changes in chromatin structure induced by AP2gamma. J Biol Chem 276: 15519–15526. [DOI] [PubMed] [Google Scholar]
- 80. Demichelis F, Attard G (2013) A step toward functionally characterized prostate cancer molecular subtypes. Nat Med 19: 966–967. [DOI] [PubMed] [Google Scholar]
- 81. Andreassen KE, Kristiansen W, Karlsson R, Aschim EL, Dahl O, et al. (2013) Genetic variation in AKT1, PTEN and the 8q24 locus, and the risk of testicular germ cell tumor. Hum Reprod 28: 1995–2002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Number of top differentially modified regions between TCam-2 and NCCIT, and their overlap between H3K27ac and H3K4me3 based on associated genes. Genes with significant differences (outside 99% confidence interval) in summed peak height per gene (ΔΣP) were identified as top-differentially modified.
(TIF)
Normalized ratios for each of the 14 loci tested in the MLPA-DNaseI assay. A threshold of <0.75 was defined for DNaseI-hypersensitivity. N = enriched in ChIP-seq analysis in NCCIT (non-seminoma cell model), S = enriched in ChIP-seq analysis in TCam-2 (seminoma model). (A) Analysis of NCCIT cells. (B) Analysis of TCam-2 cells. (C) Overview of loci and interpretation of results. DNaseI hypersensitivity is indicated if present in the cell line in which marker enrichment was also found in the ChIP-seq analysis.
(TIF)
Visualization of global methylation patterns in both cell lines. Depicted is a violin plot of the distribution of methylation values (β) for both cell lines. In general, NS are considered globally hypermethylated in comparison to SE but TCam-2 is known to show an intermediate phenotype with regard to global methylation status (see Wermann et al 2010 and Netto et al 2008 in reference list). Indeed, significantly lower methylation levels were detected in TCam-2 but the quantitative difference in methylation distribution was very moderate (p<0.01, Mann Whitney U test, medianβ (1st–3rd quantileβ): 51%TCam-2 (46%–84%) versus 63%NCCIT (58%–87%)).
(TIFF)
Venn diagrams analogous to Figure 6 corrected for gene symbols that are not represented by valid measurements in the expression or methylation data (histone modification = genome wide assessment). NCCIT: 33 genes differentially methylated were not annotated in the expression data. 17 overexpressed genes were not annotated in the methylation data. 97 genes showing differential histone modifications were not present in the expression or methylation data. For TCam-2 these numbers were 101/28/198. Based on an empirical probability distribution we assessed random overlap using 10,000 draws from simulated genelists with nexpression (EXPR) = 14,525, nhistone-modification (HM) = 22,000 and nmethylation (MEHTY) = 21,243 genes. These numbers correspond with the number of genes with valid measurements on the arrays (histone modification: genome wide proxy). Significant overlap indicates more overlapping genes identified in these venn diagrams than would we expected based on random subsets of genes. Significant overlap is indicated with a * (p<0.05). p-values TCam-2: pEXPR_HM = <0.0001, pEXPR_METHY = 0.0370–0.1604, pHM_METHY = 0.3229–0.6860, pall3 = 0.0003–0.0167. p-values NCCIT: pEXPR_HM = 0–0, pEXPR_METHY = 0.0001–0.0007, pHM_METHY = 0.3110–1.0000, pall3 = <0.0001. (P-values are ranges if in the repeated random draws used to construct the empirical cumulative distribution function a specific count of overlapping genes occurred more than once).
(TIF)
Top differentiating genes in histone modification, CpG methylation and gene expression analyses.
(XLSX)
Results of IPA/functional analysis of (differentiating) gene lists. IPA was performed using the default settings including “testis” as specific tissue of interest, and only incorporating experimentally observed evidence. Green fill indicates overlap between TCam-2 and NCCIT. Reported log(p) values are the result of IPA's internal enrichment tests.
(XLSX)
Detailed results of motif enrichment analysis (HOMER) per cell line and per histone mark.
(PDF)
ZIP file containing DMRforPairs output for significant regions. Please start from the html files.
(ZIP)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. Array and ChIP sequencing data is available at the Gene Expression Omnibus under accession number GSE56454.