Abstract
Background
Activating epidermal growth factor receptor (EGFR) mutations characterize a subgroup of non-small-cell lung cancer that benefit from first line EGFR tyrosine kinase inhibitors (EGFR-TKI). However, the existence of polyclonal cell populations may hinder personalized-medicine strategies as patients’ screening often depends upon a single tumor-biopsy sample. The purpose of this study is to clarify and to validate in clinical testing conditions the accuracy of EGFR genotyping using different tumor sites and various types of samples (transthoracic, surgical or endoscopic biopsies and cytology specimens).
Methods
We conducted a retrospective review of 357 consecutive patients addressed for EGFR mutation screening in accordance with the directive of the European Medicines Agency (stage IV NSCLC). Fifty-seven samples were EGFR mutated and 40 had adequate tumor specimens for analysis on multiple spatially separated sites. Ten wild type samples were also analyzed. A total of 153 and 39 tumor fragments, from mutated and non-mutated cases respectively, were generated to analyze tumor heterogeneity or primary-metastatic discordances. After histological review of all fragments, EGFR genotyping was assessed using the routine diagnostic tools: fragment analysis for insertions and deletions and allele specific TaqMan probes for point mutations. EGFR copy number (CN) was evaluated by qPCR using TaqMan probes.
Results
The identification of EGFR mutations was independent of localization within primary tumor, of specimen type and consistent between primary and metastases. At the opposite, for half of the samples, tumor loci showed different EGFR copy number that may affect mutation detection cut-off.
Conclusions
This is the largest series reporting multiple EGFR testing in Caucasians. It validates the accuracy of EGFR mutation screening from single tumor-biopsy samples before first line EGFR-TKI. The unpredictable variability in EGFR CN and therefore in EGFR wild type/mutant allelic ratio justifies the implementation of sensitive methods to identify patients with EGFR mutated tumors.
Keywords: EGFR mutation, Genetic heterogeneity, EGFR amplification
Introduction
Lung carcinoma is the first cause of death by cancer in the world, mostly because patients have an advanced stage disease at diagnosis [1]. Adenocarcinoma (ADC), the most frequent histological type is morphologically and biologically heterogeneous. Different architectural patterns have been described and different molecular pathways are involved in the carcinogenesis process [2]. Attention has largely been focused on proliferation pathways with the identification of mutations in oncogenes such as KRAS, EGFR, ALK, HER2, PI3KCA and BRAF that are potential or validated drug targets [3]. The first identified target in NSCLC was the EGF receptor. In 2004, EGFR activating mutations were identified in lung ADC and rapidly associated with response to EGFR-TKI [4,5]. Clinico-pathological features that correlate with these mutations include east-Asian ethnicity, adenocarcinoma histology, female sex, and never smoking history. In lung cancer the prevalence of EGFR mutations varies from 10% in Caucasians to more than 40% in Asian populations [6]. They are mainly located in the tyrosine kinase domain and 90% consist of either small deletions in exon 19 (DEL19) or a missense mutation in codon 21 that changes the leucine 858 in an arginine (p.L858R). Concerning rare alterations, about 3% of the mutations occur at codon 719, resulting in the substitution of glycine by a cysteine, alanine or serine (p.G719X) or at codon 861 (p.L861Q) [7,8]. In addition, there are rare 1 to 2% in-frame insertion mutations in exon 20 [9]. The predictive value of frequent alterations (DEL19 and p.L858R) is more or less equivalent but some studies have reported a higher sensitivity and longer PFS for patients with DEL19 mutated NSCLC [10]. Concerning rare alterations, sensitivity to EFGR-TKI and PFS are globally lower [11]. In 2010, results from large phase III trial led to the restriction of EGFR tyrosine kinase inhibitors to EGFR mutated tumors in first line treatment [12]. EGFR mutational status has therefore became mandatory to determine which therapy will be the most appropriate to patients with stage IV diseases. In this context, genetic heterogeneity is an obstacle to correct determination of EGFR status on small biopsies specimens. Previous studies showed that the EGFR mutation status was discordant in different parts of the tumor or between primary or secondary metastatic sites [13-16]. At the opposite, Yatabe et al showed in a series of Asiatic patients that discordant cases where extremely rare [17] and it was suggested that discrepancies regarding EGFR mutations distribution could be due to methodological procedures [16-18]. If EGFR genotyping results depend on sample types it will negatively impact treatment decisions. In Caucasians, EGFR molecular status at various tumor sites remains to be examined in standard testing conditions to validate EGFR molecular testing as a diagnostic tool. Finally, EGFR mutational heterogeneity could also explain the occurrence of secondary EGFR-TKI resistances. Patients undergoing EFGR-TKI treatments will ultimately relapse. Recurrences are related to various mechanisms among which the emergence of EGFR p.T790M clones seems to be the most frequent. This alteration, present as a minor sub-clone before treatment seems selected by EGFR-TKI treatments [19]. Most methods used for molecular diagnostic are not sensitive enough to detect minor p.T790M subclones (<1%) and this alteration is rarely identified in untreated patients. Indeed, the reported frequency of baseline EGFR p.T790M mutations varies widely in the literature, ranging from 1% of all EGFR-mutant lung cancers [11] to 35% of all EGFR-mutant lung cancers [20] and depends of the sensitivity of the assays used and their ability to identify minor clones within a tumor. The prognostic significance of baseline EGFR p.T790M has not been reported. In the acquired resistance setting, it has been demonstrated that the presence of p.T790M predicts a favorable prognosis and indolent progression, compared to the absence of p.T790M after TKI failure [19].
Because no large Caucasian series was tested for EGFR genetic heterogeneity, we addressed this question in clinical testing conditions thanks to a French nationwide EGFR mutation characterization program in advanced lung cancer (National Cancer Institute, INCa).
The aim of this study was to answer several questions of clinical relevance: -i- is EGFR mutation distribution, including p.T790M, heterogeneous within the primary tumor?; -ii- is EGFR mutation distribution heterogeneous between the primary tumor and thoracic metastases ?; -iii- Are microbiopsy or cytology samples suitable for EGFR screening? and iiii- is EGFR copy number heterogeneous within the tumor?
Patients and methods
Patients
We studied 357 consecutive patients with adenocarcinoma that had EGFR testing for clinical purpose at the Hôtel Dieu Hospital from January 2010 to June 2011 in accordance with the directive of the European Medicines Agency (stage IV NSCLC tested before EGFR-TKI treatment). We found that 57 patients out of 357 had EGFR mutated tumors (15.9% of the entire series). For 40 patients we had enough tissues available for a multi-localization screening. No concomitant KRAS or HER2 alteration was identified on those samples. To rule out possible genotyping errors at time of diagnosis, we also tested 10 EGFR, KRAS and HER2 wild type (WT) adenocarcinomas. This study was reviewed and approved by the local ethic committee (Comité de Protection des Personnes/CPP 2012 06-12). Patients’ characteristics are shown in Table 1 and are in accordance with previous reports on EGFR mutated Caucasian series [21,22].
Table 1.
Clinical characteristics of patients with EGFR mutated lung adenocarcinoma
| Gender | |
|---|---|
| Male |
13 |
| Female |
27 |
|
Age(years) |
Mean 64 [42 – 81] |
|
Tobacco status |
|
| ≤ 10 Pack per year |
15 |
| > 10 Pack per year |
14 |
| unknown |
11 |
|
TNM stage at initial diagnosis* |
|
| I |
4 |
| II |
4 |
| III |
9 |
| IV |
20 |
| Unknown | 3 |
Shows the clinical characteristics of patients with an EGFR mutated adenocarcinoma. Patients mentioned with stage 1 or 2 tumors have relapsed. EGFR testing was done using the available material (diagnostic samples) at time of relapse.
Samples
All EGFR mutated adenocarcinomas had thyroid transcriptional factor (TTF1) nuclear expression (Novocastra, clone SPT24). The 22 surgical resections were classified according to the IASLC/ATS/ERS recommendation [23].
A total of 153 tumor fragments from 40 patients were selected by two pathologists (MLA and DD): 5 bronchial biopsies, 2 bronchial aspirations, 5 tomography-guided needle lung biopsies, 8 pleural effusions, 33 metastatic pleural localizations, 83 pulmonary surgical samples (lobectomy and wedge resections) and 17 lymph-nodes specimens. For each patient 2 to 19 loci were generated consisting of different localizations (primary and metastatic sites) or subdivision of the tumor tissue into small parts to analyze EGFR mutation at different sites, with different tumor cell content and different architectural patterns (Table 2, Additional file 1: Table S1). We called “small specimens”: cytology (pleural effusions or bronchial aspiration) or small biopsy (tomography-guided needle lung biopsies or bronchial biopsies). Concerning wild type samples 39 fragments from 10 patients were generated, including 3 adrenal glands, 6 lymph-nodes, 2 pleural metastases and 28 fragments from the primary tumors. All contained more than 50% tumor cells to rule out possible mutant allele dilution (Additional file 2: Table S2).
Table 2.
Multi-localization EGFR genotyping screening
| |
Number and type of samples analyzed per patients |
EGFR genotyping results |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Patients | Mutation type | Primary tumor(sugical sample,lung biopsy*,bronchial biospy£) | PLEURAL BIOPSY | LYMPH NODE | Cytology: Pleural Effusion,Bronchial aspiration* | Contributive samples | Concordant EGFR status | Non contributive samples | Discordant EGFR status |
| P1 |
del19 |
7 |
0 |
0 |
0 |
7 |
7 |
0 |
|
| P2 |
del19 |
2 |
0 |
0 |
0 |
2 |
2 |
0 |
|
| P3 |
del19 |
0 |
3 |
0 |
1 |
4 |
3 |
0 |
1 (rescued by CAST PCR) |
| P4 |
del19 |
0 |
4 |
0 |
1 |
5 |
5 |
0 |
|
| P5 |
del19 |
0 |
3 |
0 |
1 |
4 |
4 |
0 |
|
| P6 |
del19 |
1* + 1* |
0 |
0 |
0 |
2 |
2 |
0 |
|
| P7 |
del19 |
0 |
0 |
0 |
2 |
2 |
2 |
0 |
|
| P8 |
del19 |
1* + 1* |
0 |
0 |
0 |
2 |
2 |
0 |
|
| P9 |
del19 |
3 |
1 |
0 |
0 |
4 |
4 |
0 |
|
| P10 |
del19 |
0 |
3 |
0 |
0 |
3 |
3 |
0 |
|
| P11 |
del19 |
0 |
3 |
0 |
0 |
3 |
3 |
0 |
|
| P12 |
del19 |
0 |
3 |
0 |
0 |
2 |
2 |
1 |
|
| P13 |
del19 |
4 |
0 |
0 |
0 |
4 |
4 |
0 |
|
| P14 |
del19 |
2 |
0 |
0 |
0 |
2 |
2 |
0 |
|
| P15 |
del19 |
0 |
2 |
0 |
1 |
3 |
3 |
0 |
|
| P16 |
del19 |
1£ + 1£ |
0 |
0 |
1* |
3 |
1 |
0 |
2 (remained WT by CAST PCR) |
| P17 |
del19 |
3 |
0 |
0 |
0 |
3 |
3 |
0 |
|
| P18 |
del19 |
0 |
0 |
1 |
1* |
1 |
1 |
1 |
|
| P19 |
del19 |
1 + 1* |
0 |
0 |
0 |
2 |
2 |
0 |
|
| P20 |
p.G719A |
1 |
0 |
4 |
0 |
5 |
5 |
0 |
|
| P21 |
p.G719A |
5 |
0 |
2 |
0 |
7 |
7 |
0 |
|
| P22 |
ins20 |
3 |
0 |
1 |
0 |
4 |
4 |
0 |
|
| P23 |
ins20 |
0 |
2 |
0 |
0 |
2 |
2 |
0 |
|
| P24 |
ins20 |
3 |
0 |
2 |
0 |
4 |
4 |
1 |
|
| P25 |
ins20 |
0 |
2 |
0 |
0 |
2 |
2 |
0 |
|
| P26 |
ins20 |
3 |
0 |
0 |
0 |
3 |
3 |
0 |
|
| P27 |
ins20 |
2 + 1 |
0 |
1 + 1 |
0 |
4 |
2 |
1 |
2 |
| P28 |
p.L858R |
4 |
0 |
2 |
0 |
6 |
6 |
0 |
|
| P29 |
p.L858R |
1£ + 1£ |
0 |
0 |
0 |
2 |
2 |
0 |
|
| P30 |
p.L858R |
2 |
0 |
0 |
0 |
2 |
2 |
0 |
|
| P31 |
p.L858R |
3 |
0 |
0 |
0 |
3 |
3 |
0 |
|
| P32 |
p.L858R |
0 |
3 |
0 |
1 |
4 |
4 |
0 |
|
| P33 |
p.L858R |
0 |
3 |
0 |
0 |
3 |
3 |
0 |
|
| P34 |
p.L858R |
1£ |
1 |
0 |
1 |
3 |
3 |
0 |
|
| P35 |
p.L858R |
4 |
0 |
2 |
0 |
6 |
6 |
0 |
|
| P36 |
p.L858R |
2 |
0 |
1 |
0 |
3 |
3 |
0 |
|
| P37 |
p.L858R |
3 |
0 |
0 |
0 |
3 |
3 |
0 |
|
| P38 |
p.L858R |
3 |
0 |
0 |
0 |
3 |
3 |
0 |
|
| P39 |
p.L858R |
3 |
0 |
0 |
0 |
3 |
3 |
0 |
|
| P40 | p.L861Q/p.T790M | 19 | 0 | 0 | 0 | 19 | 19 | 0 | |
Shows the type and number of samples tested for EGFR mutation in 40 patients. Non-contributive samples are non-amplified specimens. Discordant samples are DNAs that have been found wild type when a mutation was expected. The discordant samples are italic. Lung biopsy: *and bronchial biopsy: £.
In order to fit to the usual clinical practice of our pathology department, EGFR mutations were analyzed in a representative tumor area, without microdissection procedure. All samples (153 from EGFR mutated tumors, 39 from wild type samples) were reviewed for tumor cell content at x100 magnification (HES staining) by pathologists (MLA and DD) for 26 out of the 153 samples had low tumor cell contents (Additional file 1: Table S1 and Additional file 2: Table S2).
Molecular analysis
Genomic DNA was extracted from 20 μm-thick formalin-fixed paraffin-embedded blocks using illustra™ DNA extraction kit BACC2 (GE Healthcare), according to the manufacturer's instructions. EGFR mutations were analyzed using locally validated tests [24]. In the diagnostic setting, our screening strategy is to test EGFR exon 19 deletions (DEL19), exon 20 insertions (INS20) and the p.L858R mutation along with KRAS and HER2 exon 20 insertions. Non-mutated samples are subsequently analyzed for EGFR codons 719, 861 and mutated samples for EGFR p.T790M. The 153 fragments generated from mutated samples were analyzed for the mutation found at initial diagnosis, the 39 fragments from non-mutated cases were tested for the entire EGFR mutation panel (DEL19, INS20, p.L858R, p.L861Q and p.G719X).
Deletions and insertions were detected using fragment analysis with a FAM-labeled primer, run on an ABI 3730 XL (Applied biosystems, Foster City, CA) and analyzed with Genemapper software (Applied biosystems). Fragment analysis has a detection cut-off of 5-10% mutated/wild type allele ratio. Samples with an expected DEL19 that were found wild-type (n = 3) using fragment analysis were subsequently re-analyzed by TaqMan® Mutation Detection assays based on Competitive Allele Specific TaqMan PCR technology (CAST). Point mutations: p.L861Q, p.G719A, p.G719C, p.G719S, p.L858R and p.T790M were analyzed using similar technology. CAST probes were not available to test INS20 mutations. Allele specific assays were run in a final volume of 5 μl in 384 wells plate including 2.5 μl of 2X TaqMan® genotyping master mix (Applied Biosystems), 0.5 μl of 10X Assay Mix (Hs00000173_rf :EGFR_rf; Hs00000141_mu :EGFR_6213_mu; Hs00000104_mu: EGFR_ 6239_mu; Hs00000148_mu: EGFR_6253_mu; Hs00000146_mu: EGFR_6252_mu; Hs00000106_mu: EGFR_6240_mu; Hs00000102_mu: EGFR_6224_mu; Hs00000228_mu: EGFRex19dels_mu from Lifetechnologies) and 1μl DNA template (≤20ng/μl). Runs were performed in duplicates on an ABI Prism 7900 HT sequence detection system (Applied Biosystems) using the following thermo cycling conditions: 95°C/10 m (92°C/15 s, 58°C/1 m) for 5 cycles, then (92°C/15 s, 60°C/60 s) for 40 cycles and analyzed with the SDS 2.0 software program (Applied Biosystems). qPCR analyses have a detection cut-off of 1-2% mutated/wild type allele ratio [24]. Direct sequencing was run on a subset of samples as previously described [25].
EGFR copy number
EGFR copy number was assessed by real time quantitative PCR using TaqMan® copy number assays. Three probes were selected, in intron1, on intron7-exon8 and exon29-intron29 boundaries (Hs04960197_cn; Hs01646307_cn; Hs00458616_cn). TaqMan® Copy number reference assay RNase P was used as internal control and calibration was done using non-tumor formalin fixed paraffin embedded (FFPE) tissues. Samples were run in triplicates, the maximum difference tolerated between triplicates to calculate the average cycle threshold (Ct) was 0.3. EGFR copy number was given by the formula 2-ddct as described previously [26]. Increased copy number was defined by a CNV > 2.5. Cell lines with known copy number variations (CNV) were used as controls. FFPE samples with RNase P Ct over 30 were not used for copy number quantification and qualified as non-contributive samples.
Results
1- EGFR mutation status on multiple spatially separated samples:
From January 2010 to March 2011, 357 patients managed at the Hotel-Dieu hospital for lung cancer had EGFR testing for clinical purpose. Among the 57 EGFR mutated tumors, 40 patients had enough available tissue for multiple EGFR testing. Nineteen patients had an exon 19 deletion (DEL19), 12 a p.L858R point mutation, 6 an exon 20 insertion (INS20), 2 a p.G719A mutation and 1 a p.L861Q/p.T790M double alteration. For those 40 patients, 153 DNA samples were extracted from various tumor localizations and reanalyzed for the initial alteration. Four samples could not be amplified (2 DEL19 and 2 INS20 tumors). The initial EGFR mutation was identified in 144 of the 149 informative samples. Five samples with an expected DEL19 (n = 3) or INS20 (n = 2) were found wild type. For those 5 cases, 2 cytologic specimens (pleural effusion and bronchial aspiration), 1 small endobronchial biopsy and 2 surgical specimens, the proportion of tumor cells was low, ranging from 2% to less than 10%. One specimen (P3) was rescued by DEL19-CAST probes while both samples from P16 remained wild type (Table 2). CAST probes were not available to test INS20. There was no discordance for samples with expected p.L858R, p.L861Q/p.T790M or p.G719A mutations (Table 2 and Additional file 1: Table S1).
All specimens were tested for the p.T790M mutation. This alteration was identified in one tumor (P40). Patient was naïve of EGFR-TKI, the tumor was p.L861Q/p.T790M mutated, was divided into 19 parts and both mutations were homogeneously distributed with similar allele intensity (Additional file 1: Table S1). Samples that were initially diagnosed EGFR wild-type, were found wild-type on all sub-specimens (Additional file 2: Table S2).
2- Concordant EGFR mutation status was found within the tumor, and between primary and thoracic metastasis.
In our series, 22/40 patients had surgical resection for adenocarcinoma allowing classification according the IASLC/ATS/ERS recommendations [2]. All architectural patterns were represented except the mucinous pattern. Most tumors had a lepidic counterpart (9/22) but it was not necessary the predominant pattern (Additional file 1: Table S1). For these 22 adenocarcinomas, we analyzed different tumor area, displaying similar or different architectural patterns and no EGFR heterogeneity was seen within these specimens.
For 10 patients we analyzed both primary tumor and lymph node metastases (n = 8) or pleural metastases (n = 2). A single discordant result was found between the primary tumor (INS20 mutation) and one metastatic lymph node containing 15% of tumor cells. For this patient, the mutation was present in the other lymph node metastasis (P27, Table 2, Additional file 1: Table S1).
Two patients showed a pre-invasive lesion (atypical adenomatous hyperplasia and in situ adenocarcinoma) located at distance of the invasive adenocarcinoma. These pre-invasion lesions had the same EGFR mutation as the invasive counterpart (Additional file 1: Table S1).
3- Microbiopsy and cytology samples allow EGFR mutation analysis.
Twenty non-surgical specimens (8 pleural effusions, 2 bronchial aspirations, 5 tomography-guided needle lung biopsies, 5 bronchial biopsies) were analyzed including 8 with less than 15% tumor cells. Nineteen samples were contributive, 16 were EGFR mutated and 3 were wild type: 1 pleural effusion, 1 bronchial aspirate and 1 bronchial biopsy with 5, 5 and 10% tumor cell content respectively (Table 2).
4- Determination of EGFR copy number.
EGFR copy number (CN) was available in 132/153 samples. In this series, 26/132 samples had an EGFR CN > 2.5 and 2 an EGFR CN >5. Half of the patients (18/40) had at least one sample showing an increased EGFR CN but only one patient had homogeneous copy number increase on all fragment analyzed (CN >4 on 3 pleural biopsy fragments, P11) (Table 3).
Table 3.
Multi-localization EGFR CNV screening
| |
Number and type of samples analyzed per patients |
EGFR
CNV results |
||||||
|---|---|---|---|---|---|---|---|---|
| Patients | Mutation type | Primary tumor(sugical sample,lung biopsy*, bronchial biospy£ | Pleural biopsy | Lymph node | Cytology: Pleural effusion,Bronchial Aspiration* | EGFR CNV ≥ 2.5 | EGFR CNV < 2.5 | Non contributive samples |
|
P1 |
del19 |
7 |
0 |
0 |
0 |
5 |
2 |
|
|
P2 |
del19 |
2 |
0 |
0 |
0 |
|
2 |
|
|
P3 |
del19 |
0 |
3 |
0 |
1 |
|
3 |
1 |
|
P4 |
del19 |
0 |
4 |
0 |
1 |
1 |
3 |
1 |
|
P5 |
del19 |
0 |
3 |
0 |
1 |
1 |
3 |
|
|
P6 |
del19 |
1* + 1* |
0 |
0 |
0 |
1 |
1 |
|
|
P7 |
del19 |
0 |
0 |
0 |
2 |
1 |
1 |
|
|
P8 |
del19 |
1* + 1* |
0 |
0 |
0 |
1 |
1 |
|
|
P9 |
del19 |
3 |
1 |
0 |
0 |
1 |
3 |
|
|
P10 |
del19 |
0 |
3 |
0 |
0 |
|
1 |
2 |
|
P11 |
del19 |
0 |
3 |
0 |
0 |
3 |
|
|
|
P12 |
del19 |
0 |
3 |
0 |
0 |
1 |
2 |
|
|
P13 |
del19 |
4 |
0 |
0 |
0 |
|
4 |
|
|
P14 |
del19 |
2 |
0 |
0 |
0 |
|
2 |
|
|
P15 |
del19 |
0 |
2 |
0 |
1 |
|
3 |
|
|
P16 |
del19 |
1£ + 1£ |
0 |
0 |
1* |
|
1 |
2 |
|
P17 |
del19 |
3 |
0 |
0 |
0 |
1 |
1 |
1 |
|
P18 |
del19 |
0 |
0 |
1 |
1* |
|
1 |
1 |
|
P19 |
del19 |
1 + 1* |
0 |
0 |
0 |
|
1 |
1 |
|
P20 |
p.G719A |
1 |
0 |
4 |
0 |
1 |
2 |
2 |
|
P21 |
p.G719A |
5 |
0 |
2 |
0 |
|
6 |
1 |
|
P22 |
ins20 |
3 |
0 |
1 |
0 |
2 |
2 |
|
|
P23 |
ins20 |
0 |
2 |
0 |
0 |
|
2 |
|
|
P24 |
ins20 |
3 |
0 |
2 |
0 |
|
4 |
1 |
|
P25 |
ins20 |
0 |
2 |
0 |
0 |
1 |
1 |
|
|
P26 |
ins20 |
3 |
0 |
0 |
0 |
|
3 |
|
|
P27 |
ins20 |
2 + 1 |
0 |
1 + 1 |
0 |
|
4 |
1 |
|
P28 |
p.L858R |
4 |
0 |
2 |
0 |
|
6 |
|
|
P29 |
p.L858R |
1£ + 1£ |
0 |
0 |
0 |
1 |
1 |
|
|
P30 |
p.L858R |
2 |
0 |
0 |
0 |
|
2 |
|
|
P31 |
p.L858R |
3 |
0 |
0 |
0 |
|
3 |
|
|
P32 |
p.L858R |
0 |
3 |
0 |
1 |
2 |
2 |
|
|
P33 |
p.L858R |
0 |
3 |
0 |
0 |
1 |
1 |
1 |
|
P34 |
p.L858R |
1£ |
1 |
0 |
1 |
1 |
|
2 |
|
P35 |
p.L858R |
4 |
0 |
2 |
0 |
|
6 |
|
|
P36 |
p.L858R |
2 |
0 |
1 |
0 |
|
2 |
1 |
|
P37 |
p.L858R |
3 |
0 |
0 |
0 |
|
2 |
1 |
|
P38 |
p.L858R |
3 |
0 |
0 |
0 |
1 |
2 |
|
|
P39 |
p.L858R |
3 |
0 |
0 |
0 |
|
2 |
1 |
| P40 | p.L861Q/p.T790M | 19 | 0 | 0 | 0 | 18 | 1 | |
Shows the type and number of samples tested for EGFR copy number variation (CNV) in 40 patients. Non-contributive samples are non-amplified specimens or samples with Ct >30.
Lung biopsy: * and bronchial biopsy: £.
Concerning specimens with very low tumor cell content (≤10%, n = 25), 5 were found wild type and 2 were non informative, none of these 7 specimens had an increased EGFR CN. At the opposite, among the mutated specimens with low tumor cell content 4 /18 had a CN ≥ 2.5 suggesting that EGFR CN impacts on mutation detection for low tumor cell content specimens.
We next examined whether increased EGFR CN was associated with specific morphologic features. In our experience, there was no clear association with high-grade lesions (solid or micropapillary predominant patterns) but in one patient, high EGFR CN was found in the solid pattern and not in the lepidic counterpart (Figure 1). At the opposite, 1 out of the EGFR mutated precursor lesions (in situ adenocarcinoma and atypical adenomatous hyperplasia), had an increased EGFR CN (CN: 3.4). No EGFR copy number increase was found in EGFR wild type tumor samples.
Figure 1.
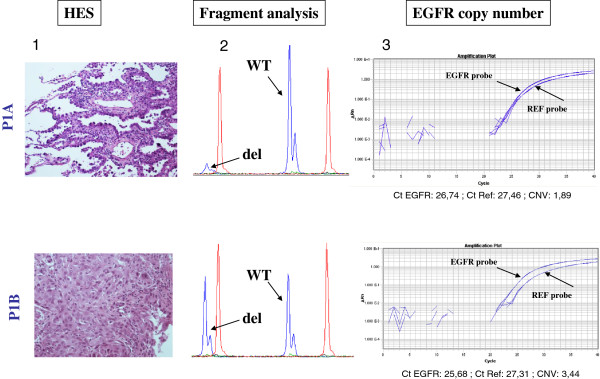
Illustration of the different patterns of EGFR DEL19 mutation, EGFR CN analyses and the associated histological features for patient P1. -1- HES staining at x100 magnification, P1A : lepidic pattern, P1B: solid pattern. -2- Fragment analysis for EGFR DEL19 mutation. -3- EGFR copy number evaluation. In sample A with lepidic pattern, mutation was validated by a cast PCR assay and there is no increased copy number of EGFR gene (CNV:1.89). In sample B with solid pattern, EGFR amplification was identified (CNV: 3.44).
Discussion
Tyrosine kinase inhibitors (TKI) have changed advanced EGFR mutated lung carcinoma clinical practice with global improved short-term survival and fewer side effects [27]. In routine clinical practice EGFR mutation screening is mandatory to decide which first line treatment would be the most appropriate [28,29]. Heterogeneous repartition of EGFR mutations within tissue or between different metastatic sites is an obstacle to accurate molecular screening. It was reported in different studies and remains an important question for clinicians [13-15,30-32]. Table 4 summarizes previous works and compared samples and technologies. Main differences are, the inclusion of various cancer types including squamous-cell cancers that are not targets for EGFR-TKI treatments or pre-treated samples, different proportion of smokers, different types of metastatic site and finally different technologies and different panels of mutations tested. It seems that higher proportion of smokers, heterogeneous tumors specimens and the use of low sensitivity methods yielded to higher rates of inconsistencies in EGFR mutation results [33,34]. Moreover lower rates of concordance are also found for rare EGFR variants [34,35]. It seems therefore important to analyze in a series of patients prospectively tested for EGFR mutation in clinical settings, the impact of heterogeneity on diagnosis. Indeed, non-surgical specimens from either the primary or metastatic site often constitute the only tissue available for molecular diagnosis in patients eligible for TKI with advanced stage cancers [13,30,32,36,37]. Are these small specimens reliable for molecular testing? We addressed this question in Caucasian patients with EGFR mutated lung tumors. Multiple samples were obtained either from different localizations or different loci within the primary tumor. As expected in Caucasians, the frequency of EGFR mutated tumor was 15% [38]. The p.T790M resistance mutation was studied in all patients (153 specimens) that were EGFR-TKI naïve. In untreated patient, the p.T790M mutation is usually described as a subclonal alteration that is difficult to identify even with highly sensitive methods [39]. Here the alteration was found once concomitantly to a p.L861Q mutation, both alleles were equally represented and the distribution was homogeneous suggesting that in this case, the p.T790M alteration is a driver mutation altogether with the p.L861Q. Routine EGFR testing gives a qualitative analysis of EGFR mutations, tumor is either mutated or non-mutated and the proportion of mutated allele is not taken into consideration. Using this definition, we found that 149/153 samples were contributive; among these 149 samples, only 5 showed a discordant genotype (WT/M). Discordant results were from low tumor cell content specimens expected to be either DEL19 or INS20. Fragment analysis, the method used to detect these alterations has a higher detection threshold as compared to allelic discrimination (10% versus 1%) [24], suggesting that these results might be false negative. Among the samples that could be tested using the DEL19 TaqMan® Mutation Detection assay one was found positive while both specimens from patient P16 remained negative. This patient had 2 primary cancers, concomitant adenocarcinoma and squamous cell cancer. Initial EGFR mutation was found on a bronchial biopsy sample showing an ADC, a second biopsy and the bronchial aspiration were found EGFR wild type even though high sensitivity CAST PCR was used. For this patient, the existence of 2 cancers might explain the presence of EGFR mutated and non-mutated samples. Indeed diagnosis on small specimens may be equivocal. To validate the accuracy of EGFR wild type status, 10 patients with a diagnosis of EGFR wild type tumor were re-analyzed at different loci. No EGFR alteration was found (mutation or increased CN). It suggests that negative results can be trusted as long as the method detection cut-off matches the tumor cell content. Although our work cannot rule out the existence of minor wild type subclones, we believe that any sample allows accurate EGFR mutations detection at initial diagnostic. This fits the results of Yatabe et al [17]. However, discrepancies may be increased by the use of low sensitivity methods such as sequencing. In this work, 38 samples were analyzed by direct sequencing. We found 23 concordances, 3 discordances and 12 informative sequences because of high background noise. We stopped the comparison as this method is not used in diagnosis in our lab and was shown to be inappropriate for biopsy or cytology FFPE samples. Indeed results depend on the estimation of the tumor cell content, on the method’s detection cut-off and on EGFR copy number. What threshold of tumor nuclei should we use? It is assumed that more than 50% of tumor cells allows accurate molecular testing and that samples under 10% may lead to false negative results [40]. Between 10% and 50% the reliability depends on the laboratory experience. Interestingly, we had detectable EGFR mutations in 18/23 contributive samples with ≤ 10% of tumor cells. It highlights the fact that if a positive result can be found in a sample with low tumor content, negative results have to be validated according to the specimen’s tumor content. Pleural metastatic evolution is frequent in lung cancer and pleural effusion and/or tissue are convenient materials for molecular biology.
Table 4.
Summary of previously published series
| Article | Histological subtype | No of samples | Smoking status | Concordance PT/M | Analysis within PT | Concordance within PT | Method | Mutation type | Metastatic sites |
|---|---|---|---|---|---|---|---|---|---|
| Yatabe et al. 2011
[17] |
ADC (77) |
77 |
|
100 |
|
|
qPCR |
L858R |
Lymph nodes |
| Fragment analysis |
DEL19 |
||||||||
| INS20 | |||||||||
| 719X | |||||||||
| Sun et al. 2011
[37] |
ADC (39), SCC (31), ADSQ (6), LCC (4) |
80 |
Ever (49) |
|
|
|
Direct sequencing |
all |
Lymph nodes |
| |
Never (31) |
|
|
|
|||||
| |
Global |
92,5% (74/80) |
|
|
|||||
| Wei et al. 2014
[41] |
ADC (49) |
50 |
Ever (10) |
80% |
|
|
qPCR (commercial kit) |
45 hotspots |
Lymph nodes |
| SCC (1) | |||||||||
| |
Never (40) |
97,5% |
|
|
|||||
| |
Global |
93% (47/50) |
|
|
|||||
| |
|
|
|
|
|||||
| Bai et al. 2013
[36] |
ADC (63) |
85 (45 EGFRmt 40 EGFRwt) |
|
|
1431 foci |
87,1% |
ARMS |
DXS EGFR mutation Kit |
|
| SCC (10) |
1238 foci (foci : capture with laser microdissection 0,1cm2) |
4 cases with 5% - 8% of foci showing mutations |
|||||||
| |
ADSQ (5) |
||||||||
| |
Other (7) |
||||||||
| | |||||||||
| Chang et al. 2011
[42] |
ADC (34) |
56 (27 EGFRmt) |
Ever (29) |
62% |
|
|
Direct sequencing |
all |
Lymph nodes |
| SCC (17) | |||||||||
| |
ADSQ (1) |
Never (23) |
70% |
||||||
| |
Other (1) |
Unknown (4) |
|
||||||
| |
Global |
68% (38/56) |
|||||||
| Schmid et al. 2009
[33] |
ADC (96) |
96 (7 EGFRmt) |
Ever (74) |
|
|
|
Direct sequencing |
L858R (3) |
Lymph nodes |
| DEL19 (3) | |||||||||
| |
Never (22) |
|
INS20 (1) |
||||||
| |
Global |
14% (1/7) |
|||||||
| Gow et al 2009
[34] |
ADC (42) |
67 (35 EGFRmt) |
Ever (26) |
|
|
|
Direct sequencing and ARMS for discordant results |
all |
Brain (25) |
| SCC (21) |
Bone (20) |
||||||||
| |
ADSQ (0) |
(19 with adjuvant treatment before molecular analysis on metastatic site) |
Never (41) |
|
Other (22) |
||||
| |
Other (4) |
Global |
26% (9/35) seq and 57 %(20/35) ARMS |
||||||
| Mattsson et al. 2012
[18] |
ADC (6) |
6 |
|
|
3 foci per tumor (distinct morphologies) |
100% |
Direct sequencing |
L858R and DEL19 |
|
| |
|
|
|
|
|
|
|
|
|
| Kalikaki et al. 2008
[35] |
ADC (20) |
25 (7 EGFRmt) |
Ever (22) |
|
|
|
Direct sequencing |
all |
Brain (3) |
| SCC (2) |
Pleura (5) |
||||||||
| |
ADSQ (0) |
(17 with adjuvant treatment before molecular analysis on metastatic site) |
Never (3) |
|
Lung (9) |
||||
| |
Other (3) |
Global |
14% (1/7) 5 mutations are rare alterations (codons 692-847-746-857) |
Adrenal gland (3) |
|||||
| | |||||||||
| |
Bone (2) |
||||||||
| |
Skin (1) |
||||||||
| |
Liver (1) |
||||||||
| Matsumoto et al. 2006
[43] |
ADC (19) |
19 (12 EGFRmut) |
|
100% |
|
|
Direct sequencing |
L858R, DEL19 |
Brain (19) |
| Yatabe et al. 2011
[17] |
ADC (50) |
50 EGFRmt |
|
|
3 foci per tumor (50) |
100% |
qPCR |
L858R |
|
| Fragment analysis |
DEL19 |
||||||||
| 100 foci per tumor (5) | 100% |
Schematic review of previously published series comparing primary tumor and metastasis or different loci within primary tumor. Tumor type, smoking status, detection methods, mutation tested and metastatic sites are given. PT : primary Tumor, M: metastasis, ADC: adenocarcinoma, SCC: squamous cell carcinoma, ADSQ : adenosquamous carcinoma, LCC: large cell carcinoma mt: mutated, wt : wild type, ARMS: amplification refractory mutation system.
We secondly tested whether EGFR CN increase could explain why EGFR mutation assessment, remain possible for cases with very few tumor cells. We found that EGFR copy number was heterogeneous between different fragments from the same tumor. Thus, the allelic ratio of each specimen depends not only on the tumor cell content but also on the EGFR copy number and on the number of mutated allele in tumor cells. In clinical settings the proportion of EGFR mutated cells or the presence of an associated increased copy number, which is frequent in EGFR mutated tumors, is not taken into account. In samples with various tumor cell contents and various EGFR gene copy number the quantification of the mutated allele/wild type allele ratio is difficult. It is clear to us that this ratio varies between tumor sites and that low tumor cell content specimens are rescued by the presence of a high mutant/wild type allele ratio. At the opposite, high tumor cell content specimens may show low mutant/wild type allele ratio estimated by the intensity of the mutant probe signal. This variability may explain discrepancies between series using different detection strategies or using microdissection of very few tumor cells as PCR amplifications in those conditions may lead to amplification errors [44]. However, it would be important to set up assays to analyze the allelic ratio’s impact on response to treatment. One other question was the link between histology and EGFR mutation or CN. In our experience, no difference in EGFR mutation status was found according to adenocarcinoma architectural patterns. This is in accordance with a report on a small series of samples (n = 11) from a Swedish group [18]. As already reported EGFR copy number was variable within the same tumor and may vary according to the architectural pattern [17,45]. Here, for one tumor, EGFR increased copy number was restricted to the solid counterpart as compared to lepidic pattern. But this could not be generalized as other solid or micropapillary predominant pattern tumors had no EGFR CN increase and at the opposite a preinvasive lesion showed a high EGFR copy number. Finally, our results suggested that the role of EGFR amplification in cancer progression might not be directly linked to the histological tumor grade.
Patients undergoing EGFR TKI treatment will develop resistance after several months. Heterogeneity has been described as a phenomenon that could drive secondary resistance. Issues are either the development of a p.T790M or a wild type subclone [15,30,39]. In our series, there was no p.T790M heterogeneity. Considering that our detection method (CAST-PCR) can detect approximately 1 to 2% of mutated alleles in a background of wild type this might not have been sufficient for p.T790M subclone detection [24]. It was also suggested that secondary resistances might be due to EGFR mutation loss. Our study was not designed to detect the presence of EGFR wild type subclones however, if this wild type tumor cell population exists, it is not an obstacle to initial EGFR molecular diagnosis defined by the qualitative presence of the EGFR mutation.
Conclusions
This retrospective series reports multiple EGFR testing in lung cancer in routine diagnostic conditions and validates that molecular testing from single tumor-biopsy sample before first line EGFR-TKI may be conducted on any specimens. This is an important result for clinical practice, it indicates that EGFR testing is relevant on biopsies, cytological samples, lymph nodes and metastatic sites using standardized and validated procedures with a defined detection cut-off. This existence of multiple primary tumors with possible distinct genotypes needs to be considered and the development of sensitive methods is recommended because the wild type/mutated allele ratio is unpredictable. Finally, the clinical impact of the associated EGFR copy number increase in a subset of samples remains to be evaluated.
Competing interest
Following authors have declared that they do not have any conflict of interests: Mansuet-Lupo A, Zouiti F, Alifano M, Charpentier MC, Ducruit V, Devez F, Lemaitre F, Damotte D and Blons H Pierre Laurent-Puig declared being a consultant or advisory role for Amgen, Roche, Merck-Serono, Genomic Health, Astellas, Sanofi, Integragen and Raindance.
Authors’ contribution
AL carried out the molecular genetic studies, carried out tumor classification according to the IASLC/ATS/ERS recommendation, analyzed the data, and drafted the manuscript. FZ, FL carried out the molecular genetic studies, analyzed molecular data. MA recruited the patients, drafted the manuscript. CMC carried out tumor classification according to the IASLC/ATS/ERS recommendation. DV, DF carried out the pathology laboratory work. DM, PLP participated in the design of the study, performed analysis and helped to draft the manuscript. HB conceived of the study participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Detailed results showing multilocalisation screening for EGFR mutation and EGFR CNV.
Wild type sample results.
Contributor Information
Audrey Mansuet-Lupo, Email: audrey.lupo@htd.aphp.fr.
Fouzia Zouiti, Email: fouziaz@hotmail.fr.
Marco Alifano, Email: marco.alifano@cch.aphp.fr.
Anne Tallet, Email: anne_tallet@hotmail.com.
Marie-Christine Charpentier, Email: marie-christine.charpentier@htd.aphp.fr.
Véronique Ducruit, Email: ducruit.veronique@htd.aphp.fr.
Fabrice Devez, Email: fabrice.devez@htd.aphp.fr.
Fanny Lemaitre, Email: lemaitre.fanny@gmail.com.
Pierre Laurent-Puig, Email: pierre.laurent-puig@parisdescartes.fr.
Diane Damotte, Email: diane.damotte@htd.aphp.fr.
Hélène Blons, Email: helene.blons@parisdescartes.fr.
Acknowledgements
We thank The French National Cancer Institute (INCa) for its financial support (Plan cancer 2009-2013).
References
- Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa A, Motoi N, Riely GJ, Sima CS, Gerald WL, Kris MG, Park BJ, Rusch VW, Travis WD. Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage I cases. Mod Pathol. 2011;24:653–664. doi: 10.1038/modpathol.2010.232. [DOI] [PubMed] [Google Scholar]
- Coate LE, John T, Tsao MS, Shepherd FA. Molecular predictive and prognostic markers in non-small-cell lung cancer. Lancet Oncol. 2009;10:1001–1010. doi: 10.1016/S1470-2045(09)70155-X. [DOI] [PubMed] [Google Scholar]
- Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Subramanian J, Govindan R. Lung cancer in never smokers: a review. J Clin Oncol. 2007;25:561–570. doi: 10.1200/JCO.2006.06.8015. [DOI] [PubMed] [Google Scholar]
- Mitsudomi T, Yatabe Y. Mutations of the epidermal growth factor receptor gene and related genes as determinants of epidermal growth factor receptor tyrosine kinase inhibitors sensitivity in lung cancer. Cancer Sci. 2007;98:1817–1824. doi: 10.1111/j.1349-7006.2007.00607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatabe Y, Mitsudomi T. Epidermal growth factor receptor mutations in lung cancers. Pathol Int. 2007;57:233–244. doi: 10.1111/j.1440-1827.2007.02098.x. [DOI] [PubMed] [Google Scholar]
- Arcila ME, Nafa K, Chaft JE, Rekhtman N, Lau C, Reva BA, Zakowski MF, Kris MG, Ladanyi M. EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther. 2013;12:220–229. doi: 10.1158/1535-7163.MCT-12-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman DM, Yeap BY, Sequist LV, Lindeman N, Holmes AJ, Joshi VA, Bell DW, Huberman MS, Halmos B, Rabin MS, Haber DA, Lynch TJ, Meyerson M, Johnson BE, Janne PA. Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non-small cell lung cancer patients treated with gefitinib or erlotinib. Clin Cancer Res. 2006;12:3908–3914. doi: 10.1158/1078-0432.CCR-06-0462. [DOI] [PubMed] [Google Scholar]
- Wu JY, Yu CJ, Chang YC, Yang CH, Shih JY, Yang PC. Effectiveness of tyrosine kinase inhibitors on "uncommon" epidermal growth factor receptor mutations of unknown clinical significance in non-small cell lung cancer. Clin Cancer Res. 2011;17:3812–3821. doi: 10.1158/1078-0432.CCR-10-3408. [DOI] [PubMed] [Google Scholar]
- Fukuoka M, Wu YL, Thongprasert S, Sunpaweravong P, Leong SS, Sriuranpong V, Chao TY, Nakagawa K, Chu DT, Saijo N, Duffield EL, Rukazenkov Y, Speake G, Jiang H, Armour AA, To KF, Yang JC, Mok TS. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS) J Clin Oncol. 2011;29:2866–2874. doi: 10.1200/JCO.2010.33.4235. [DOI] [PubMed] [Google Scholar]
- Jiang SX, Yamashita K, Yamamoto M, Piao CJ, Umezawa A, Saegusa M, Yoshida T, Katagiri M, Masuda N, Hayakawa K, Okayasu I. EGFR genetic heterogeneity of nonsmall cell lung cancers contributing to acquired gefitinib resistance. Int J Cancer. 2008;123:2480–2486. doi: 10.1002/ijc.23868. [DOI] [PubMed] [Google Scholar]
- Sakurada A, Lara-Guerra H, Liu N, Shepherd FA, Tsao MS. Tissue heterogeneity of EGFR mutation in lung adenocarcinoma. J Thorac Oncol. 2008;3:527–529. doi: 10.1097/JTO.0b013e318168be93. [DOI] [PubMed] [Google Scholar]
- Taniguchi K, Okami J, Kodama K, Higashiyama M, Kato K. Intratumor heterogeneity of epidermal growth factor receptor mutations in lung cancer and its correlation to the response to gefitinib. Cancer Sci. 2008;99:929–935. doi: 10.1111/j.1349-7006.2008.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen JN, Sorensen JB. Intratumor heterogeneity and chemotherapy-induced changes in EGFR status in non-small cell lung cancer. Cancer Chemother Pharmacol. 2012;69:289–299. doi: 10.1007/s00280-011-1791-9. [DOI] [PubMed] [Google Scholar]
- Yatabe Y, Matsuo K, Mitsudomi T. Heterogeneous distribution of EGFR mutations is extremely rare in lung adenocarcinoma. J Clin Oncol. 2011;29:2972–2977. doi: 10.1200/JCO.2010.33.3906. [DOI] [PubMed] [Google Scholar]
- Mattsson JS, Imgenberg-Kreuz J, Edlund K, Botling J, Micke P. Consistent mutation status within histologically heterogeneous lung cancer lesions. Histopathology. 2011;61:744–751. doi: 10.1111/j.1365-2559.2012.04245.x. [DOI] [PubMed] [Google Scholar]
- Oxnard GR, Arcila ME, Sima CS, Riely GJ, Chmielecki J, Kris MG, Pao W, Ladanyi M, Miller VA. Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant lung cancer: distinct natural history of patients with tumors harboring the T790M mutation. Clin Cancer Res. 2011;17:1616–1622. doi: 10.1158/1078-0432.CCR-10-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosell R, Molina MA, Costa C, Simonetti S, Gimenez-Capitan A, Bertran-Alamillo J, Mayo C, Moran T, Mendez P, Cardenal F, Isla D, Provencio M, Cobo M, Insa A, Garcia-Campelo R, Reguart N, Majem M, Viteri S, Carcereny E, Porta R, Massuti B, Queralt C, De Aguirre I, Sanchez JM, Sanchez-Ronco M, Mate JL, Ariza A, Benlloch S, Sanchez JJ, Bivona TG. et al. Pretreatment EGFR T790M mutation and BRCA1 mRNA expression in erlotinib-treated advanced non-small-cell lung cancer patients with EGFR mutations. Clin Cancer Res. 2011;17:1160–1168. doi: 10.1158/1078-0432.CCR-10-2158. [DOI] [PubMed] [Google Scholar]
- Calvo E, Baselga J. Ethnic differences in response to epidermal growth factor receptor tyrosine kinase inhibitors. J Clin Oncol. 2006;24:2158–2163. doi: 10.1200/JCO.2006.06.5961. [DOI] [PubMed] [Google Scholar]
- Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12:175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, Beer DG, Powell CA, Riely GJ, Van Schil PE, Garg K, Austin JH, Asamura H, Rusch VW, Hirsch FR, Scagliotti G, Mitsudomi T, Huber RM, Ishikawa Y, Jett J, Sanchez-Cespedes M, Sculier JP, Takahashi T, Tsuboi M, Vansteenkiste J, Wistuba I, Yang PC, Aberle D, Brambilla C, Flieder D. et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot A, Le Corre D, Luscan A, Cazes A, Pallier K, Emile JF, Laurent-Puig P, Blons H. Competitive allele specific TaqMan PCR for KRAS, BRAF and EGFR mutation detection in clinical formalin fixed paraffin embedded samples. Exp Mol Pathol. 2012;92:275–280. doi: 10.1016/j.yexmp.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Pallier K, Houllier AM, Le Corre D, Cazes A, Laurent-Puig P, Blons H. No somatic genetic change in the paxillin gene in nonsmall-cell lung cancer. Mol Carcinog. 2009;48:581–585. doi: 10.1002/mc.20538. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Sequist LV, Martins RG, Spigel D, Grunberg SM, Spira A, Janne PA, Joshi VA, McCollum D, Evans TL, Muzikansky A, Kuhlmann GL, Han M, Goldberg JS, Settleman J, Iafrate AJ, Engelman JA, Haber DA, Johnson BE, Lynch TJ. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- Santarpia M, De Pas TM, Altavilla G, Spaggiari L, Rosell R. Moving towards molecular-guided treatments: erlotinib and clinical outcomes in non-small-cell lung cancer patients. Future Oncol. 2013;9:327–345. doi: 10.2217/fon.13.6. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Hagiwara K. Epidermal growth factor receptor (EGFR) mutation and personalized therapy in advanced nonsmall cell lung cancer (NSCLC) Target Oncol. 2013;8:27–33. doi: 10.1007/s11523-013-0258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZY, Zhong WZ, Zhang XC, Su J, Yang XN, Chen ZH, Yang JJ, Zhou Q, Yan HH, An SJ, Chen HJ, Jiang BY, Mok TS, Wu YL. EGFR mutation heterogeneity and the mixed response to EGFR tyrosine kinase inhibitors of lung adenocarcinomas. Oncologist. 2012;17:978–985. doi: 10.1634/theoncologist.2011-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda Y, Takigawa N, Fushimi S, Ochi N, Kubo T, Ozaki S, Tanimoto M, Kiura K. Disappearance of an activated EGFR mutation after treatment with EGFR tyrosine kinase inhibitors. Lung Cancer. 2012;78:121–124. doi: 10.1016/j.lungcan.2012.07.003. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Yukawa T, Hirami Y, Okita R, Saisho S, Maeda A, Yasuda K, Nakata M. Heterogeneity of the EGFR mutation status between the primary tumor and metastatic lymph node and the sensitivity to EGFR tyrosine kinase inhibitor in non-small cell lung cancer. Target Oncol. 2012;8:237–242. doi: 10.1007/s11523-012-0241-x. [DOI] [PubMed] [Google Scholar]
- Schmid K, Oehl N, Wrba F, Pirker R, Pirker C, Filipits M. EGFR/KRAS/BRAF mutations in primary lung adenocarcinomas and corresponding locoregional lymph node metastases. Clin Cancer Res. 2009;15:4554–4560. doi: 10.1158/1078-0432.CCR-09-0089. [DOI] [PubMed] [Google Scholar]
- Gow CH, Chang YL, Hsu YC, Tsai MF, Wu CT, Yu CJ, Yang CH, Lee YC, Yang PC, Shih JY. Comparison of epidermal growth factor receptor mutations between primary and corresponding metastatic tumors in tyrosine kinase inhibitor-naive non-small-cell lung cancer. Ann Oncol. 2009;20:696–702. doi: 10.1093/annonc/mdn679. [DOI] [PubMed] [Google Scholar]
- Kalikaki A, Koutsopoulos A, Trypaki M, Souglakos J, Stathopoulos E, Georgoulias V, Mavroudis D, Voutsina A. Comparison of EGFR and K-RAS gene status between primary tumours and corresponding metastases in NSCLC. Br J Cancer. 2008;99:923–929. doi: 10.1038/sj.bjc.6604629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai H, Wang Z, Wang Y, Zhuo M, Zhou Q, Duan J, Yang L, Wu M, An T, Zhao J, Wang J. Detection and clinical significance of intratumoral EGFR mutational heterogeneity in Chinese patients with advanced non-small cell lung cancer. PLoS One. 2013;8:e54170. doi: 10.1371/journal.pone.0054170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Zhang Q, Luan H, Zhan Z, Wang C, Sun B. Comparison of KRAS and EGFR gene status between primary non-small cell lung cancer and local lymph node metastases: implications for clinical practice. J Exp Clin Cancer Res. 2011;30:30. doi: 10.1186/1756-9966-30-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Satouchi M, Ishii G, Nishio K, Hagiwara K, Mitsudomi T, Whiteley J, Donald E, McCormack R, Todo T. An evaluation study of EGFR mutation tests utilized for non-small-cell lung cancer in the diagnostic setting. Ann Oncol. 2012;23:2914–2919. doi: 10.1093/annonc/mds121. [DOI] [PubMed] [Google Scholar]
- Ma C, Wei S, Song Y. T790M and acquired resistance of EGFR TKI: a literature review of clinical reports. J Thorac Dis. 2011;3:10–18. doi: 10.3978/j.issn.2072-1439.2010.12.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison G, Zhu G, Moulis A, Dearden S, Speake G, McCormack R. EGFR mutation testing in lung cancer: a review of available methods and their use for analysis of tumour tissue and cytology samples. J Clin Pathol. 2012;66:79–89. doi: 10.1136/jclinpath-2012-201194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei B, Yang K, Zhao J, Chang Y, Ma Z, Dong B, Guo Y, Ma J. Quantification of EGFR mutations in primary and metastatic tumors in non-small cell lung cancer. J Exp Clin Cancer Res. 2014;33:5. doi: 10.1186/1756-9966-33-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YL, Wu CT, Shih JY, Lee YC. Comparison of p53 and epidermal growth factor receptor gene status between primary tumors and lymph node metastases in non-small cell lung cancers. Ann Surg Oncol. 2011;18:543–50. doi: 10.1245/s10434-010-1295-6. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Takahashi K, Iwakawa R, Matsuno Y, Nakanishi Y, Kohno T, Shimizu E, Yokota J. Frequent EGFR mutations in brain metastases of lung adenocarcinoma. Int J Cancer. 2006;119:1491–4. doi: 10.1002/ijc.21940. [DOI] [PubMed] [Google Scholar]
- Akbari M, Hansen MD, Halgunset J, Skorpen F, Krokan HE. Low copy number DNA template can render polymerase chain reaction error prone in a sequence-dependent manner. J Mol Diagn. 2005;7:36–39. doi: 10.1016/S1525-1578(10)60006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob TJ, Hoenig T, Clauditz TS, Atanackovic D, Koenig AM, Vashist YK, Klose H, Simon R, Pantel K, Izbicki JR, Bokemeyer C, Sauter G, Wilczak W. Frequent intratumoral heterogeneity of EGFR gene copy gain in non-small cell lung cancer. Lung Cancer. 2012;79:221–227. doi: 10.1016/j.lungcan.2012.11.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed results showing multilocalisation screening for EGFR mutation and EGFR CNV.
Wild type sample results.


