Abstract
Sphingosine 1-phosphate (S1P), a potent bioactive phospholipid, has been reported to regulate a broad spectrum of biological processes. However, little is known regarding S1P’s effects on mitochondrial function. In this study, we investigated the S1P’s effects on the Peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) signaling pathway and mitochondrial biogenesis in Hep G2 cells. Our results indicate that administration of S1P leads to a significant upregulation of mitochondrial DNA replication and transcription, increased mitochondrial mass, and elevated adenosine triphosphate synthesis. In addition, we found that treatment with S1P stimulates expression of PGC-1α, a master regulator of mitochondrial biogenesis, as well as its downstream targets: nuclear respiratory factor 1 (NRF1) and mitochondrial transcription factor A (TFAM). Moreover, our data demonstrate that S1P’s effects on PGC-1α and mitochondrial biogenesis are mediated by the protein kinase A/cAMP response element-binding protein (PKA/CREB) pathway. Importantly, we also revealed that S1P’s effects on mitochondrial biogenesis are dependent on its type 2 receptor (S1P2), though not on either its type 1 (S1P1) or type 3 (S1P3) receptors. Based on these observations, we concluded that S1P activates the PKA/CREB pathway through S1P2, which then promotes expression of PGC-1α/NRF1/TFAM and subsequent mitochondrial biogenesis in Hep G2 cells.
Keywords: Sphingosine 1-phosphate (S1P), Peroxisome proliferator-activated receptor c coactivator 1α (PGC-1α), Mitochondrial biogenesis, Mitochondrial DNA, cAMP response element-binding protein (CREB)
Introduction
Sphingosine 1-phosphate (S1P), a bioactive phospholipid, is derived from sphingosine through activation of sphingosine kinase. It has been shown to be a key signaling molecule involved in various intracellular physiological activities such as proliferation, survival, migration, inflammation, and angiogenesis (Pitson 2011). S1P is known to play a crucial role in both healthy and disease states and has been reported to affect cardiac function, immune cell function, vascular development and permeability, and inflammation (Pyne and Pyne 2011). S1P’s effects are mediated through certain S1P-specific receptors (Chun et al. 2010). S1P-induced activation of these receptors results in stimulation of several downstream effector enzymes, such as extracellular-signal-regulated kinases 1 and 2 (ERK-1/2), protein kinase B (AKT), and phospholipase C (Yester et al. 2010). Moreover, it has been demonstrated that S1P increases production of cyclic adenosine monophosphate (cAMP), a second messenger, through activation of adenylyl cyclase (AC) (Ammit et al. 2001). Consistently, the cAMP response element-binding protein (CREB) transcription factor was activated by treatment with S1P through phosphorylation at Ser133 (Che et al. 2012).
Mitochondria perform multiple functions within the cell environment. Their main function is to produce adenosine triphosphate (ATP). The life cycles of mitochondria are highly dynamic and mitochondrial biogenesis plays an important role in maintaining a sufficient population of healthy mitochondria which is needed to meet the physiological needs of eukaryotic cells (Ferramosca and Zara 2013; Scarpulla et al. 2012). Peroxisome proliferator-activated receptor c coactivator 1α (PGC-1α) has emerged as a master regulator of mitochondrial biogenesis and function (Fernandez-Marcos and Auwerx 2011). PGC-1α modulates a number of genes associated with mitochondrial biogenesis pathways, including nuclear respiratory factor 1 (NRF1) and mitochondrial transcription factor A (TFAM). NRF1 directly regulates expression of several nuclear-encoded electron transport chain (ETC) proteins and indirectly regulates the three mitochondrial-encoded cytochrome c oxidase (COX) subunit genes. Previous studies have demonstrated that the PGC-1α gene possesses a binding site for CREB, which is well conserved between humans (spanning the region 2133–2116 in PGC-1α promoter) and mice (region 2146–2129) and renders PGC-1α transcription highly reactive to CREB activation (Handschin et al. 2003; Herzig et al. 2001).
Mitochondrial dysfunction is strongly associated with liver disease (Grattagliano et al. 2011). In light of this, regulation of production of mitochondria and their chaperones has become a subject of great interest in furthering our understanding of how energy metabolism and susceptibility to hepatotoxicity are controlled. However, the effects of S1P on mitochondrial biogenesis in hepatocytes and on the PGC-1α pathway are still unknown. Actually, both S1P (Strub et al. 2011) and PGC-1α (Aquilano et al. 2010) are localized inside the mitochondria. The effects of S1P on CREB activation lead us to speculate that S1P might promote mitochondrial biogenesis through activation of the PGC-1α pathway in Hep G2 cells and, in our study, we verified this hypothesis. Our findings indicate that treatment with S1P promotes mitochondrial biogenesis through activating the PGC-1α/NRF1/TFAM pathway. These findings expand our understanding on the physiological function of S1P.
Materials and methods
Cell culture and treatment
Human hepatocellular carcinoma cells (Hep G2) were purchased from ATCC and maintained in RMPI 1640 medium (Hyclone) plus 10 % fetal bovine serum (FBS), 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells were treated with 5 μM S1P for 48 h. H89 (15 μM, Sigma), compound C (10 μM, Sigma), JTE-013 (10 μM; Tocris Bioscience), pertussis toxin (PTX) (100 ng/ml; Merck Chemicals), and suramin (5 μM; Sigma-Aldrich) were used to treat Hep G2 cells for 48 h at the same time when S1P was added. Human WRO cells were grown in RPMI medium with 0.1 mM non-essential amino acids, 2 mM l-glutamine, 10 % (v/v) FBS, 50 units/ml of penicillin, and 50 units/ml streptomycin.
Real-time polymerase chain reaction for mtDNA transcripts quantification
Total RNA was extracted using an RNeasy Mini Kit (Qiagen Australia, Australia) following the manufacturer’s instructions. A 2-μg total RNA was used for reverse transcription by employing the RevertAid First Strand cDNA Synthesis Kit (Fermentas Life Sciences, USA). Target messenger RNA (mRNA) levels were measured using real-time reverse transcriptase (RT)-polymerase chain reaction (PCR) on an ABI Prism 7500 (Applied Biosystems, USA). The primers for COX subunit I (COX I) were 5′-AGCAG GGATACCTCGTCGTTAC-3′ and 5′-GTTTTGATGCGA AGGCTTCTCAAA-3′ (fragment length, 140 bp). The primers for β-actin, which were chosen as an internal standard, were 5′-TCCCAGCACACTTAACTTAGC-3′ and 5′-AGCCACAAGAAACACTCAGG-3′ (fragment length, 98 bp). The ddCt (COX I to β-actin) represents the mitochondrial DNA (mtDNA) transcripts in a cell.
Determination of mitochondrial DNA copy number
QIAamp DNA mini kit (QIAGEN, USA) was used to extract total intracellular DNA. mtDNA copy number was measured by real-time PCR, as mentioned above. ND1, a human electron transport chain gene, was used to represent mtDNA. The gene 18S was used to represent nuclear DNA (nDNA) copy numbers. The ddCt (ND1 to 18S) represents the mtDNA copy number in a cell. The following primers were used in this study: ND1: forward, 5′-ATGGCCAACCTCCTACTCCT-3′; reverse: 5′-GCGGTGATG TAGAGGGTGAT-3′; nuclear 18S: forward, 5′-ACGGACCAGAGCGAAAGCA-3′and reverse, 5′-GACATCTAAGGGCATCACAGAC-3′.
Reverse transcriptase polymerase chain reaction to determine expression of S1P receptors
cDNA, synthesized by RT-PCR as described in “Real-time polymerase chain reaction for mtDNA transcripts quantification”, was used to study expression of S1P receptors in Hep G2 cells.
The following primers were used in this study:
S1P1: forward, 5′-GGCTGGAACTGCATCAGTGCG-3′; reverse, 5′-GAGCAGCGCCACATTCTCAGAGC-3′;
S1P2: forward, 5′-CCGAAACAGCAAGTTCCACT-3′; reverse, 5′-CCAGGAGGCTGAAGACAGAG-3′;
S1P3: forward, 5′-AAGGCTCAGTGGTTCATCGT-3′; reverse, 5′-GCTATTGTTGCTGCTGCTTG-3′.
The primers used for b-actin are: forward, 5′-CTGTCGAGTCGCGTCCACCC-3′, and reverse, 5′-GCTTTGCACATGCCGGAGCC-3′.
Small interfering RNA knockdown
Small interfering RNA (siRNA) oligonucleotide targeting S1P2 RNA was used in this study. A scrambled siRNA was used as the negative control. siRNAs were transfected into Hep G2 cells using lipofectamine RNAiMAX (Invitrogen, USA) according to the manufacturer’s recommendations. The degree of knockdown was determined by quantitative RT-PCR 48 h after transfection.
Western blot analysis
Whole-cell lysates were prepared using cell lysis buffer (Cell signaling, USA) following the manufacturer’s instructions. Cells were sonicated and centrifuged at 4 °C for 15 min. Protein concentration was assayed using a BCA kit (Sigma, USA). Proteins were size-fractionated by SDS–polyacrylamide gel electrophoresis (PAGE). After transferring proteins onto PVDF membranes, blots were blocked with 5 % non-fat milk for 2 h at RT and incubated with primary antibodies for 3 h at RT (Sheng et al. 2012). Following incubation with secondary antibodies, immunocomplexes were developed using chemiluminescence.
Measurement of intracellular ATP
ATP levels in Hep G2 cells were determined using a bioluminescence somatic cell assay kit (Invitrogen, USA) according to the manufacturer’s instructions; 2 × 104 cells were used for each sample. Chemiluminescence signals were acquired using Victor X3 Multilabel Plate Readers (PerkinElmer, USA).
Mitochondria staining with MitoTracker red
Cells were stained with MitoTracker red (Invitrogen, USA) (Sheng et al. 2009) to investigate mitochondrial density. Briefly, cells were plated on cover-slips and subjected to the indicated transfection and incubation. Cells were then washed with HBSS and stained with 20 nM MitoTracker red. 4′,6-diamidino-2-phenylindole (DAPI) was used to counterstain cell nuclei. Staining was observed at ×60 oil immersion using a Zeiss fluorescence microscope. Intracellular integrated optical density (IOD) of red fluorescence, as analyzed with Image-Pro Plus software (Version 5.0), was used to index mitochondrial mass.
Statistical analysis
Significant differences between samples were determined using one-way analysis of variance (ANOVA) along with the Student/Newman–Keuls multiple comparison test. P ≤ 0.05 was considered statistically significant.
Results
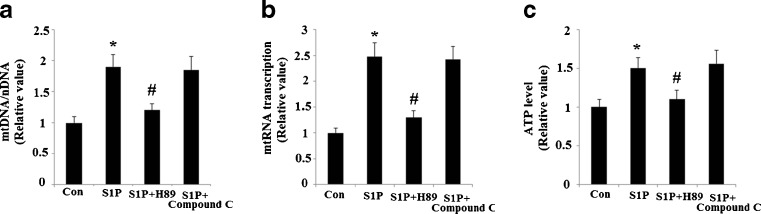
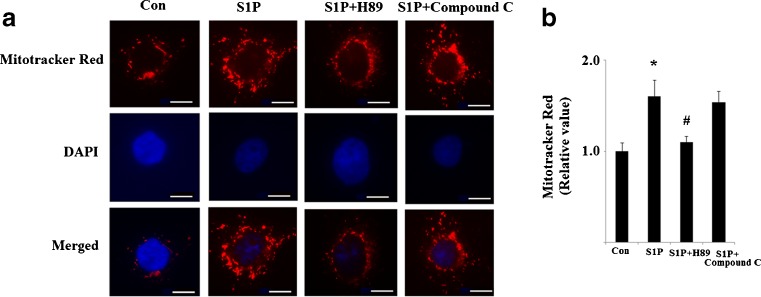
In order to test the effects of S1P on mitochondrial biogenesis, we evaluated the abundance of mtDNA present in cells following administration of S1P. As is shown in Fig. 1a, administration of S1P led to significant upregulation of mtDNA copy numbers. An increase in mtDNA transcription was also observed following a 48-h treatment with S1P (Fig. 1b). When ATP synthesis was normalized over protein content, a significant change was observed (Fig. 1c). This data suggests an overall increase in mitochondrial mass. MitoTracker red staining experiments confirmed the presence of an overall increase in mitochondrial mass (Fig. 2). Both the PKA/CREB and the AMP-activated protein kinase (AMPK) pathways are reported to affect PGC-1α transcriptional activity and sequent mitochondrial biogenesis (Scarpulla. 2011). Therefore, we investigated whether these pathways were involved in the effects induced by S1P. Our results indicate that S1P’s effects on mitochondrial biogenesis were completely abolished by 15 μM H89, a specific inhibitor of the PKA/CREB pathway. However, compound C, a potent AMPK inhibitor, did not affect the patterns of mitochondrial biogenesis induced by S1P (Figs. 1 and 2). This data suggests that the PKA pathway, though not the AMPK pathway, plays a critical role in the observed increase in mitochondrial biogenesis following S1P treatment.
Fig. 1.
The effects of S1P on the replication, transcription of mitochondrial DNA and ATP levels. A 5 μM S1P leads to the induction of mitochondrial biogenesis in Hep G2 cells, which is inhibited by the PKA inhibitor, H89, but not the AMPK inhibitor, compound C. a Relative mtDNA copy number (n = 4, ANOVA, *P < 0.01 versus non-treatment group; # P < 0.01 versus S1P-treated group); b relative mtRNA transcription (n = 4, ANOVA, *P < 0.01 versus non-treatment group; # P < 0.01 versus S1P-treated group); c relative ATP levels (n = 4, ANOVA, *P < 0.01 versus non-treatment group; # P < 0.01 versus S1P-treated group)
Fig. 2.
S1P stimulates mitochondrial mass. A 5 μM S1P leads to the induction of mitochondrial biogenesis in Hep G2 cells, which is inhibited by the PKA inhibitor, H89, but not the AMPK inhibitor, compound C. a The representative picture of mitochondrial mass stained by MitoTracker red dye and DAPI as described in methods. Thirty individual cells were randomly selected from different visual fields in each group. The intracellular integrated optical density (IOD) of red fluorescence was analyzed with Image-Pro Plus software (Version 5.0). Scale bar, 20 μm. b Quantitative analysis of fluorescence intensity (ANOVA, *P < 0.01 versus non-treatment group; # P < 0.01 versus S1P-treated group)
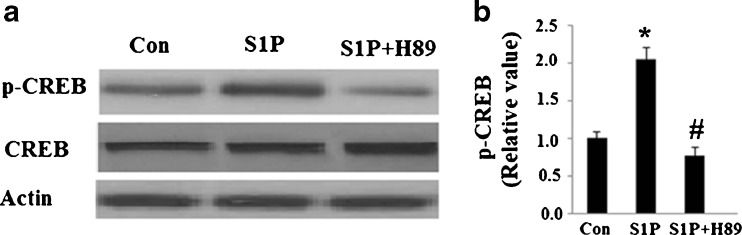
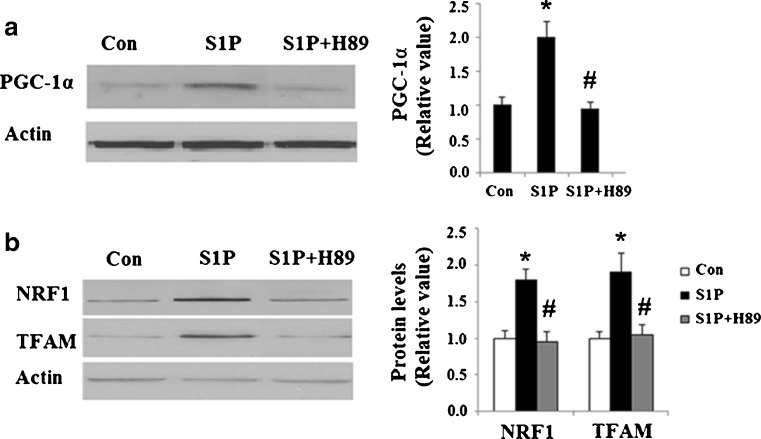
Next, we set out to further explore the underlying mechanisms behind S1P’s effects on mitochondrial biogenesis. The principal action of S1P is to elevate intracellular cAMP levels by activating adenylyl cyclase (AC). cAMP is known to be a main activator of the PKA pathway, which has the ability to increase phosphorylated CREB and its activity. Additionally, phosphorylated CREB may be able to activate transcription of PGC-1α. By combining this information with the results described above, we speculated that activated CREB was involved in S1P’s effects on mitochondrial biogenesis. Indeed, S1P treatment led to an increase in phosphorylated levels of CREB at Ser133 (Fig. 3), which was successfully abolished by H89. However, total levels of CREB were consistent throughout the course of our study. Importantly, S1P treatment increased levels of PGC-1α (Fig. 4a) along with its downstream targets NRF1 and TFAM (Fig. 4b). The increased expression of these functional proteins was also abolished by administration of H89.
Fig. 3.
The PKA/CREB pathway is activated by S1P. Hep G2 cells were treated with 5 μM S1P with or without the presence of the PKA inhibitor H89 (15 μM). Expression levels of proteins were determined by western blot analysis. a Representative western blot analysis of phosphorylated CREB at Ser133; b Quantitative analysis of the relative level of p-CREB (n = 4, ANOVA, *P < 0.01 versus non-treatment group; # P < 0.01 versus S1P-treated group)
Fig. 4.
S1P treatment stimulates the activation of PGC-1α/NRF1/TFAM pathway. Hep G2 cells were treated with 5 μM S1P with or without the presence of the PKA inhibitor H89 (15 μM). Expression levels of proteins were determined by western blot analysis. Con control group, S1P S1P-treated group; S1P + H89 cells were treated with S1P and H89. a Protein levels of PGC-1α were significantly increased in S1P-treated group, which was abolished by H89 (n = 4, ANOVA, *P < 0.01 versus non-treatment group; # P < 0.01 versus S1P-treated group); b protein levels of NRF1 and TFAM were significantly increased in S1P-treated group, which was abolished by H89 (n = 4, ANOVA, *P < 0.01 versus non-treatment group; # P < 0.01 versus S1P-treated group)
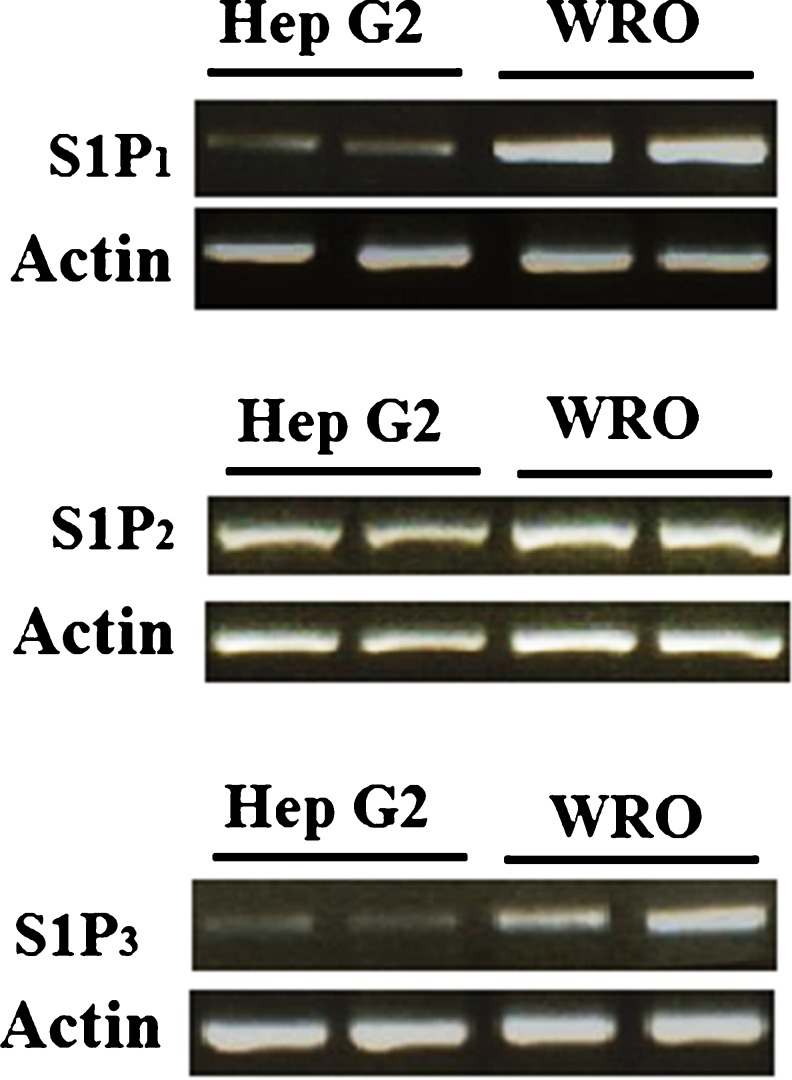
By binding to specific G protein-coupled receptors, S1P is able to carry out its various functions. S1P receptors types 1, 2, and 3 (S1P1, 2, 3) have been reported to be expressed in human liver cells (Li et al. 2011). To investigate expression patterns of S1P receptors in Hep G2 cells, an RT-PCR experiment was performed. The results indicate that S1P1, S1P2, and S1P3 are all expressed in Hep G2 cells (Fig. 5). WRO cells, which also express all three of these receptors, were used as a positive control (Balthasar et al. 2006). Our results successfully verified that S1P2 levels are highest in Hep G2 cells.
Fig. 5.
Hep G2 cells express S1P receptors. a RT-PCR for expression of S1P1, with WRO cells used as a positive control and actin serving as the housekeeping gene; b RT-PCR for expression of S1P2, with WRO cells used as a positive control and actin serving as the housekeeping gene; c RT-PCR for expression of S1P3, with WRO cells used as a positive control and actin serving as the housekeeping gene
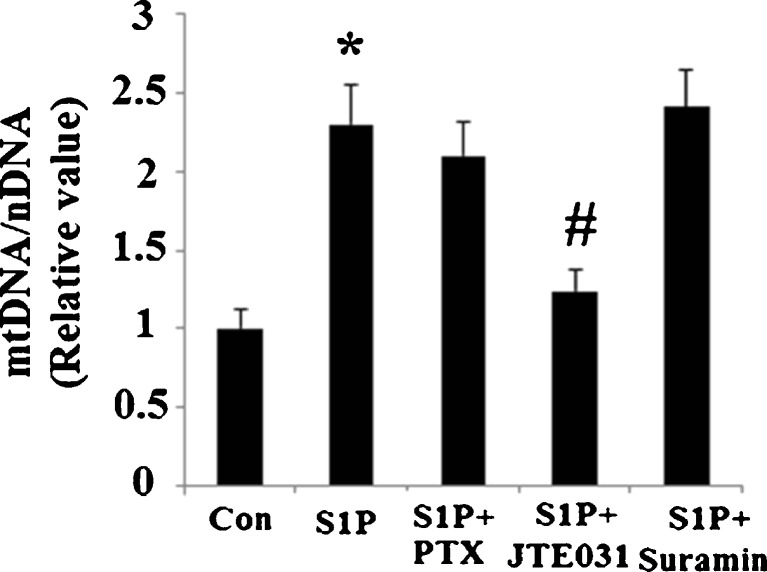
Next, we further investigated whether these receptors are involved in S1P’s effects on mitochondrial biogenesis. In Hep G2 cells incubated with JTE-013, an inhibitor of S1P2 receptor signaling, S1P’s positive effects on upregulation of mitochondrial DNA were fully reversed (Fig. 6a). Neither incubation with Suramin, which selectively inhibits S1P3, nor incubation with PTX, an inhibitor of Gαi signaling (required for functional S1P1), reversed the effects brought on by administration of S1P.
Fig. 6.
S1P receptor type 2 (S1P2), but not S1P1 or S1P3, mediates the effects of S1P on mitochondrial biogenesis. a Hep G2 cells were treated with 5 μM sphingosine 1-phosphate (S1P), 100 ng/ml pertussis toxin (PTX) (inhibitor of Gαi/S1P1), 10 μM JTE-013 (inhibitor of S1P2), or 5 μM suramin (inhibitor of S1P3). Relative mtDNA copy numbers were measured to index mitochondrial biogenesis (n = 4, ANOVA, *P < 0.01 versus non-treatment group; # P < 0.01 versus S1P-treated group)
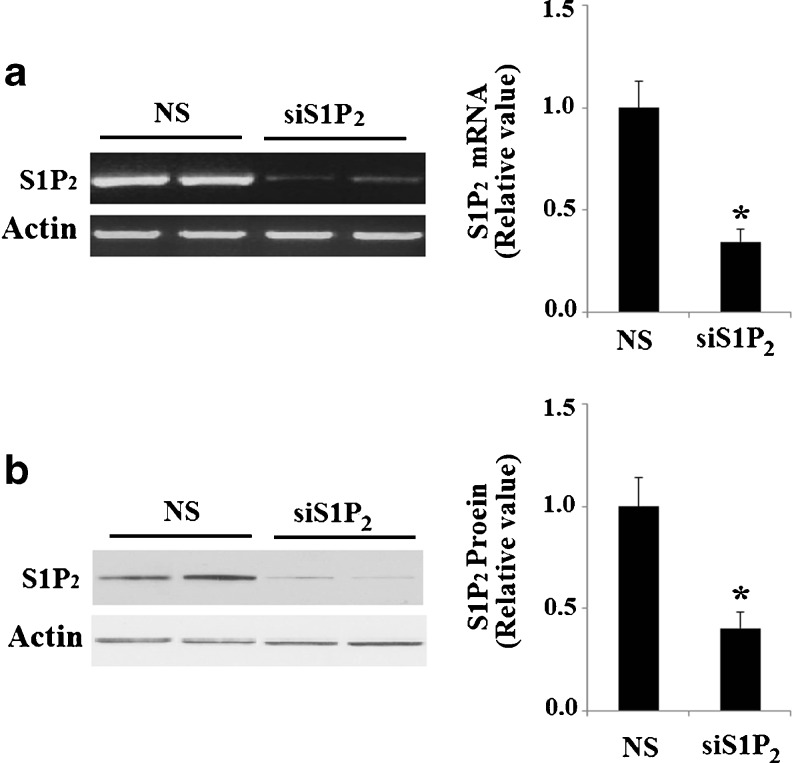
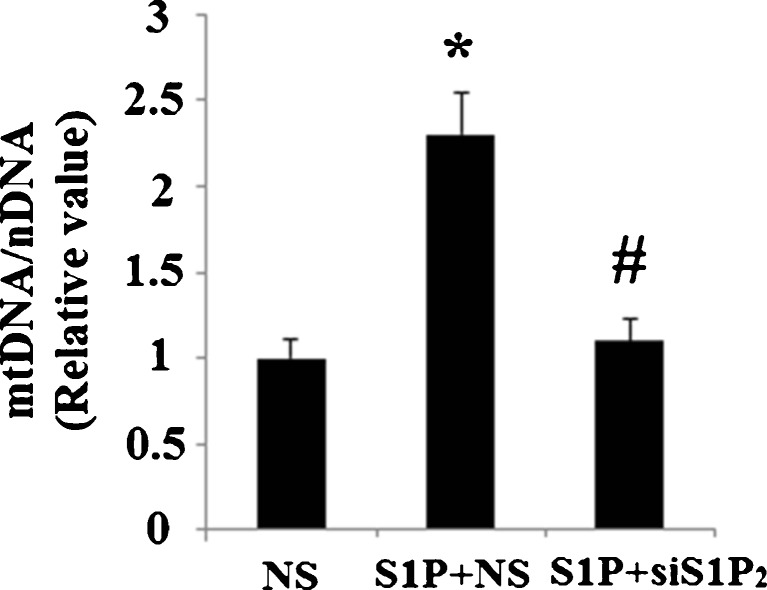
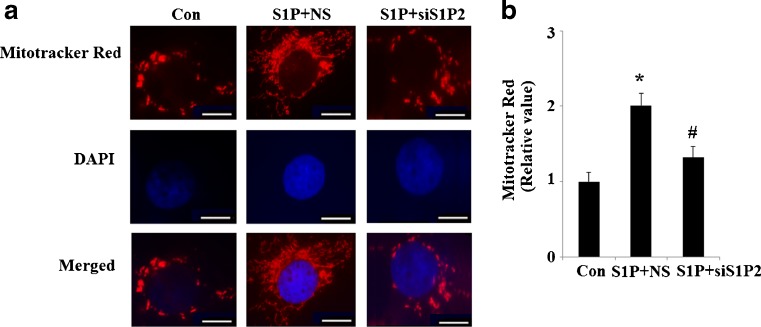
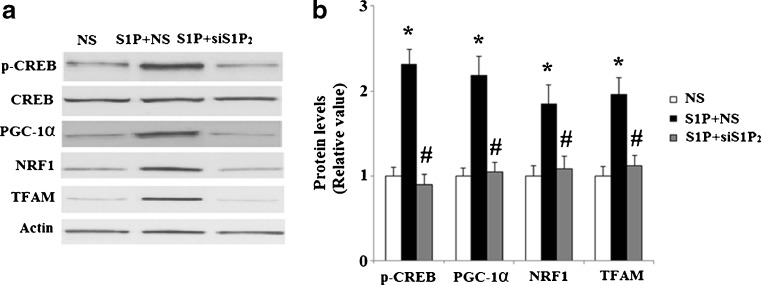
To confirm these results, we knocked down S1P2 using siRNA technology. Efficacy of S1P2-knockdown in Hep G2 cells was confirmed by a 65.7 % reduction in S1P2 mRNA expression (Fig. 7a). Successful knockdown of S1P2 was also verified at the protein level using western blot analysis (Fig. 7b). In Hep G2 cells transfected with non-sense siRNA, mitochondrial DNA was observed to be significantly increased after treatment with S1P (P < 0.01). However, in cells transfected with S1P2 siRNA, S1P’s effects were diminished (P < 0.01) (Fig. 8). MitoTracker red dye was used to stain mitochondria in Hep G2 cells. Importantly, successful silencing of S1P2 abolished the increase in mitochondrial mass induced by treatment with S1P (Fig. 9a, b). Consistently, transfection with S1P2 siRNA abolished the effects of S1P on CREB phosphorylation and upregulation of PGC-1α, NRF 1, and TFAM (Fig. 10). These results indicate that S1P2, though not S1P1 or S1P3, plays a role in S1P’s effects on mitochondrial biogenesis.
Fig. 7.
Successful knockdown of in Hep G2 cells. NS non-specific RNA; siS1P 2 S1P2 siRNA. a RT-PCR revealed the successful knockdown of S1P2 at mRNA levels (*P < 0.01 vs. NS group); b Western blot analysis revealed the successful knockdown of S1P2 at protein levels (*P < 0.01 vs. NS group)
Fig. 8.
Knockdown of S1P2 abolished the effect of S1P on mitochondrial biogenesis. After transfection with small interfering RNA (siRNA) against S1P2 (siS1P 2) or a non-specific control (NS), cells were treated with 5 μM S1P. Relative mtDNA copy number was measured to index mitochondrial biogenesis (n = 4, ANOVA, *P < 0.01 versus non-treatment group; # P < 0.01 versus NS + S1P group)
Fig. 9.
Knockdown of S1P2 abolished the effect of S1P on mitochondrial mass. After transfection with small interfering RNA (siRNA) against S1P2 (siS1P 2) or a non-specific control (NS), cells were treated with 5 μM S1P. a The representative pictures of MitoTracker red staining and nuclear staining. Thirty individual cells were randomly selected from different visual fields in each group. The intracellular integrated optical density (IOD) of red fluorescence was analyzed with Image-Pro Plus software (Version 5.0). Scale bar 20 μm. b Quantitative analysis of fluorescence intensity (ANOVA, *P < 0.01 versus non-treatment group; # P < 0.01 versus S1P + NS group)
Fig. 10.
Knockdown of S1P2 abolished the effect of S1P on CREB/PGC-1α pathway. After transfection with small interfering RNA (siRNA) against S1P2 or a non-specific control (NS), cells were treated with 5 μM S1P. Protein levels of phosphorylated CREB at Ser133, PGC-1α, NRF1, and TFAM were measured by western blot analysis. NS non-specific RNA, siS1P 2 S1P2 RNA interference. a Representative western blot bands; b Quantitative analysis of relative protein levels (n = 4, ANOVA, *P < 0.01 versus non-treatment group; # P < 0.01 versus NS + S1P group)
Discussion
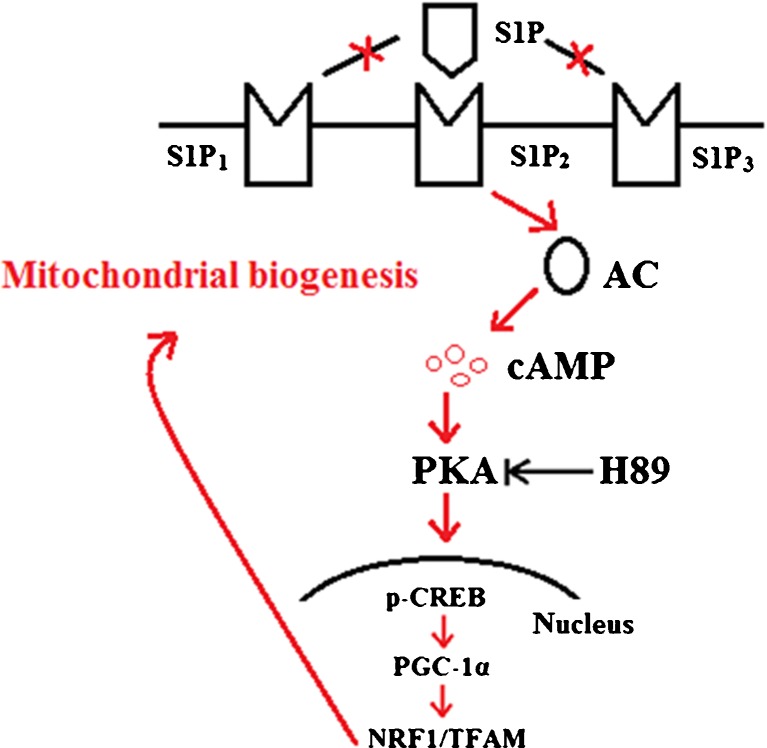
It has been found that mitochondrial damage and dysfunction occur in several acute and chronic liver diseases, such as Alcoholic and Non-Alcoholic Fatty Liver Disease (NAFLD), drug-induced steatohepatitis, viral hepatitis, biliary cirrhosis, hepatocellular carcinoma, ischemia/reperfusion injury, and transplant rejection, as well as various other diseases (Fellman and Kotarsky 2011). Identification of the mechanisms promoting mitochondrial biogenesis in hypocytes is helpful in developing improved pharmacological approaches to the prevention of mitochondrial dysfunction in liver disease (Piantadosi and Suliman 2012). In this study, we demonstrated that S1P can promote mitochondrial biogenesis in Hep G2 cells by increasing ATP levels, mtDNA/nDNA ratios, and mitochondrial contents. To the best of our knowledge, this is the first time that the effects of S1P on mitochondrial biogenesis have been reported. Consistent with this notion, we also found that S1P treatment upregulates expression of PGC-1α as well as its downstream target molecules, NRF 1 and TFAM. Moreover, we demonstrated that S1P-induced mitochondrial biogenesis and induction of PGC-1α is prevented by H89, a PKA inhibitor. We also revealed that S1P’s effects on mitochondrial biogenesis are dependent on its type 2 receptor (S1P2), though not on its types 1 or 3 receptors (S1P1, S1P3). Based on these observations, we concluded that S1P activates the PKA/CREB pathway through S1P2, which then promotes expression of PGC-1α and subsequent mitochondrial biogenesis. A schematic representation of the underlying mechanism is shown in Fig. 11.
Fig. 11.
Schematic drawing of the effects of S1P on mitochondrial biogenesis in Hep G2 cells
It has been found that hepatic expression of PGC-1α is decreased in patients with cholesterol cholelithiasis (Bertolotti et al. 2006). Moreover, it has also been found that hepatic PGC-1α expression is elevated in type 2 diabetes mouse models (Koo et al. 2004). PGC-1α is able to increase both the number of mitochondria and the oxidative phosphorylation capacity of each mitochondrion. Previous observations suggest that PGC-1α coordinates the process of metabolic adaptation that occurs in the liver, including both glucose homeostasis and lipid homeostasis (Zhang et al. 2004). In agreement with our findings, another study has demonstrated that PKA-mediated activation of CREB enhances hepatocyte nuclear factor-4α (HNF-4α) transcriptional activity via induction of the PGC-1α gene in Hep G2 cells (Dankel et al. 2010; Yamamoto et al. 2004). It is likely that increased hepatic PGC-1α expression can stimulate hepatic glucose output. Sphingosine kinase 2 (SphK2) plays an important role in producing S1P. A recent study demonstrated that SphK2 is localized on the inner mitochondrial membrane, where it regulates levels of mitochondrial S1P. S1P binds with high affinity and specificity to prohibitin 2 (PHB2), a highly conserved protein that regulates mitochondrial assembly and function. S1P’s interaction with homomeric PHB2 is important for cytochrome c oxidase assembly and mitochondrial respiration (Strub et al. 2011). In addition, a previous study has reported that PGC-1α is localized within mitochondria. More importantly, mitochondrial PGC-1α is associated with nucleoids and forms a multiprotein complex with TFAM, thereby suggesting their possible involvement in regulation of mitochondrial biogenesis and metabolism (Aquilano et al. 2010). Thus, we speculate that treatment with S1P might increase levels of mitochondrial PGC-1α in order to promote mitochondrial biogenesis. On the other hand, whether or not S1P and PGC-1α are able to interact directly in mitochondria is still an enigma. Future study will provide a complete picture of the underlying mechanisms involved in the effects of S1P on PGC-1α and mitochondrial biogenesis.
Previous studies have demonstrated that S1P can improve mitochondrial function under different types of stress. Mitochondria are also central to intracellular Ca2+ homeostasis, free radicals, and pro- and anti-apoptotic balance. As such, maintaining mitochondrial homeostasis is important for maintaining cell survival. It has been reported that S1P treatment reduces oxygen-glucose deprivation (OGD)-induced membrane depolarization and also reduces the increase of Ca2+ in mitochondria that occurs during OGD (Agudo-López et al. 2010). Moreover, S1P was demonstrated to have the ability to exert its cytoprotective effects on mitochondrial events during apoptosis through regulation of both pro-apoptotic Bad and Bax proteins in an MEK-dependent manner (Betito and Cuvillier 2006). The data attained in this study serve to expand our knowledge of S1P’s protective effects on mitochondrial function. These findings suggest that S1P could potentially have therapeutic effects on mitochondrial disorders through stimulating expression of PGC-1α and mitochondrial biogenesis in liver diseases.
References
- Agudo-López A, Miguel BG, Fernández I, et al. Involvement of mitochondria on neuroprotective effect of sphingosine-1-phosphate in cell death in an in vitro model of brain ischemia. Neurosci Lett. 2010;470(2):130–133. doi: 10.1016/j.neulet.2009.12.070. [DOI] [PubMed] [Google Scholar]
- Ammit AJ, Hastie AT, Edsall LC, et al. Sphingosine 1-phosphate modulates human airway smooth muscle cell functions that promote inflammation and airway remodeling in asthma. FASEB J. 2001;15:1212–1214. doi: 10.1096/fj.00-0742fje. [DOI] [PubMed] [Google Scholar]
- Aquilano K, Vigilanza P, Baldelli S, et al. Peroxisome proliferator-activated receptor gamma co-activator 1alpha (PGC-1alpha) and sirtuin 1 (SIRT1) reside in mitochondria: possible direct function in mitochondrial biogenesis. J Biol Chem. 2010;285:21590–21599. doi: 10.1074/jbc.M109.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balthasar S, Samulin J, Ahlgren H, et al. Sphingosine 1-phosphate receptor expression profile and regulation of migration in human thyroid cancer cells. Biochem J. 2006;398(3):547–556. doi: 10.1042/BJ20060299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti M, Gabbi C, Anzivino C, et al. Decreased hepatic expression of PPAR-gamma coactivator-1 in cholesterol cholelithiasis. Eur J Clin Invest. 2006;36(3):170–175. doi: 10.1111/j.1365-2362.2006.01607.x. [DOI] [PubMed] [Google Scholar]
- Betito S, Cuvillier O. Regulation by sphingosine 1-phosphate of Bax and Bad activities during apoptosis in a MEK-dependent manner. Biochem Biophys Res Commun. 2006;340(4):1273–1277. doi: 10.1016/j.bbrc.2005.12.138. [DOI] [PubMed] [Google Scholar]
- Che W, Manetsch M, Quante T, et al. Sphingosine 1-phosphate induces MKP-1 expression via p38 MAPK- and CREB-mediated pathways in airway smooth muscle cells. Biochim Biophys Acta. 2012;1823(10):1658–1665. doi: 10.1016/j.bbamcr.2012.06.011. [DOI] [PubMed] [Google Scholar]
- Chun J, Hla T, Lynch KR, et al. International Union of Basic and Clinical Pharmacology. LXXVIII. Lysophospholipid receptor nomenclature. Pharmacol Rev. 2010;62:579–587. doi: 10.1124/pr.110.003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankel SN, Hoang T, Flågeng MH, et al. CAMP-mediated regulation of HNF-4α depends on the level of coactivator PGC-1α. Biochim Biophys Acta. 2010;1803:1013–1019. doi: 10.1016/j.bbamcr.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Fellman V, Kotarsky H. Mitochondrial hepatopathies in the newborn period. Semin Fetal Neonatal Med. 2011;16(4):222–228. doi: 10.1016/j.siny.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Fernandez-Marcos PJ, Auwerx J. Regulation of PGC-1α, a nodal regulator of mitochondrial biogenesis. Am J Clin Nutr. 2011;93:884S–890S. doi: 10.3945/ajcn.110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferramosca A, Zara V. Biogenesis of mitochondrial carrier proteins: molecular mechanisms of import into mitochondria. Biochim Biophys Acta. 2013;1833:494–502. doi: 10.1016/j.bbamcr.2012.11.014. [DOI] [PubMed] [Google Scholar]
- Grattagliano I, Russmann S, Diogo C, et al. Mitochondria in chronic liver disease. Curr Drug Targets. 2011;12(6):879–893. doi: 10.2174/138945011795528877. [DOI] [PubMed] [Google Scholar]
- Handschin C, Rhee J, Lin J, et al. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc Natl Acad Sci U S A. 2003;100:7111–7113. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig S, Long F, Jhala US, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- Koo SH, Satoh H, Herzig S, et al. PGC-1 promotes insulin resistance in liver through PPAR-alpha-dependent induction of TRB-3. Nat Med. 2004;10(5):530–534. doi: 10.1038/nm1044. [DOI] [PubMed] [Google Scholar]
- Li C, Zheng S, You H, et al. Sphingosine 1-phosphate (S1P)/S1P receptors are involved in human liver fibrosis by action on hepatic myofibroblasts motility. J Hepatol. 2011;54(6):1205–1213. doi: 10.1016/j.jhep.2010.08.028. [DOI] [PubMed] [Google Scholar]
- Piantadosi CA, Suliman HB. Transcriptional control of mitochondrial biogenesis and its interface with inflammatory processes. Biochim Biophys Acta. 2012;1820(4):532–541. doi: 10.1016/j.bbagen.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitson SM. Regulation of sphingosine kinase and sphingolipid signaling. Trends Biochem Sci. 2011;36:97–107. doi: 10.1016/j.tibs.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Pyne S, Pyne NJ. Translational aspects of sphingosine 1-phosphate biology. Trends Mol Med. 2011;17:463–472. doi: 10.1016/j.molmed.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim Biophys Acta. 2011;1813(7):1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC, Vega RB, Kelly DP. Transcriptional integration of mitochondrial biogenesis. Trends Endocrinol Metab. 2012;2:459–466. doi: 10.1016/j.tem.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng B, Niu Y, Zhou H, et al. The mitochondrial function was impaired in APP knockout mouse embryo fibroblast cells. Chin Sci Bull. 2009;54:1725–1731. doi: 10.1007/s11434-009-0239-7. [DOI] [Google Scholar]
- Sheng B, Wang X, Su B, et al. Impaired mitochondrial biogenesis contributes to mitochondrial dysfunction in Alzheimer’s disease. J Neurochem. 2012;120(3):419–429. doi: 10.1111/j.1471-4159.2011.07581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub GM, Paillard M, Liang J, et al. Sphingosine-1-phosphate produced by sphingosine kinase 2 in mitochondria interacts with prohibitin 2 to regulate complex IV assembly and respiration. FASEB J. 2011;25(2):600–612. doi: 10.1096/fj.10-167502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Shimano H, Nakagawa Y, et al. SREBP-1 interacts with hepatocyte nuclear factor-4α and interferes with PGC-1 recruitment to suppress hepatic gluconeogenic genes. J Biol Chem. 2004;279(13):12027–12035. doi: 10.1074/jbc.M310333200. [DOI] [PubMed] [Google Scholar]
- Yester JW, Tizazu E, Harikumar KB, et al. Extracellular and intracellular actions of sphingosine-1-phosphate. Adv Exp Med Biol. 2010;688:141–155. doi: 10.1007/978-1-4419-6741-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ma K, Song S, et al. Peroxisomal proliferator-activated receptor-gamma coactivator-1 alpha (PGC-1 alpha) enhances the thyroid hormone induction of carnitine palmitoyltransferase I (CPT-I alpha) J Biol Chem. 2004;279(52):53963–53971. doi: 10.1074/jbc.M406028200. [DOI] [PubMed] [Google Scholar]