Abstract
NF-κB is a transcription factor that integrates pro-inflammatory and pro-survival responses in diverse cell types. The activity of NF-κB is regulated in part by acetylation of its p65 subunit at lysine 310,which is required for transcription complex formation. De-acetylation at this site is performed by sirtuin 1(SIRT1) and possibly other sirtuins in an NAD+ dependent manner, such that SIRT1 inhibition promotes NF-κB transcriptional activity. It is unknown, however, whether changes in NAD+ levels can influence p65 acetylation and cellular inflammatory responses. Poly(ADP-ribose)-1 (PARP-1) is an abundant nuclear enzyme that consumes NAD+ in the process of forming (ADP-ribose)polymers on target proteins, and extensive PARP-1 activation can reduce intracellular NAD+ concentrations. Here we tested the idea that PARP-1 activation can regulate NF-κB transcriptional activity by reducing NAD+ concentrations and thereby inhibiting de-acetylation of p65. Primary astrocyte cultures were treated with the alkylating agent N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) to induce PARP-1 activation. This resulted in sustained acetylation of p65 and increased NF-κB transcriptional activity as monitored by a κB-driven eGFP reporter gene. These effects of MNNG were negated by a PARP-1 inhibitor, in PARP-1−/− cells, and in PARP-1−/− cells transfected with a catalytically inactive PARP-1 construct, thus confirming that these effects are mediated by PARP-1 catalytic activity. The effects of PARP-1 activation were replicated by a SIRT1 inhibitor, EX-527, and were reversed by exogenous NAD+. These findings demonstrate that PARP-1-induced changes in NAD+ levels can modulate NF-κB transcriptional activity through effects on p65 acetylation.
Keywords: Acetylation, Astrocyte, NAD+, NF-κB, PARP-1, SIRT1
1. Introduction
NF-κB is a transcription factor that integrates pro-inflammatory and pro-survival responses in diverse cell types. NF-κB is activated by numerous stimuli including DNA damage, which can occur as a direct secondary effect of infection, ischemia, trauma, and other stressors [1]. The NF-κB family includes five members, p65 c-Rel, RelB, p105/p50 and p100/p52 that form homo- or heterodimers [2]. In most cell types, the most abundant heterodimer consists of p65 and p50. The regulation of NF-κB activity is complex, and occurs at multiple levels [2]. One level of regulation in canonical NF-κB activation involves the release of p65/p50 dimers from the IκB subunit in the cytosol, and subsequent translocation to the nucleus. A second layer of regulation occurs within the nucleus, with the formation of an active transcription complex. p300/CBP mediated acetylation of various lysine sites (at least 5 sites reported) of the p65 subunit has been established as a key regulatory event in this process. In particular, acetylation at lysine 310 of p65 is required for full transcriptional activity of p65, while acetylation at lysine 218/221 enhances DNA binding and impairs assembly with IκBα [3,4]. Acetylation of these lysine sites can be reversed by histone deacetylase (HDAC) corepressor proteins [5,6]. Deacetylation of p65 lysine 218/221 does not require NAD, but Lysine 310 of p65 is deacetylated in the nucleus by the atypical class III HDACs, SIRT1 and possibly other sirtuins in a reaction that requires NAD+ [3,4,7,8].
NAD+ is also required by the abundant nuclear enzyme, poly(ADP-ribose) polymerase-1 (abbreviated as PARP-1). Upon activation by DNA damage, cytokines, or other signals, PARP-1 consumes NAD+ to generate branched ADP-ribose polymers on chromatin and other acceptor proteins [9,10]. These polymers have diverse functions in DNA repair and other nuclear processes [11,12]. Extensive PARP-1 activation can deplete the cytosolic (non-mitochondrial) NAD+ pool ([13–16]. PARP-1 also regulates NF-κB activity [17,18], but there remains uncertainty as to the role of PARP-1 activity in this process. Several studies indicate that inhibition of PARP-1 enzymatic activity impairs NF-κB-mediated responses [19–24], whereas others have found that PARP-1 influences NF-κB-mediated responses independent of its enzymatic activity [17,25–27].
Given that both SIRT1 and PARP-1 require NAD+, one mechanism by which PARP-1 activation could influence NF-κB activity is by reducing the amount of NAD+ available for SIRT1 de-acetylation of p65 [28]. Here we tested this hypothesis using primary astrocyte cultures in which PARP-1 activation was induced by the DNA alkylating agent N-methyl-N′-nitro-N-nitrosoguanidine (MNNG). Astrocytes are experimentally convenient because they are non-neoplastic cells that can be prepared in homogeneous cultures, and because prior work has established key aspects of PARP-1 activation and regulation in this cell type. Results of the present studies confirm that extensive PARP-1 activation promotes NF-κB transcriptional activation by a mechanism involving p65 acetylation.
2. Materials and methods
2.1. Materials
Cell culture reagents were obtained from Cellgro/Mediatech (Herndon, VI). 3,4-dihydro-5-[4-(1-piperidinyl)butoxy]-1(2h)-isoquinolinone (DPQ)was obtained from Calbiochem (San Diego, CA). All other reagents were from Sigma/Aldrich (St. Louis, MO) except where otherwise stated.
2.2. Cell cultures
All animal studies were approved by the San Francisco Veterans Affairs Medical Center animal studies committee and follow the NIH guidelines for humane care of animals. PARP-1−/− mice were obtained from the Jackson Labs and out bred for more than 10 generations to the CD1 background. Wild type mice were from the closely related Swiss Webster strain and were obtained from Simonsen Labs (Gilroy, CA). Astrocyte cultures were prepared from mouse pups of both sexes in 24-well plates as described previously [29]. At confluency (12–14 days in vitro), the cultures were treated with 22 µM cytosine β-D-arabinofuranoside for 2 days to inhibit the proliferation of microglia. The cultures were subsequently maintained in Eagle's minimal essential medium (MEM) supplemented with 3% fetal bovine serum (FBS).
2.3. Cell culture experiments
Studies were initiated by placing the astrocyte cultures in a physiological balanced salt solution (BSS) containing 3.1 mMKCl, 134 mM NaCl, 1.2 mM CaCl2, 1.2 mM MgSO4, 0.5 mM KH2PO4, 15.7 mM NaHCO3 and 2 mM glucose, pre-equilibrated to pH 7.2 in a 5% CO2 atmosphere. Experiments were performed at 37 °C in 5% CO2 with test compounds prepared as concentrated stock solutions in BSS. Stimulations with 100 µM MNNG for 10 min were terminated by complete medium exchange and replacement with BSS. Control wells received medium exchanges only.
2.4. Immunostaining
Fixation and immunostaining of cell cultures were performed as previously described [23]. Incubations were performed with a 1:1000 dilution of rabbit anti-poly(ADP-ribose) (Trevigen, Gaithersburg, MD), or with 1:30 dilution of rabbit polyclonal anti-NF-κB p65 (Cell Signaling Technology, Inc., Danvers, MA) at 4 °C for 24 h, followed by incubation with Alexa Fluor 594-conjugated anti-rabbit IgG (Molecular Probes, Eugene, OR), 1:500 dilution, for 2 h. For evaluation of NF-κB nuclear translocation, nuclei were stained with propidium iodide. Confocal photomicrographs were obtained from single layer scan with pinhole of 1 Airy (~100 µm). Controls prepared in the absence of primary antibody showed no staining under the conditions described (not shown).
2.5. Western blotting
Western blots from whole cell samples were prepared and quantified as described [23]. Membranes were incubated with 1:200 dilutions of rabbit anti-NF-κB Ac-p65 (acetyl K310; Abcam, Inc. Cambridge, MA), 1:2500 dilution of rabbit polyclonal anti-NF-κB (p65; Calbiochem/EMD Bioscience, Inc., La Jolla, CA) or with a 1:500 dilution of mouse monoclonal anti-poly(ADP-ribose) (clone 10HA; Trevigen, Gaithersburg, MD). After washing, the membranes were incubated for 2 h with peroxidase-conjugated anti-rabbit or anti mouse IgG (Vector Laboratories, Burlingame, CA) diluted 1:7500. The protein bands were visualized using ECL™ Plus WB Detection kit (Amersham-Pharmacia Biotech) and X-OMAT AR film (Kodak). To quantify protein loading, the membranes were re-probed with mouse monoclonal anti β-actin at a 1:10,000 dilution, followed by peroxidase-conjugated anti-mouse IgG (1:10,000 dilution). Controls performed in the absence of primary or secondary Abs showed no signal (data not shown). Band densities were quantified with the NIH Image J program.
2.6. Cell transfections
Wild-type and mutant human PARP-1 constructs were prepared as previously described [30]. PARP-1−/− mouse cells were transfected at 8–9 days in vitro, at about 90% confluency, using Lipofectamine 2000 (Invitrogen) at a 2:1 ratio with DNA. Cells were used for experiments 40–48 h after transfection. The lentivirus κB-dEGFP construct was prepared as described previously [31] and astrocytes were treated with the lentivirus 2–3 days prior to experiments. eGFP expression was evaluated by fluorescence microscopy at the designated time points after MNNG exposure by counting the number of eGFP-expressing cells in 5 randomly selected fields within each culture well.
2.7. Statistics
Each “n” denotes an independent experiment comprised of 3–4 parallel treatments per condition. Results are presented as a mean ± standard error. Statistical significance was evaluated by one-way ANOVA followed by the Dunnett's test for multiple comparisons against a common control group.
3. Results
3.1. PARP-1 inhibition prevents NF-κB p65 acetylation
Astrocyte cultures treated for 10 min with the alkylating agent MNNG (100 µM) developed a robust activation of PARP-1 (Fig. 1). PAR immunoreactivity appears as a smear in these Western blots, indicating poly(ADP-ribosylation) of many different protein targets [32,33]. This signal was completely blocked by the addition of the PARP inhibitor DPQ, but was unaffected by the addition of NAD+ (Fig. 1). MNNG treatment also induced acetylation of p65 NF-κB subunit (Fig. 2A). The increase in p65 acetylation was blocked by both pharmacological PARP inhibition (DPQ) and by genetic PARP-1 deletion (Fig. 2B, C, D).
Fig. 1.
MNNG-induced PARP-1 activation is blocked by DPQ, but not NAD+. Immunoblots show formation of poly(ADP-ribose)-conjugated proteins (PAR). A, Ten-minute incubations with 100 µM MNNG induce robust PAR formation that is prevented by the PARP-1 inhibitor, DPQ (25 µM). Time points denote intervals following washout of MNNG. B, NAD+ (2.5 mM) does not prevent PAR formation. Representative of n = 3.
Fig. 2.
NF-κB subunit p65 acetylation is affected by PARP-1 activation and NAD+ levels. A, MNNG (100 µM for 10 min) induces acetylation of NF-κB p65 in wild-type (wt) astrocytes. B, MNNG does not induce p65 acetylation in PARP-1 deficient astrocytes. C, MNNG induced p65 acetylation in wt astrocytes is blocked by both the PARP inhibitor DPQ (25 µM) and by medium supplementation with NAD+ (2.5 mM). Time points denote intervals following washout of MNNG. D, Graph shows quantified data of p65 acetylation in wt astrocytes (A and C) (#p < 0.05 compared to control, *p < 0.05 compared to MNNG at each time point, n = 3–4.).
3.2. NAD+ influences NF-κB acetylation
Our prior studies have established that MNNG-induced PARP-1 activation under the conditions of these experiments causes a rapid and sustained depletion of cytosolic NAD+ (from 9.51 ± 0.72 to 2.85 ± 0.93 nmol/mg protein) [14]. We also previously demonstrated that this depletion can be reversed by medium supplementation with 2.5 mM NAD+, which gains entry into astrocytes through P2×7 receptor-gated channels [15,32,34]. Since NAD+ is a requisite substrate for SIRT1 de-acetylation of p65, we evaluated the possibility that PARP-1 activation increases p65 acetylation and resultant NF-κB transcriptional activation by impairing the ability of SIRT1 or other NAD+-dependent sirtuins to de-acetylate p65 (Fig. 3A). Consistent with this idea, 2.5 mM NAD+ was found to prevent the MNNG-induced increase in p65 acetylation (Fig. 2C, D). This effect cannot be attributed to PARP-1 inhibition by NAD+ or NAD+ metabolites because NAD+ supplementation did not block PARP-1 activity (Fig. 1B). Moreover, NAD+ was found to have no effect on p65 acetylation when SIRT1 activity was blocked by 10 µM EX-527 [35,36] (Fig. 3B).
Fig. 3.
NF-κB subunit p65 acetylation is increased by SIRT1 inhibition. A, Diagram showing the proposed relationships between PARP-1, SIRT1, NAD+, and NF-κB. NF-κB is normally sequestered in the cytosol. In canonical NF-κB activation [2], phosphorylation of the IκB subunit permits the p65/p50 dimer to translocate to the nucleus, where it is acetylated and binds with other proteins to form an activated transcription complex on gene promoter regions. The activated transcription complex is normally deactivated by NAD+-dependent deacetylation of the p65 subunit, catalyzed by SIRT-1. NAD+ levels are reduced by PARP-1 activation, thereby preventing this de-acetylation step and promoting gene transcription. This effect can be negated by either PARP-1 inhibition (DPQ) or by NAD+ repletion. The SIRT-1 inhibitor EX-527 also blocks de-acetylation of NF-κB, but this effect cannot be negated by NAD+. B, Astrocytes treated with the SIRT1 inhibitor EX-572 (10 µM) show accumulation of acetylated p65, and this effect is not blocked by NAD+. Representative of n = 3. C, Photomicrographs show NF-κB transcriptional activity in astrocytes transfected with a κB reporter gene driving eGFP expression. eGFP expression is increased by EX-527, and this effect is not blocked by NAD+. D, Graph shows quantification of eGFP expression. (*p < 0.05, n = 3).
3.3. PARP-1 enzymatic activity is required for NF-κB transcriptional activation
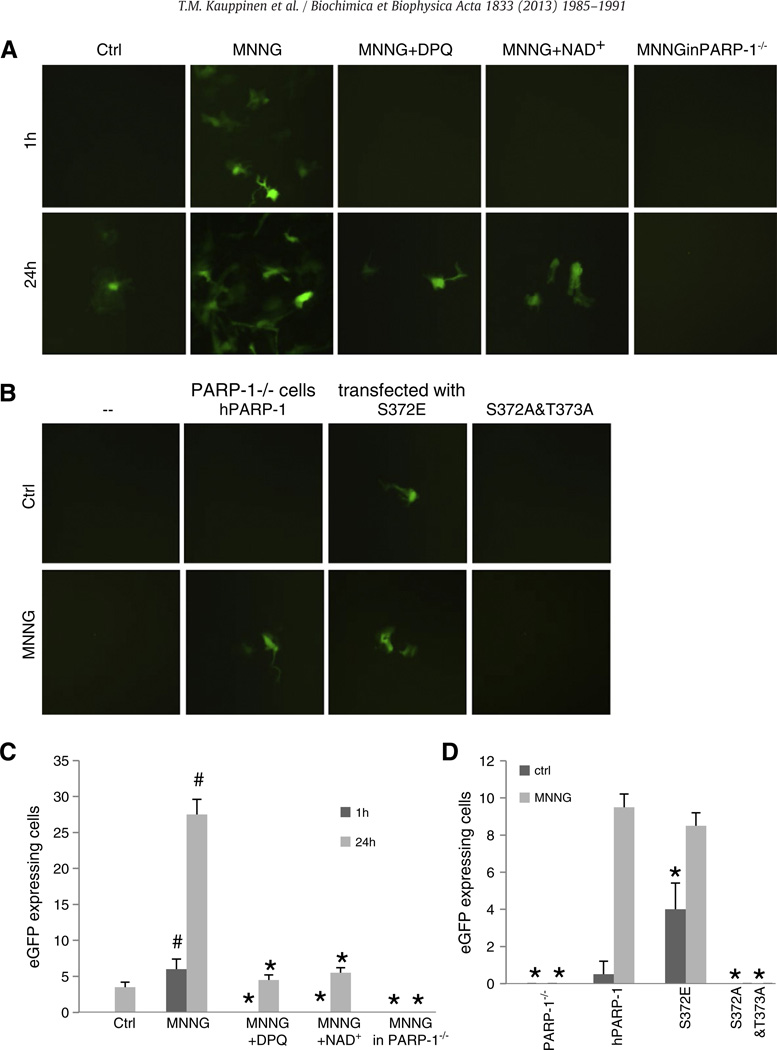
Activation of NF-κB signaling was further evaluated in astrocytes transfected with a κB reporter gene that responds to NF-κB (p65) transcriptional activation by expressing enhanced green fluorescent protein (eGFP) [31]. Inhibition of SIRT-1 activity with EX-527 increased eGFP expression in concert with increased p65 acetylation (Fig. 3C, D). MNNG exposure similarly increased NF-κB transcriptional activity in wild-type astrocytes, but not in PARP-1−/− astrocytes or wild-type astrocytes treated with the PARP inhibitor DPQ. MNNG also failed to induce NF-κB activation in cultures supplemented with 2.5 mM NAD+ (Fig. 4A, C).
Fig. 4.
PARP-1 enzymatic activity is required for NF-κB transcriptional activation. A, NF-κB transcription activity detected in astrocytes transfected with an eGFP-expressing κB reporter gene. Photomicrographs were prepared at 1 and 24 h after MNNG exposures (100 µM for 10 min). MNNG triggers NF-κB activation in wild-type cells, but not in PARP-1−/− cells or in wild-type cells treated with 25 µMDPQ or 2.5 mM NAD+. B, PARP-1−/− cells exhibit NF-κB transcriptional activation when transfected normal human PARP-1 (hPARP-1) or with hPARP-1 containing the S372E phosphomimetic mutation, but not when transfected with hPARP-1 containing the S372A and T373A mutations that prevent enzymatic activation. C and D, eGFP expression quantification. For C, #p < 0.05 compared to control, *p < 0.05 compared to MNNG, n = 3. For D, *p < 0.05 compared to PARP-1−/− cultures transfected with hPARP-1, n = 3. Where no bar is visible, there was no detectable eGFP expression.
The role of PARP-1 enzymatic activity in NF-κB activation was further evaluated by using previously developed PARP-1 mutants containing modifications at phosphorylation sites that influence enzymatic activity [30]. PARP-1−/− cells transfected with wild-type PARP-1 and treated with MNNG showed eGFP expression in about 30% of the cells. Prior studies have shown comparable transfection efficiency attained with all 3 constructs used [30]. PARP-1−/− cells transfected with S372A & T373A PARP-1, which lacks phosphorylation sites required for enzymatic activation, failed to show NF-κB activation after MNNG stimulus. Conversely, PARP-1−/− cells transfected with the S372E PARP-1 mutant, which mimics constitutive phosphorylation at this site, showed increased NF-κB activation even in the absence of MNNG stimulation (Fig. 4B, D). Additional studies confirmed that the impaired NF-κB transcription activity in PARP-1−/− cells is not due to deficient p65 protein expression (Fig. 5A), and that PARP-1 depletion does not either interfere with NF-κB p65 nuclear translocation (Fig. 5B).
Fig. 5.
PARP-1 depletion does not affect NF-κB p65 expression or nuclear translocation. A, Immunoblots showing p65 expression in wild-type and PARP−/− cells. B, Confocal images demonstrate NF-κB p65 subunit nuclear translocation 30 min after MNNG stimulation (100 µM for 10 min) in both wild-type and PARP-1−/− cells. Representative of n = 3.
4. Discussion
These results show that extensive activation of PARP-1 can promote the transcriptional activity of NF-κB by a mechanism involving p65 acetylation. Reversal of this effect by exogenous NAD+, in conjunction with previously demonstrated effects of PARP-1 and exogenous NAD+ on intracellular NAD+ levels, identify this as an NAD+ dependent mechanism. At present, the only enzymes known to perform NAD+-dependent de-acetylation of lysine residues are the class III histone de-acetylases, also known as sirtuins [37]. SIRT1 in particular can catalyze de-acetylation of the lysine 310 of p65 [31]. SIRT2 has also been reported to do so [8], but SIRT2 is not usually localized to the nucleus. However, there is presently no way to directly evaluate in situ SIRT1 activity (other than by p65 acetylation), so we cannot exclude the possibility that NAD+ levels influence p65 acetylation by some other, as yet unrecognized mechanism.
PARP-1 activation not only consumes NAD+, but also produces nicotinamide as a product of NAD+ cleavage. The production of nicotinamide has been suggested as another possible way that PARP-1 and sirtuins may interact, because many sirtuins (including SIRT1) are inhibited by nicotinamide [37,38]. The expected effect of nicotinamide in this instance would be prolonged p65 acetylation and enhanced NF-κB transcriptional activation, as was observed here. However, the additional observation that exogenous NAD+ supplementation did not inhibit PARP-1 activity while was still able to reverse the effects of PARP-1 on both p65 acetylation and NF-κB transcriptional activity argues against the idea that cleavage of NAD+ to nicotinamide or other metabolites is a significant regulatory factor under these conditions. This notion is further supported by our previously published data, in which NAD+ supplementation (5 mM) failed to increase intracellular levels required for PARP-1 inhibition [15].
Canonical NF-κB transcriptional activation requires both translocation of NF-κB dimers to the nucleus and the subsequent formation and maintenance of a DNA-bound transcriptional complex. Under the conditions of MNNG-induced DNA damage employed here, NF-κB translocation is likely induced by the ATM/NEMO pathway leading to IκB phosphorylation [39–41]. The role of PARP-1 activation in NF-κB signaling has been demonstrated in many levels, PARP-1 promotes complex formation [19,22,42], participates in DNA binding of p65 subunit [26], is involved in IKK complex activation [43], and regulates NF-κB translocation between cytosol and nucleus [44–46]. In our model PARP-1 depletion did not prevent cytosol-to-nucleus translocation of NF-κB (p65) but NF-κB transcriptional activity was prevented. This finding underscores the fact that p65 nuclear translocation is not functionally equivalent to transcriptional activation. Our results point to the ability of PARP-1 to affect activity of other enzymes regulating NF-κB signaling. This finding is consistent with the model proposed (Fig. 3A), and underscores the importance of NAD+ in PARP-1 mediated NF-κB regulation.
The role of PARP-1 enzymatic activity in the regulation of NF-κB is also addressed by these studies. Astrocytes transfected with the catalytically inactive PARP-1 construct failed to show increased NF-κB transcriptional activity in the presence of MNNG, and wild type astrocytes treated with the PARP inhibitor, DPQ, similarly failed to show increased NF-κB transcriptional activity. These results agree with those of several prior studies that demonstrate that catalytic PARP-1 inhibition suppresses variety of NF-κB-mediated responses [17,20,22,23,45,47–51]. However, these results do not exclude the possibility that PARP-1 may also interact with NF-κB by non-enzymatic mechanisms. Strong evidence for this possibility exists [46,52], including evidence for a physical association between PARP-1 and the p65 subunit [26]. Such a mechanism has already been demonstrated with p53 [53], which shares intriguing similarities with PARP-1 mediated NF-κB regulation [45].
The present studies were performed under conditions of extensive PARP-1 activation, such as those that occur during oxidative stress or excitotoxicity. As such, they provide proof of principle that PARP-1 mediated depletion of NAD+, as occurs during genotoxic stress, is capable of influencing NF-κB transcriptional activity. However, it remains to be established whether this is an active mechanism during more physiological PARP-1 activation. The SIRT1Km for NAD+ is near the estimated cytosolic NAD+ concentration [54], suggesting that even modest levels of PARP-1 activation could influence NF-κB transcriptional activity by this mechanism. Recent work evaluating the effects of PARP-1 and SIRT1 on fat and muscle metabolism supports this contention [55].
5. Conclusions
PARP-1 activation can regulate NF-κB transcriptional activity by reducing NAD+ concentrations and thereby inhibiting deacetylation of p65. This observation identifies a novel mechanism by which DNA damage can promote NF-κB transcriptional activity.
Abbreviations
- BSS
balanced salt solution
- HDAC
histone deacetylases
- MNNG
N-methyl-N′-nitro-N-nitrosoguanidine
- PAR
poly(ADP-ribose)
- PARP-1
poly(ADP-ribose) polymerase-1
- SIRT1
sirtuin 1
References
- 1.McCool KW, Miyamoto S. DNA damage-dependent NF-kappaB activation: NEMO turns nuclear signaling inside out. Immunological reviews. 2012;246:311–326. doi: 10.1111/j.1600-065X.2012.01101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann A, Baltimore D. Circuitry of nuclear factor kappaB signaling. Immunol Rev. 2006;210:171–186. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- 3.Chen LF, Mu Y, Greene WC. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 2002;21:6539–6548. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. Embo J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashburner BP, Westerheide SD, Baldwin AS., Jr The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol Cell Biol. 2001;21:7065–7077. doi: 10.1128/MCB.21.20.7065-7077.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 7.Buck SW, Gallo CM, Smith JS. Diversity in the Sir2 family of protein deacetylases. J Leukoc Biol. 2004;75:939–950. doi: 10.1189/jlb.0903424. [DOI] [PubMed] [Google Scholar]
- 8.Rothgiesser KM, Erener S, Waibel S, Luscher B, Hottiger MO. SIRT2 regulates NF-kappaB dependent gene expression through deacetylation of p65 Lys310. J Cell Sci. 2010;123:4251–4258. doi: 10.1242/jcs.073783. [DOI] [PubMed] [Google Scholar]
- 9.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. The Biochemical journal. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 10.Davidovic L, Vodenicharov M, Affar EB, Poirier GG. Importance of poly(ADP-ribose) glycohydrolase in the control of poly(ADP-ribose) metabolism. Experimental cell research. 2001;268:7–13. doi: 10.1006/excr.2001.5263. [DOI] [PubMed] [Google Scholar]
- 11.Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Molecular cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji Y, Tulin AV. The roles of PARP1 in gene control and cell differentiation. Curr Opin Genet Dev. 2010;20:512–518. doi: 10.1016/j.gde.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger NA. Poly(ADP-ribose) in the cellular response to DNA damage. Radiation research. 1985;101:4–15. [PubMed] [Google Scholar]
- 14.Alano CC, Tran A, Tao R, Ying W, Karliner JS, Swanson RA. Differences among cell types in NAD(+) compartmentalization: a comparison of neurons, astrocytes, and cardiac myocytes. J Neurosci Res. 2007;85:3378–3385. doi: 10.1002/jnr.21479. [DOI] [PubMed] [Google Scholar]
- 15.Alano CC, Garnier P, Ying W, Higashi Y, Kauppinen TM, Swanson RA. NAD+ depletion is necessary and sufficient for poly(ADP-ribose) polymerase-1-mediated neuronal death. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2010;30:2967–2978. doi: 10.1523/JNEUROSCI.5552-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zong WX, Ditsworth D, Bauer DE, Wang ZQ, Thompson CB. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev. 2004;18:1272–1282. doi: 10.1101/gad.1199904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ha HC, Hester LD, Snyder SH. Poly(ADP-ribose) polymerase-1 dependence of stress-induced transcription factors and associated gene expression in glia. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:3270–3275. doi: 10.1073/pnas.052712399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol. 2008;20:294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang WJ, Alvarez-Gonzalez R. The sequence-specific DNA binding of NF-kappa B is reversibly regulated by the automodification reaction of poly (ADP-ribose) polymerase 1. The Journal of biological chemistry. 2001;276:47664–47670. doi: 10.1074/jbc.M104666200. [DOI] [PubMed] [Google Scholar]
- 20.Chiarugi A, Moskowitz MA. Poly(ADP-ribose) polymerase-1 activity promotes NF-kappaB-driven transcription and microglial activation: implication for neurodegenerative disorders. J Neurochem. 2003;85:306–317. doi: 10.1046/j.1471-4159.2003.01684.x. [DOI] [PubMed] [Google Scholar]
- 21.Hasko G, Mabley JG, Nemeth ZH, Pacher P, Deitch EA, Szabo C. Poly(ADP-ribose) polymerase is a regulator of chemokine production: relevance for the pathogenesis of shock and inflammation. Mol Med. 2002;8:283–289. [PMC free article] [PubMed] [Google Scholar]
- 22.Nakajima H, Nagaso H, Kakui N, Ishikawa M, Hiranuma T, Hoshiko S. Critical role of the automodification of poly(ADP-ribose) polymerase-1 in nuclear factor-kappaB-dependent gene expression in primary cultured mouse glial cells. The Journal of biological chemistry. 2004;279:42774–42786. doi: 10.1074/jbc.M407923200. [DOI] [PubMed] [Google Scholar]
- 23.Kauppinen TM, Swanson RA. Poly(ADP-ribose) polymerase-1 promotes microglial activation, proliferation, and matrix metalloproteinase-9-mediated neuron death. Journal of immunology. 2005;174:2288–2296. doi: 10.4049/jimmunol.174.4.2288. [DOI] [PubMed] [Google Scholar]
- 24.Chiarugi A. Inhibitors of poly(ADP-ribose) polymerase-1 suppress transcriptional activation in lymphocytes and ameliorate autoimmune encephalomyelitis in rats. Br J Pharmacol. 2002;137:761–770. doi: 10.1038/sj.bjp.0704934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hassa PO, Hottiger MO. The functional role of poly(ADP-ribose)polymerase 1 as novel coactivator of NF-kappaB in inflammatory disorders. Cell Mol Life Sci. 2002;59:1534–1553. doi: 10.1007/s00018-002-8527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hassa PO, Covic M, Hasan S, Imhof R, Hottiger MO. The enzymatic and DNA binding activity of PARP-1 are not required for NF-kappa B coactivator function. The Journal of biological chemistry. 2001;276:45588–45597. doi: 10.1074/jbc.M106528200. [DOI] [PubMed] [Google Scholar]
- 27.Kraus WL, Lis JT. PARP goes transcription. Cell. 2003;113:677–683. doi: 10.1016/s0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J. Are poly(ADP-ribosyl)ation by PARP-1 and deacetylation by Sir2 linked? Bioessays. 2003;25:808–814. doi: 10.1002/bies.10317. [DOI] [PubMed] [Google Scholar]
- 29.Swanson RA, Farrell K, Stein BA. Astrocyte energetics, function, and death under conditions of incomplete ischemia: a mechanism of glial death in the penumbra. Glia. 1997;21:142–153. doi: 10.1002/(sici)1098-1136(199709)21:1<142::aid-glia16>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 30.Kauppinen TM, Chan WY, Suh SW, Wiggins AK, Huang EJ, Swanson RA. Direct phosphorylation and regulation of poly(ADP-ribose) polymerase-1 by extracellular signal-regulated kinases 1/2. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7136–7141. doi: 10.1073/pnas.0508606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen J, Zhou Y, Mueller-Steiner S, Chen LF, Kwon H, Yi S, Mucke L, Gan L. SIRT1 protects against microglia-dependent amyloid-beta toxicity through inhibiting NF-kappaB signaling. The Journal of biological chemistry. 2005;280:40364–40374. doi: 10.1074/jbc.M509329200. [DOI] [PubMed] [Google Scholar]
- 32.Alano CC, Ying W, Swanson RA. Poly(ADP-ribose) polymerase-1-mediated cell death in astrocytes requires NAD+ depletion and mitochondrial permeability transition. The Journal of biological chemistry. 2004;279:18895–18902. doi: 10.1074/jbc.M313329200. [DOI] [PubMed] [Google Scholar]
- 33.Ying W, Sevigny MB, Chen Y, Swanson RA. Poly(ADP-ribose) glycohydrolase mediates oxidative and excitotoxic neuronal death. Proc Natl Acad Sci U S A. 2001;98:12227–12232. doi: 10.1073/pnas.211202598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagasawa K, Escartin C, Swanson RA. Astrocyte cultures exhibit P2X7 receptor channel opening in the absence of exogenous ligands. Glia. 2009;57:622–633. doi: 10.1002/glia.20791. [DOI] [PubMed] [Google Scholar]
- 35.Napper AD, Hixon J, McDonagh T, Keavey K, Pons JF, Barker J, Yau WT, Amouzegh P, Flegg A, Hamelin E, Thomas RJ, Kates M, Jones S, Navia MA, Saunders JO, DiStefano PS, Curtis R. Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J Med Chem. 2005;48:8045–8054. doi: 10.1021/jm050522v. [DOI] [PubMed] [Google Scholar]
- 36.Peck B, Chen CY, Ho KK, Di Fruscia P, Myatt SS, Coombes RC, Fuchter MJ, Hsiao CD, Lam EW. SIRT inhibitors induce cell death and p53 acetylation through targeting both SIRT1 and SIRT2. Mol Cancer Ther. 2010;9:844–855. doi: 10.1158/1535-7163.MCT-09-0971. [DOI] [PubMed] [Google Scholar]
- 37.Smith BC, Hallows WC, Denu JM. Mechanisms and molecular probes of sirtuins. Chem Biol. 2008;15:1002–1013. doi: 10.1016/j.chembiol.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- 39.Miyamoto S. Nuclear initiated NF-kappaB signaling: NEMO and ATM take center stage. Cell Res. 2011;21:116–130. doi: 10.1038/cr.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mabb AM, Wuerzberger-Davis SM, Miyamoto S. PIASy mediates NEMO sumoylation and NF-kappaB activation in response to genotoxic stress. Nat Cell Biol. 2006;8:986–993. doi: 10.1038/ncb1458. [DOI] [PubMed] [Google Scholar]
- 41.Wu ZH, Shi Y, Tibbetts RS, Miyamoto S. Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science. 2006;311:1141–1146. doi: 10.1126/science.1121513. [DOI] [PubMed] [Google Scholar]
- 42.Hassa PO, Buerki C, Lombardi C, Imhof R, Hottiger MO. Transcriptional coactivation of nuclear factor-kappaB-dependent gene expression by p300 is regulated by poly(ADP)-ribose polymerase-1. The Journal of biological chemistry. 2003;278:45145–45153. doi: 10.1074/jbc.M307957200. [DOI] [PubMed] [Google Scholar]
- 43.Stilmann M, Hinz M, Arslan SC, Zimmer A, Schreiber V, Scheidereit C. A nuclear poly(ADP-ribose)-dependent signalosome confers DNA damage-induced IkappaB kinase activation. Molecular cell. 2009;36:365–378. doi: 10.1016/j.molcel.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 44.Oumouna-Benachour K, Hans CP, Suzuki Y, Naura A, Datta R, Belmadani S, Fallon K, Woods C, Boulares AH. Poly(ADP-ribose) polymerase inhibition reduces atherosclerotic plaque size and promotes factors of plaque stability in apolipoprotein E-deficient mice: effects on macrophage recruitment, nuclear factor-kappaB nuclear translocation, and foam cell death. Circulation. 2007;115:2442–2450. doi: 10.1161/CIRCULATIONAHA.106.668756. [DOI] [PubMed] [Google Scholar]
- 45.Zerfaoui M, Errami Y, Naura AS, Suzuki Y, Kim H, Ju J, Liu T, Hans CP, Kim JG, Abd Elmageed ZY, Koochekpour S, Catling A, Boulares AH. Poly(ADP-ribose) polymerase-1 is a determining factor in Crm1-mediated nuclear export and retention of p65 NF-kappa B upon TLR4 stimulation. Journal of immunology. 2010;185:1894–1902. doi: 10.4049/jimmunol.1000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zerfaoui M, Suzuki Y, Naura AS, Hans CP, Nichols C, Boulares AH. Nuclear translocation of p65 NF-kappaB is sufficient for VCAM-1, but not ICAM-1, expression in TNF-stimulated smooth muscle cells: differential requirement for PARP-1 expression and interaction. Cellular signalling. 2008;20:186–194. doi: 10.1016/j.cellsig.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kauppinen TM, Higashi Y, Suh SW, Escartin C, Nagasawa K, Swanson RA. Zinc triggers microglial activation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:5827–5835. doi: 10.1523/JNEUROSCI.1236-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kauppinen TM, Suh SW, Higashi Y, Berman AE, Escartin C, Won SJ, Wang C, Cho SH, Gan L, Swanson RA. Poly(ADP-ribose)polymerase-1 modulates microglial responses to amyloid beta. Journal of neuroinflammation. 2011;8:152. doi: 10.1186/1742-2094-8-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCourtie AS, Farivar AS, Woolley SM, Merry HE, Wolf PS, Szabo C, Mulligan MS. Poly (ADP) ribose synthetase inhibition in alveolar macrophages undergoing hypoxia and reoxygenation. Experimental and molecular pathology. 2008;84:141–144. doi: 10.1016/j.yexmp.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 50.Ullrich O, Diestel A, Eyupoglu IY, Nitsch R. Regulation of microglial expression of integrins by poly(ADP-ribose) polymerase-1. Nature cell biology. 2001;3:1035–1042. doi: 10.1038/ncb1201-1035. [DOI] [PubMed] [Google Scholar]
- 51.Veuger SJ, Hunter JE, Durkacz BW. Ionizing radiation-induced NF-kappaB activation requires PARP-1 function to confer radio resistance. Oncogene. 2009;28:832–842. doi: 10.1038/onc.2008.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hassa PO, Haenni SS, Buerki C, Meier NI, Lane WS, Owen H, Gersbach M, Imhof R, Hottiger MO. Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-kappaB-dependent transcription. J Biol Chem. 2005;280:40450–40464. doi: 10.1074/jbc.M507553200. [DOI] [PubMed] [Google Scholar]
- 53.Kanai M, Hanashiro K, Kim SH, Hanai S, Boulares AH, Miwa M, Fukasawa K. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nature cell biology. 2007;9:1175–1183. doi: 10.1038/ncb1638. [DOI] [PubMed] [Google Scholar]
- 54.Houtkooper RH, Canto C, Wanders RJ, Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev. 2010;31:194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, Schoonjans K, Schreiber V, Sauve AA, Menissier-de Murcia J, Auwerx J. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 2011;13:461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]