Immunoglobulin heavy chain (IGH) gene-associated translocations occur frequently in different subtypes of B-cell lymphomas.1 These translocations result in deregulated expression of partner genes and play pivotal roles in the pathogenesis of lymphomas. Molecular characterization of IGH-associated translocation breakpoints has been instrumental in the identification of genes important to the development of normal and malignant B cells.
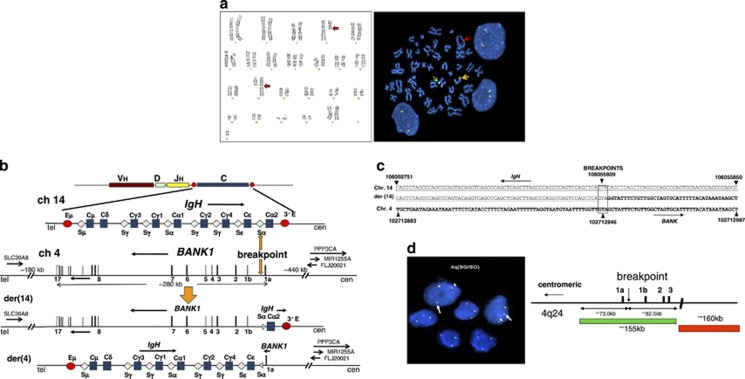
We identified a novel balanced reciprocal translocation, t(4;14)(q24;q32), by conventional karyotyping (Figure 1a) in a case of post-transplant lymphoproliferative disorder (PTLD). The patient underwent stem cell transplantation for classical Hodgkin's lymphoma and developed a gastric mass that was diagnosed as Epstein–Barr virus (EBV)-positive polymorphic PTLD with monomorphic areas. Fluorescence in situ hybridization (FISH) with IGH dual-color probe demonstrated that the translocation involved the IGH locus (Figure 1b). The breakpoint sequence was cloned by long-distance inverse PCR2, 3, 4 and sequence analysis identified the partner gene as BANK1 (B-cell scaffold protein with ankyrin repeats 1) (Figure 1c). The breakpoints were located in the switch region (Sα) proximal to the Cα2 gene segment near the 3′ end of IGH, and in intron 1 of the BANK1 gene ∼830 bp telomeric to the 3′ end of exon 1a. The juxtaposed IGH and BANK1 have opposite transcription orientations with a head-to-head configuration.
Figure 1.
Identification of a novel translocation t(4;14)(q24;q32) and identification of BANK1 as an IGH-associated partner. (a, left) Karyotype showing the t(4;14)(q23;q32) translocation and trisomy 9. (a, right) Metaphase and interphase FISH assay showing rearrangement of the IGH gene. Fusion (yellow) signal indicates the normal IGH allele and the two arrows show the split signals of IGH on the der(4) and der(14) indicating the rearrangement of the gene. (b) The structures of the germline IGH and BANK1 alleles, as well as the translocated IGH/BANK1 allele in der(14) and der(4), are depicted. Vertical boxes denote exons of the BANK1 gene. Sequence analysis of the breakpoint region showed that the BANK1 gene is juxtaposed to IGH. The breakpoint is located at the switch (S) region proximal to the Cα2 gene segment at the 3′ portion of the IGH gene, and in intron 1 of the BANK1 gene, ∼830 bp telomeric to the 3′ end of exon 1a. IGH and BANK1 are arranged in a head-to-head transcription orientation. This IGH/BANK1 translocation results in removal of the major BANK1 promoter from the rest of the gene. Four neighboring coding genes (PPP3CA, MIR1255A, FLJ20021 and SLC39A8) are shown by horizontal arrows, the direction and length of which represent the orientation and size of the transcription units. The approximate genomic distance between the 5′ or 3′ end of BANK1 and the closest exon of the neighboring genes are indicated. C, constant; cen, centromeric; E, enhancer; S, switch; tel, telomeric. (c) The sequence alignment of the translocation breakpoint is shown. IGH sequence is underlined and BANK1 sequence is in bold. The breakpoint is boxed (the exact nucleotide location cannot be determined). The translocated BANK1 sequence shows a C to G mutation (in italics) in very close vicinity to the breakpoint. (d) Interphase nuclei were hybridized with BAC RP11-96J17 (green, centromeric) and BAC RP11-138I19 (red, telomeric) in 4q24 to confirm BANK1 rearrangement. The arrow indicates the green signal present on derivative chromosome 4 generated by a split within RP11-96J17. The arrowhead points to the signal generated from the remainder of BAC RP11-96J17 and the adjacent RP11-138I19, present on derivative chromosome 14. The yellow signal (not marked) in the cells corresponds to the intact, untranslocated BANK1 allele. Only exons 1a, 1b, 2 and 3 of BANK1 are shown.
IGH/BANK1 translocation was further confirmed by FISH using double-color break-apart assay with two BAC probes, RP11-96J17 and RP11-138I19, performed on cells isolated from the PTLD case (Figure 1d). We screened an additional 68 diffuse large B-cell lymphomas (DLBCLs) and 15 PTLDs by FISH using the same probes. None of them were found to harbor any translocations affecting the BANK1 locus (data not shown), suggesting that translocation involving BANK1 is a rare event.
BANK1 acts as an important adaptor molecular to link B-cell receptor (BCR)-mediated signaling to the generation of intracellular secondary messengers in B cells. Phosphorylated BANK1 enhanced BCR-induced calcium mobilization by binding to LYN, thus promoting LYN-mediated tyrosine phosphorylation of 1,4,5-triphosphate receptor (IP3R) that leads to release of calcium from intracellular stores.5 BANK1 has recently been shown to physically interact with B-cell lymphoid kinase (BLK),6 another B-cell specific Src family kinase, and phospholipase C γ2 (PLCg2),7 a major molecular switch in B-cell signal transduction, upon BCR engagement. In addition to an enhancing function in BCR signaling, BANK1 may have an inhibitory role in the CD40 signaling pathway. Studies on BANK1-deficient mice suggested that BANK1 attenuates CD40-mediated AKT activation to prevent hyperactive B-cell responses.8 BANK1 single-nucleotide polymorphic variants have also been implicated in autoimmune diseases such as systemic lupus erythematosus.9 Although evidence of an important function of BANK1 in B-cell physiology is emerging, a role for BANK1 in lymphoma development has not been reported previously.
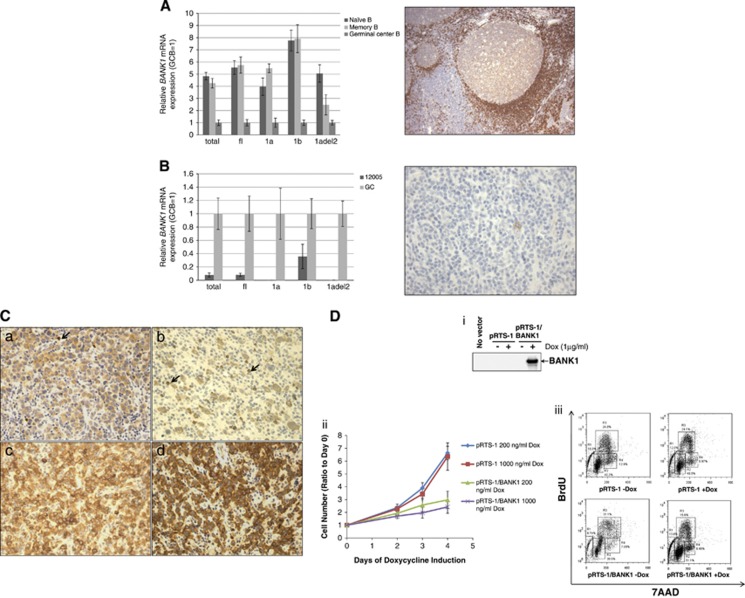
BANK1 encodes three RNA isoforms, 1a, 1b and Δ2, with 1a being the major isoform.5, 9 The two full-length isoforms, 1a and 1b, are derived from alternative promoter usage that results in the generation of exon 1a or 1b. The Δ2 isoform contains an in-frame deletion of exon 2. BANK1 is mainly expressed in immature and mature B cells with very low expression in pro B cells and absent expression in T cells. To determine whether BANK1 expression is differentially expressed during B-cell differentiation, B-cell subsets including naive, germinal center (GC) and memory B cells were isolated and BANK1 mRNA expression was determined by real-time quantitative reverse transcriptase-PCR (qRT-PCR). All BANK1 isoforms were expressed at higher levels (2.5–8 fold) in the naive and memory B cells compared with the GC B cells (Figure 2A). In line with this, immunohistochemistry (IHC) on reactive tonsils showed a distinctly lower level of expression in the GCs and higher levels in the mantle zones (Figure 2A). These findings imply a physiological downregulation of BANK1 expression during GC transit.
Figure 2.
IgH translocation of BANK1 resulted in its downregulation and may contribute to lymphoma development by promoting cell proliferation. (A, left) Levels of different BANK1 mRNA isoforms: total, full-length (fl), isoform 1a (1a), isoform 1b (1b) and Δ2 isoform (1adel2) were measured by qRT-PCR and expressed relative to the normal GC B cells. BANK1 mRNA expression in naive and memory B cells is significantly higher compared with that in GC B cells. (A, right) IHC was performed on paraffin sections of a reactive tonsil using a polyclonal antibody against BANK1. BANK1 is weakly expressed in GC but strongly expressed in the mantle zones (MZ). T cells are negative for BANK1. (B, left) BANK1 mRNA isoforms were determined in the lymphoma case (12005) harboring BANK1 translocation by qRT-PCR as described in (A). BANK1 expression is decreased compared with GC B-cells. In particular, isoforms 1a as well as Δ2 are virtually absent, mostly likely because of dissociation of exon 1-initiated promoter from the rest of the BANK1 gene in the translocated allele and silencing of the nontranslocated allele. (B, right) IHC was performed on paraffin sections of the lymphoma case (12005) using a polyclonal antibody against BANK1. The tumor cells are negative for BANK1. A rare BANK1-positive tumor cell is shown as an internal control. (C) Variable expressions of BANK1 in PTLD and DLBCL. (a) Weak expression in a monomorphic PTLD; (b) weak expression in a polymorphic PTLD; (c) moderate expression in a monomorphic PTLD; (d) strong expression in a DLBCL-NOS. Arrows indicate normal reactive B cells that show distinctly higher BANK1 expression compared with the weak BANK1-expressing lymphoma cells. (D) BANK1 has a negative effect on cell cycle. (Di) BC3 primary effusion lymphoma (PEL) cells stably transfected with pRTS-1/BANK1 expressed BANK1 in the presence of doxycycline (Dox). (Dii) Overexpression of BANK1 in BC3 cells upon Dox addition resulted in a slower growth rate compared with controls. (Diii) Bromodeoxyuridine (BrdU) incorporation assay showed that BANK1 caused a decrease in cell proliferation by 50%.
To determine the effect of the t(4;14) translocation on BANK1 expression, real-time qRT-PCR was performed to quantitate total BANK1, total full-length BANK1 (that is, 1a plus 1b) and the three individual BANK1 isoforms (1a, 1b and Δ2) mRNA levels. Interestingly, we did not detect evidence of activation of BANK1 expression by the IGH translocation. Instead, we observed downregulation of BANK1 expression as a result of the translocation. The expression for total and full-length BANK1 mRNAs was greatly reduced (>90%) in the PTLD case compared with GC B cells (Figure 2B). BANK1 1a and Δ2 transcript isoforms, both initiated from the exon 1a promoter, were only barely detectable. The level of BANK1 1b transcript isoform was ∼40% of the normal GC B cells. These results are consistent with the loss of expression of the major BANK1 1a isoform because of dissociation of exon 1 from the rest of the BANK1 gene, and concurrent silencing of the nontranslocated BANK1 allele. The latter allele is grossly intact, as implied by FISH, and we have excluded the possibility of hypermethylation (data now shown), suggesting other unknown mechanisms are responsible for repressing BANK1 transcription from the nontranslocated allele. It is noteworthy that although the translocated IGH locus is in a favorable position to do so, we did not observe activation of transcription initiated from the adjacent alternative BANK1 exon 1b promoter or from an intronic cryptic promoter. IHC confirmed the lack of BANK1 in the tumor cells (Figure 2B). In contrast, we detected variable BANK1 expression by IHC in other (8 of 8) PTLDs, 4 of 4 EBV-positive DLBCLs of the elderly and 48 of 54 DLBCL, not otherwise specified (DLBCL-NOS), all of which did not harbor BANK1 rearrangements (Supplementary Table 1 and Figure 2C). Thus, we believe that the main consequence of IGH translocation to the BANK1 locus is most likely downregulation of BANK1 through dissociation of the major promoter. Of the 54 DLBCLs, 6 also lacked BANK1 expression, suggesting the presence of alternative mechanism(s) of BANK1 downregulation in B-cell lymphomas.
Target gene downregulation is a highly unusual consequence of IGH translocations, as they have been shown to lead to overexpression of the target gene in almost all circumstances. IGH-mediated gene inactivation has been implicated only once before in hematopoietic malignancies for the t(14;16)(q32;q23) translocation that is found in <5% of plasma cell myelomas.10 All the breakpoints of this translocation are located in the last intron of the tumor suppressor gene WWOX (WW domain containing oxidoreductase).11, 12 This intron is nearly 1-Mb long and contains the fragile site FRA16D.13 IGH in this translocation is thought to have a dual pathogenetic function. It is in a position to disrupt the WWOX gene and prevent the generation of a full-length protein and, at the same time, causes overexpression of the neighboring MAF proto-oncogene located telomeric to WWOX via the IGH enhancer.14
We have also considered the remote possibility that the IGH translocation activates a gene in the vicinity of BANK1. Three coding genes, PPP3CA, MIR1255A and FLJ20021, reside ∼443 kb centromeric to the BANK1 breakpoint, and one coding gene, SLC39A8, is located ∼460 kb telomeric to the BANK1 breakpoint (http://www.ncbi.nlm.nih.gov/gene/55024) (Figure 1b). PPP3CA, MIR1255A and SLC39A8 have the same orientation as the translocated IGH, whereas FLJ20021 has the opposite orientation to the translocated IGH. It is unlikely that the translocated IGH on der(4) drives expression of any of these three genes using the germline I transcript promoter because of their considerable distance from the IgH and/or incompatible relative transcriptional orientation. Furthermore, the configurations of the translocated IGH relative to these genes, either upstream/same orientations for PPP3CA and MIR1255A or downstream/opposite orientations for FLJ20021, are not considered favorable for an enhancer activation mechanism. Although IgH is located downstream of SL39A8 in the same orientation and can conceivably activate the latter through its 3′ enhancer on the der(14) chromosome, SLC39A8 mRNA level in this case was not significantly higher compared with EBV-positive B-cell lymphomas without the IGH/BANK1 translocation (data not shown). Thus, there is no definitive evidence for activation of SLC39A8 by the translocated IGH locus.
The inactivation of BANK1 in a IGH-associated translocation suggests that abnormal downregulation of BANK1 can promote lymphoma development. To show directly that BANK1 has a negative inhibitory effect on lymphomagenesis, we transfected BC3 primary effusion lymphoma cells with an all-in-one doxycycline-inducible expression vector pRTS-1,15 carrying BANK1 complementary DNA to establish an inducible BANK1-expressing cell line. The BC3 cell line was chosen because it has no detectable endogenous BANK1. Addition of doxycycline resulted in efficient induction of BANK1 and a significant retardation in the rate of cell number increase compared with the control transfectants (Figure 2Di and ii). Bromodeoxyuridine staining demonstrated a decreased incorporation from 31.1 to 15.6% upon BANK1 expression, implying a negative effect of BANK1 on cell cycle (Figure 2Diii).
This study identified BANK1 as a novel IGH translocation partner and demonstrated a rare mechanism of gene inactivation by IGH through promoter dissociation. Our initial functional studies demonstrating a negative effect of BANK1 on cell proliferation suggest that BANK1 inactivation may contribute to lymphoma by facilitating cell proliferation. Unlike the myeloma-associated t(14;16)(q32;q23), which has a dual impact on two neighboring genes (WWOX and MAF), the impact of (4;14) translocation described here appears to be restricted to BANK1. Our study underscores the utility of cloning IGH-associated translocations, even the rare ones, to identify genes with important functions in normal and neoplastic B-cell biology. We provide, for the first time, genetic and functional data suggesting a possible tumor suppressor role of BANK1 in B-cell lymphomagenesis. Further studies are warranted to further investigate the involvement of BANK1 in mature B-cell malignancies.
Acknowledgments
This work was supported by a generous departmental grant to WT.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Blood Cancer Journal website (http://www.nature.com/bcj)
Supplementary Material
References
- Robbiani DF, Nussenzweig MC. Chromosome translocation, B cell lymphoma, and activation-induced cytidine deaminase. Annu Rev Pathol. 2013;8:79–103. doi: 10.1146/annurev-pathol-020712-164004. [DOI] [PubMed] [Google Scholar]
- Karran EL, Sonoki T, Dyer MJS. Cloning of immunoglobulin chromosomal translocations by long-distance inverse polymerase chain reaction. Methods Mol Med. 2005;115:217–230. doi: 10.1385/1-59259-936-2:217. [DOI] [PubMed] [Google Scholar]
- Willis TG, Jadayel DM, Coignet LJ, Abdul-Rauf M, Treleaven JG, Catovsky D, et al. Rapid molecular cloning of rearrangements of the IGHJ locus using long-distance inverse polymerase chain reaction. Blood. 1997;90:2456–2464. [PubMed] [Google Scholar]
- Sonoki T, Willis TG, Oscier DG, Karran EL, Siebert R, Dyer MJ. Rapid amplification of immunoglobulin heavy chain switch (IGHS) translocation breakpoints using long-distance inverse PCR. Leukemia. 2004;18:2026–2031. doi: 10.1038/sj.leu.2403500. [DOI] [PubMed] [Google Scholar]
- Yokoyama K, I-h SuIh, Tezuka T, Yasuda T, Mikoshiba K, Tarakhovsky A, et al. BANK regulates BCR-induced calcium mobilization by promoting tyrosine phosphorylation of IP(3) receptor. EMBO J. 2002;21:83–92. doi: 10.1093/emboj/21.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo-Lopez C, Delgado-Vega AM, Wojcik J, Kozyrev SV, Thavathiru E, Wu YY, et al. Genetic and physical interaction of the B-cell systemic lupus erythematosus-associated genes BANK1 and BLK. Ann Rheum Dis. 2012;71:136–142. doi: 10.1136/annrheumdis-2011-200085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal-Quiros M, Wu YY, Alarcon-Riquelme ME, Castillejo-Lopez C. BANK1 and BLK act through phospholipase C gamma 2 in B-cell signaling. PLoS One. 2013;8:e59842. doi: 10.1371/journal.pone.0059842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiba Y, Yamazaki T, Okada T, Gotoh K, Sanjo H, Ogata M, et al. BANK negatively regulates Akt activation and subsequent B cell responses. Immunity. 2006;24:259–268. doi: 10.1016/j.immuni.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Kozyrev SV, Abelson A-K, Wojcik J, Zaghlool A, MVPL Reddy, Sanchez E, et al. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:211–216. doi: 10.1038/ng.79. [DOI] [PubMed] [Google Scholar]
- Avet-Loiseau H, Malard F, Campion L, Magrangeas F, Sebban C, Lioure B, et al. Translocation t(14;16) and multiple myeloma: is it really an independent prognostic factor. Blood. 2011;117:2009–2011. doi: 10.1182/blood-2010-07-295105. [DOI] [PubMed] [Google Scholar]
- Walker BA, Wardell CP, Johnson DC, Kaiser MF, Begum DB, Dahir NB, et al. Characterization of IGH locus breakpoints in multiple myeloma indicates a subset of translocations appear to occur in pregerminal center B cells. Blood. 2013;121:3413–3419. doi: 10.1182/blood-2012-12-471888. [DOI] [PubMed] [Google Scholar]
- Paige AJ, Taylor KJ, Taylor C, Hillier SG, Farrington S, Scott D, et al. WWOX: a candidate tumor suppressor gene involved in multiple tumor types. Proc Natl Acad Sci USA. 2001;98:11417–11422. doi: 10.1073/pnas.191175898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krummel KA, Roberts LR, Kawakami M, Glover TW, Smith DI. The characterization of the common fragile site FRA16D and its involvement in multiple myeloma translocations. Genomics. 2000;69:37–46. doi: 10.1006/geno.2000.6321. [DOI] [PubMed] [Google Scholar]
- Chesi M, Bergsagel PL, Shonukan OO, Martelli ML, Brents LA, Chen T, et al. Frequent dysregulation of the c-maf proto-oncogene at 16q23 by translocation to an Ig locus in multiple myeloma. Blood. 1998;91:4457–4463. [PubMed] [Google Scholar]
- Bornkamm GW, Berens C, Kuklik-Roos C, Bechet J-M, Laux G, Bachl J, et al. Stringent doxycycline-dependent control of gene activities using an episomal one-vector system. Nucleic Acids Res. 2005;33:e137. doi: 10.1093/nar/gni137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.