Abstract
Objective
Studies of neutralizing antibodies in HIV-1 infected individuals provide insights into the quality of the response that should be possible to elicit with vaccines and ways to design effective immunogens. Some individuals make high titres of exceptional broadly reactive neutralizing antibodies that are of particular interest; however, more modest responses may be a reasonable goal for vaccines. We performed a large cross-sectional study to determine the spectrum of neutralization potency and breadth that is seen during chronic HIV-1 infection.
Design
Neutralization potency and breadth were assessed with genetically and geographically diverse panels of 205 chronic HIV-1 sera and 219 Env-pseudotyped viruses representing all major genetic subtypes of HIV-1.
Methods
Neutralization was measured by using Tat-regulated luciferase reporter gene expression in TZM-bl cells. Serum-neutralizing activity was compared with a diverse set of human mAbs that are widely considered to be broadly neutralizing.
Results
We observed a uniform continuum of responses, with most sera displaying some level of cross-neutralization, and approximately 50% of sera neutralizing more than 50% of viruses. Titres of neutralization (potency) were highly correlated with breadth. Many sera had breadth comparable to several of the less potent broadly neutralizing human mAbs.
Conclusion
These results help clarify the spectrum of serum-neutralizing activity induced by HIV-1 infection and that should be possible to elicit with vaccines. Importantly, most people appear capable of making low to moderate titres of broadly neutralizing antibodies. Additional studies of these relatively common responses might provide insights for practical and feasible vaccine designs.
Keywords: HIV-1, immunity, neutralizing antibodies, serum, vaccines
Introduction
Efforts to elicit broadly neutralizing antibodies (bnAbs) with HIV-1 vaccine immunogens face daunting challenges, owing to extraordinary genetic diversity of the gp120 and gp41 viral envelope glycoproteins (Env) and the poor immunogenicity of conserved Env epitopes [1]. For many years, only a small number of bnAbs were known; these recognized the CD4-binding site (CD4bs) (b12), a cluster of N-linked glycans on gp120 (2G12) and three adjacent epitopes in the membrane proximal external region (MPER) of gp41 (2F5, 4E10, Z13) [2–4]. Cautious optimism arose when it was shown that sera from some antiretroviral-naive chronically HIV-1 infected individuals neutralized a majority of viruses tested [5–9], suggesting it should be possible to design vaccines that elicit similar responses [10]. Since then, studies of infected individuals who possess the best bnAb activity have yielded a number of important advances. A plethora of new bnAbs were isolated and characterized, and together, these categories of antibodies explain much of the bnAb activity in HIV-1 sera. These new bnAbs permitted detailed analyses of additional epitopes in the CD4bs and MPER, as well as novel epitopes in the V1V2-glycan, V3/C3-glycan and C2-glycan regions of gp120 [11–13]. Some of the newer bnAbs are much better neutralizers than the earlier ones and are gaining more attention, although any of these bnAbs would be desirable to induce with HIV-1 vaccines.
Notably, the best bnAbs exhibit one or more unusual features, including high levels of somatic mutation in the complementary determining regions (CDRs) and framework regions (FWRs), long heavy-chain third CDRs (HCDR3 s) and polyreactivity, all of which may contribute to the relatively uncommon occurrence of such superior bnAbs during natural infection [12,14,15]. These features may also pose substantial potential barriers to the elicitation of similar bnAbs by vaccination. A more plausible vaccine approach might aim to elicit neutralizing antibodies that exhibit more moderate breadth and potency but are easier to induce because of their less restrictive maturation requirements. Polyvalent immunogens that induce several neutralizing antibody specificities, each with moderate breadth, could be used to generate a protective antibody response with increased breadth [16].
The feasibility of eliciting cross-reactive neutralizing antibodies via immunization would be better understood if more accurate information was available on the breadth of neutralization that is seen in the majority of infected individuals. Previous studies have estimated that only 10–25% of people can make antibodies that cross-neutralize many of the HIV-1 strains tested [5–9]; however, cross-reactive neutralizing antibodies with more modest potency and breadth may be a reasonable goal for HIV-1 vaccines. Due to the limited size and diversity of previous datasets, the level of breadth that is seen most often during chronic infection has remained unclear. We therefore assessed the neutralization profiles of a large multisubtyped panel of chronic sera from HIV-1 infected individuals assayed against an equally large multisubtype panel of Env-pseudotyped viruses. Our results demonstrate that broadly neutralizing antibody responses of moderate potency are a common feature of HIV-1 infection.
Materials and methods
HIV-1 positive sera
Serum samples were obtained from chronically infected individuals who were antiretroviral drug-naive and infected with HIV-1 subtypes A (n = 8), B (n = 59), C (n = 58), D (n = 3), CRF01-AE (n = 15), CRF07_BC (n = 16), CRF02_AG (n = 2), CRF10_CD (n = 1), AC (n = 4), AD (n = 3) and ABCD (n = 2); 34 were infected with subtypes that could not be determined due to low levels of plasma viraemia (Supplementary Tables 1–3, http://links.lww.com/QAD/A426). When possible, sets of sera collected from particular sites were prescreened to exclude sera with little or no neutralizing activity, as at the time of collection (prior to 2009), there was a concern that such sera might be frequent and would be noninformative. In such cases, six Env-pseudotyped viruses commonly used in neutralization assay panels, mostly from clades B and C, were used to select 88 sera after prescreening for neutralization activity. The other 117 sera were collected from sites where no prescreening was performed. Prescreening would bias the data set towards individuals with higher levels of neutralizing activity, and one of our goals was to estimate the frequency of individuals with varying levels of neutralizing activity in the infected population, including low levels. Thus, for such estimates, we excluded prescreened sample sets. However, we were also interested in exploring the relationship between serum potency and breadth, and for this comparison, the 88 prescreened samples were a useful addition. HIV-1 genetic subtypes in the samples were determined by single genome amplification and sequencing of a single serum gp160 gene as described [17].
Env-pseudotyped viruses
Molecularly cloned env-rev cassettes containing full-length gp160 were used to produce Env-pseudotyped viruses by cotransfection with an Env-defective backbone plasmid (pSG3Δenv) in 293T cells as described [17]. The panel of 219 Env-pseudotyped viruses included subtypes A (n = 10), B (n = 54),C (n = 67), D (n = 5), G (n = 8), CRF01_AE (n = 21), CRF02_AG (n = 16), CRF06 (n = 1), CRF07_BC (n = 14), AC (n = 6), AD (n = 5), ACD (n = 1), BC (n = 4), BG (n = 1) and CD (n = 6). Assay stocks of Env-pseudotyped viruses were titrated in TZM-bl cells. None of the viruses included in this panel were among the readily neutralizing tier 1 category [18].
Neutralization assay
Neutralization was measured in 96-well culture plates by using Tat-regulated firefly luciferase (Luc) reporter gene expression to quantify reductions in virus infection in TZM-bl [17]. Heat-inactivated (56°C, 1 h) serum samples were assayed at three-fold dilutions starting at 1 : 20. Human mAbs were assayed at three-fold dilutions starting at 25–50µg/ml. Neutralization titres are either the serum dilution (ID50) or mAb concentration (IC50) at which relative luminescence units (RLUs) were reduced by 50% compared with RLU in virus control wells after subtraction of background RLU in cell control wells. Titres that reduced 80% of the virus signal were also recorded.
Data analyses/statistical methods
R (version 2.15.2) was used to analyse and visualize neutralization data. Spearman’s nonparametric correlation test was used with α equal to 0.05 on each of two tests of independent data. Cumulative distribution of 117 serum neutralization breadths were compared with a uniform distribution with the one-sample Kolmogorov test, though tied values prevent computing an exact P value. Serum percentile scores were computed as the complement of the cumulative distribution function from neutralization breadths of 117 nonprescreened sera (Serum percentile=100% − Cumulative breadth). All analyses were repeated with neutralization data for 80% neutralization titres and, as expected, this more stringent criterion yielded fewer quantified results within current assay sensitivity limits (57.2% of 44 758 assays quantified by ID50s versus 23.2% from ID80s). When most assay results fell below the sensitivity limit, the censored data caused intractable difficulties for statistical inference when using the ID80s. To mitigate this, and because we think they are most relevant to the discussion at hand, we emphasize analysis of 50% neutralization titres, but still report salient 80% neutralization results in Table 1.
Table 1.
Neutralization breadth, and proportion of sera with comparable breadth, for 10 human bnAbs tested against a multisubtype panel of 119 tier 2 Env-pseudotyped viruses.
| 10E8 | 4E10 | VRC01 | PG9 | PGT145 | PGT128 | 2F5 | CH01 | b12 | 2G12 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 50% neutralization | ||||||||||
| Breadth (50)a | 97.3 | 96.6 | 89.2 | 86.2 | 75.7 | 59.1 | 58.0 | 49.5 | 29.4 | 20.2 |
| Serum %ileb | 2.6 | 3.4 | 9.4 | 12.0 | 19.7 | 33.3 | 37.6 | 50.4 | 77.8 | 83.8 |
| Breadth (5)a | 92.8 | 42.9 | 82.9 | 80.2 | 62.6 | 53.9 | 26.1 | 37.8 | 7.6 | 11.8 |
| Serum %ileb | 6.0 | 61.5 | 12.8 | 14.5 | 32.5 | 41.0 | 78.6 | 67.5 | 97.4 | 94.0 |
| Breadth (1)a | 59.5 | 11.8 | 58.6 | 69.8 | 56.5 | 48.7 | 5.9 | 16.2 | 0.8 | 3.4 |
| Serum %ileb | 33.3 | 94.0 | 34.2 | 27.4 | 39.3 | 51.3 | 98.3 | 90.6 | 100 | 100 |
| 80% neutralization | ||||||||||
| Breadth (50)a | 97.2 | 73.9 | 87.7 | 80.6 | 63.0 | 55.6 | 42.9 | 13.5 | 16.0 | 12.6 |
| Serum %ileb | 0 | 3.4 | 0.9 | 1.7 | 5.1 | 6.8 | 12.8 | 39.3 | 37.6 | 41.9 |
| Breadth (5)a | 65.1 | 8.4 | 69.8 | 70.8 | 47.2 | 48.6 | 5.0 | 6.7 | 1.7 | 3.4 |
| Serum %ileb | 5.1 | 51.3 | 5.1 | 4.3 | 11.1 | 10.3 | 65.8 | 61.5 | 84.6 | 73.5 |
| Breadth (1)a | 15.1 | 0 | 36.8 | 55.6 | 41.7 | 45.8 | 0 | 3.8 | 0 | 0 |
| Serum %ileb | 37.6 | 94.0 | 13.7 | 6.8 | 13.7 | 11.1 | 94.0 | 71.8 | 94.0 | 94.0 |
bnAbs, broadly neutralizing antibodies.
Percentage of bnAb IC50 and IC80 assay results below the concentration (µg/ml) shown in parentheses.
Percentage of nonprescreened sera with equal or greater breadth than bnAbs (shown in boldface type).
Study approval
This study utilized preexisting, de-identified specimens and was conducted under the approval of the Duke University Medical Center Institutional Review Board. The data were analysed anonymously.
Results and discussion
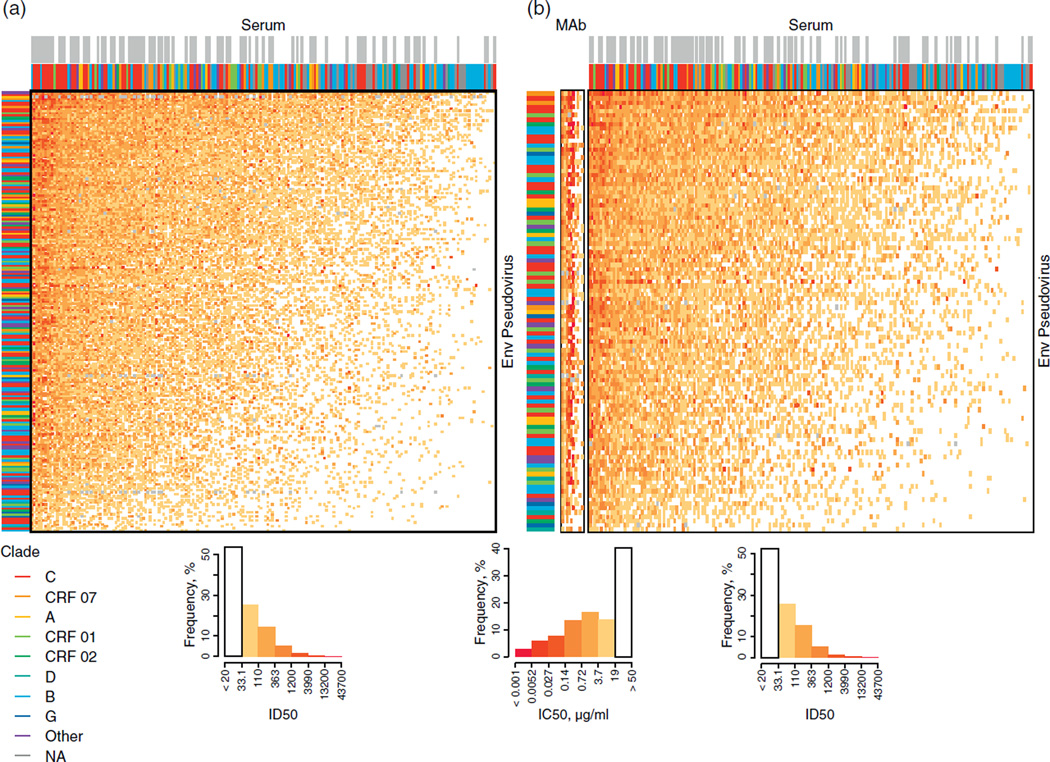
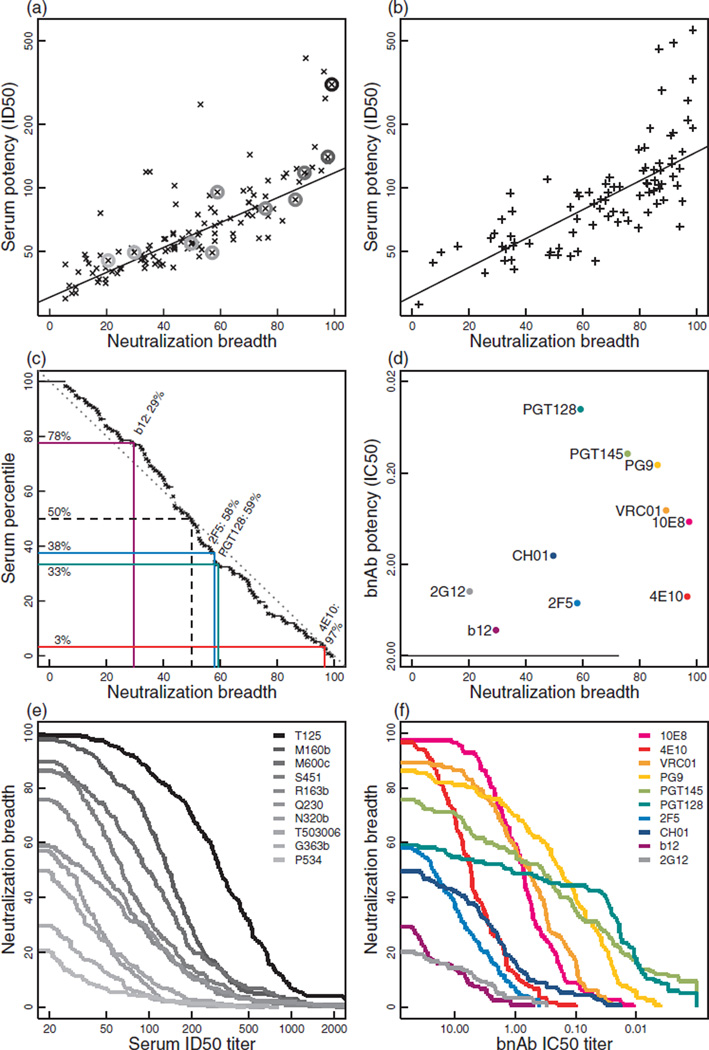
The neutralization profiles of 205 chronic sera from HIV-1 infected individuals were assessed against a panel of 219 Env-pseudotyped viruses that exhibit a tier 2 neutralization phenotype resembling most circulating strains [18] (Fig. 1). Both the viruses and sera were sampled globally from diverse geographic locations and together represented five HIV-1 M-group subtypes (A, B, C, D and G), and three common circulating recombinant forms (CRFs 01, 02 and 07), as well as additional unique recombinants (Supplementary Tables 1–3, http://links.lww.com/QAD/A426). Neutralization was assessed with molecularly cloned Env-pseudotyped viruses expressing the entire gp160 of the designated strain. Among the serum samples, 117 out of 205 were selected randomly from chronically infected individuals, and these were analysed separately in Fig. 2a, enabling an assessment of the range of breadth of neutralizing activity found among randomly selected HIV-1 chronically infected individuals. Essentially, all sera could neutralize at least a few of the tier 2 pseudoviruses, and most had activity against a substantial proportion of the diverse Envs that were tested in our panel (Fig. 1a, Fig. 2a). In hindsight, because of the unexpected high frequency of positive neutralization seen in the randomly selected group, prescreening was not necessary. The cumulative distribution of neutralization breadths for the 117 randomly selected sera in this large panel revealed a strikingly uniform continuum of the response (Fig. 2c). Although only a small fraction of these sera neutralized most viruses (10% neutralized 90% of viruses), 50% of them neutralized 50% of the virus panel. Neutralization potency (geometric mean ID50 titre) was highly correlated with breadth; this was true of both the randomly selected (Fig. 2a) and prescreened samples (Fig. 2b).
Fig. 1. Variation in serum neutralization potency and virus susceptibility.
(a) Heatmap displaying ID50 titres from 205 chronic donor sera (columns) tested against 229 phylogenetically diverse Env-pseudotyped viruses (rows). (b) Heatmaps comparing 10 bnAbs tested against 103 diverse viruses, and the 205 chronic donor sera assayed with the same set of 103 viruses. Columns in both heatmaps are ordered with greatest potencies to the left. Rows are ordered with greatest virus susceptibilities at the top. Neutralization titres of bnAbs and sera are presented separately because their different units are not directly comparable (i.e. concentration for bnAbs, dilution for sera). For bnAbs, lower values represent more potent neutralization, whereas for serum samples, higher values represent more potent neutralization. The column order of bnAbs follows that of columns in Table 1 (10E8 to 2G12 from left to right). Colour-key histograms (left and right histograms are for serum samples, middle histogram is for bnAbs) summarize the percentage of the total number of reactions that fell within a given range of neutralization titres. Annotation bands indicate virus clades (left column), serum clades (coloured bars, immediately above heatmaps for all 205 samples) and the 88 prescreened sera (grey bars above the colored bars). An ‘x’ indicates missing data.
Fig. 2. Serum neutralization breadth and potency continuum.
(a) Breadth and potency of neutralization were highly correlated among the 117 nonprescreened sera (Spearman rank correlation, P = 2.8 × 10−34, ρ = 0.85), though a few sera had exceptionally high potency. Circles indicate sera profiled in (e), selected for having breadths that match representative human bnAbs. (b) Breadth and potency were highly correlated among 88 sera selected by prescreening (Spearman rank correlation, P = 4.1× 10−21, ρ = 0.80); a few sera again had exceptionally high potency. (c) Serum percentiles on the y-axis showing the proportion of 117 nonprescreened sera with equal or greater breadth than the breadth shown along the x-axis. Breadths of four representative bnAbs are superimposed for comparison (b12, 2F5, PGT128, 4E10). Serum breadths were consistent with a uniform distribution (Kolmogorov test, P = 0.31, approximated due to 27 ties). (d) Breadth and potency of 10 bnAbs. (e) Magnitude-breadth functions showing proportion of viruses neutralized versus ID50 titre for 10 sera having breadths that match representative bnAbs. (f) Magnitude-breadth functions of 10 representative bnAbs.
This activity was compared with the breadth of a set of 10 human mAbs selected because they are considered broadly neutralizing and represent the known major neutralization epitopes on tier 2 viruses (Fig. 2d). We included the CD4bs bnAbs VRC01 [19,20], CH01 [16,21] and b12 [2], the glycan-dependent V1V2 bnAbs PG9 [22–24] and PGT145 [23], the V3/C3 glycan-dependent bnAb PGT128 [23], the glycan-specific bnAb 2G12 [3] and the MPER bnAbs 2F5, 4E10 [4] and 10E8 [25]. This includes the older set of bnAbs that have lower potency and breadth but are still considered broadly neutralizing (b12, 2G12, 2F5, 4E10), as well as some of the more potent recently isolated bnAbs (VRC01, PGT128, 10E8). The bnAbs were tested against a panel of 119 tier 2 viruses, 103 of which were also in the panel of 219 used to evaluate the sera (Fig. 1b). At the highest concentration tested (50µg/ml), 4E10, 10E8, VRC01, PG9 and PGT145 demonstrated the greatest breadth (75.7–97.3%of viruses), corresponding to 2.6–19.7% of sera using 50% neutralization cut-off (Fig. 2d–f, Table 1). Other bnAbs (e.g. PGT128, 2F5, CH01, b12, 2G12) neutralized 20.2–59.1% of the virus panel and this level of breadth was seen among 33.3–83.8% of the sera. Indeed, 77% of the sera exhibited greater breadth than the prototypic CD4bs bnAb, b12. The sera also exhibited substantial breadth when compared with several bnAbs using more stringent 80% neutralization values, especially when compared with more moderate concentrations of 1–5µg/ml of the bnAbs (Table 1).
Notably, our results with b12 and 2G12 suggest that these two bnAbs may not be as broadly neutralizing as previously thought. We found that b12 (50µg/ml) neutralized only 29.4% of all 119 viruses tested, including 57.1 and 33% of subtype B and C viruses, respectively. A previous large study estimated these values to be 50% (all), 72% (subtype B) and 67% (subtype C) [26]. We also found that 2G12 (50µg/ml) neutralized 20.2% of the 119 viruses tested, including 57.1 and 7.7% of subtype B and C viruses, respectively, whereas previous estimates were 41, 72 and 0%. These differences may reflect greater genetic diversity among the viruses in our panel than in previous panels.
Although it was not possible to directly compare the inhibitory concentration of monoclonal bnAbs with the inhibitory dilution of sera, we note that many serum neutralization titres were moderate in magnitude, often near or below the threshold shown to prevent infection after experimental challenge with R5, tier 2 simian HIV (SHIV) in nonhuman primates [27,28]. Nonetheless, our results demonstrate the potential to elicit measurable cross-neutralizing activity in most people at levels that exceed the best responses seen in HIV-1 vaccine efficacy trials [29,30]. Although it is not known what titres will be needed to protect against natural HIV-1 transmission, relatively low serum neutralization titres in the range of 1 : 20 to 1 : 50, which were often exceeded here, are protective against other viruses [31] and have been shown to impede HIV-1 replication and drive neutralization escape in humans [32], suggesting that they may have adequate potency for vaccines that aim to prevent HIV-1 infection.
We have not determined whether the serum neutralization breadth seen here is monoclonal or polyclonal in nature. In most cases examined, broad serum neutralization is explained by the presence of one or very few antibody specificities, each of which exhibits substantial breadth against tier 2 viruses [16,19,22,23,25,33–35]. Strain-specific neutralizing antibodies to other epitopes are also present in these sera; however, polyvalent mixtures of such strain-specific antibodies exhibit little breadth [36]. Thus, detection of even moderate breadth against tier 2 viruses usually indicates the presence of antibodies to one or more of a small number of conserved vulnerable regions on the virus.
We note thatmoderate cross-neutralizing activity generally is not observed prior to 3 years of HIV-1 infection [37]. Because we do not know the length of time of infection in our study participants, some individuals might have been infected for less than 3 years, which could have resulted in an underestimation of neutralization breadth. Also, serum neutralization potency and breadth can fluctuate over time during chronic infection. In a study of 155 chronically infected individuals sampled at two time points and tested against a panel of 12 tier-2 viruses, 30% had geometric mean titres that varied more than two-fold, and 6% had geometric mean titres that varied between three and six-fold (unpublished data). This dynamic nature of the bnAb response suggests that cross-sectional studies based on single samples may underestimate the potential of infected individuals to make bnAbs at some point during infection.
Overall, these observations provide a more accurate picture of the spectrum of neutralizing antibody responses that are possible in HIV-1 infected individuals. Sera with greatest breadth of neutralization are certainly of particular interest for detailed study. Still, most people are capable of making antibodies with more moderate breadth of coverage that would be deemed of value if elicited in a vaccine context. Studies of the relatively common HIV-1 infected individuals who have moderate breadth of neutralization could be valuable for understanding how to elicit similar responses via vaccination. An example of particular interest is the recent description of a clonal lineage of CD4bs-specific bnAbs that exhibits less breadth of neutralization but is also less mutated than other CD4bs bnAbs, and that may serve as a template to identify suitable immunogens that will re-elicit this lineage and generate neutralizing antibodies with similar breadth [38].
Supplementary Material
Acknowledgements
We thank Francine McCutchan, Carolyn Williamson, Beatrice Hahn, Ronald Swanstrom, Feng Gao, Jerome Kim and Miguel Thompson for molecular Env clones. We also thank Lynn Morris, Guy de Bruyn, Ramesh Paranjape, Pachamuthu Balakrishnan, Yiming Shao, Kunxue Hong, Hao Wu, Ning Li, Linqi Zhang, Hong Shang, Aine McKnight, Ruengpung Sutthent, Esper Kallas, Center For HIV/AIDS Vaccine Immunology, Centre for the AIDS Programme of Research in South Africa, International AIDS Vaccine Initiative, HIV Vaccine Trials Network, HIV Prevention Trials Network, Southern African National Blood Services, US Military HIV Research Program, Zambia-Emory HIV Research Project and the Bill and Melinda Gates Foundation’s Collaboration for AIDS Vaccine Discovery for serum specimens from HIV-1 infected individuals. We thank Dennis Burton for b12,PG9,PGT128 andPGT145,Mark Connors for 10E8, Herman Katinger for 2G12, 2F5 and 4E10, and Barton Haynes for CH01 and longitudinal neutralization data from 155 chronic HIV-1 infected individuals. Finally, we thank Kelli Greene and Hongmei Gao for coordinating the acquisition of reagents and the activities involved in performing assays and quality assurance of the final data.
D.C.M. designed and led the CAVD Neutralization Serotype Discovery Project (NSDP) that provided the data for analysis. B.T.K. and P.H. designed and conducted the data analyses. All three wrote the initial draft of the manuscript. M.S.S., R.T.B. and J.R.M. supervised the majority of NSDP neutralization assays and assisted with the interpretation of results and manuscript writing.
This work was funded by a grant from the Bill & Melinda Gates Foundation (Collaboration for AIDS Vaccine Discovery) and by the Intramural Research Program of the Vaccine Research Center, NIAID, NIH.
Footnotes
Conflicts of interest
There are no conflicts of interest.
References
- 1.Mascola JR, Montefiori DC. The role of antibodies in HIV vaccines. Ann Rev Immunol. 2010;28:413–444. doi: 10.1146/annurev-immunol-030409-101256. [DOI] [PubMed] [Google Scholar]
- 2.Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 3.Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, et al. Antibody domain exchange is an immunologic solution to carbohydrate cluster recognition. Science. 2003;300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- 4.Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, et al. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2001;75:10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binley JM, Lybarger EA, Crooks ET, Seaman MS, Gray E, Davis KL, et al. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J Virol. 2008;82:11651–11668. doi: 10.1128/JVI.01762-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doria-Rose NA, Klein RM, Manion MM, O’Dell S, Phogat A, Chakrabarti B, et al. Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J Virol. 2009;83:188–199. doi: 10.1128/JVI.01583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Svehla K, Louder MK, Wycuff D, Phogat S, Tang M, et al. Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J Virol. 2009;83:1045–1059. doi: 10.1128/JVI.01992-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sather DN, Armann J, Ching LK, Mavrantoni A, Sellhorn G, Caldwell Z, et al. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J Virol. 2009;83:757–769. doi: 10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, et al. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol. 2009;83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stamatatos L, Morris L, Burton DR, Mascola JR. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat Med. 2009;15:866–870. doi: 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- 11.Burton DR, Ahmed R, Barouch DH, Butera ST, Crotty S, Godzik A, et al. A blueprint for HIV vaccine discovery. Cell Host Microbe. 2012;12:396–407. doi: 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.West AP, Jr, Scharf L, Horwitz J, Klein F, Nussenzweig MC, Bjorkman PJ. Computational analysis of anti-HIV-1 antibody neutralization panel data to identify potential functional epitope residues. Proc Nat Acad Sci U S A. 2013;110:10598–10603. doi: 10.1073/pnas.1309215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verkoczy L, Kelsoe G, Moody MA, Haynes BF. Role of immune mechanisms in induction of HIV-1 broadly neutralizing anti-bodies. Curr Opin Immunol. 2011;23:383–390. doi: 10.1016/j.coi.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013;153:126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonsignori M, Montefiori DC, Wu X, Chen X, Hwang KK, Tsao CY, et al. Two distinct broadly neutralizing antibody specificities of different clonal lineages in a single HIV-1-infected donor: implications for vaccine design. J Virol. 2012;86:4688–4692. doi: 10.1128/JVI.07163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessments of neutralizing antibodies. J Virol. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien J-P, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2010;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, et al. Comprehensive cross-clade neutralization analysis of a panel of antihuman immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78:13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moldt B, Rakasz EG, Schultz N, Chan-Hui P-Y, Swiderek K, Weisgrau KL, et al. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc Natl Acad Sci U S A. 2012;109:18921–18925. doi: 10.1073/pnas.1214785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hessell AJ, Poignard P, Hunter M, Hangartner L, Tehrani DM, Bleeker WK, et al. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med. 2009;15:951–955. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilbert P, Wang M, Wrin T, Petropoulos C, Gurwith M, Sinangil F, et al. Magnitude and breadth of a nonprotective neutralizing antibody response in an efficacy trial of a candidate HIV-1 gp120 vaccine. J Infect Dis. 2010;202:595–605. doi: 10.1086/654816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montefiori DC, Karnasuta C, Huang Y, Ahmed H, Gilbert P, de Souza MS, et al. Magnitude and breadth of the neutralizing antibody response in the RV144 and Vax003 HIV-1 vaccine efficacy trials. J Infec Dis. 2012;206:431–441. doi: 10.1093/infdis/jis367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plotkin SA. Correlates of protection induced by vaccination. Clin Vaccine Immunol. 2010;17:1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bar KJ, Tsao CY, Iyer SS, Decker JM, Yang Y, Bonsignori M, et al. Early low-titer neutralizing antibodies impede HIV-1 replication and select for virus escape. PLoS Pathog. 2012;8:e1002721. doi: 10.1371/journal.ppat.1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A. 2012;109:E3268–E3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein F, Gaebler C, Mouquet H, Sather DN, Lehmann C, Scheid JF, et al. Broad neutralization by a combination of antibodies recognizing the CD4 binding site and a new conformational epitope on the HIV-1 envelope protein. J Exp Med. 2012;209:1469–1479. doi: 10.1084/jem.20120423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corti D, Langedijk JP, Hinz A, Seaman MS, Vanzetta F, Fernandez-Rodriguez BM, et al. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One. 2010;5:e8805. doi: 10.1371/journal.pone.0008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johannes F, Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 37.Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, et al. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4 T cell decline and high viral load during acute infection. J Virol. 2011;85:4828–4840. doi: 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao H-X, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.