Abstract
Background/Aim
Cancer cells have an essential demand for vitamin B12 (cobalamin) to enable cellular replication. The present pilot study quantified the immunohistochemical expression of vitamin B12 transport protein (Transcobalamin II; TCII), cell surface receptor (Transcobalamin II-R; TCII-R) and proliferation protein (Ki-67) in human tumor xenografts.
Materials and Methods
Tissue microarray slides containing 34 xenograft tumor tissues were immunohistochemically stained using TCN2 (anti-TCII), CD320 (anti-TCII-R) and MIB-1 (anti-Ki-67) antibodies. Representatively stained areas of all slides were digitally imaged and protein expression was quantified using ImageJ software plugins.
Results
All xenograft tumor tissues stained positively for TCII, TCII-R and Ki-67 proteins; expression varied both within and between tumor types. Correlation between TCII/TCII-R and Ki-67 expression was not significant in xenograft tissues.
Conclusion
Proliferating cancer cells express measurable levels of TCII and TCII-R. Immunohistochemical quantification of these markers may be useful as a tool for detection of tumors, tailored selection of anti-tumor therapies and surveillance for evidence of recurrent disease.
Keywords: Vitamin B12, immunohistochemistry, transcobalamin II, transport protein, receptor, Ki-67, xenograft
Despite numerous advances in medical research, cancer remains one of the leading causes of death in the United States. Therefore, use of biological markers to more accurately diagnose and predict effective therapy is receiving increased interest in clinical cancer medicine. The ability to direct an individualized diagnostic and therapeutic plan can increase the probability of diagnosis, increase likelihood of treatment success, maximize use of valuable treatment time and minimize financial costs associated with ineffective diagnostics or drugs. Development of new, accurate biomarkers is critical to increasing our ability to appropriately select diagnostic and therapeutic modalities unique to each patient.
Vitamin B12 (cobalamin) is an essential micronutrient that was first isolated and crystallized in 1948 (1). Since then, studies have shown that vitamin B12 plays an important role in the differentiation, proliferation and metabolic stability of cells (2–4). Once absorbed into the bloodstream, vitamin B12 binds to transcobalamin II (TCII), a non-glycosylated transport protein found in blood (5, 6). The vitamin B12-TCII complex is then transported to cells and undergoes receptor-mediated endocytosis using the transcobalamin II receptor (TCII-R), a specific vitamin B12 cell surface receptor (5, 6). While all living cells require vitamin B12, rapidly dividing tumor cells have a highly increased need for this vitamin, which is critical in the methylation process associated with DNA synthesis. In order to supply this substantial demand for vitamin B12, proliferative tumor cells: i) increase production of TCII that is able to scavenge, bind, transport and deliver vitamin B12 (7–9), and ii) express far greater numbers of cell surface TCII receptors than normal cells do, enabling them to take up and accumulate vitamin B12 in large quantities (10–14). Vitamin B12 coupling offers an effective means of targeting diagnostic and therapeutic agents to cancer cells using the TCII/TCII-R delivery system.
Imaging of primary and metastatic tumors using radio-labeled vitamin B12 analogs to target the TCII receptors of tumor cells is gaining momentum. The dependence of image quality on target to non-target ratio, especially with molecular imaging modalities such as single photon emission computed tomography (SPECT), positron emission tomography (PET) and magnetic resonance imaging (MRI), highlights the critical need for tumor targeting (15). Most recently, the vitamin B12 analog diethylenetriaminepentaacetate (DTPA)-adenosylcobalamin has been used in SPECT imaging to successfully detect a variety of high-grade primary and metastatic tumors (16). TCII-R-targeted imaging agents such as DTPA-adenosylcobalamin show promise for early detection of both primary and metastatic tumors, including those eluding detection by standard diagnostic means. Furthermore, targeted imaging may play an important role in assessing tumor response to conventional radiation and chemotherapeutic treatments, and in monitoring patients for development of recurrent or metastatic disease.
The TCII/TCII-R delivery system may also be used to effectively promote selective uptake of chemotherapeutic agents by cancer cells, thereby increasing the efficacy of therapy and decreasing adverse effects on normal tissues (15, 17–20). Studies have demonstrated the potential use of vitamin B12 as a carrier to target cisplatin (15) and doxorubicin-loaded liposomes (20) to tumor cells. Nitrosylcobalamin (NO-Cbl), a novel vitamin B12 analog comprised of nitric oxide (NO) bound to hydroxocobalamin, was developed as a potential anti-tumor agent (21), and has been shown to inhibit tumor growth both in the laboratory and in the clinical treatment of canine malignancies (12, 22–24). NO-Cbl functions as a biological ‘Trojan horse’, using TCII transport and TCII-R-mediated vitamin B12 uptake to specifically target NO-Cbl to cancer cells (12). Following internalization of NO-Cbl into tumor cells through TCII-R-mediated endocytosis, NO is liberated from cobalamin resulting in decreased cellular metabolism, activation of apoptotic mechanisms and inhibition of survival pathways (12, 22, 23, 25). Recent work suggests that NO-Cbl may also function as a folate antagonist (26). The anti-tumor activity of NO-Cbl has been shown to directly correlate with the expression of TCII-R on the plasma membranes of tumor cells (12).
Uptake of vitamin B12-based imaging and chemotherapeutic analogs has been shown to be increased in highly proliferative tumors (16, 27). Determination of the proliferative status of a tumor is most commonly estimated by the immunohistochemical assessment of Ki-67, a nuclear antigen that is strictly associated with cellular proliferation (28). Ki-67 is present during all active phases of the cell cycle (G1, S, G2 and M phases) but is absent from resting cells (G0) (29). Because Ki-67 protein expression is required for progression through the cell division cycle, it provides an excellent marker of the proliferation status of a given cell population (28, 29). Ki-67 expression has been measured immunohistochemically in a wide variety of tumor types, and is typically conveyed as the percentage of positively stained tumor cells out of the total number of cells. Ki-67 estimation has been used to help differentiate benign from malignant tissue (30–32), establish tumor grade (33–35), identify malignant transformation of benign tumors (36), assess extent of peritumoral vascular invasion (37, 38), evaluate lymphatic invasion (39–41), aid in selection of treatment protocols (42–44), measure treatment response (45–47), determine prognosis (31, 48–50) and monitor for recurrent (51–53) or metastatic (54–56) disease. Because of its wide utility, Ki-67 has become one of the most frequently assessed tumor biomarkers in use today.
Immunohistochemical (IHC) staining is currently the gold standard for the quantitative measurement of protein biomarkers, especially when biomarker numbers are increased in proliferative tumor tissues (57). Several studies have demonstrated a significant correlation between results of IHC protein quantification and quantification using established methods including enzyme-linked immunosorbent assay (ELISA) (58–60) and western blot (61, 62). IHC staining uses specific antibodies to differentially stain for the antigens of choice, and relies on the principle that greater antigen content leads to increased precipitation of immune complexes coupled to a chromophore such as 3,3’ diaminobenzidine (DAB) (60). Quantitative analysis of IHC-stained slides has traditionally relied on manual counting of positively immunostained cells. However, manual counts may be associated with interpretation subjectivity, intra- and inter-observer variation, reproducibility issues, and labor intensive protocols (63–66). Computer-assisted analysis of digitally imaged IHC-stained tissues has been introduced as a means of overcoming the shortfalls associated with manual counts (60, 67). ImageJ is a public domain, Java-based image processing program, developed at the National Institutes of Health (Bethesda, MD, USA) and released in 1997 (68). Designed with an open architecture to enable expandability using Java plugins, it has become the standard in scientific image analysis. The color deconvolution plugin utilizes an algorithm to calculate the contribution of multiple stains (such as DAB and hematoxylin) on each tissue slide based on stain-specific red-green-blue (RGB) absorption, and generates a unitless value corresponding to the amount of chromophore-bound antigen present (69–73). The Immunoratio plugin utilizes the color deconvolution algorithm to analyze nuclear immunostains such as Ki-67, and generates a value expressed as a percentage of positively immunostained area out of total nuclear area (33, 66). ImageJ, as well as the color deconvolution and Immunoratio plugins, are freely available web-based applications, enabling their routine use for quantitative IHC analysis.
The aim of this pilot study was to develop a standardized IHC staining, digital imaging and computer-assisted software protocol to permit objective quantification and comparison of expression of TCII, TCII-R and Ki-67 in a variety of human tumor xenografts. Results from this study may be used to evaluate the potential usefulness of IHC quantification of these proteins for detecting tumors and for selection and monitoring of vitamin B12-based anti-tumor therapies.
Materials and Methods
Tissue samples
Commercial tissue microarray slides were purchased from the Tissue Acquisition and Cellular/Molecular Analysis Shared Service at the University of Arizona Cancer Center (Tucson, AZ, USA). Expression of TCII, TCII-R and Ki-67 were measured in thirty-four human cancer cell line xenografts. Cancer cell lines evaluated included: (1) breast carcinoma MCF7/S; (2) breast carcinoma MDA-MB-231; (3) breast carcinoma ZR-75-1; (4) cervical carcinoma C-33A; (5) cervical carcinoma Ca Ski; (6) cervical carcinoma HeLa; (7) colonic carcinoma HCT-116; (8) colonic adenocarcinoma HT-29; (19) epidermoid carcinoma A431NS; (10) Ewing’s sarcoma CHP-100; (11) glioblastoma multiforme SF-767; (12) leukemia MDS/SP1; (13) B cell lymphoma Granta-4; (14) Burkitt’s lymphoma Ramos; (15) Burkitt’s lymphoma Raji; (16) melanoma A375; (17) non-small cell lung cancer (NSCLC) adenocarcinoma H1975; (18) NSCLC carcinoma MV-522; (19) NSCLC carcinoma NCI: H460; (20) ovarian carcinoma 2780AD; (21) ovarian carcinoma NIH-OVCAR-3; (22) pancreatic adenocarcinoma AsPC-1; (23) pancreatic adenocarcinoma BxPC-3; (24) pancreatic adenocarcinoma CAPAN-2; (25) pancreatic adenocarcinoma CFPAC-1; (26) pancreatic adenocarcinoma HPAF-II; (27) pancreatic adenocarcinoma MIA PaCa-2; (28) pancreatic carcinoma Panc-1; (29) pancreatic carcinoma SU.86.86; (30) prostatic carcinoma DU-145; (31) prostatic adenocarcinoma PC-3; (32) renal adenocarcinoma ACHN; (33) renal carcinoma Caki; and (34) uterine epithelial carcinoma RL 95-2.
Commercial microarray preparation
Tumor xenografts were developed by injecting 10×106 cancer cells into the flanks of severe combined immunodeficient mice. When the tumor volume of each xenograft approximated 500 mm3, mice were sacrificed by carbon dioxide asphyxiation and tumors were harvested. Tissues were fixed for 24 h in 10% neutral buffered formalin, transferred to 70% ethanol, processed overnight in a Tissue Tek V Tissue Processor (Sakura Finetek, Torrence, CA, USA), and embedded into paraffin blocks. A hematoxylin and eosin slide was cut and stained from each individual xenograft block and was reviewed by a board-certified pathologist, who marked optimal areas for coring. A tissue microarray (TMA) block was made by removing a 2 mm core from each individual paraffin block and re-embedding multiple donor cores into a recipient TMA paraffin block. Three-micron thick sections were cut from the donor TMA block, placed on positively charged glass slides and baked overnight for 8–15 h at 65°C to minimize potential loss of core samples during automated staining. Slides were then allowed to cool at room temperature.
Immunohistochemical staining procedure for TCII and TCII-R
Determination of a staining protocol and selection of antibody dilutions were first determined by automated staining of formalin-fixed, paraffin-embedded NIH-OVCAR 3 xenograft tissues (Tissue Acquisition and Cellular/Molecular Analysis Shared Service, University of Arizona Cancer Center, Tucson, AZ, USA) using incremental antibody dilutions and comparing results to those obtained and used in a previous study (12). Tissue microarray slides were processed in two changes of xylene for 5 min each, and two changes of 100% histology grade alcohol (anhydrous ethyl alcohol 90±1% v/v, methyl alcohol approximately 5% v/v, 2-propanol approximately 5% v/v) for 1 min each. Slides were then processed with one change of 95% histology grade alcohol for 30 s and rinsed. Once deparaffinization was complete, endogenous peroxidase activity of the sections was quenched by incubation in 3% hydrogen peroxide for 15 min at room temperature. Following rinsing and antigen retrieval in a sodium citrate buffer (pH 6.0), heat-induced epitope retrieval was accomplished using a Dako Pascal pressure chamber (Dako North America, Carpinteria, CA, USA) at 120°C for 30 s. Slides were cooled, placed into Supersensitive™ Wash Buffer 20× (Biogenex, Fremont, CA, USA) and loaded onto an automated stainer (Dako Autostainer Plus; Dako North America, Carpinteria, CA, USA). In order to reduce nonspecific background staining, slides were blocked by sequential incubation with avidin and biotin for 25 min each (Avidin/Biotin Blocking System; Covance Inc., Princeton, NJ, USA), followed by 10 min with a protein block (Background Sniper; Biocare Medical, Concord, CA, USA). Slides were then incubated for 1 h at room temperature with the following primary antibodies: rabbit polyclonal antibody against TCII (TCN2; 1:50 dilution; Proteintech, Chicago, IL, USA) and rabbit polyclonal antibody against TCII-R (CD320; 1:50 dilution; Proteintech, Chicago IL, USA). After washing, slides were incubated with a biotinylated secondary antibody for 20 min, followed by an enzyme-streptavidin-horse radish peroxidase conjugate for 20 min (Supersensitive™ Link-Label IHC Detection System; Biogenex, Fremont, CA, USA). Immunostaining visualization was achieved by immersion of slides in diaminobenzidine (DAB) (Betazoid DAB Chromagen Kit; Biocare Medical, Concord, CA, USA) for 5 min, followed by counterstaining with Mayer’s hematoxylin for 30 s. Slides were then air-dried. To ensure the specificity of immunostaining, negative control TMA slides were prepared by omitting the primary antibody.
Immunohistochemical staining procedure for Ki-67
Tissue microarray slides were processed in two changes of 70% ethanol, two changes of 0.03% hydrogen peroxide in ethanol, and rinsed three times with phosphate-buffered saline (PBS). Slides were placed into a citrate buffer (pH 6.0) antigen-retrieval solution. Following antigen retrieval in a sodium citrate buffer (pH 6.0). heat-induced epitope retrieval was accomplished using a Dako Pascal pressure chamber (Dako North America, Carpinteria, CA, USA) at 120°C for 30 s. Slides were cooled, rinsed three times with PBS and blocked with 10% normal horse serum (Quad Five, Ryegate, MT, USA) for 20 min. One TMA slide was incubated with a mouse monoclonal primary antibody to Ki-67 (Clone MIB-1; Dako North America, Carpinteria, CA, USA) at a 1:40 dilution for 1 h; the second TMA slide was not exposed to the primary antibody in order to serve as a negative control. Both slides were rinsed three times with PBS and incubated with a horse anti-mouse secondary antibody (Biocare Medical, Concord, CA, USA) for 10 min. Slides were rinsed three times with PBS and incubated with streptavidinhorse radish peroxidase (Biocare Medical, Concord, CA, USA) for 10 minutes. Slides were rinsed three times with PBS, developed with Nova Red (Vector Labs, Burlingame, CA, USA), counterstained in Mayers hematoxylin for 15 s and air-dried.
Microscopic evaluation and imaging
Slides were examined by light microscopy using an Olympus BH-2 microscope (Olympus, Center Valley, PA, USA), fitted with an Olympus DP71 12.5-megapixel, 12-bit digital color camera with Peltier cooling and DP Manager and Controller software (version 3.0; Olympus, Center Valley, PA, USA). An ultra-high resolution mode (4080×3072) was used, and light settings were standardized for all imaging sessions. TCII- and TCII-R-positive cells exhibited DAB-positive (brown) staining, and Ki-67-positive cells exhibited Nova Red-positive (red-brown) staining; negative cells stained with the hematoxylin counterstain only. Stained slides were reviewed by a board-certified pathologist. A representative area of solid tumor devoid of connective tissue and ischemic necrosis and with even distribution of cells was selected and digitally photographed for each tissue sample. Digital images for TCII- and TCII-R analysis were obtained at 10× magnification (200 micron scale bar), and images for Ki-67 analysis were obtained at 40× magnification (50 micron scale bar). For Ki-67 analysis, slide labels were omitted in order to minimize extraneous background staining. JPEG format was used to capture all digital images.
Digital image analysis for TCII and TCII-R
For each image, the color deconvolution method was used to isolate TCII- and TCII-R positive DAB-stained cells from TCII- and TCII-R negative hematoxylin-stained cells. DAB and hematoxylin were digitally separated using ImageJ software (version 1.46c; WS Rasband, National Institutes of Health, Bethesda, MD, USA, http://rsb.info.nih.gov/ij/) and an ImageJ plugin for color deconvolution (70), which calculated the contribution of DAB and hematoxylin, based on stain-specific red-green-blue (RGB) absorption (69). Following deconvolution, scale was set to the 200 micron scale bar on each image. The deconvoluted image was subjected to histogram analysis, with the lower threshold set at 10, and the upper threshold set at 100. For each image, three fields of consistent staining were selected and measured using 200×200 pixel boxes. A value was assigned to each field using ImageJ software, and the average value of all three fields was used to assign a staining value (arbitrary units) to each image.
Digital image analysis for Ki-67
Staining was evaluated using ImageJ software and the ImmunoRatio plugin (66) optimized for Ki-67 nuclear staining and utilizing the color deconvolution method described above (69). Background correction was performed using an image from an empty slide background (blankfield image) to address image color balance and uneven illumination. The number of Ki-67-positive NovaRed-stained cells over the total number of cells was calculated and used to assign a staining value (%) to each image.
Statistical analysis
The Spearman rank correlation co-efficient was used as a non-parametric measure of statistical dependence between TCII, TCII-R and Ki-67 expression. Correlation co-efficient values were calculated using an online computer software program (Free Statistics Software, Office for Research Development and Education, version 1.1.23-r7, URL http://www.wessa.net). Significance was set at p≤0.05.
Results
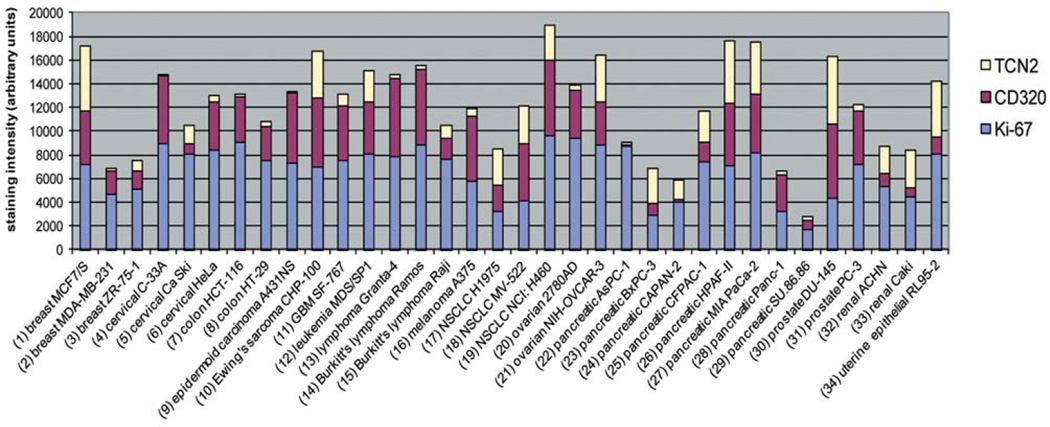
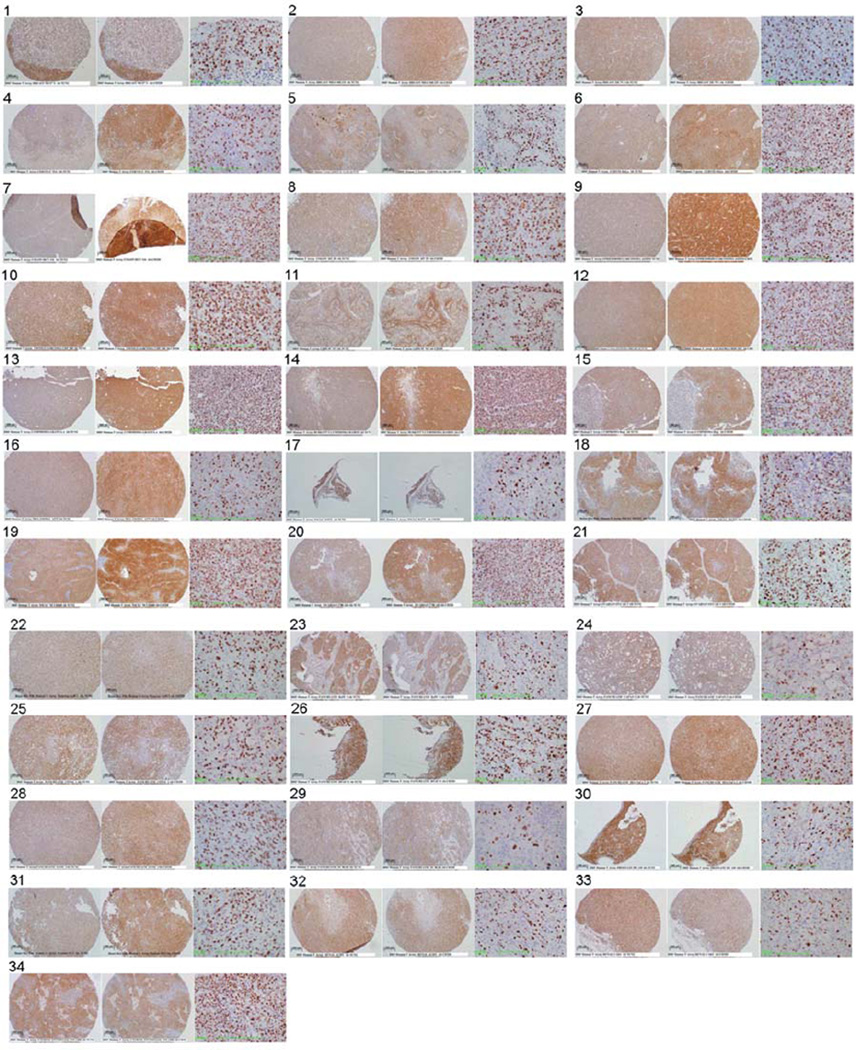
Immunohistochemical staining values for all xenograft tissues are summarized in Table 1 and tabulated in Figure 1. Digital images of TCII, TCII-R and Ki-67-stained slides are included in Figure 2.
Table I.
Immunohistochemical staining scores of human tumor xenografts.
| Tumor type | Cell line | TCII (arbitrary units) |
TCII-R (arbitrary units) |
Ki-67 (% positive cells) |
|---|---|---|---|---|
| Breast carcinoma | MCF7/S | 5444 | 4543 | 72 |
| Breast carcinoma | MDA-MB-231 | 239 | 2039 | 46 |
| Breast carcinoma | ZR-75-1 | 918 | 1551 | 51 |
| Cervical carcinoma | C-33A | 125 | 5674 | 90 |
| Cervical carcinoma | Ca Ski | 1598 | 842 | 81 |
| Cervical carcinoma | HeLa | 601 | 4059 | 84 |
| Colonic carcinoma | HCT-116 | 263 | 3869 | 90 |
| Colonic adenocarcinoma | HT-29 | 491 | 2828 | 75 |
| Epidermoid carcinoma | A431NS | 98 | 5874 | 73 |
| Ewing's sarcoma | CHP-100 | 3905 | 5878 | 69 |
| Glioblastoma multiforme | SF-767 | 947 | 4606 | 76 |
| Leukemia | MDS/SP1 | 2650 | 4319 | 81 |
| B-cell lymphoma | Granta-4 | 332 | 6514 | 79 |
| Burkitt's lymphoma | Ramos | 329 | 6413 | 88 |
| Burkitt's lymphoma | Raji | 1022 | 1779 | 77 |
| Melanoma | A375 | 579 | 5500 | 58 |
| NSCLC adenocarcinoma | H1975 | 3043 | 2184 | 33 |
| NSCLC carcinoma | MV-522 | 3284 | 4747 | 42 |
| NSCLC carcinoma | NCI:H460 | 2951 | 6346 | 96 |
| Ovarian carcinoma | 2780 AD | 459 | 4124 | 94 |
| Ovarian carcinoma | NIH-OVCAR-3 | 3859 | 3625 | 89 |
| Pancreatic adenocarcinoma | AsPC-1 | 190 | 149 | 87 |
| Pancreatic adenocarcinoma | BxPC-3 | 3030 | 960 | 29 |
| Pancreatic adenocarcinoma | CAPAN-2 | 1612 | 267 | 40 |
| Pancreatic adenocarcinoma | CFPAC-1 | 2619 | 1618 | 74 |
| Pancreatic adenocarcinoma | HPAF-II | 5235 | 5243 | 71 |
| Pancreatic adenocarcinoma | MIA-PaCa-2 | 4405 | 4999 | 81 |
| Pancreatic carcinoma | Panc-1 | 369 | 3090 | 32 |
| Pancreatic carcinoma | SU.86.86 | 337 | 772 | 17 |
| Prostatic carcinoma | DU-145 | 5793 | 6221 | 44 |
| Prostatic adenocarcinoma | PC-3 | 572 | 4497 | 72 |
| Renal adenocarcinoma | ACHN | 2281 | 1102 | 53 |
| Renal carcinoma | Caki | 3204 | 827 | 44 |
| Uterine epithelial carcinoma | RL95-2 | 4721 | 1496 | 81 |
Immunohistochemical staining was performed using TCN2 (anti-TCII), CD320 (anti-TCII-R) and MIB-1 (anti-Ki-67)-specific antibodies. TCII=Transcobalamin II transport protein; TCII-R=transcobalamin II cell surface receptor; NSCLC=non-small cell lung cancer.
Figure 1.
Immunohistochemical expression of the transcobalamin II transport protein (TCII), transcobalamin II cell surface receptor (TCII-R) and proliferative index (Ki-67) in human cancer xenografts. Immunohistochemical staining was performed using TCN2 (anti-TCII), CD320 (anti-TCII-R) and MIB-1 (anti-Ki-67)-specific antibodies. Ki-67 percentages were multiplied by 100 to enable use of the y axis scale.
Figure 2.
Tumor tissue sections, immunohistochemically stained for TCII (TCN2 antibody, 10× objective), TCII-R (CD320 antibody, 10× objective) and Ki-67 (MIB-1 antibody, 40× objective), are shown from left to right for each xenograft tissue sample: (1) breast carcinoma MCF7/S; (2) Breast carcinoma MDA-MB-231; (3) breast carcinoma ZR-75-1; (4) cervical carcinoma C-33A; (5) cervical carcinoma Ca Ski; (6) cervical carcinoma HeLa; (7) colonic carcinoma HCT-116; (8) colonic adenocarcinoma HT-29; (19) epidermoid carcinoma A431NS; (10) Ewing’s sarcoma CHP-100; (11) glioblastoma multiforme SF-767; (12) leukemia MDS/SP1; (13) B-cell lymphoma Granta-4; (14) Burkitt’s lymphoma Ramos; (15) Burkitt’s lymphoma Raji; (16) melanoma A375; (17) non-small cell lung cancer (NSCLC) adenocarcinoma H1975; (18) NSCLC carcinoma MV-522; (19) NSCLC carcinoma NCI: H460; (20) ovarian carcinoma 2780AD; (21) ovarian carcinoma NIH-OVCAR-3; (22) pancreatic adenocarcinoma AsPC-1; (23) pancreatic adenocarcinoma BxPC-3; (24) pancreatic adenocarcinoma CAPAN-2; (25) pancreatic adenocarcinoma CFPAC-1; (26) pancreatic adenocarcinoma HPAF-II; (27) pancreatic adenocarcinoma MIA PaCa-2; (28) pancreatic carcinoma Panc-1; (29) pancreatic carcinoma SU.86.86; (30) prostatic carcinoma DU-145; (31) prostatic adenocarcinoma PC-3; (32) renal adenocarcinoma ACHN; (33) renal carcinoma Caki; and (34) uterine epithelial carcinoma RL 95-2.
All tumor xenografts stained positively for TCII, TCII-R and Ki-67. Average/median staining values of xenograft tumor tissues were 1985/1310 (TCII), 3487/3964 (TCII-R) and 67%/74% (Ki–67). The range of staining values was 98–5793 (TCII), 149–6413 (TCII-R) and 17–96% (Ki-67).
There was no statistically significant correlation between TCII and TCII-R expression (Spearman rank coefficient: 0.03, p=0.87). Grouping of tumors by type did not identify any correlation between these proteins. Thirty-two percent of tumor tissues had staining values that were higher for TCII than for TCII-R; in these tissues, staining values for TCII were 2 times greater on average than those for TCII-R. Sixty-eight percent of tissues had higher staining values for TCII-R; in these tissues, staining values for TCII-R were 10 times greater on average than those for TCII. Immunohistochemical staining values for TCII and TCII-R varied both between and within xenograft tumor types.
Overall, there was no statistically significant correlation between TCII and Ki-67 expression (Spearman rank coefficient: −0.22, p=0.21), or between TCII-R and Ki-67 expression (Spearman rank coefficient: 0.32, p=0.07). Furthermore, there were no statistically significant correlations between expression of these proteins when tumors were grouped by type. Staining values for Ki-67 also varied both between and within tumor types.
Serial passage number for each xenograft tumor included in the TMA block was available for 19 out of the 34 xenograft tissues. Average passage number was 36, and median passage number was 18.5, with a range of 3 to 169. There was no statistically significant correlation between passage number and TCII expression (Spearman rank coefficient: 0.29, p=0.22), TCII-R expression (Spearman rank coefficient: –0.17, p=0.48) or Ki-67 expression (Spearman rank coefficient: −0.41, p=0.09).
Discussion
This pilot study is one of the first studies attempting to quantify TCII, TCII-R and Ki-67 expression in a variety of xenograft tumor tissues using a standardized IHC staining, imaging and analysis protocol. All xenograft tissues were developed, passaged, processed and fixed in a single laboratory, thus minimizing variables such as specimen handling, fixative selection, fixation duration, paraffin embedding, storage and sectioning that often complicate comparison of xenograft tissues from multiple sources (74–76). Use of TMA slides facilitated identical processing, staining, imaging and analysis of all xenograft tissues, enabling an objective and comparative quantitative analysis of protein expression.
All xenograft tumor tissues stained positively for TCII and TCII-R expression, suggesting an intracellular demand for cobalamin during DNA synthesis. Of interest was the fact that in over half the xenograft tissues, staining values for TCII-R were on average 10 times greater than for TCII. While TCII-R expression is regulated in part by intracellular cobalamin levels (77, 78), its expression is reported to be more critically regulated by the proliferation and differentiation status of the cells (10, 11). Studies have shown that cellular proliferation may be increased in serially passaged xenograft tumor tissues compared to the original tumor tissues from which the xenografts were derived (79, 80). It is possible that in the xenograft tumor tissues of this study, increased cellular proliferation in some serially passaged tissue samples prompted up-regulation of TCII-R expression. Variations in protein expression in passaged xenograft tissue cell lines may have also influenced the ability to detect discrete staining patterns within specific tumor types.
All xenograft tumor tissues exhibited positive nuclear staining for Ki-67, indicating that xenograft tissues were obtained from actively proliferating tumors. Staining values obtained for Ki-67 in this study were within ranges similar to those determined in previous IHC studies utilizing xenograft tumor tissues and computer-assisted image analysis methods (81, 82). Lack of significant correlation between Ki-67 and TCII/TCII-R expression was somewhat surprising, especially considering that vitamin B12 is an essential requirement for DNA synthesis during rapid cellular proliferation. Physiological and/or technical variables may have contributed to this finding. While it has been shown that Ki-67 expression increases during the latter half of the S and G2 phases and peaks in the M phase of the cell cycle (28), it is unknown whether expression of TCII and TCII-R are linked to specific phases of the cell cycle during active cellular proliferation. Furthermore, the half lives of Ki-67 and TCII are relatively short at 60–90 min (28, 83) in comparison to a half life of 8 h for TCII-R (84). Due to the dynamic process of protein expression, it may have been difficult to uniformly capture timing of maximal expression for each protein. From a technical standpoint, Ki-67 expression has been shown to vary between serial passages of xenograft tissues (79, 80); effects of passaging on TCII and TCII-R expression are less well known. It is possible that the range of passage numbers between xenograft tissues in this study may have had a more pronounced effect on Ki-67 expression than on TCII /TCII-R expression. In addition, comparison of expression between proteins using TMA slides is best performed with serial tissue sections from the core TMA paraffin block. In this study, the commercial TMA slides used for IHC staining were obtained from as close to serial cuts as possible: the slide made from section 20 of the TMA block was used to stain for TCII, the slide made from section 21 of the TMA block was used to stain for TCII-R, and the slide made from section 32 of the TMA block was used to stain for Ki-67. Use of a non-consecutive TMA section for Ki-67 evaluation may have influenced the ability to identify a correlation between Ki-67 and TCII /TCII-R expression.
TCII, TCII-R and Ki-67 expression varied both between and within tumor types. Variation in expression of these proteins between tumor types was not unexpected due to the diversity of biological characteristics that are unique to each tumor type. Protein expression within tumor types may have been influenced by the range of serial passages within cell lines of each tumor type, as well as by small sample size in the 9 of 15 tumor types that were represented by more than one cell line in the TMA. However, variations in TCII, TCII-R and Ki-67 expression were evident even within 8 pancreatic tumor cell lines, suggesting a significant individual component to protein expression and adding strength to the rationale for personalized cancer medicine.
The use of digital imaging and color deconvolution-based computer image analysis provided an efficient and effective way to objectively assign a staining value to each image. With the ImageJ color deconvolution plugin, upper and lower thresholds for histogram analysis of the deconvoluted images were chosen such that immunostaining was minimal within fragments of normal tissues apparent in some of the TMA cores. While selection of upper and lower thresholds was somewhat subjective, these thresholds were standardized for every image, enabling direct comparison of staining values between xenograft tumor tissues.
Subjective bias in this study may have been introduced by manual selection of the region of interest for both TMA tumor coring as well as stained slide imaging. However, recent work has suggested that use of computer-assisted image analysis may be robust enough to overcome the variability associated with manual selection of the region of interest for measurement of protein expression (85). Furthermore, microscopic evaluation of tumor tissues by a pathologist was beneficial in this study as it provided the opportunity to exclude normal stromal and inflammatory cells from tissue samples, thus overcoming one of the reported shortfalls of computer-assisted image analysis (85, 86).
This pilot study established a standardized method to comparatively detect and quantify TCII, TCII-R and Ki-67 protein expression in a variety of xenograft tumor tissues. Results from this study suggest that proliferative tumors expressing TCII and TCII-R may be good candidates for diagnostic imaging with agents such as DTPA-adenosylcobalamin, and for treatment with vitamin B12-based anti-tumor agents such as NO-Cbl. Furthermore, measurable changes in TCII and TCII-R expression may be useful in assessing response to anti-tumor therapy and in monitoring for evidence of disease recurrence. These preliminary results will be used as a starting point to aid in the design of additional studies evaluating the IHC expression of these proteins in tumor tissues. Future considerations include: i) measurement of TCII, TCII-R and Ki-67 in naturally occurring tumors; ii) evaluation of whole tissue sections to address the heterogeneous nature of tumor tissues that may not be accurately represented in TMA tissue cores; iii) quantification of TCII, TCII-R and Ki-67 expression in adjacent malignant and benign tissues to identify and measure biomarker up-regulation in proliferative tumor tissues; and iv) quantification of TCII, TCII-R and Ki-67 in patient tumors and correlation with outcome, prognosis and monitoring following therapy with cobalamin analogs such as NO-Cbl in a clinical trial setting. Results of these studies may support the use of immunohistochemical quantification of TCII, TCII-R and Ki-67 protein expression as a screening tool to detect tumors and to help tailor treatment selection and monitoring of anti-tumor therapies.
Acknowledgements
The Authors would like to acknowledge Gillian Paine-Murrieta and Kathleen McDaniel of the Tissue Acquisition and Cellular/Molecular Analysis Shared Service of the University of the Arizona Cancer Center for provision of the xenograft tissue microarray slides; these services are supported by the Cancer Center Support Grant # NIH CA023074. The authors would also like to thank Melanie Walker (VDx Research Services, Davis, CA), and Barry Puget and Michael Manzer (Histopathology Laboratory - UC Davis, Davis, CA) for their technical assistance.
This project was funded in part by the Bauer Research Foundation, Inc. and by contributions from David C. Falk, Sr. (Kingsmill Farm II, Wake Forest, NC).
References
- 1.Rickes EL, Brink NG, Koniuszy FR, et al. Crystalline Vitamin B12. Science. 1948;107(2781):396–397. doi: 10.1126/science.107.2781.396. [DOI] [PubMed] [Google Scholar]
- 2.Linnell JC, Matthews DM. Cobalamin metabolism and its clinical aspects. Clin Sci (Lond) 1984;66(2):113–121. doi: 10.1042/cs0660113. [DOI] [PubMed] [Google Scholar]
- 3.Seetharam B, Bose S, Li N. Cellular import of cobalamin (Vitamin B-12) J Nutr. 1999;129(10):1761–1764. doi: 10.1093/jn/129.10.1761. [DOI] [PubMed] [Google Scholar]
- 4.Weir DG, Scott JM. Vitamin B12 "cobalamin". In: Shils ME, Olseon JA, Shike M, Ross AC, editors. Modern Nutrition in Health and Disease. Baltimore: Lippincott Williams & Wilkins; 1999. pp. 447–458. [Google Scholar]
- 5.Seetharam B, Li N. Transcobalamin II and its cell surface receptor. Vitam Horm. 2000;59:337–366. doi: 10.1016/s0083-6729(00)59012-8. [DOI] [PubMed] [Google Scholar]
- 6.Seetharam B, Yammani RR. Cobalamin transport proteins and their cell-surface receptors. Expert Rev Mol Med. 2003;5(18):1–18. doi: 10.1017/S1462399403006422. [DOI] [PubMed] [Google Scholar]
- 7.Rabinowitz R, Rachmilewitz B, Rachmilewitz M, et al. Production of transcobalamin II by various murine and human cells in culture. Isr J Med Sci. 1982;18(7):740–745. [PubMed] [Google Scholar]
- 8.Hall CA, Green-Colligan PD, Begley JA. Synthesis of transcobalamin II by cultured human hepatocytes. Biochim Biophys Acta. 1985;838(3):387–389. doi: 10.1016/0304-4165(85)90239-9. [DOI] [PubMed] [Google Scholar]
- 9.Schohn H, Gueant JL, Girr M, et al. Synthesis and secretion of a cobalamin-binding protein by HT 29 cell line. Biochem J. 1991;280(Pt 2):427–430. doi: 10.1042/bj2800427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindemans J, Kroes AC, van Geel J, et al. Uptake of transcobalamin II-bound cobalamin by HL-60 cells: effects of differentiation induction. Exp Cell Res. 1989;184(2):449–460. doi: 10.1016/0014-4827(89)90343-1. [DOI] [PubMed] [Google Scholar]
- 11.Amagasaki T, Green R, Jacobsen DW. Expression of transcobalamin II receptors by human leukemia K562 and HL-60 cells. Blood. 1990;76(7):1380–1386. [PubMed] [Google Scholar]
- 12.Bauer JA, Morrison BH, Grane RW, et al. Effects of interferon beta on transcobalamin II-receptor expression and antitumor activity of nitrosylcobalamin. J Natl Cancer Inst. 2002;94(13):1010–1019. doi: 10.1093/jnci/94.13.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russell-Jones G, McTavish K, McEwan J, et al. Vitamin-mediated targeting as a potential mechanism to increase drug uptake by tumours. J Inorg Biochem. 2004;98(10):1625–1633. doi: 10.1016/j.jinorgbio.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 14.Russell-Jones G, McTavish K, McEwan J. Preliminary studies on the selective accumulation of vitamin-targeted polymers within tumors. J Drug Target. 2011;19(2):133–139. doi: 10.3109/10611861003734027. [DOI] [PubMed] [Google Scholar]
- 15.Ruiz-Sanchez P, Konig C, Ferrari S, et al. Vitamin B12 as a carrier for targeted platinum delivery: in vitro cytotoxicity and mechanistic studies. J Biol Inorg Chem. 2011;16(1):33–44. doi: 10.1007/s00775-010-0697-z. [DOI] [PubMed] [Google Scholar]
- 16.Collins DA, Hogenkamp HP, O’Connor MK, et al. Biodistribution of radiolabeled adenosylcobalamin in patients diagnosed with various malignancies. Mayo Clin Proc. 2000;75(6):568–580. doi: 10.4065/75.6.568. [DOI] [PubMed] [Google Scholar]
- 17.Zee-Cheng RK, Cheng CC. Delivery of anticancer drugs. Methods Find Exp Clin Pharmacol. 1989;11(7–8):439–529. [PubMed] [Google Scholar]
- 18.Duncan R. Drug-polymer conjugates: potential for improved chemotherapy. Anticancer Drugs. 1992;3(3):175–210. doi: 10.1097/00001813-199206000-00001. [DOI] [PubMed] [Google Scholar]
- 19.Bagnato JD, Eilers AL, Horton RA, et al. Synthesis and characterization of a cobalamin-colchicine conjugate as a novel tumor-targeted cytotoxin. J Org Chem. 2004;69(26):8987–8996. doi: 10.1021/jo049953w. [DOI] [PubMed] [Google Scholar]
- 20.Gupta Y, Ganesh N, Kohli DV, et al. Development and characterization of doxorubicin bearing vitamin B12 coupled sterically stabilized liposomes for tumor targeting. Current Nanoscience. 2011;7(3):427–235. [Google Scholar]
- 21.Bauer JA. Synthesis, characterization and nitric oxide release profile of nitrosylcobalamin: a potential chemotherapeutic agent. Anticancer Drugs. 1998;9(3):239–244. doi: 10.1097/00001813-199803000-00006. [DOI] [PubMed] [Google Scholar]
- 22.Chawla-Sarkar M, Bauer JA, Lupica JA, et al. Suppression of NF-kappa B survival signaling by nitrosylcobalamin sensitizes neoplasms to the anti-tumor effects of Apo2L/TRAIL. J Biol Chem. 2003;278(41):39461–39469. doi: 10.1074/jbc.M306111200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Bauer JA, Lupica JA, Schmidt H, et al. Nitrosylcobalamin potentiates the anti-neoplastic effects of chemotherapeutic agents via suppression of survival signaling. PLoS One. 2007;2(12):e1313. doi: 10.1371/journal.pone.0001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bauer JA, Frye G, Bahr A, et al. Anti-tumor effects of nitrosylcobalamin against spontaneous tumors in dogs. Invest New Drugs. 2010;28(5):694–702. doi: 10.1007/s10637-009-9282-0. [DOI] [PubMed] [Google Scholar]
- 25.Tang Z, Bauer JA, Morrison B, et al. Nitrosylcobalamin promotes cell death via S nitrosylation of Apo2L/TRAIL receptor DR4. Mol Cell Biol. 2006;26(15):5588–5594. doi: 10.1128/MCB.00199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sysel AM, Horne WI, Steiner JM, et al. Serum folate: a pharmacodynamic biomarker of intracellular nitrosylcobalamin activity following intravenous administration in dogs. Anticancer Res. 2012;32(10):4307–4312. [PubMed] [Google Scholar]
- 27.Collins DA, Hogenkamp HP, Gebhard MW. Tumor imaging via indium 111-labeled DTPA-adenosylcobalamin. Mayo Clin Proc. 1999;74(7):687–691. doi: 10.4065/74.7.687. [DOI] [PubMed] [Google Scholar]
- 28.Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182(3):311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 29.Gerdes J, Lemke H, Baisch H, et al. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133(4):1710–1715. [PubMed] [Google Scholar]
- 30.Ohsie SJ, Sarantopoulos GP, Cochran AJ, et al. Immunohistochemical characteristics of melanoma. J Cutan Pathol. 2008;35(5):433–444. doi: 10.1111/j.1600-0560.2007.00891.x. [DOI] [PubMed] [Google Scholar]
- 31.Heeran MC, Hogdall CK, Kjaer SK, et al. Prognostic value of tissue protein expression levels of MIB-1 (Ki-67) in Danish ovarian cancer patients. From the ‘MALOVA’ ovarian cancer study. APMIS. 2013 Apr 18; doi: 10.1111/apm.12071. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 32.Kim BH, Bae YS, Kim SH, et al. Usefulness of Ki-67 (MIB-1) immunostaining in the diagnosis of pulmonary sclerosing hemangiomas. APMIS. 2013;121(2):105–110. doi: 10.1111/j.1600-0463.2012.02945.x. [DOI] [PubMed] [Google Scholar]
- 33.Remes SM, Tuominen VJ, Helin H, et al. Grading of neuroendocrine tumors with Ki-67 requires high-quality assessment practices. Am J Surg Pathol. 2012;36(9):1359–1363. doi: 10.1097/PAS.0b013e3182632038. [DOI] [PubMed] [Google Scholar]
- 34.Canlorbe G, Laas E, Bendifallah S, et al. Contribution of immunohistochemical profile in assessing histological grade of endometrial cancer. Anticancer Res. 2013;33(5):2191–2198. [PubMed] [Google Scholar]
- 35.Nadler A, Cukier M, Rowsell C, et al. Ki-67 is a reliable pathological grading marker for neuroendocrine tumors. Virchows Arch. 2013;462(5):501–505. doi: 10.1007/s00428-013-1410-8. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Yang QX, Tian XT, et al. Malignant transformation of nasal chondromesenchymal hamartoma in adult: a case report and review of the literature. Histol Histopathol. 2013;28(3):337–344. doi: 10.14670/HH-28.337. [DOI] [PubMed] [Google Scholar]
- 37.Colleoni M, Rotmensz N, Maisonneuve P, et al. Prognostic role of the extent of peritumoral vascular invasion in operable breast cancer. Ann Oncol. 2007;18(10):1632–1640. doi: 10.1093/annonc/mdm268. [DOI] [PubMed] [Google Scholar]
- 38.Ladstein RG, Bachmann IM, Straume O, et al. Ki-67 expression is superior to mitotic count and novel proliferation markers PHH3, MCM4 and mitosin as a prognostic factor in thick cutaneous melanoma. BMC Cancer. 2010;10:140. doi: 10.1186/1471-2407-10-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gasparini G, Pozza F, Meli S, et al. Breast cancer cell kinetics: immunocytochemical determination of growth fractions by monoclonal antibody Ki-67 and correlation with flow cytometric S-phase and with some features of tumor aggressiveness. Anticancer Res. 1991;11(6):2015–2021. [PubMed] [Google Scholar]
- 40.Bader AA, Tio J, Petru E, et al. T1 breast cancer: identification of patients at low risk of axillary lymph node metastases. Breast Cancer Res Treat. 2002;76(1):11–17. doi: 10.1023/a:1020231300974. [DOI] [PubMed] [Google Scholar]
- 41.Margulis V, Shariat SF, Ashfaq R, et al. Ki-67 is an independent predictor of bladder cancer outcome in patients treated with radical cystectomy for organ-confined disease. Clin Cancer Res. 2006;12(24):7369–7373. doi: 10.1158/1078-0432.CCR-06-1472. [DOI] [PubMed] [Google Scholar]
- 42.Miyake H, Sakai I, Muramaki M, et al. Prediction of response to combined immunotherapy with interferon-alpha and low-dose interleukin-2 in metastatic renal cell carcinoma: expression patterns of potential molecular markers in radical nephrectomy specimens. Int J Urol. 2009;16(5):465–471. doi: 10.1111/j.1442-2042.2009.02262.x. [DOI] [PubMed] [Google Scholar]
- 43.Takei H, Kurosumi M, Yoshida T, et al. Neoadjuvant endocrine therapy of breast cancer: which patients would benefit and what are the advantages? Breast Cancer. 2011;18(2):85–91. doi: 10.1007/s12282-010-0239-0. [DOI] [PubMed] [Google Scholar]
- 44.Lazzeroni M, Guerrieri-Gonzaga A, Botteri E, et al. Tailoring treatment for ductal intraepithelial neoplasia of the breast according to Ki-67 and molecular phenotype. Br J Cancer. 2013;108(8):1593–1601. doi: 10.1038/bjc.2013.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mao JT, Roth MD, Fishbein MC, et al. Lung cancer chemoprevention with celecoxib in former smokers. Cancer Prev Res (Phila) 2011;4(7):984–993. doi: 10.1158/1940-6207.CAPR-11-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dede DS, Gumuskaya B, Guler G, et al. Evaluation of changes of biologic markers ER, PR, HER 2 and Ki-67 in breast cancer with administration of neoadjuvant dose-dense doxorubicin, cyclophosphamide followed by paclitaxel. J BUON. 2013;18(1):57–63. [PubMed] [Google Scholar]
- 47.Brandao RD, Veeck J, Van de Vijver KK, et al. A randomised controlled phase II trial of pre-operative celecoxib treatment reveals anti-tumour transcriptional response in primary breast cancer. Breast Cancer Res. 2013;15(2):R29. doi: 10.1186/bcr3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fisher G, Yang ZH, Kudahetti S, et al. Prognostic value of Ki-67 for prostate cancer death in a conservatively managed cohort. Br J Cancer. 2013;108(2):271–277. doi: 10.1038/bjc.2012.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reyal F, Hajage D, Savignoni A, et al. Long-term prognostic performance of Ki67 rate in early stage, pT1-pT2, pN0, invasive breast carcinoma. PLoS One. 2013;8(3):e55901. doi: 10.1371/journal.pone.0055901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanchez-Munoz A, Plata-Fernandez YM, Fernandez M, et al. The role of immunohistochemistry in breast cancer patients treated with neoadjuvant chemotherapy: an old tool with an enduring prognostic value. Clin Breast Cancer. 2013;13(2):146–152. doi: 10.1016/j.clbc.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 51.Wang Y, Yin Q, Yu Q, et al. A retrospective study of breast cancer subtypes: the risk of relapse and the relations with treatments. Breast Cancer Res Treat. 2011;130(2):489–498. doi: 10.1007/s10549-011-1709-6. [DOI] [PubMed] [Google Scholar]
- 52.Yamashita S, Moroga T, Tokuishi K, et al. Ki-67 labeling index is associated with recurrence after segmentectomy under video-assisted thoracoscopic surgery in stage I non-small cell lung cancer. Ann Thorac Cardiovasc Surg. 2011;17(4):341–346. doi: 10.5761/atcs.oa.10.01573. [DOI] [PubMed] [Google Scholar]
- 53.Ramirez C, Cheng S, Vargas G, et al. Expression of Ki-67, PTTG1, FGFR4, and SSTR 2, 3, and 5 in nonfunctioning pituitary adenomas: a high throughput TMA, immunohistochemical study. J Clin Endocrinol Metab. 2012;97(5):1745–1751. doi: 10.1210/jc.2011-3163. [DOI] [PubMed] [Google Scholar]
- 54.Li R, Heydon K, Hammond ME, et al. Ki-67 staining index predicts distant metastasis and survival in locally advanced prostate cancer treated with radiotherapy: an analysis of patients in radiation therapy oncology group protocol 86–10. Clin Cancer Res. 2004;10(12 Pt 1):4118–4124. doi: 10.1158/1078-0432.CCR-1052-03. [DOI] [PubMed] [Google Scholar]
- 55.Gimotty PA, Van Belle P, Elder DE, et al. Biologic and prognostic significance of dermal Ki67 expression, mitoses, and tumorigenicity in thin invasive cutaneous melanoma. J Clin Oncol. 2005;23(31):8048–8056. doi: 10.1200/JCO.2005.02.0735. [DOI] [PubMed] [Google Scholar]
- 56.Lusby K, Savannah KB, Demicco EG, et al. Uterine leiomyosarcoma management, outcome, and associated molecular biomarkers: A single institution’s experience. Ann Surg Oncol. 2013;20(7):2364–2372. doi: 10.1245/s10434-012-2834-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dunstan RW, Wharton KA, Jr, Quigley C, et al. The use of immunohistochemistry for biomarker assessment – can it compete with other technologies? Toxicol Pathol. 2011;39(6):988–1002. doi: 10.1177/0192623311419163. [DOI] [PubMed] [Google Scholar]
- 58.Lehr HA, Mankoff DA, Corwin D, et al. Application of photoshop-based image analysis to quantification of hormone receptor expression in breast cancer. J Histochem Cytochem. 1997;45(11):1559–1565. doi: 10.1177/002215549704501112. [DOI] [PubMed] [Google Scholar]
- 59.Bhatnagar J, Tewari HB, Bhatnagar M, et al. Comparison of carcinoembryonic antigen in tissue and serum with grade and stage of colon cancer. Anticancer Res. 1999;19(3B):2181–2187. [PubMed] [Google Scholar]
- 60.Helps SC, Thornton E, Kleinig TJ, et al. Automatic nonsubjective estimation of antigen content visualized by immunohistochemistry using color deconvolution. Appl Immunohistochem Mol Morphol. 2012;20(1):82–90. doi: 10.1097/PAI.0b013e31821fc8cd. [DOI] [PubMed] [Google Scholar]
- 61.Podhajsky RJ, Bidanset DJ, Caterson B, et al. A quantitative immunohistochemical study of the cellular response to crush injury in optic nerve. Exp Neurol. 1997;143(1):153–161. doi: 10.1006/exnr.1996.6354. [DOI] [PubMed] [Google Scholar]
- 62.Dias P, Chen B, Dilday B, et al. Strong immunostaining for myogenin in rhabdomyosarcoma is significantly associated with tumors of the alveolar subclass. Am J Pathol. 2000;156(2):399–408. doi: 10.1016/S0002-9440(10)64743-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jacobs JJ, Lehe C, Cammans KD, et al. An automated method for the quantification of immunostained human Langerhans cells. J Immunol Methods. 2001;247(1–2):73–82. doi: 10.1016/s0022-1759(00)00328-8. [DOI] [PubMed] [Google Scholar]
- 64.Bloom K, Harrington D. Enhanced accuracy and reliability of HER-2/neu immunohistochemical scoring using digital microscopy. Am J Clin Pathol. 2004;121(5):620–630. doi: 10.1309/Y73U-8X72-B68T-MGH5. [DOI] [PubMed] [Google Scholar]
- 65.Lopez C, Lejeune M, Salvado MT, et al. Automated quantification of nuclear immunohistochemical markers with different complexity. Histochem Cell Biol. 2008;129(3):379–387. doi: 10.1007/s00418-007-0368-5. [DOI] [PubMed] [Google Scholar]
- 66.Tuominen VJ, Ruotoistenmaki S, Viitanen A, et al. ImmunoRatio: a publicly available web application for quantitative image analysis of estrogen receptor (ER), progesterone receptor (PR), and Ki-67. Breast Cancer Res. 2010;12(4):R56. doi: 10.1186/bcr2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lejeune M, Jaen J, Pons L, et al. Quantification of diverse subcellular immunohistochemical markers with clinicobiological relevancies: validation of a new computer-assisted image analysis procedure. J Anat. 2008;212(6):868–878. doi: 10.1111/j.1469-7580.2008.00910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol. 2001;23(4):291–299. [PubMed] [Google Scholar]
- 70.Landini G. Color deconvolution using ImageJ. 2013 URL http://www.dentistry.bham.ac.uk/landinig/software/software.html. [Google Scholar]
- 71.Vrekoussis T, Chaniotis V, Navrozoglou I, et al. Image analysis of breast cancer immunohistochemistry-stained sections using ImageJ: an RGB-based model. Anticancer Res. 2009;29(12):4995–4998. [PubMed] [Google Scholar]
- 72.Safadi RA, Musleh AS, Al-Khateeb TH, et al. Analysis of immunohistochemical expression of k19 in oral epithelial dysplasia and oral squamous cell carcinoma using color deconvolution-image analysis method. Head Neck Pathol. 2010;4(4):282–289. doi: 10.1007/s12105-010-0210-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hirano H, Yonezawa H, Yunoue S, et al. Immunoreactivity of Wnt5a, Fzd2, Fzd6, and Ryk in glioblastoma: evaluative methodology for DAB chromogenic immunostaining. Brain Tumor Pathol. 2013 Jun 9; doi: 10.1007/s10014-013-0153-1. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 74.Taylor CR, Levenson RM. Quantification of immunohistochemistry – issues concerning methods, utility and semiquantitative assessment II. Histopathology. 2006;49(4):411–424. doi: 10.1111/j.1365-2559.2006.02513.x. [DOI] [PubMed] [Google Scholar]
- 75.Walker RA. Quantification of immunohistochemistry – issues concerning methods, utility and semiquantitative assessment I. Histopathology. 2006;49(4):406–410. doi: 10.1111/j.1365-2559.2006.02514.x. [DOI] [PubMed] [Google Scholar]
- 76.Dowsett M, Nielsen TO, A’Hern R, et al. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. J Natl Cancer Inst. 2011;103(22):1656–1664. doi: 10.1093/jnci/djr393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hall CA. The uptake of vitamin B12 by human lymphocytes and the relationships to the cell cycle. J Lab Clin Med. 1984;103(1):70–81. [PubMed] [Google Scholar]
- 78.Fiskerstrand T, Riedel B, Ueland PM, et al. Disruption of a regulatory system involving cobalamin distribution and function in a methionine-dependent human glioma cell line. J Biol Chem. 1998;273(32):20180–20184. doi: 10.1074/jbc.273.32.20180. [DOI] [PubMed] [Google Scholar]
- 79.Abe T, Tada M, Shinohara N, et al. Establishment and characterization of human urothelial cancer xenografts in severe combined immunodeficient mice. Int J Urol. 2006;13(1):47–57. doi: 10.1111/j.1442-2042.2006.01220.x. [DOI] [PubMed] [Google Scholar]
- 80.Flatmark K, Reed W, Halvorsen T, et al. Pseudomyxoma peritonei--two novel orthotopic mouse models portray the PMCA-I histopathologic subtype. BMC Cancer. 2007;7:116. doi: 10.1186/1471-2407-7-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stoyanova R, Hachem P, Hensley H, et al. Antisense-MDM2 sensitizes LNCaP prostate cancer cells to androgen deprivation, radiation, and the combination in vivo. Int J Radiat Oncol Biol Phys. 2007;68(4):1151–1160. doi: 10.1016/j.ijrobp.2007.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fatema CN, Zhao S, Zhao Y, et al. Monitoring tumor proliferative response to radiotherapy using (18)F-fluorothymidine in human head and neck cancer xenograft in comparison with Ki-67. Ann Nucl Med. 2013;27(4):355–362. doi: 10.1007/s12149-013-0693-9. [DOI] [PubMed] [Google Scholar]
- 83.Schneider RJ, Burger RL, Mehlman CS, et al. The role and fate of rabbit and human transcobalamin II in the plasma transport of vitamin B12 in the rabbit. J Clin Invest. 1976;57(1):27–38. doi: 10.1172/JCI108265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Youngdahl-Turner P, Mellman IS, Allen RH, et al. Protein mediated vitamin uptake. Adsorptive endocytosis of the transcobalamin II-cobalamin complex by cultured human fibroblasts. Exp Cell Res. 1979;118(1):127–134. doi: 10.1016/0014-4827(79)90590-1. [DOI] [PubMed] [Google Scholar]
- 85.Fasanella S, Leonardi E, Cantaloni C, et al. Proliferative activity in human breast cancer: Ki-67 automated evaluation and the influence of different Ki-67 equivalent antibodies. Diagn Pathol. 2011;6(Suppl 1):S7. doi: 10.1186/1746-1596-6-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pathmanathan N, Balleine RL. Ki67 and proliferation in breast cancer. J Clin Pathol. 2013;66(6):512–516. doi: 10.1136/jclinpath-2012-201085. [DOI] [PubMed] [Google Scholar]