Abstract
The cardiac transcription factor Tbx20 has a critical role in the proper morphogenetic development of the vertebrate heart, and its misregulation has been implicated in human congenital heart disease. Although it is established that Tbx20 exerts its function in the embryonic heart through positive and negative regulation of distinct gene programs, it is unclear how Tbx20 mediates proper transcriptional regulation of its target genes. Here, using a combinatorial proteomic and bioinformatic approach, we present the first characterization of Tbx20 transcriptional protein complexes. We have systematically investigated Tbx20 protein-protein interactions by immunoaffinity purification of tagged Tbx20 followed by proteomic analysis using GeLC-MS/MS, gene ontology classification, and functional network analysis. We demonstrate that Tbx20 is associated with a chromatin remodeling network composed of TLE/Groucho co-repressors, members of the Nucleosome Remodeling and Deacetylase (NuRD) complex, the chromatin remodeling ATPases RUVBL1/RUVBL2, and the T-box repressor Tbx18. We determined that the interaction with TLE co-repressors is mediated via an eh1 binding motif in Tbx20. Moreover, we demonstrated that ablation of this motif results in a failure to properly assemble the repression network and disrupts Tbx20 function in vivo. Importantly, we validated Tbx20-TLE interactions in the mouse embryonic heart, and identified developmental genes regulated by Tbx20:TLE binding, thereby confirming a primary role for a Tbx20-TLE repressor complex in embryonic heart development. Together, these studies suggest a model in which Tbx20 associates with a Gro/TLE-NuRD repressor complex to prevent inappropriate gene activation within the forming heart.
Keywords: Tbx20, repression complex, T-box, label-free MS/MS, Groucho, TLE3, heart development, cardiac, Tbx18
Introduction
The development and maturation of a functional heart is a complex process that involves distinct but overlapping phases of specification, proliferation, migration, differentiation, and morphogenesis. Disturbances in any of these processes can lead to a number of congenital heart defects. Currently, congenital heart defects affect nearly 1% of all newborns and are a significant cause of infant death 1. Recent studies have demonstrated that human patients with dilated cardiomyopathy, atrial septal defects, or mitral valve disease carry mutations in the transcription factor Tbx20, while upregulation of Tbx20 gene expression has been reported in patients with tetralogy of Fallot 2. Tbx20 is a member of the T-box family of transcription factors, all of which share a well-conserved DNA binding domain known as the T-box and have diverse roles in embryonic development. Tbx20 has been identified in many organisms, including Drosophila, zebrafish, Xenopus, and mouse, and in all species examined Tbx20 transcripts are strongly expressed throughout the developing heart 3. Results from genetic analysis and protein depletion studies are consistent with a role for Tbx20 during the early stages of vertebrate heart development; hearts lacking Tbx20 show a progressive loss of cardiomyocytes, a failure of the heart to undergo looping and chamber formation, and defects in cardiomyocyte maturation 4. Collectively, these studies suggest that the sequence, expression, and function of Tbx20 are evolutionarily conserved from flies to human.
Similar to other T-box factors, Tbx20 is localized to the nucleus, binds DNA in a sequence-specific manner, and modulates transcription of downstream target genes 4a-e, 5. Results from a number of studies have shown that Tbx20 can act to both promote and repress target gene expression in the heart; however, it is unclear how Tbx20 initiates a transcriptional repressive program within the same cells in which it also acts as a potent transcriptional activator. It has been proposed that protein co-factors may act to specify Tbx20 transcriptional activity 5c. A model in which protein co-factors act as determinants of Tbx20 activity has several unresolved issues because few in vivo Tbx20 co-factors have been identified. Additionally, there is uncertainty about the precise mechanism by which binding of Tbx20 to DNA results in either activation or repression of a target gene. In vitro assays have been used to demonstrate interactions between Tbx20 and a suite of cardiac transcription factors that include Tbx5, Nkx2.5, Gata4, Gata5, and Islet1 4a, 5a, although none of these interactions have been shown to occur in vivo in the embryonic heart. Indeed, the presence of DNA-binding motifs for Nkx2.5, Gata4, and Tbx5 in the promoter regions of Tbx20 target genes, in combination with evidence that these transcription factors act combinatorially to promote target gene expression suggest that cardiac transcription factors are important co-factors for Tbx20 to activate gene expression in the developing heart 4e, 5a, 5c. However, it is not well understood how Tbx20 functions as a transcriptional repressor as co-factors that may act as functional co-repressors have not been identified. Therefore, the precise mechanisms by which Tbx20 regulates distinct gene programs in the heart remains unclear.
To begin to address these questions, we have undertaken, to our knowledge, the first proteomic study aimed at identifying Tbx20 protein interactions. Using affinity purification mass spectrometry (AP-MS) 6, we have systematically characterized Tbx20-containing transcriptional complexes. With this approach, we have identified a unique Tbx20 chromatin remodeling network that includes the Groucho-related proteins Transducin-like Enhancer of Split 1 and 3 (TLE1/3), Metastasis-associated Protein 1 (MTA1), the histone-binding proteins RBBP4 and RBBP7, RUVB-like 1 and 2, Nucleolin, Nucleophosmin, Histone Deacetylase 2 (HDAC2), and the T-box repressor Tbx18. We provide evidence that Tbx20 recruits TLE1/3 through an evolutionarily conserved N-terminal engrailed homology 1 (eh1) binding motif, and demonstrate that recruitment of NuRD complex components requires binding of TLE3 to Tbx20. We find that TLE family members are expressed in mouse embryonic heart tissue, and that Tbx20 interacts with both TLE1 and TLE3 in vivo during heart development, representing the first endogenous Tbx20 interactions identified in embryonic heart tissue to date. Finally, we observe that the Tbx20-TLE interaction is essential for Tbx20 transcriptional activity in the embryo, and we define a unique set of genes that are repressed by the Tbx20-TLE transcriptional complex. We propose a model in which Tbx20 binds to TLE factors to assemble a chromatin remodeling and deacetylase complex on target gene loci to repress distinct genetic programs in the forming heart.
Materials and Methods
DNA constructs
Mouse Tbx20a cDNA was fused to EGFP and cloned into the pMONO-neo-mcs plasmid (Invitrogen) for expression in HEK293 cells. The Tbx20eh1mut-EGFP construct was generated by site-direct mutagenesis (Stratagene) of phenylalanine 18 (F18L) and serine 19 (S19I) using the primers 5’-CTCTCGAGCCAATGCCTTAATCATCGCCGCGCTTATGTC -3’ and 5’- GACATAAGCGCGGCGATGATTAAGGCATTGGCTCGAGAG -3’ according to manufacturer's instructions. To generate the Tbx20-HA construct, mouse Tbx20a cDNA was fused to an HA epitope and cloned into pMONO-neo-mcs. The pCMV2-TLE1-Flag construct was generously provided by Dr. Stefano Stifani 7. The pCMX-TLE3 plasmid was kindly provided by Dr. Peter Tontonoz 8. Tbx18-Flag was generously provided by Dr. Chen-Leng Cai 9.
Xenopus injections and animal cap isolation
Xenopus laevis embryos were staged according to Nieuwkoop and Faber 10 and injected with 1 ng Tbx20 or Tbx20-EGFP mRNA at the one-cell stage using established protocols 11. Animal caps were excised at stage 8-9 and cultured in 1X modified Barth's saline (MBS) until sibling embryos reached stage 13. Activin-treated caps were cultured in 8 Units/mL Activin in 1X MBS.
Isolation of Tbx20-EGFP protein complexes
pMONO-Tbx20-EGFP or Tbx20eh1mut-EGFP plasmids were transfected into HEK293 cells using FuGENE (Roche Applied Science). Tbx20-EGFP complexes and GFP complexes were immunoaffinity purified from cells using in-house developed rabbit polyclonal anti-GFP antibodies conjugated to magnetic beads, as previously described 12. Briefly, HEK293 cells expressing Tbx20-EGFP or GFP alone were washed with cold PBS, harvested from the plate by scraping with a plastic spatula and pelleted at 1500 rpm for 10 min at 4°C. The cell pellet was resuspended in 100 μL/1 μg 20 mM HEPES, pH 7.4, containing 1.2% polyvinylpyrrolidone and protease inhibitors and snap frozen in liquid nitrogen. Cells were lysed by cryogenic grinding using a Retsch MM 301 Mixer Mill (10 cycles × 2.5 min at 30 Hz) (Retsch, Newtown, PA) and the frozen cell powder was resuspended in optimized lysis buffer (5 mL/1 g cells) (20 mM K-HEPES pH 7.4, 0.1 M KOAc, 2 mM MgCl2, 0.1% Tween-20, 1 μM ZnCl2, 1 μM CaCl2, 150 mM NaCl, 0.5% Triton X-100 containing protease and phosphatase inhibitors). Cell lysates were homogenized using a Polytron (Kinematica) step (2 × 15 sec) and pelleted at 8000 rpm at 4°C. Cleared lysates were rotated with 7 mg magnetic beads (M270 Epoxy Dynabeads, Invitrogen) coupled to anti-GFP antibodies for 1 hr at 4°C. The magnetic beads were then washed in lysis buffer (6 × 1 mL) (without protease and phosphatase inhibitors) and eluted from the beads in 40 μL 1× LDS Sample Buffer (Invitrogen) at 70°C for 15 min. Eluted proteins were alkylated with 100 mM iodoacetamide for 1 hr at room temperature and subjected to mass spectrometry analysis.
Mass spectrometry analysis of Tbx20-EGFP protein complexes
Immunoisolates were analyzed by mass spectrometry as previously described 13 with minor differences. Briefly, reduced and alkylated eluates were partially resolved by SDS-PAGE on 4-12% Bis-Tris NuPAGE gels (Invitrogen) and stained using SimplyBlue Coomassie stain (Invitrogen). Each lane was divided into 1 mm slices and binned into 8 wells of a 96-well plate. Gel slices were destained in 50 mM ammonium bicarbonate (ABC) containing 50% acetonitrile (ACN). Proteins were digested in-gel with 20 μL of 12.5 ng/μL trypsin in 50 mM ABC for 5 hrs at 37°C. Tryptic peptides were extracted in 0.5% formic acid for 4 hrs at room temperature, followed by 0.5% formic acid/50% ACN for 2 hrs at room temperature. The extracted peptides were concentrated by vacuum centrifugation to 10 μL and either desalted online (trap column, Magic C18 AQ, 100 μm × 2.5cm) or offline using StageTips. Desalted peptides (4 μL) were separated online by reverse phase C18 (Acclaim PepMap RSLC, 1.8 μm, 75 μm × 25 cm) over 90 min at 250 nL/min using a Dionex Ultimate 300 nanoRSLC and detected by an LTQ Orbitrap Velos or XL mass spectrometer (Thermofisher Scientific, San Jose, CA).
The mass spectrometer was operated in data-dependent acquisition mode with dynamic exclusion enabled. A single acquisition cycle comprised a single full-scan mass spectrum (m/z = 350–1700) in the Orbitrap (r = 30,000 at m/z = 400), followed by collision-induced dissociation (CID) fragmentation in the linear ion trap of the top 10 (XL) or 20 (Velos) most intense precursor ions. FT full scan target value was 1E6 with a max. injection time of 300 ms. IT tandem MS target values were 5E3 (XL) or 1E4 (Velos) with a maximum injection time of 100 ms. CID fragmentation was performed at an isolation width of 2.0 Th, normalized collision energy of 30, and activation time of 30 (XL) or 10 ms (Velos).
Data processing and functional protein analyses
MS/MS spectra were extracted from Thermo RAW files and searched by Proteome Discoverer/SEQUEST (version 1.3, Thermo Fisher Scientific) against the UniProt SwissProt protein sequence database (release 2010-11) containing forward and reverse entries from human and the mouse Tbx20a sequence plus common contaminants (20,324 forward sequences). SEQUEST search parameters were as follows: full enzyme specificity with 2 missed cleavages, precursor and fragment tolerances, 10 ppm and 0.5 Da, fixed modification, carbamidomethylation of cysteine, and variable modifications, oxidized methionine and phosphorylation of STY. SEQUEST peptide spectrum matches (MSF files) were loaded into Scaffold software (ver. 3.5.1, Proteome Software, Inc), subjected to an X! Tandem refinement search, and then analyzed by PeptideProphet and ProteinProphet algorithms to determine peptide and protein probabilities. The high mass accuracy option for probability scoring was enabled. The following peptide modifications were included in the X!Tandem refinement search: deamidation of NQ, and acetylation of K and amino-terminus. Protein groups were assembled by Scaffold and filtered by a minimum of 2 unique peptides. Probability thresholds were empirically defined to achieve < 1% peptide and protein FDR as assessed by matches to the reverse database. Proteins descriptions, accession numbers, and their respective unique peptides and unweighted spectrum counts were exported to Excel for further analysis.
Specific Tbx20 protein interactions were identified by spectral counting enrichment analysis 13 comparing the Tbx20-EGFP versus EGFP alone conditions. The following criteria were applied to each individual replicate (N=3): 1) only proteins with ≥ 5 spectrum counts were retained, 2) only those proteins that had a spectral count enrichment of ≥ 2.5-fold versus GFP alone were retained, and 3) using GO annotations, proteins assigned a “nuclear” localization ontology term were retained (Supplemental Table 1; see Supplemental Table 2 for proteins excluded for not being associated with a nuclear localization ontology term). The proteins that fulfilled these criteria in all three replicates were imported into Cytoscape 14 for classification into functional subgroups according to biological processes using the plugin ClueGO (Supplemental Table 3) 15. Proteins within GO term clusters were analyzed in STRING using protein accessions as input 16.
Comparison of interaction protein abundance versus estimated average proteome abundance, defined here as an enrichment index, was used to identify prominent interaction candidates from the GO-classified nuclear proteins, as described in 13. To calculate this enrichment index, first, a protein's spectral counts was normalized within each biological replicate (N = 3) by the Tbx20 spectral count ratio of each individual replicate / average. Then, normalized spectral counts were converted to NSAF (Normalized Spectral Abundance Factor) values 17 and further normalized by estimated proteome abundance from the human subset of the PAX (protein abundance across organisms) database 18 (Supplemental Table 4). Tbx20 spectral counts were excluded from this analysis. We have previously shown that calculating this enrichment index aids in identifying high confidence interactions that prominently associate with the bait 13.
Analysis of Tbx20-EGFP and Tbx20eh1mut-EGFP protein complexes using mass spectrometry-based label-free analysis
Tbx20-EGFP and Tbx20eh1mut-EGFP complexes were immunoisolated and analyzed by LC-MS/MS as described above. Peptide and protein identifications were filtered using the criteria described above, including the requirement of at least 5 spectra per protein identified for either wild-type or mutant isolations and the requirement that all proteins identified in the wild-type isolation be at least 2.5-fold enriched over isolations from EGFP-expressing cells. To determine differences in interacting proteins between wild-type and mutant Tbx20 isolations, a combination of spectral counting and MS1 label-free approaches were employed. For spectral counting analysis, the spectral counts for putative protein interactions in the Tbx20eh1mut isolation were normalized by the Tbx20 spectral count ratio of eh1mut/wild-type isolation. Then, the fold-difference in spectral counts of putative interactions (Tbx20eh1mut isolation relative to wild-type Tbx20) was used to identify differential interacting proteins. For selected Tbx20 interactions, label-free quantification was performed by MS1 peak area measurements using the MaxQuant software (ver 1.4.0.8) 19. Relative quantification was performed using the MaxQuant “intensity” measurements and the “Match between runs” feature enabled, which quantified non-sequenced peptide features based on accurate mass, retention time, and sequence information collected from other data-dependent LC-MS/MS runs. Only unmodified peptides designated as unique at the protein group level were used to calculate summed protein intensities. The intensity ratio of Tbx20 in the eh1mut/wild type isolation was used to normalize protein interaction ratios. The normalized fold-differences of putative interactions (Tbx20eh1mut isolation relative to the wild-type Tbx20) were used as a measure of differential interaction. Interacting proteins with fold-differences ≤ 0.5 were considered to be significantly reduced in mutant isolations (Supplemental Tables 5, 6).
Construction of a HEK293 HDAC2-EGFP stable cell line
The HDAC2 ORF was amplified from an HDAC2 plasmid (gift from E. Seto, Moffitt Cancer Center), and inserted into the pLXSN-C-EGFP-FLAG vector to create the HDAC2-EGFP-flag fusion, as in 20. The Phoenix™ retrovirus expression system (Orbigen, San Diego, CA) was used to transduce HEK293 cells to express the HDAC2-EGFP-FLAG fusion according to the manufacturer's instructions. The transduced cells were selected in 300 mg/L G418 (EMD, Gibbstown, NJ) and sorted by FACS (Vantage S.E. with TurboSort II, Becton Dickinson, Franklin Lakes, NJ) to obtain a stable cell line. The nuclear localization and deacetylation activity of the GFP-tagged HDAC2 were confirmed.
Isolation of endogenous Tbx20 from mouse embryonic hearts
Pregnant CD1 females were sacrificed on embryonic day 10.5 (E10.5) and the embryos removed. Embryonic hearts (n=25) were dissected from the embryos in cold PBS and snap-frozen in liquid nitrogen. Embryonic hearts were cryogenically lysed, and endogenous Tbx20 protein complexes were immunoaffinity purified as described using 5 mg magnetic beads (M270 Epoxy Dynabeads, Invitrogen) conjugated to anti-Tbx20 antibodies (Santa Cruz Biotechnology). The isolated proteins were analyzed by Western blotting.
Immunofluorescence and immunoblotting
For immunofluorescence of HEK293 cells, cells were cultured in 8-well chamber slides pretreated with poly-D-lysine. For live imaging of EGFP fluorescence, cells were transfected with pMONO-Tbx20-EGFP. Forty-eight hours later, the cells were rinsed with 1X PBX and DAPI added (200 ng/mL in 1X PBS) for 30 min. Cells were imaged by confocal microscopy on a Zeiss 710.
Antibodies used for immunoblotting include mouse anti-GFP (JL8) (Clontech Living Colors Monoclonal), mouse anti-Flag (M2) (Sigma), goat anti-TLE1 (N-18) (Santa Cruz Biotechnology), rabbit anti-TLE3 (M-201) (Santa Cruz Biotechnology), mouse anti-GAPDH (Millipore), and goat anti-Tbx20 (Santa Cruz Biotechnology).
RNA extraction and RT-PCR
RNA was extracted using Trizol (Invitrogen) and purified on RNeasy columns (Qiagen). cDNA synthesis was performed from 0.5-1 μg of RNA using random primers and SuperScript II reverse transcriptase (Invitrogen). Expression levels were assessed using GoTaq Green Master Mix (Promega) and Taq polymerase on a GeneAmp PCR System (Applied Biosystems). PCR products were analyzed by 2.5% agarose gel electrophoresis.
In situ hybridization
E10.5 embryos were fixed whole in 4% paraformaldehyde and 16 μm tissue sections were cut for in situ hybridization. Fragments corresponding to the 5’ UTR of Tbx20, TLE1, and TLE3 transcripts were cloned into a pStrataclone cloning vector (Strataclone Blunt PCR Cloning Kit, Agilent Technologies), then digoxigenin-labeled antisense riboprobes were generated and hybridized to the tissue sections using standard techniques. Sections were imaged on an Olympus BX61 Upright Wide Field microscope.
RNA-seq and differential expression analyses
RNA was extracted from stage 13 Xenopus laevis embryos injected with Tbx20-EGFP or Tbx20eh1mut-EGFP mRNA. As a control, RNA was also extracted from uninjected stage 13 siblings. The Illumina Tru-seq RNA sample prep kit was used for cDNA library preparation. Libraries were sequenced using an Illumina HiSeq 2500 system (Vanderbilt Technologies for Advanced Genomics, Vanderbilt University). Each sample (two replicates per condition) generated between 32 million and 38.5 million single-end, 50-bp reads. These were mapped to a set of 8,879 X. laevis transcripts using the default options of Bowtie2 (version 2.1.0) 21 as described previously22, with a unique-mapping rate of 45-47%. Bowtie2 output files were converted to binary forms and sorted using Samtools (version 0.1.19)23. Transcript assemblies and expression level quantification were performed on each sample using the default settings of Cufflinks (version 2.0.2)24. The transcript lists were subsequently merged using the default settings of the Cuffmerge script in Cufflinks to produce a comprehensive list of transcripts represented in all our Cufflinks assemblies (Supplemental Tables 7, 8). Differential expression analysis was performed on all samples using the default settings of the Cuffdiff program in Cufflinks, with the bias detection and correction option (-b) and the multi-mapping correction option (-u) (Supplemental Tables 7, 8). Gene ontology analysis was performed using GOrilla25 (Supplemental Table 9, 10). Heat maps were generated using the heatmap.2 function of the gplots package (R package version 2.11.3, R Foundation for Statistical Computing, Vienna, Austria) in R (version 3.0.1, R Foundation for Statistical Computing, Vienna, Austria).
Results
Tbx20-EGFP is localized to the nucleus and transcriptionally active
Identification of critical Tbx20 protein co-factors in a high throughput manner has been hampered by a lack of antibodies against Tbx20 that are suitable for directed proteomics analyses. Additionally, there are no cell lines that recapitulate endogenous Tbx20 expression and thus could provide sufficient material for large-scale proteomics studies of Tbx20 protein complexes. Since the main goal of this set of studies was to determine the general transcriptional mechanisms by which Tbx20 functions, we generated human embryonic kidney (HEK293) cells expressing Tbx20 tagged at the C-terminus with EGFP (Figure 1A). HEK293 cells have been used as a cell culture model for studies on the transcriptional activity of Tbx20 4a, b, indicating that this cell line contains the necessary cohort of transcriptional co-factors required for Tbx20-mediated transcriptional regulation. In agreement with its known role in transcription, Tbx20-EGFP localizes to the nucleus when expressed in HEK293 cells, as shown by live GFP fluorescence microscopy (Figure 1B).
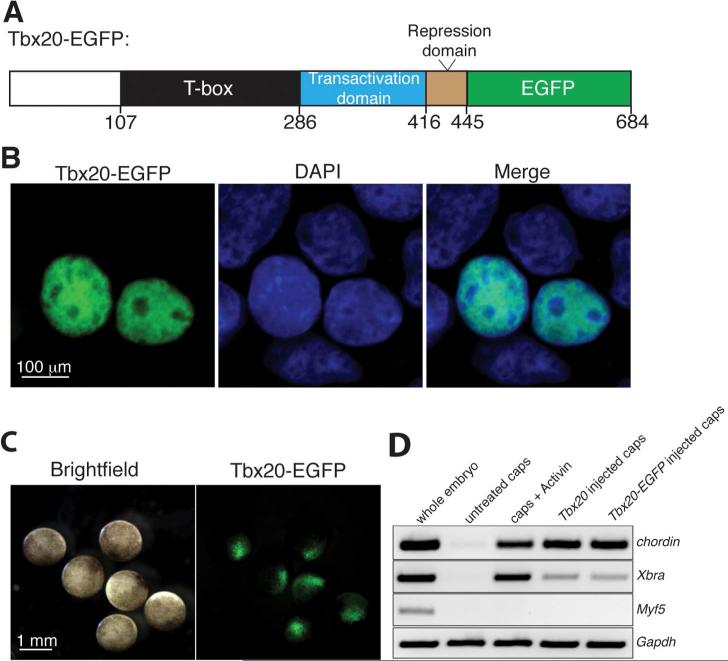
Figure 1.
Tbx20-EGFP is nuclear-localized and transcriptionally active. (A) Schematic of EGFP-tagged (green) Tbx20 expression construct, showing the N-terminal (white), T-box (black), transactivation (blue), and repression (brown) domains. A putative Tbx5 protein-protein interaction (PPI) domain lies within the N-terminus and T-box. Numbers denote amino acid residues. (B) Tbx20-EGFP is localized to the nucleus in HEK293 cells, as confirmed by live GFP fluorescence and colocalization with DAPI. (C) Tbx20-EGFP mRNA was injected at the 1-cell stage into Xenopus embryos. Expression of Tbx20-EGFP in the animal pole of stage 9 Xenopus embryos was confirmed by live GFP fluorescence. (D) RT-PCR analysis of the mesodermal genes chordin and Xbra and the skeletal muscle gene Myf5 in stage 13 whole embryos, stage-matched untreated animal caps, Activin-treated animal caps, Tbx20-injected animal caps, and Tbx20-EGFP-injected animal caps. The housekeeping gene Gapdh was used as a loading control for all RT-PCR reactions.
To confirm that the EGFP-tagged Tbx20 is transcriptionally active, we made use of a Xenopus animal cap assay. The animal cap is a region of the Xenopus blastula and early gastrula stage embryo (stages 8/9) that consists of naïve pluripotent cells. Animal caps normally contribute to skin and nervous tissue; however, a recent study demonstrated that animal caps excised from embryos injected with Tbx20 mRNA express the early mesoderm marker Xbra, but not the skeletal muscle marker Myf5, indicating that Tbx20 can induce cell fate changes in the early embryo 5a. To assess whether EGFP-tagged Tbx20 has the same ability to induce gene expression changes as untagged Tbx20, we injected Tbx20 and Tbx20-EGFP mRNA into one-cell stage Xenopus laevis embryos. By stage 9, Tbx20-EGFP protein is localized throughout the animal pole of the embryo, as shown by live GFP fluorescence microscopy (Figure 1C). At this stage, animal caps were excised from Tbx20- and Tbx20-EGFP-injected embryos and cultured in isolation until uninjected sibling whole embryos reached stage 13. After extraction of RNA, we assessed the expression of the early mesodermal markers chordin and Xbra, and as a negative control, Myf5, by RT-PCR. The levels of induction were compared to expression of these genes within whole embryos, untreated caps, and, as a positive control, caps treated with the mesoderm-inducing factor Activin. Both EGFP-tagged Tbx20 and untagged Tbx20 induce chordin expression to the same degree as Activin (Figure 1D). Further, Tbx20 and Tbx20-EGFP induce moderate levels of Xbra compared to Activin-induced caps; however, there was no induction of the muscle marker Myf5 by either version of Tbx20 or by Activin treatment. Untreated caps did not express any of the tissue-specific markers tested. These data indicate that EGFP-tagged Tbx20 is transcriptionally active and retains the ability to modulate downstream gene expression.
Proteomic analysis of Tbx20-EGFP interactions reveals association with a unique chromatin remodeling network
To systematically identify Tbx20-associated proteins, we performed immunoaffinity purifications of Tbx20-EGFP complexes from HEK293 cells using a high affinity in-house developed antibody against GFP 12. In parallel, as controls, we performed immunoaffinity purifications from cells expressing EGFP alone (Supplemental Tables 1, 2). Immunopurified proteins were partially resolved by SDS-PAGE, digested in-gel with trypsin, and analyzed by nLC-tandem MS (MS/MS) on an LTQ Orbitrap XL or an LTQ Orbitrap Velos. Three independent biological replicates were performed (Supplemental Tables 1,2); two of these immunopurifications were performed in the presence of DNase to eliminate interactions mediated by binding of factors on adjacent DNA sequences. Raw MS/MS spectra from each experiment were analyzed by SEQUEST database searches (Proteome Discoverer) and loaded into Scaffold for further analysis. Protein identifications from all three replicates were filtered using stringent confidence parameters (see Materials and Methods). A spectrum counting approach was employed to assess enrichment of protein interactions with Tbx20-EGFP relative to EGFP alone. First, proteins were required to be reproducibly present, detected by at least 5 spectrum counts. Second, proteins showing less than 2.5-fold spectrum count enrichment over proteins co-isolated with EGFP alone were deemed non-specific and were excluded. Further, given the nuclear localization of Tbx20-EGFP, proteins lacking a nuclear gene ontology term were excluded as likely non-specific associations occurring during whole-cell lysis (Supplemental Table 2).
In total, 114 proteins with annotated nuclear localization passed our spectrum count and fold enrichment criteria, 97% of which occur in all three biological replicates. There were no proteins identified as unique to a single Tbx20-EGFP isolation; however, 3 proteins, while detected in all three isolations, failed to pass our stringent criteria in one of the three experiments (Supplemental Table 1). To assess the Tbx20 nuclear interaction network, we classified the proteins passing our spectrum count and fold enrichment criteria into functional subgroups by Cytoscape, using the ClueGO plugin 14-15. Specifically, proteins were assigned into gene ontology (GO) term clusters according to biological function ontologies. When we examined Tbx20 nuclear interactions, the most prominent biological function category contained 53 proteins related to RNA processing (Figure 2A and Supplemental Table 3). This is expected, as it is well established that the DNA template that is being actively transcribed is often closely associated with the RNA processing machinery and suggests that a portion of Tbx20 binding is closely associated with active transcription. Nuclear interactions also included 13 proteins involved in nucleosome assembly (Figure 2A and Supplemental Table 3). Nucleosome assembly functions were represented by such GO terms as chromatin remodeling, DNA conformation change, and nucleosome organization , indicating a crucial role for Tbx20 in modification of chromatin architecture. Sixteen proteins were assigned to DNA repair/synthesis, and 10 proteins were assigned to nuclear transport (Figure 2A and Supplemental Table 3). Collectively, these putative Tbx20 interactions represent different complexes and functions of Tbx20 throughout the nucleus.
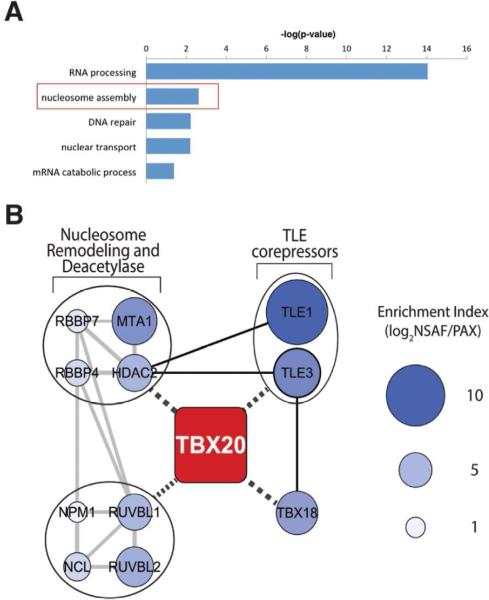
Figure 2.
Shotgun proteomics of Tbx20-EGFP protein complexes reveals association with a chromatin remodeling and Groucho transcriptional protein network. (A) GO enrichment analysis of Tbx20 interactions using ClueGO clustering according to biological function ontologies. (B) The Cytoscape network was assembled from automated retrieval and manual curation of protein functional associations using STRING analysis (grey lines) and literature curation (solid black lines), respectively. Potential interactions/functional associations with Tbx20 are indicated by black dashed lines. Nodes are labeled with respective gene symbols and enrichment index values (see Materials and Methods) are represented by node size and blue color intensity. Functional protein groupings are indicated in closed circles.
Given the lack of knowledge regarding the molecular mechanisms of Tbx20-mediated transcriptional regulation, we reasoned that the specific protein functions represented within the ‘nucleosome assembly’ category could provide new insight on the potential roles of Tbx20 in gene regulation. Moreover, since chromatin remodeling and transcription repression complexes are often large multi-protein complexes, we speculated that the specific proteins represented within the ‘nucleosome assembly’ category were likely to be interconnected. To test this hypothesis, we analyzed the proteins within this functional cluster (Table 1 and Supplemental Table 3) using STRING, a knowledge database of known and predicted protein-protein interactions16, with the aim of generating a predictive Tbx20 interaction network. Eight of these proteins form a highly interconnected network containing chromatin remodeling and deacetylase functions and include the nucleolar proteins Nucleophosmin (NPM1) and Nucleolin (NCL), as well as core components of the Nucleosome Remodeling and Deacetylase (NuRD) complex (MTA1, RBBP4, RBBP7, and HDAC2)—a major ATP-dependent chromatin remodeling complex with important roles in transcription and chromatin assembly (reviewed in 26) (Table 1 and Figure 2B, gray lines). NCL and MTA1 were integrated into this network based upon their known or predicted functional association with components of the chromatin remodeling network 27. Additionally, this network includes two members of the INO80 chromatin remodeling complex, the ATPases RUVBL1 and RUVBL2. The INO80 complex is a highly conserved, multisubunit, ATP-dependent chromatin remodeling complex that contributes to both transcriptional activation and repression 28.
Table 1.
Tbx20-associated proteins identified by LC-MS/MS. Numbers represent an average across three experimental replicates.
| Protein description | Gene name | GFP | Tbx20-EGFP | ||
|---|---|---|---|---|---|
| Spectrum counts | Spectrum counts | Unique peptides | Percent coverage | ||
| T-Box transcription factor Tbx20 | Tbx20 | 1 | 707 | 54 | 73 |
| Green fluorescent protein | GFP | 429 | 411 | 23 | 63 |
| T-box transcription factor Tbx18 | Tbx18 | 1 | 11 | 2 | 5 |
| Transducin-like enhancer of split 1 | TLE1 | 1 | 17 | 5 | 15 |
| Transducin-like enhancer of split 3 | TLE3 | 2 | 29 | 13 | 23 |
| Histone-binding protein Rbbp4 | RBBP4 | 3 | 15 | 7 | 21 |
| Histone-binding protein Rbbp7 | RBBP7 | 1 | 10 | 2 | 16 |
| Histone deacetylase 2 | HDAC2 | 3 | 11 | 3 | 14 |
| Metastasis-associated protein MTA1 | MTA1 | 1 | 7 | 4 | 7 |
| Nucleophosmin | NPM1 | 7 | 40 | 10 | 51 |
| Nucleolin | NCL | 14 | 65 | 14 | 22 |
| RuvB-like 1 | RUVBL1 | 3 | 23 | 8 | 23 |
| RuvB-like 2 | RUVBL2 | 4 | 24 | 10 | 24 |
As this is the first demonstration that Tbx20 associates with chromatin remodeling proteins, it was unclear how Tbx20 might be functionally linked to this chromatin modification network. There is evidence that MTA family proteins interact directly with transcription factors at target gene loci 29; however, different transcription factors have been shown to bind to different regions of individual subunits of the NuRD complex 30. To attempt to identify the functional link between Tbx20 and the chromatin remodeling network, we first examined the list of nuclear-enriched proteins for additional components of a chromatin modification network that may have been excluded in our original analysis due to incomplete GO or functional annotation. Surprisingly, this search uncovered the transcriptional co-repressors Transducin-like Enhancer of split 1 and 3 (TLE1 and TLE3) and the T-box transcriptional repressor Tbx18. TLE family members are orthologs of the Drosophila Groucho protein and have been previously demonstrated to bind directly to T-box factors, including Tbx18 and Tbx15, through an engrailed homology 1 (eh1) binding motif and achieve transcriptional repression by recruiting histone deacetylases 31. Based upon the reported functional relationships between TLE proteins, T-box factors, and HDACs, we incorporated TLE1/3 and Tbx18 into the STRING network (Table 1 and Figure 2B, black lines).
To examine if Gro/TLE factors could serve as proximal interacting partners, linking Tbx20 to chromatin remodeling complexes, we estimated the relative enrichment of proteins within the interaction network. As previously described 6, 13, the relative protein enrichment within the immunoisolates was estimated by normalizing their relative protein abundances (NSAF values) 17 to their proteome abundances from the PAX database (pax-db.org) (Supplemental Table 4). This relative enrichment analysis was performed for the 114 proteins enriched in Tbx20 immunoisolations and in the “nuclear” GO subcellular localization (Supplementary Table 3). Respective enrichment index values were then illustrated in the Gro/TLE-chromatin remodeling network (Figure 2B). We hypothesized that proteins within the interaction network having greater enrichment indices would represent more proximal and perhaps essential Tbx20 interactions. Indeed, TLE1 and TLE3 comprised two of the most highly enriched components of the interaction network. This supports a critical role for these proteins in regulating Tbx20 function, suggesting they may be directly linked to Tbx20 (Figure 2B, black dashed lines).
Tbx20 forms protein complexes with TLE1/3, Tbx18, and HDAC2
While our AP-MS analysis of Tbx20 complexes does not establish which proteins are direct interactions (Figure 2B, black dashed lines), other T-box proteins, Tbx15 and Tbx18, have been reported to bind directly to TLE3 via N-terminal eh1 binding motifs 32, indicating that the eh1 motif may represent a common motif used by T-box transcription factors to bind Gro/TLE family members. Therefore, to determine whether Tbx20 directly recruits Groucho co-repressors via an eh1 binding motif, we next investigated the interaction between Tbx20 and TLE1/3. Tbx20 contains an N-terminal eh1 binding motif that is fully conserved in all vertebrate orthologs of Tbx20 (Figure 3A). To confirm an interaction between Tbx20 and TLE1/3 and determine whether these interactions require the eh1 motif, we generated a Tbx20eh1mut-EGFP expression construct in which the eh1 motif has been ablated by site-directed mutation of phenylalanine 18 and serine 19 to an isoleucine and a leucine, respectively (Tbx20F18I; S19L-EGFP). Reciprocal immunoisolations of TLE1 and TLE3 complexes were performed in the presence of wildtype Tbx20-EGFP or Tbx20eh1mut-EGFP. Mutation of the eh1 motif significantly reduced the ability of Tbx20-EGFP to be co-isolated with either TLE1 or TLE3, suggesting that Tbx20 binds TLE1/3 directly through this motif (Figure 3B,C).
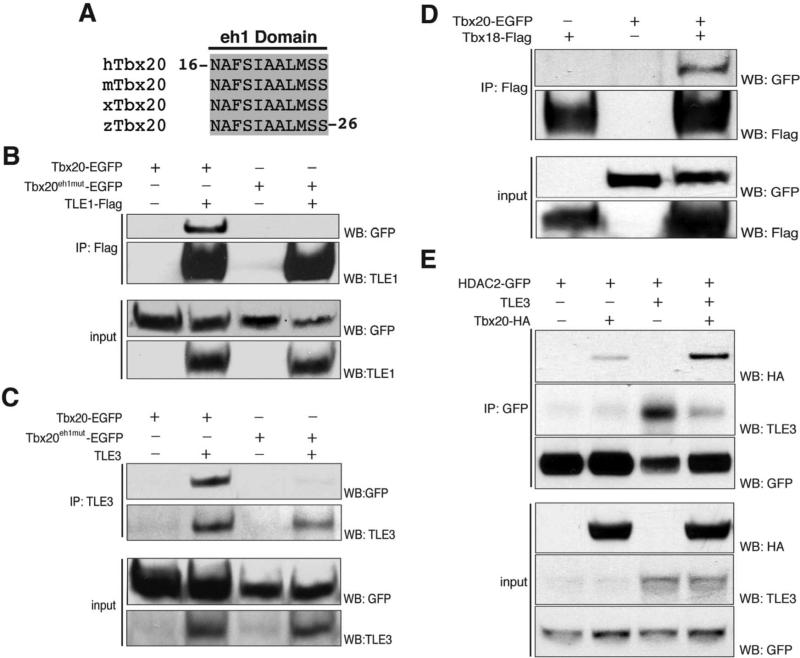
Figure 3.
Tbx20 interacts with TLE1/3, HDAC2, and Tbx18. (A) Protein sequence alignment of an N-terminal eh1 binding motif in Tbx20 demonstrating complete conservation of the eh1 motif across all vertebrate homologs of Tbx20. h, human; m, mouse; x, Xenopus; z, zebrafish. (B) Reciprocal immunoisolations of TLE1-Flag complexes from HEK293 cells expressing either Tbx20-EGFP or Tbx20eh1mut -EGFP. (C) Reciprocal immunoisolations of TLE3 complexes from HEK293 cells expressing either Tbx20-EGFP or Tbx20eh1mut-EGFP. (D) Reciprocal immunoisolations of Tbx18-Flag complexes from HEK293 cells expressing Tbx18-Flag in the presence or absence of Tbx20-EGFP. (E) Reciprocal immunoisolations of HDAC2-GFP from HEK293 cells expressing Tbx20-HA and/or TLE3. IP, immunoprecipitation; WB, western blot.
The finding that Tbx20 interacts with both a Gro/TLE complex and the Groucho dependent repressor Tbx18 32 suggests that Tbx20 and Tbx18 may heterodimerize to regulate a common set of targets in a Groucho-dependent manner. To further investigate the interaction between Tbx20 and Tbx18, we transfected HEK293 cells with Tbx18-Flag alone or in the presence of Tbx20-EGFP. Immunopurification with an anti-Flag antibody and Western blot analysis revealed efficient co-isolation of Tbx20 (Figure 3D). Collectively, these data imply a role for a Tbx20-Tbx18 repressor complex during vertebrate development.
To investigate the link between the Tbx20-TLE1/3 complex and HDAC2, we generated an HEK293 cell line stably expressing HDAC2 tagged at the C-terminus with EGFP. To confirm an interaction between Tbx20 and HDAC2, we performed reciprocal isolations of HDAC2-EGFP in the presence or absence of Tbx20-HA. Tbx20 was successfully co-isolated with HDAC2-EGFP (Figure 3E). As a number of studies indicate that binding to Groucho co-factors results in recruitment of deacetylase machinery 31, 33, we also performed isolations of HDAC2-EGFP in the presence of over-expressed TLE3 and Tbx20. Interestingly, excess TLE3 results in a substantial increase in the amount of Tbx20 associated with HDAC2, suggesting that TLE3 bridges an interaction between Tbx20 and HDAC2 (Figure 3E). However, less TLE3 associates with HDAC2 when Tbx20 is overexpressed; therefore, an alternate explanation is that Tbx20 competes with TLE3 for binding to HDAC2.
Label-free mass spectrometry analysis reveals that Tbx20:TLE binding triggers recruitment of NuRD components
To distinguish between these possibilities and to assess precisely which components of the chromatin modification network (Figure 2B) are dependent on the Tbx20-TLE3 interaction, we used label-free mass spectrometry to assess the differences between Tbx20-EGFP and Tbx20eh1mut-EGFP protein complexes. To do this, we expressed Tbx20-EGFP and Tbx20eh1mut-EGFP in HEK293 cells and performed parallel isolations of EGFP-tagged Tbx20 complexes, as described above. Changes in the relative abundance of interacting chromatin remodeling factors between wild-type and mutant Tbx20 complexes were assessed using two label-free quantification approaches: 1) spectral counting, and 2) MS1 peak area quantification (see Materials and Methods). To correct for the total amount of isolated Tbx20 complexes between conditions, we normalized the quantitative values for associated proteins identified in mutant Tbx20 complexes by the ratio of wild-type/eh1mut Tbx20-EGFP. Also, Tbx18 was excluded from label-free quantification due to the presence of only one tryptic peptide that was exclusive to Tbx18 (the other peptide was isobaric with a tryptic peptide from Tbx20).
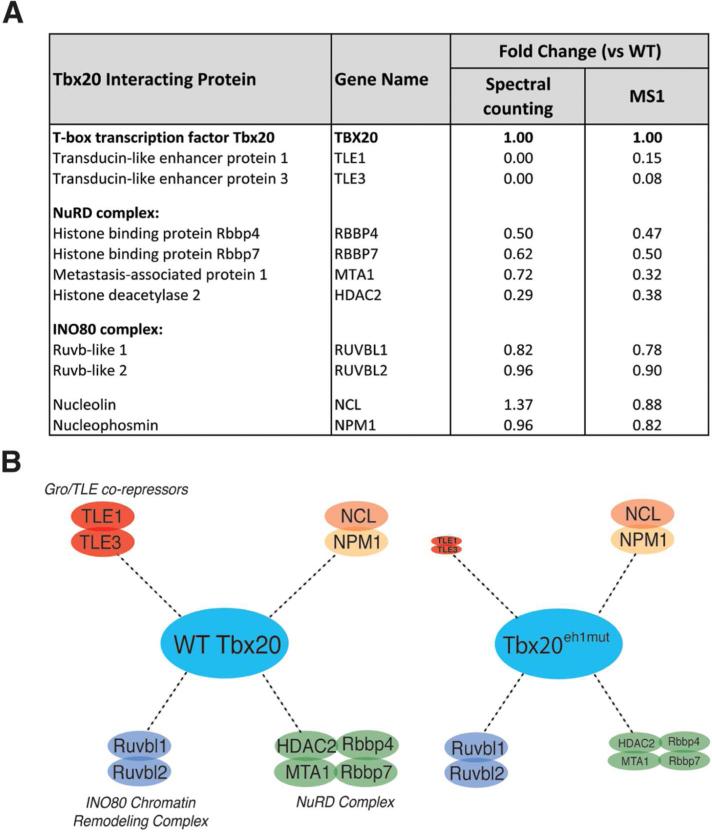
Consistent with our previous Western blot analysis (Figure 3B-C), we no longer identified TLE1 or TLE3 in the Tbx20eh1mut mutant isolation (Figure 4A and Supplementary Table 5). We also observed an eh1mut-dependent spectral count fold-decrease that was specific to components of the NuRD complex, though with variation in the magnitude of the decrease between proteins (Figure 4A). To address this variation, we also employed MS1-based quantification, which allows more precise measurement of relative abundance, particularly for proteins with few spectral counts (e.g. in the mutant isolation, Supplemental Table 5) and/or with shared tryptic sequences (e.g., RBBP4 and RBBP7). MS1-based analysis confirmed the spectral counting result for TLE1/3, suggesting a relative decrease of at least 10-fold in the eh1mut vs wild-type Tbx20 (Supplemental Table 6). Further, we observed that association with HDAC2, MTA1, and the histone binding proteins RBBP4/RBBP7 was significantly reduced and to a similar extent in the eh1mut compared to wild-type Tbx20, indicating that the interaction of Tbx20 with NuRD complex components is partially Groucho-dependent (Figure 4A,B and Supplemental Table 5, 6). These data suggest that TLE co-repressors play a central role in the formation of a Tbx20 transcriptional repressive complex by recruiting core components of the NuRD complex including HDAC2. Overall, these results demonstrate that this AP-MS approach can efficiently isolate and identify specific Tbx20 interactions, which may help to define the mechanisms involved in Tbx20-mediated gene regulation.
Figure 4.
Tbx20 assembles a Gro/TLE-NuRD repression complex via the eh1 binding motif. Wild-type Tbx20-EGFP and Tbx20eh1mut-EGFP were immunoaffinity purified from HEK293 cells with associated proteins and analyzed by mass spectrometry. Fold changes for each interaction illustrated in (A) is shown for the isolated Tbx20eh1mut mutant versus wild-type Tbx20. (B) The relative size of the circles indicates increased or decreased relative abundance of each interaction as determined by the MS1-based fold-change for mutant versus wild-type Tbx20 isolations.
Endogenous Tbx20 interacts with TLE factors in mouse embryonic hearts
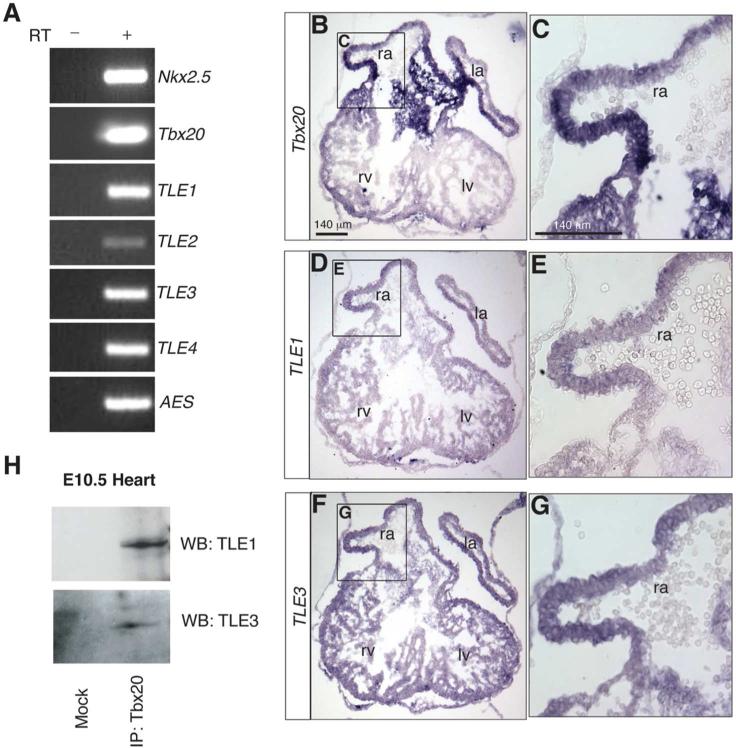
Our proteomic analysis suggests that Tbx20 is linked to a transcriptional repressive complex via direct interaction with a TLE family member. To assess the endogenous Tbx20 association with TLE family members in vivo at the time in embryogenesis at which Tbx20 has been shown to function, we first examined the expression of all of the TLE family members in mouse embryonic day 10.5 (E10.5) heart tissue by RT-PCR. At this stage of development, Tbx20 is uniformly expressed throughout the four-chambered embryonic heart 34 where it is required for proper transcriptional regulation of cardiac chamber-specific genes4b-e. All of the TLE family members are highly expressed at this time, with the exception of TLE2, which is expressed at relatively low levels (Figure 5A). To further confirm that Tbx20 co-localizes with TLE1/3 within the same cells of the embryonic heart, we performed in situ hybridization for Tbx20, TLE1, and TLE3 transcripts on serial tissue sections from an E10.5 heart. This analysis revealed that TLE1 and TLE3 are expressed broadly with Tbx20 throughout the myocardium (Figure 5B-G).
Figure 5.
Endogenous Tbx20 interacts with TLE1/3 in mouse embryonic hearts. (A) RT-PCR analysis of TLE family members and cardiac-specific markers Nkx2.5 and Tbx20 in E10.5 heart tissue. All samples derived from embryonic hearts dissected at E10.5. (B,C) Section in situ hybridization analysis for Tbx20 expression on a cryosection of an E10.5 heart. Neighboring sections from the same embryonic heart were used to assess TLE1 (D,E) and TLE3 (F,G) expression. (H) 25 hearts were dissected, and endogenous Tbx20 complexes were isolated with an antibody against Tbx20, analyzed by SDS-PAGE, and immunoblotted with antibodies against TLE1 and TLE3. In parallel and as a control, a mock immunoprecipitation was performed in the absence of Tbx20 antibody. ra, right atrium; la, left atrium; rv, right ventricle; lv, left ventricle.
Importantly, we were able to detect an interaction between endogenous Tbx20 and both TLE1 and TLE3 in the embryonic heart, indicating that Tbx20 binds both TLE factors in vivo during heart development (Figure 5H). This finding represents the first demonstration of endogenous Tbx20 protein-protein interactions from an embryonic heart. These data further validate our proteomic approach and demonstrate that Tbx20 assembles a TLE repressor complex in the embryonic heart at the time at which Tbx20 functions in cardiac development.
Ablation of Tbx20:TLE binding disrupts Tbx20-mediated transcriptional repression in Xenopus embryos
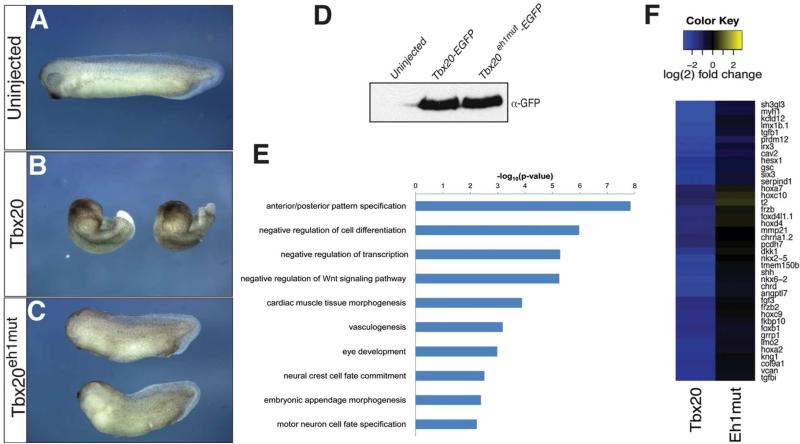
Having established that Tbx20 assembles a TLE co-repressor complex in the forming heart, we next sought to establish the consequences of Tbx20:TLE binding on Tbx20 biological activity. To address this issue, we examined whether Tbx20eh1mut mutant protein could promote morphogenetic changes in the embryo when ectopically expressed in developing Xenopus laevis embryos. Xenopus embryos were injected at the one-cell stage with Tbx20-EGFP or Tbx20eh1mut-EGFP mRNA and allowed to develop until control uninjected embryos reached tailbud stages (stage 23/33). Ectopic expression of Tbx20-EGFP drastically disturbs cell movements during gastrulation, leading to a marked shortening of the anterior/posterior axis, consistent with a previous report 5a (Figure 6A, B). Strikingly, these defects are partially rescued by ablation of the eh1 binding motif, indicating that recruitment of TLE factors is critical for the ability of Tbx20 to promote changes in morphogenesis (Figure 6C). This finding is supported by the observation that Tbx20-EGFP and Tbx20eh1mut-EGFP are expressed at comparable levels from injected mRNAs, as shown by Western blotting for GFP (Figure 6D).
Figure 6.
RNA-sequencing reveals disrupted transcriptional output in Tbx20eh1mut-expressing embryos. Xenopus embryos were injected at the one-cell stage with Tbx20 (B) or Tbx20eh1mut (C) mRNA and allowed to develop until sibling uninjected embryos (A) reached tadpole stages. (D) Total protein equivalent to one uninjected, Tbx20-EGFP, or Tbx20eh1mut embryo (stage 12.5) was analyzed by SDS-PAGE and immunoblotted with an antibody against GFP. Western blot shows comparable protein levels from injected mRNAs. (E) Gene ontology grouping (GOrilla) of the 173 genes downregulated at least 2-fold by wild-type Tbx20, but not by Tbx20eh1mut. GO terms enriched in this gene set were plotted by the log of their FDR-adjusted p-values. (F) Heat map depicting differential regulation of genes by wild-type Tbx20 and Tbx20eh1mut. The 173 genes downregulated at least 2-fold only by wild-type Tbx20 were ranked by the fold change ratio between the mutant and wild-type. The 40 genes with the greatest disparity between the mutant and wild-type are shown.
Our findings are consistent with an essential role for Tbx20-TLE repressor complexes in embryonic heart formation. To gain insights into what molecular processes and genes are regulated by Tbx20-TLE complexes, we used high-throughput sequence analysis to examine the transcriptomes in Tbx20-EGFP and Tbx20eh1mut-EGFP-injected embryos. Specifically, we sought to identify genes downregulated by Tbx20-TLE repressor complexes. We chose stage 12.5 (early neurula-stage) embryos for analysis because misexpressed Tbx20 leads to gene expression changes at this stage (Figure 1D), and to eliminate the detection of gene expression changes induced by endogenous Tbx20 transcripts, which can be detected as early as stage 16 3e. A total of 201 genes were significantly downregulated at least 2-fold by ectopic Tbx20 when compared to control uninjected embryos (Supplemental Figure 1). Of these 201 genes, 173 were not repressed by ectopic expression of Tbx20eh1mut protein (86% of genes downregulated by wild-type Tbx20; Supplemental Figure 1 and Supplemental Table 9), indicating a role for Tbx20-TLE complexes in negative regulation of this group of genes. Gene ontology enrichment analysis revealed a trend whereby the 173 genes not repressed by Tbx20eh1mut were significantly enriched for functions involving anterior/posterior pattern specification, regulation of cell differentiation and transcription, regulation of the Wnt signaling pathway, and tissue patterning (Figure 6E and Supplemental Table 10), cellular processes known to be associated with Gro/TLE repressive function in the embryo 35. Indeed, upon ranking the 173 genes uniquely repressed by Tbx20 overexpression, the 40 genes most differentially regulated between Tbx20- and Tbx20eh1mut-expressing embryos included hox genes involved in anterior/posterior patterning (hoxa7, hoxc10, hoxd4, hoxc9, hoxa2)36, transcription factors important for control of cellular differentiation (irx3, tbx3, gata3, and nkx6.2), and a number of Wnt signaling pathway components (frzb, dkk1, frzb2) (Figure 6F). Notably, Irx3 has recently been identified as a direct ChIP-seq genomic target of Tbx20 in the adult heart, and a recent study finds that Tbx20 directly regulates Wnt pathway genes in the endocardial cushions, suggesting that our RNA-seq analysis identified primary mediators of Tbx20 cardiac function 5b, 37. Additionally, early, ectopic expression of Tbx20 leads to downregulation of a number of genes with important roles in cardiac muscle tissue morphogenesis, including the cardiac transcription factors nkx2.5, isl1, pitx2, and foxc1 (Figure 6E, F and Supplemental Table 10). Interestingly, these cardiac genes are not downregulated by Tbx20eh1mut protein (Figure 6E, F and Supplemental Table 10), implying that in the absence of other cues (ie. transcriptional coactivators), Tbx20 may act in combination with TLE repressor complexes to repress cardiac promoters. Collectively, these results provide additional evidence that Tbx20-mediated repression is facilitated by Gro/TLE complexes.
Conclusions
Despite the critical role of Tbx20 in cardiac development, the precise mechanisms by which Tbx20 regulates distinct gene programs in the heart are not understood. Studies in mouse knockout models of Tbx20 indicate that Tbx20 is required for proper patterning and morphogenesis of working myocardium 4b-e. Thus, activating and/or repressive activity of Tbx20 on target genes underlies the primary cardiomyocyte lineage split into specialized chamber and non-chamber myocardium. To identify and characterize the determinants of Tbx20 transcriptional activity in the heart, it is essential to identify Tbx20 interacting proteins. Our study constitutes the first unbiased analysis of Tbx20 protein interactions. Using a proteomics/bioinformatics approach, we identified a unique transcriptional repression network that includes Groucho co-repressors, components of the NuRD complex, components of the INO80 chromatin remodeling complex (RUVBL1 and RUVBL2), and the T-box repressor Tbx18.
A Tbx20-TLE-NuRD repressor complex
The Tbx20 homolog Midline was recently demonstrated to bind Groucho directly in an eh1-dependent manner in Drosophila whole embryos, and this interaction was required for proper transcriptional repression of the wingless gene during segmentation of the ectoderm 38. Our studies confirm and expand upon this work by demonstrating that 1) vertebrate Tbx20 interacts with Groucho homologs, 2) vertebrate Tbx20 interacts with at least two members of the Groucho-related TLE family, TLE1 and TLE3, in human cells and in the mouse embryonic heart through the eh1 motif, and 3) Tbx20-TLE interactions directly result in the recruitment of components of the chromatin-remodeling NuRD complex including HDAC2. These data suggest that recruitment of Gro/TLE co-repressor complexes and subsequent deacetylation of target loci represent an evolutionarily conserved mechanism by which Tbx20 functions and thus, one mode by which Tbx20 promotes inactive chromatin states during development. A thorough expression analysis of TLE factors in the developing heart has not been published; although, it has been reported that TLE1 and TLE3 transcripts were not detectable in the mouse embryonic heart by in situ hybridization 39. Our data, however, suggests that most members of the mouse TLE family are expressed in the heart at stage E10.5 of embryogenesis. Further, we find that TLE1 and TLE3 transcripts are broadly expressed throughout the E10.5 myocardium by in situ hybridization. Finally, TLE1 and TLE3 proteins co-isolate with endogenous Tbx20 at E10.5. The availability of other TLE factors in the heart, and our results showing that Tbx20 interacts with both TLE1 and TLE3, suggests that Tbx20 may interact with multiple members of this family to cooperatively regulate genes in the heart. As such, it will be interesting to determine whether other TLE family members have distinct temporal and spatial expression patterns in the forming heart or whether they can act redundantly on Tbx20 target genes.
Our proteomic and biochemical analyses of Tbx20 complexes indicate that binding of TLE factors by Tbx20 results in the recruitment of core components of the NuRD complex40, specifically, the histone binding proteins RBBP4 and RBBP7, MTA1, and HDAC2. Historically, requirements for the NuRD complex in early cell fate decisions in the embryo41 have prevented the identification of a role for NuRD specifically in the heart; however, HDAC2 is ubiquitously expressed in developing myocardium 42, and mice that are mutant for both HDAC1 and -2 die neonatally of cardiac arrhythmias and dilated cardiomyopathy43. Taken together, these studies implicate an important role for a Tbx20-NuRD association in developing cardiomyocytes.
Interestingly, interactions with the remainder of the chromatin remodeling network were unaffected by the eh1 mutation, suggesting that they are recruited independently of the eh1 binding motif and TLE recruitment. Of note, RUVBL2 (also called Reptin) was demonstrated to co-occupy the Hesx1 promoter with TLEs and HDAC1 to silence Hesx1 expression during mouse pituitary development 44, indicating that this protein, although still present within Tbx20eh1mut mutant complexes, may be part of an intact TLE corepressor complex via recruitment by a as yet unidentified binding motif within Tbx20.
The finding that Tbx20 represses cardiac genes when ectopically expressed in early Xenopus embryos indicates that Tbx20 activating or repressive activity is likely context dependent. Previous studies have reported that Tbx20, in combination with other cardiac transcription factors, primarily acts to promote cardiac gene expression while repressing genetic programs of other tissues 4e, 5a, 5c. In accordance with this, we identified genes essential for vasculogenesis, eye development, neural crest development, and motor neuron development downregulated by Tbx20 misexpression. Given that Tbx20 misexpression also downregulates cardiac transcription factors, one possibility is that additional tissue-specific factors or cues are needed to antagonize default repression achieved by Tbx20. Indeed, we observed that Tbx20eh1mut mutant protein fails to repress these genetic programs, implying that displacement of the TLE corepressor complex is sufficient to abolish Tbx20 repressive activity on these genes. As such, it would be interesting to determine whether TLE-mediated repression can be out-competed by cardiac coactivators in the context of a cardiac progenitor or myocyte. This principle has been illustrated previously by the role Gro/TLE plays in cells that do not receive Wg/Wnt signaling. Binding of Gro/TLE to Tcf/Lef repressors on Wg/Wnt target genes blocks recruitment of beta-catenin, an obligatory Tcf/Lef coactivator. Effective Wg/Wnt signaling leads to accumulation of beta-catenin, which subsequently can out-compete Gro/TLE for binding to Tcf/Lef 45. Thus, the level of Wn/Wnt signaling via beta-catenin accumulation dictates which cofactor binds Tcf/Lef, leading to transcription activation or repression. Our data therefore highlight a unique mechanism by which TLE-mediated repression may function as a context-dependent regulatory switch required for Tbx20 to alternate between activator and repressor.
A Tbx20-INO80 complex
The INO80 chromatin remodeling complex is a very large protein complex with 11 to 16 members including the ATP-dependent helicases Ino80 and SRCAP, the DNA helicases Pontin (Ruvbl1, Tip49, Tip49a) and Reptin (Ruvbl2, Tip48, Tip49b), actin, and various actin-related proteins (Arp4, Arp5, Arp8, β-actin, Arp7, Arp9) 28. The INO80 complex alters chromatin accessibility resulting in activation or repression of target genes 46. Two members of the complex, Pontin and Reptin, have opposing acitivities within the INO80 complex, and this antagonistic relationship has been shown to play a role in cardiac growth in zebrafish 47. An ENU-induced mutation in Reptin leads to cardiac hyperplasia and embryonic lethality. This mutation is an activating mutation in Reptin, increasing the ATPase activity of Reptin complexes, thus overriding Pontin inhibition of Reptin activity, and increasing the transcriptional repressor activity of the complex. Mutant zReptin was subsequently shown to have a stronger repressive effect on beta-catenin/TCF-mediated transactivation, leading to the hypothesis that Pontin/Reptin complexes regulate beta-catenin-mediated activation of cell cycle genes such as cyclin D and c-Myc to control the balance between proliferation and differentiation in the developing zebrafish heart 47. Our data suggests that Tbx20 interacts with a RUVBL1-RUVBL2 protein complex independently of its association with a TLE repressor complex, suggesting that Tbx20 may also recruit the INO80 complex for gene regulation. Tbx20 has a well-documented role in cardiomyocyte proliferation4b and seems to exert opposite effects on cell proliferation in embryonic versus fetal cardiomyocytes48. Therefore, it is tempting to speculate that association of Tbx20 with the INO80 complex in fetal cardiomyocytes underlies this switch in the activity of Tbx20 transcriptional complexes.
A Tbx20-Tbx18 complex
An unexpected finding of these studies is that Tbx20 interacts with the tissue-specific transcription factor Tbx18. In the developing heart, Tbx18 expression overlaps with that of Tbx20 in a subset of cardiomyocytes within the interventricular septum and a portion of the left ventricle at a stage when the heart is undergoing chamber specialization and expansion, processes that are both dependent on proper Tbx20 function 3f, g, 49. Thus, interaction with Tbx18 in this subset of cardiomyocytes provides one potential mechanism through which Tbx20 may function to regulate regionally distinct gene programs in the heart. Tbx18 is also expressed in the epicardium, an epithelial monolayer that covers the myocardium and is a critical source of signals and cells for the underlying myocardium 34. Recently, a microarray analysis of isolated epicardial cells revealed Tbx20 as an epicardium-enriched transcription factor 50, opening the possibility for a Tbx18-Tbx20 transcriptional complex within the epicardium. Tbx18 also plays a prominent role in formation of the myocardial sinus horns that make up the venous pole of the heart; however, it is not clear whether Tbx20 is co-expressed with Tbx18 within this tissue 51. Additionally, our lab has previously reported that Tbx20 interacts physically with Tbx5 4a, suggesting that heterodimerization with other T-box factors may represent an important mechanism by which Tbx20 regulates gene expression in the embryo.
Interestingly, Tbx18 has also been shown to interact with TLE3 via an eh1 binding motif within the N-terminus of the protein 32. In this study, transcriptional assays demonstrated that Tbx18 can repress activation of the Nppa/ANF promoter by the cardiac transcription factors Gata4, Nkx2.5, and Tbx5. This finding was interpreted as Tbx18 abrogation of Nppa/ANF expression through competition with Tbx5, a second T-box protein, for T-box binding sites (TBEs) within the Nppa/ANF promoter. Similarly, Tbx18 is predicted to repress Tbx6-mediated activation of the Notch ligand Delta-like 1 (Dll1) in anterior somites through competition with Tbx6 32. Collectively, these data imply a model in which Tbx18 competes with other T-box activators for occupancy of TBEs and subsequently achieves repression by recruiting a TLE co-repressor complex to the target gene. Here, we have demonstrated the Tbx20 and Tbx18 physically interact, and Tbx20 recruits a TLE repressor complex similar to that reported for Tbx18. Therefore, a second model predicts that Tbx18 and Tbx20 may be acting cooperatively as repressors on a common set of target genes. Further studies are needed to distinguish between these possibilities.
In summary, the goal of our study was to expand upon the current knowledge of the Tbx20 transcriptional network. By combining immunoaffinity purification with proteomic and functional network analysis, we have identified a Tbx20 transcription repression network with chromatin remodeling and deacetylase functions. In particular, our results reveal a crucial role for Gro/TLE-NuRD corepressor complexes in facilitating Tbx20-mediated transcriptional repression. We also identified Tbx18 as a Tbx20 interaction, raising the question of whether Tbx20 transcriptional repression relies on cooperative activity of Tbx20 and other cardiac transcription factors, similar to what has been shown for Tbx20 transcriptional activation in the presence of the activators Gata4, Nkx2.5, and Tbx5 4e, 5a. Future studies will aim to delineate the biological role of these repressive interactions, particularly as they relate to regulation of the cardiogenic program.
Supplementary Material
Supplemental Figure 1. Venn diagram summarizing overlap of regulatory capabilities between wild-type Tbx20 an Tbx20eh1mut. Genes considered were restricted to those significantly up- or down-regulated at 2-fold compared to uninjected control dataset, in addition to having an FDR-adjusted p-value <0.05. Genes downregulated at least 2-fold only by wild-type Tbx20 were used for gene ontology analysis to investigate transcriptional repression mediated specifically by the Tbx20-TLE complex.
ACKNOWLEDGEMENTS
These studies were funding by grants to F.L.C from NIH/NHLBI (RO1 DE018825 and RO1 HL089641), to I.M.C. from NIH/NIDA (DP1DA026192) and HFSPO (RGYoo79/2009-C), to F.L.C and I.M.C from NIH/NICHD (R21 HD073044), and by an American Heart Association Pre-Doctoral Fellowship and a UNC Dissertation Completion Fellowship awarded to E.K. We also thank Panna Tandon in the Conlon Lab for critical reading of the manuscript.
ABBREVIATIONS
- NuRD
Nucleosome Remodeling and Deacetylase
- TLE
Transducin-like Enhancer of Split
- Gro
Groucho
- HEK293
Human Embryonic Kidney 293
- HDAC2
Histone Deacetylase 2
- AP-MS
Affinity Purification-Mass Spectrometry
- MTA1
Metastasis-associated protein 1
- eh1
engrailed homology 1
- NCL
Nucleolin
- NPM1
Nucleophosmin
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
ASSOCIATED CONTENT
Supporting Information Available: This material is free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
References
- 1.a van der Linde D, Konings EE, Slager MA, Witsenburg M, Helbing WA, Takkenberg JJ, Roos-Hesselink JW. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J Am Coll Cardiol. 2011;58(21):2241–7. doi: 10.1016/j.jacc.2011.08.025. [DOI] [PubMed] [Google Scholar]; b Reller MD, Strickland MJ, Riehle-Colarusso T, Mahle WT, Correa A. Prevalence of congenital heart defects in metropolitan Atlanta, 1998-2005. J Pediatr. 2008;153(6):807–13. doi: 10.1016/j.jpeds.2008.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a Kirk EP, Sunde M, Costa MW, Rankin SA, Wolstein O, Castro ML, Butler TL, Hyun C, Guo G, Otway R, Mackay JP, Waddell LB, Cole AD, Hayward C, Keogh A, Macdonald P, Griffiths L, Fatkin D, Sholler GF, Zorn AM, Feneley MP, Winlaw DS, Harvey RP. Mutations in cardiac T-box factor gene TBX20 are associated with diverse cardiac pathologies, including defects of septation and valvulogenesis and cardiomyopathy. Am J Hum Genet. 2007;81(2):280–91. doi: 10.1086/519530. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Hammer S, Toenjes M, Lange M, Fischer JJ, Dunkel I, Mebus S, Grimm CH, Hetzer R, Berger F, Sperling S. Characterization of TBX20 in human hearts and its regulation by TFAP2. J Cell Biochem. 2008;104(3):1022–33. doi: 10.1002/jcb.21686. [DOI] [PubMed] [Google Scholar]; c Liu C, Shen A, Li X, Jiao W, Zhang X, Li Z. T-box transcription factor TBX20 mutations in Chinese patients with congenital heart disease. Eur J Med Genet. 2008;51(6):580–7. doi: 10.1016/j.ejmg.2008.09.001. [DOI] [PubMed] [Google Scholar]; d Qian L, Mohapatra B, Akasaka T, Liu J, Ocorr K, Towbin JA, Bodmer R. Transcription factor neuromancer/TBX20 is required for cardiac function in Drosophila with implications for human heart disease. Proc Natl Acad Sci U S A. 2008;105(50):19833–8. doi: 10.1073/pnas.0808705105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a Ahn DG, Ruvinsky I, Oates AC, Silver LM, Ho RK. tbx20, a new vertebrate T-box gene expressed in the cranial motor neurons and developing cardiovascular structures in zebrafish. Mech Dev. 2000;95(1-2):253–8. doi: 10.1016/s0925-4773(00)00346-4. [DOI] [PubMed] [Google Scholar]; b Griffin KJ, Stoller J, Gibson M, Chen S, Yelon D, Stainier DY, Kimelman D. A conserved role for H15-related T-box transcription factors in zebrafish and Drosophila heart formation. Dev Biol. 2000;218(2):235–47. doi: 10.1006/dbio.1999.9571. [DOI] [PubMed] [Google Scholar]; c Meins M, Henderson DJ, Bhattacharya SS, Sowden JC. Characterization of the human TBX20 gene, a new member of the T-Box gene family closely related to the Drosophila H15 gene. Genomics. 2000;67(3):317–32. doi: 10.1006/geno.2000.6249. [DOI] [PubMed] [Google Scholar]; d Iio A, Koide M, Hidaka K, Morisaki T. Expression pattern of novel chick T-box gene, Tbx20. Dev Genes Evol. 2001;211(11):559–62. doi: 10.1007/s00427-001-0187-y. [DOI] [PubMed] [Google Scholar]; e Brown DD, Binder O, Pagratis M, Parr BA, Conlon FL. Developmental expression of the Xenopus laevis Tbx20 orthologue. Dev Genes Evol. 2003;212(12):604–7. doi: 10.1007/s00427-002-0276-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Kaltenbrun E, Tandon P, Amin NM, Waldron L, Showell C, Conlon FL. Xenopus: An emerging model for studying congenital heart disease. Birth Defects Res A Clin Mol Teratol. 2011;91(6):495–510. doi: 10.1002/bdra.20793. [DOI] [PMC free article] [PubMed] [Google Scholar]; g Conlon FL, Yutzey KE. T-Box Factors. In Heart Development and Regeneration. In: Rosenthal N, Harvey RP, editors. Vol. Volume 2. Elsevier; 2010. pp. 651–665. [Google Scholar]
- 4.a Brown DD, Martz SN, Binder O, Goetz SC, Price BM, Smith JC, Conlon FL. Tbx5 and Tbx20 act synergistically to control vertebrate heart morphogenesis. Development. 2005;132(3):553–63. doi: 10.1242/dev.01596. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Cai CL, Zhou W, Yang L, Bu L, Qyang Y, Zhang X, Li X, Rosenfeld MG, Chen J, Evans S. T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development. 2005;132(10):2475–87. doi: 10.1242/dev.01832. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Singh MK, Christoffels VM, Dias JM, Trowe MO, Petry M, Schuster-Gossler K, Burger A, Ericson J, Kispert A. Tbx20 is essential for cardiac chamber differentiation and repression of Tbx2. Development. 2005;132(12):2697–707. doi: 10.1242/dev.01854. [DOI] [PubMed] [Google Scholar]; d Stennard FA, Costa MW, Lai D, Biben C, Furtado MB, Solloway MJ, McCulley DJ, Leimena C, Preis JI, Dunwoodie SL, Elliott DE, Prall OW, Black BL, Fatkin D, Harvey RP. Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development. 2005;132(10):2451–62. doi: 10.1242/dev.01799. [DOI] [PubMed] [Google Scholar]; e Takeuchi JK, Mileikovskaia M, Koshiba-Takeuchi K, Heidt AB, Mori AD, Arruda EP, Gertsenstein M, Georges R, Davidson L, Mo R, Hui CC, Henkelman RM, Nemer M, Black BL, Nagy A, Bruneau BG. Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development. 2005;132(10):2463–74. doi: 10.1242/dev.01827. [DOI] [PubMed] [Google Scholar]; f Mandel EM, Kaltenbrun E, Callis TE, Zeng XX, Marques SR, Yelon D, Wang DZ, Conlon FL. The BMP pathway acts to directly regulate Tbx20 in the developing heart. Development. 2010;137(11):1919–29. doi: 10.1242/dev.043588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a Stennard FA, Costa MW, Elliott DA, Rankin S, Haast SJ, Lai D, McDonald LP, Niederreither K, Dolle P, Bruneau BG, Zorn AM, Harvey RP. Cardiac T-box factor Tbx20 directly interacts with Nkx2-5, GATA4, and GATA5 in regulation of gene expression in the developing heart. Dev Biol. 2003;262(2):206–24. doi: 10.1016/s0012-1606(03)00385-3. [DOI] [PubMed] [Google Scholar]; b Shen T, Aneas I, Sakabe N, Dirschinger RJ, Wang G, Smemo S, Westlund JM, Cheng H, Dalton N, Gu Y, Boogerd CJ, Cai CL, Peterson K, Chen J, Nobrega MA, Evans SM. Tbx20 regulates a genetic program essential to adult mouse cardiomyocyte function. J Clin Invest. 2011;121(12):4640–54. doi: 10.1172/JCI59472. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Sakabe NJ, Aneas I, Shen T, Shokri L, Park SY, Bulyk ML, Evans SM, Nobrega MA. Dual transcriptional activator and repressor roles of TBX20 regulate adult cardiac structure and function. Hum Mol Genet. 2012;21(10):2194–204. doi: 10.1093/hmg/dds034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miteva YV, Budayeva HG, Cristea IM. Proteomics-based methods for discovery, quantification, and validation of protein-protein interactions. Anal Chem. 2013;85(2):749–68. doi: 10.1021/ac3033257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buscarlet M, Hermann R, Lo R, Tang Y, Joachim K, Stifani S. Cofactor-activated phosphorylation is required for inhibition of cortical neuron differentiation by Groucho/TLE1. PLoS One. 2009;4(12):e8107. doi: 10.1371/journal.pone.0008107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Villanueva CJ, Waki H, Godio C, Nielsen R, Chou WL, Vargas L, Wroblewski K, Schmedt C, Chao LC, Boyadjian R, Mandrup S, Hevener A, Saez E, Tontonoz P. TLE3 is a dual-function transcriptional coregulator of adipogenesis. Cell Metab. 2011;13(4):413–27. doi: 10.1016/j.cmet.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nie X, Sun J, Gordon RE, Cai CL, Xu PX. SIX1 acts synergistically with TBX18 in mediating ureteral smooth muscle formation. Development. 2010;137(5):755–65. doi: 10.1242/dev.045757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nieuwkoop P. D. a. F., J. Normal table of Xenopus laevis (Daudin) North Holland; Amsterdam, The Netherlands: 1974. [Google Scholar]
- 11.a Smith JC, Slack JM. Dorsalization and neural induction: properties of the organizer in Xenopus laevis. J Embryol Exp Morphol. 1983;78:299–317. [PubMed] [Google Scholar]; b Goetz SC, Brown DD, Conlon FL. TBX5 is required for embryonic cardiac cell cycle progression. Development. 2006;133(13):2575–84. doi: 10.1242/dev.02420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cristea IM, Williams R, Chait BT, Rout MP. Fluorescent proteins as proteomic probes. Mol Cell Proteomics. 2005;4(12):1933–41. doi: 10.1074/mcp.M500227-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Tsai YC, Greco TM, Boonmee A, Miteva Y, Cristea IM. Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Mol Cell Proteomics. 2012;11(5):60–76. doi: 10.1074/mcp.A111.015156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–2. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–3. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von Mering C. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39(Database issue):D561–8. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zybailov BL, Florens L, Washburn MP. Quantitative shotgun proteomics using a protease with broad specificity and normalized spectral abundance factors. Mol Biosyst. 2007;3(5):354–60. doi: 10.1039/b701483j. [DOI] [PubMed] [Google Scholar]
- 18.Wang M, Weiss M, Simonovic M, Haertinger G, Schrimpf SP, Hengartner MO, von Mering C. PaxDb, a database of protein abundance averages across all three domains of life. Mol Cell Proteomics. 2012;11(8):492–500. doi: 10.1074/mcp.O111.014704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 20.Greco TM, Yu F, Guise AJ, Cristea IM. Nuclear import of histone deacetylase 5 by requisite nuclear localization signal phosphorylation. Mol Cell Proteomics. 2011;10(2):M110, 004317. doi: 10.1074/mcp.M110.004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.a Amin NM, Tandon P, Osborne Nishimura E, Conlon FL. RNA-seq in the tetraploid Xenopus laevis enables genome-wide insight in a classic developmental biology model organism. Methods. 2013 doi: 10.1016/j.ymeth.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Tandon P, Miteva YV, Kuchenbrod LM, Cristea IM, Conlon FL. Tcf21 regulates the specification and maturation of proepicardial cells. Development. 2013;140(11):2409–21. doi: 10.1242/dev.093385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowen NJ, Fujita N, Kajita M, Wade PA. Mi-2/NuRD: multiple complexes for many purposes. Biochim Biophys Acta. 2004;1677(1-3):52–7. doi: 10.1016/j.bbaexp.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 27.a Li YP, Busch RK, Valdez BC, Busch H. C23 interacts with B23, a putative nucleolar-localization-signal-binding protein. Eur J Biochem. 1996;237(1):153–8. doi: 10.1111/j.1432-1033.1996.0153n.x. [DOI] [PubMed] [Google Scholar]; b Sardiu ME, Cai Y, Jin J, Swanson SK, Conaway RC, Conaway JW, Florens L, Washburn MP. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc Natl Acad Sci U S A. 2008;105(5):1454–9. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102(4):463–73. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]; d Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2(6):851–61. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 28.a Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406(6795):541–4. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]; b Jin J, Cai Y, Yao T, Gottschalk AJ, Florens L, Swanson SK, Gutierrez JL, Coleman MK, Workman JL, Mushegian A, Washburn MP, Conaway RC, Conaway JW. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J Biol Chem. 2005;280(50):41207–12. doi: 10.1074/jbc.M509128200. [DOI] [PubMed] [Google Scholar]
- 29.Roche AE, Bassett BJ, Samant SA, Hong W, Blobel GA, Svensson EC. The zinc finger and C-terminal domains of MTA proteins are required for FOG-2-mediated transcriptional repression via the NuRD complex. J Mol Cell Cardiol. 2008;44(2):352–60. doi: 10.1016/j.yjmcc.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.a Fujita N, Jaye DL, Geigerman C, Akyildiz A, Mooney MR, Boss JM, Wade PA. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell. 2004;119(1):75–86. doi: 10.1016/j.cell.2004.09.014. [DOI] [PubMed] [Google Scholar]; b Li R, Zhang H, Yu W, Chen Y, Gui B, Liang J, Wang Y, Sun L, Yang X, Zhang Y, Shi L, Li Y, Shang Y. ZIP: a novel transcription repressor, represses EGFR oncogene and suppresses breast carcinogenesis. Embo J. 2009;28(18):2763–76. doi: 10.1038/emboj.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen G, Fernandez J, Mische S, Courey AJ. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 1999;13(17):2218–30. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farin HF, Bussen M, Schmidt MK, Singh MK, Schuster-Gossler K, Kispert A. Transcriptional repression by the T-box proteins Tbx18 and Tbx15 depends on Groucho corepressors. J Biol Chem. 2007;282(35):25748–59. doi: 10.1074/jbc.M703724200. [DOI] [PubMed] [Google Scholar]
- 33.a Choi CY, Kim YH, Kwon HJ, Kim Y. The homeodomain protein NK-3 recruits Groucho and a histone deacetylase complex to repress transcription. J Biol Chem. 1999;274(47):33194–7. doi: 10.1074/jbc.274.47.33194. [DOI] [PubMed] [Google Scholar]; b Yochum GS, Ayer DE. Pf1, a novel PHD zinc finger protein that links the TLE corepressor to the mSin3A-histone deacetylase complex. Mol Cell Biol. 2001;21(13):4110–8. doi: 10.1128/MCB.21.13.4110-4118.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kraus F, Haenig B, Kispert A. Cloning and expression analysis of the mouse T-box gene Tbx18. Mech Dev. 2001;100(1):83–6. doi: 10.1016/s0925-4773(00)00494-9. [DOI] [PubMed] [Google Scholar]
- 35.Buscarlet M, Stifani S. The ‘Marx’ of Groucho on development and disease. Trends Cell Biol. 2007;17(7):353–61. doi: 10.1016/j.tcb.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Gaunt SJ, Miller JR, Powell DJ, Duboule D. Homoeobox gene expression in mouse embryos varies with position by the primitive streak stage. Nature. 1986;324(6098):662–4. doi: 10.1038/324662a0. [DOI] [PubMed] [Google Scholar]
- 37.Cai X, Zhang W, Hu J, Zhang L, Sultana N, Wu B, Cai W, Zhou B, Cai CL. Tbx20 acts upstream of Wnt signaling to regulate endocardial cushion formation and valve remodeling during mouse cardiogenesis. Development. 2013;140(15):3176–87. doi: 10.1242/dev.092502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Formaz-Preston A, Ryu JR, Svendsen PC, Brook WJ. The Tbx20 homolog Midline represses wingless in conjunction with Groucho during the maintenance of segment polarity. Dev Biol. 2012;369(2):319–29. doi: 10.1016/j.ydbio.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 39.Santisteban P, Recacha P, Metzger DE, Zaret KS. Dynamic expression of Groucho-related genes Grg1 and Grg3 in foregut endoderm and antagonism of differentiation. Dev Dyn. 2010;239(3):980–6. doi: 10.1002/dvdy.22217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joshi P, Greco TM, Guise AJ, Luo Y, Yu F, Nesvizhskii AI, Cristea IM. The functional interactome landscape of the human histone deacetylase family. Mol Syst Biol. 2013;9:672. doi: 10.1038/msb.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.a Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15(6):710–23. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Marino S, Nusse R. Mutants in the mouse NuRD/Mi2 component P66alpha are embryonic lethal. PLoS One. 2007;2(6):e519. doi: 10.1371/journal.pone.0000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hang CT, Yang J, Han P, Cheng HL, Shang C, Ashley E, Zhou B, Chang CP. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466(7302):62–7. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, Hill JA, Richardson JA, Olson EN. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21(14):1790–802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olson LE, Tollkuhn J, Scafoglio C, Krones A, Zhang J, Ohgi KA, Wu W, Taketo MM, Kemler R, Grosschedl R, Rose D, Li X, Rosenfeld MG. Homeodomain-mediated beta-catenin-dependent switching events dictate cell-lineage determination. Cell. 2006;125(3):593–605. doi: 10.1016/j.cell.2006.02.046. [DOI] [PubMed] [Google Scholar]
- 45.Cinnamon E, Paroush Z. Context-dependent regulation of Groucho/TLE-mediated repression. Curr Opin Genet Dev. 2008;18(5):435–40. doi: 10.1016/j.gde.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 46.a Cai Y, Jin J, Yao T, Gottschalk AJ, Swanson SK, Wu S, Shi Y, Washburn MP, Florens L, Conaway RC, Conaway JW. YY1 functions with INO80 to activate transcription. Nat Struct Mol Biol. 2007;14(9):872–4. doi: 10.1038/nsmb1276. [DOI] [PubMed] [Google Scholar]; b Ford J, Odeyale O, Eskandar A, Kouba N, Shen CH. A SWI/SNF- and INO80-dependent nucleosome movement at the INO1 promoter. Biochem Biophys Res Commun. 2007;361(4):974–9. doi: 10.1016/j.bbrc.2007.07.109. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Klopf E, Paskova L, Sole C, Mas G, Petryshyn A, Posas F, Wintersberger U, Ammerer G, Schuller C. Cooperation between the INO80 complex and histone chaperones determines adaptation of stress gene transcription in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 2009;29(18):4994–5007. doi: 10.1128/MCB.01858-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rottbauer W, Saurin AJ, Lickert H, Shen X, Burns CG, Wo ZG, Kemler R, Kingston R, Wu C, Fishman M. Reptin and pontin antagonistically regulate heart growth in zebrafish embryos. Cell. 2002;111(5):661–72. doi: 10.1016/s0092-8674(02)01112-1. [DOI] [PubMed] [Google Scholar]
- 48.Chakraborty S, Yutzey KE. Tbx20 regulation of cardiac cell proliferation and lineage specialization during embryonic and fetal development in vivo. Dev Biol. 2012;363(1):234–46. doi: 10.1016/j.ydbio.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.a Christoffels VM, Grieskamp T, Norden J, Mommersteeg MT, Rudat C, Kispert A. Tbx18 and the fate of epicardial progenitors. Nature. 2009;458(7240):E8–9. doi: 10.1038/nature07916. discussion E9-10. [DOI] [PubMed] [Google Scholar]; b Zeng B, Ren XF, Cao F, Zhou XY, Zhang J. Developmental patterns and characteristics of epicardial cell markers Tbx18 and Wt1 in murine embryonic heart. J Biomed Sci. 2011;18:67. doi: 10.1186/1423-0127-18-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang GN, Thatcher JE, McAnally J, Kong Y, Qi X, Tan W, DiMaio JM, Amatruda JF, Gerard RD, Hill JA, Bassel-Duby R, Olson EN. C/EBP transcription factors mediate epicardial activation during heart development and injury. Science. 2012;338(6114):1599–603. doi: 10.1126/science.1229765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Christoffels VM, Mommersteeg MT, Trowe MO, Prall OW, de Gier-de Vries C, Soufan AT, Bussen M, Schuster-Gossler K, Harvey RP, Moorman AF, Kispert A. Formation of the venous pole of the heart from an Nkx2-5-negative precursor population requires Tbx18. Circ Res. 2006;98(12):1555–63. doi: 10.1161/01.RES.0000227571.84189.65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Venn diagram summarizing overlap of regulatory capabilities between wild-type Tbx20 an Tbx20eh1mut. Genes considered were restricted to those significantly up- or down-regulated at 2-fold compared to uninjected control dataset, in addition to having an FDR-adjusted p-value <0.05. Genes downregulated at least 2-fold only by wild-type Tbx20 were used for gene ontology analysis to investigate transcriptional repression mediated specifically by the Tbx20-TLE complex.