Abstract
Release of bud dormancy in perennial plants resembles vernalization in Arabidopsis thaliana and cereals. In both cases, a certain period of chilling is required for accomplishing the reproductive phase, and several transcription factors with the MADS-box domain perform a central regulatory role in these processes. The expression of DORMANCY-ASSOCIATED MADS-box (DAM)-related genes has been found to be up-regulated in dormant buds of numerous plant species, such as poplar, raspberry, leafy spurge, blackcurrant, Japanese apricot, and peach. Moreover, functional evidence suggests the involvement of DAM genes in the regulation of seasonal dormancy in peach. Recent findings highlight the presence of genome-wide epigenetic modifications related to dormancy events, and more specifically the epigenetic regulation of DAM-related genes in a similar way to FLOWERING LOCUS C, a key integrator of vernalization effectors on flowering initiation in Arabidopsis. We revise the most relevant molecular and genomic contributions in the field of bud dormancy, and discuss the increasing evidence for chromatin modification involvement in the epigenetic regulation of seasonal dormancy cycles in perennial plants.
Keywords: bud dormancy, chilling, DAM gene, chromatin, histone modifications
SEASONALITY OF BUD DORMANCY FOR ADAPTATION
The vegetative and reproductive meristems of many perennial plants in temperate climates remain in a non-growing latent state during the cold period of autumn and winter, within buds, which ensure an optimal protection against low temperatures and drought. The cessation of meristem growth and bud set are controlled by photoperiod, temperature, or a combination of both in different species (Olsen, 2010; Cooke et al., 2012); followed by the induction of bud dormancy, which precludes the growth resumption even under favorable environmental conditions (Rohde and Bhalerao, 2007). Bud dormancy is dynamically modulated by intrinsic, environmental, and hormonal factors (Arora et al., 2003; Horvath et al., 2003; Allona et al., 2008; Cooke et al., 2012), which in several aspects, resemble those factors regulating dormancy in seeds (Powell, 1987; Leida et al., 2012b). Paradoxically, in species from the Rosaceae family among others, bud dormancy is released by the same low-temperature conditions that induce dormancy, differing in the cumulative and quantitative perception of chilling required for an effective dormancy release (Coville, 1920; Couvillon and Erez, 1985).
The genotype specificity of chilling requirements for dormancy release, in addition to fruit maturation dates, are two major factors limiting the geographical distribution of temperate species (Chuine, 2010). Under a perspective of global warming, phenological issues may have a critical impact on the distribution and performance of cultivars and species. For instance, a later fulfillment of the chilling requirements of the meadow and steppe vegetation of the Tibetan Plateau from the mid-1990s, due to a warmer environment, has been found associated with a delay in the beginning of the growing season (Yu et al., 2010). The length of the growth season, limited by the growth cessation and bud-burst dates, is nevertheless expected to increase or decrease in different species according to their particular interaction with the environment (Hänninen and Tanino, 2011). This significant impact on yield from established orchards could alter the range of both native and invasive plants, and it also represents a challenge for tree breeders involved in improving varieties for better adaptability to a particular environment.
REGULATORY FACTORS IMPINGING ON BUD DORMANCY
The study of the mechanisms involved in bud dormancy initiation and release, and chilling perception, has been approached by genetic, genomic, and physiological analyses. Numerous transcriptomic studies have arisen during the last few years, addressing the changes in gene expression triggered by bud dormancy events in poplar (Populus spp.; Rohde et al., 2007; Ruttink et al., 2007), raspberry (Rubus idaeus; Mazzitelli et al., 2007), leafy spurge (Euphorbia esula; Horvath et al., 2008), Japanese apricot (Prunus mume; Yamane et al., 2008; Zhong et al., 2013), grapevine (Vitis spp.; Mathiason et al., 2009; Díaz-Riquelme et al., 2012), peach (Prunus persica; Jiménez et al., 2010a; Leida et al., 2010), blackcurrant (Ribes nigrum; Hedley et al., 2010), white spruce (Picea glauca; El Kayal et al., 2011), and pear (Pyrus pyrifolia; Liu et al., 2012; Bai et al., 2013) among other perennial species. These studies highlight the existence of common and specific gene expression programs affecting cell cycle regulation, light perception, hormonal signaling and stress response, and novel putative regulators of the dormancy stage of buds (Horvath et al., 2003; Cooke et al., 2012; Leida et al., 2012a; Doğramaci et al., 2013).
The identification and characterization of non-dormant mutants, such as the evergrowing (evg) mutant of peach (Rodriguez et al., 1994), in addition to several functional studies using transgenic poplar, have contributed to renew the dormancy field with increasing molecular works at the gene level. A deletion affecting several members of a series of six tandemly repeated MADS-box genes (DAM1-6, for DORMANCY-ASSOCIATED MADS-box) has been proposed to cause the non-dormant phenotype of the evg mutant (Bielenberg et al., 2004,2008). The six DAM genes of peach were presumably originated by serial tandem duplications from an ancestor related to the flowering transition regulator SHORT VEGETATIVE PHASE (SVP) of Arabidopsis thaliana (Jiménez et al., 2009). In addition to their tight linkage to the evg locus, several recent molecular studies support the involvement of DAM genes in promoting dormancy. DAM genes are expressed in buds following different developmental patterns and are distinctly affected by photoperiod and chilling signals (Li et al., 2009). However, the expression of two of them, DAM5 and DAM6, correlates particularly well with the dormancy status of buds, being high in dormant buds and low after the fulfillment of chilling requirements prior to bud dormancy release (Jiménez et al., 2010b; Yamane et al., 2011). To acquire further genetic evidences about DAM function, DAM6 has been constitutively expressed in transgenic plums (Prunus domestica), showing some degree of dwarfing and increased branching (Fan, 2010). Moreover, several independent quantitative trait loci (QTL) analyses dissecting the flowering time and chilling requirement traits in peach (P. persica), apricot (Prunus armeniaca), and almond (Prunus dulcis) have found a common QTL in linkage group 1 coincident with the genomic location of the DAM locus (Quilot et al., 2004; Olukolu et al., 2009; Fan et al., 2010; Sánchez-Pérez et al., 2012; Romeu et al., 2014). Interestingly, the low chilling requirement trait evaluated in one of these QTL segregating populations in peach, is associated with the presence of large insertions within intronic sequences in DAM5 and DAM6 genes (Zhebentyayeva et al., 2014). However, none of the poplar homologs of DAM genes colocalizes with QTLs associated with bud set in this species (Rohde et al., 2011), suggesting a somehow different role or a reduced natural variability of DAM genes in poplar.
Related DAM-like genes with dormancy-dependent expression have been found in other perennial species such as raspberry (Mazzitelli et al., 2007), leafy spurge (Horvath et al., 2008,2010), Japanese apricot (Yamane et al., 2008), pear (Ubi et al., 2010; Saito et al., 2013), blackcurrant (Hedley et al., 2010), and kiwifruit (Actinidia deliciosa; Wu et al., 2012). The heterologous expression of some of these genes in transgenic plants has offered additional clues about their role in flowering and dormancy. Arabidopsis plants expressing DAM1 from leafy spurge (Horvath et al., 2010) and SVP1 and SVP3 from kiwifruit (Wu et al., 2012) show a delay in flowering time; whereas the expression of PmDAM6 from Japanese apricot in poplar causes growth cessation and bud set under conditions favorable for growth (Sasaki et al., 2011).
Other regulatory genes belonging to different clades within the MADS-box family have also been found to be related to bud dormancy events. Thus, the overexpression of the birch (Betula pendula) FRUITFULL (FUL)-like gene BpMADS4 delays senescence and winter dormancy in Populus tremula (Hoenicka et al., 2008), and the presence of different allelic variants in a SUPPRESSOR OF OVEREXPRESSION OF CONSTANS (CO) 1 (SOC1)-like gene in apricot is found associated with different chilling requirements for dormancy break (Trainin et al., 2013). Interestingly, FUL and SOC1 are activated in the shoot apex of Arabidopsis by the mobile florigen protein produced by the FLOWERING LOCUS T (FT) gene, and mediate the effect of FT and CO on the photoperiod-dependent induction of flowering (Yoo et al., 2005; Melzer et al., 2008). In poplar, a regulatory pathway including two genes homologous to CO and FT controls the growth cessation induced by short days, and also seasonal bud set (Böhlenius et al., 2006). Moreover, transgenic plums overexpressing FT1 from poplar do not enter dormancy and experience continuous flowering among other developmental alterations (Srinivasan et al., 2012). A deep study about the role of the paralogs FT1 and FT2 in poplar, using heat-inducible constructs, has shown that FT1 determines the reproductive onset in winter, whereas FT2 promotes the vegetative growth in spring and summer (Hsu et al., 2011). This work postulates the seasonal succession of reproductive, vegetative, and dormant phases in poplar through the divergent expression and function of FT1 and FT2 genes.
The role of abscisic acid (ABA) and ABA-responsive factors in dormancy maintenance in buds has been mainly addressed in poplar. The overexpression and downregulation of PtABI3, an homolog of ABSCISIC ACID INSENSITIVE 3 (ABI3) of Arabidopsis involved in seed dormancy regulation by ABA signaling, cause developmental alterations in bud formation and misregulation of gene expression during bud dormancy processes (Rohde et al., 2002; Ruttink et al., 2007). Furthermore, the ectopic expression of the mutant gene abscisic acid insensitive 1 (abi1) in poplar modifies the dormant response of buds to exogenous ABA (Arend et al., 2009).
PIECES OF AN EPIGENETIC “ALARM CLOCK” FOR BUD DORMANCY AND AWAKENING
Several reviews postulate the participation of different epigenetic mechanisms involving histone modification, DNA methylation, and the synthesis of small non-coding RNAs in regulating bud dormancy events, based on evident similarities between the environmental and molecular control of dormancy in buds and other well-known processes, such as flowering initiation, vernalization, and seed development (Horvath et al., 2003; Horvath, 2009; Hemming and Trevaskis, 2011; Cooke et al., 2012). However, only few recent works have provided experimental support for these postulates.
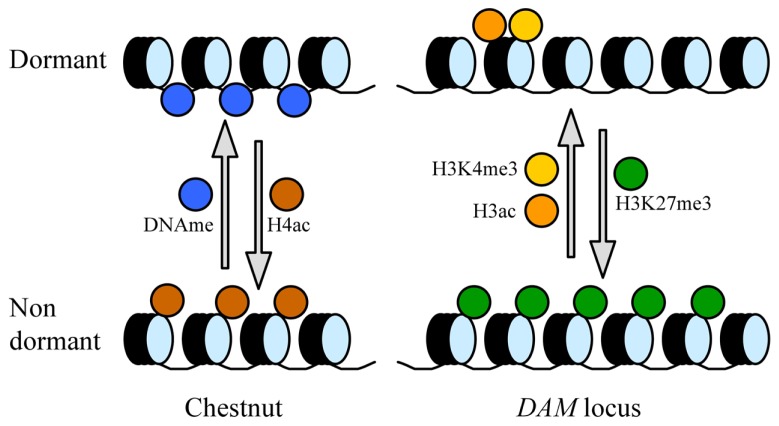
The global levels of genomic DNA methylation and acetylated histone 4 (H4) show cyclic and opposite variations during the seasonal development of chestnut (Castanea sativa), with higher DNA methylation ratios and lower H4 acetylation levels in dormant buds with respect to actively growing tissues (Santamaría et al., 2009; Figure 1). These data support a significant silencing of bulk gene expression concomitant with bud dormancy; however, the epigenetic regulation of particular genes with a relevant role in regulatory issues may differ from this global tendency to gene repression. Indeed, in a subsequent work, the authors have found the gene CsAUR3 encoding an H3 kinase-like expressed in growing tissue, and genes CsHUB2 and CsGCN5L coding for putative histone mono-ubiquitinase and histone acetyltransferase with higher expression in dormant buds (Santamar et al., 2011). Interestingly, the hub2 mutant in Arabidopsis displays reduced seed dormancy (Liu et al., 2007). In agreement with overall DNA methylation changes in chestnut, a decrease in DNA methylation at 5′-CCGG-3′ sites precedes dormancy release, transcriptional activation, and meristem growth in potato tubers (Law and Suttle, 2003).
FIGURE 1.
General and specific modifications of chromatin in dormant and dormancy-released buds. The following chromatin modifications have been identified in chestnut at the genome level (left), and specifically in the DAM locus in leafy spurge and peach (right): DNA methylation (DNAme), acetylation of histone H4 (H4ac), acetylation of H3 (H3ac), trimethylation of H3 at K4 (H3K4me3), and trimethylation of H3 at K27 (H3K27me3).
Some transcriptomic studies have contributed to the identification of genes involved in epigenetic regulation differentially expressed in dormant and growing samples. Two genes similar to FERTILIZATION INDEPENDENT ENDOSPERM (FIE) and PICKLE (PKL) are strongly up-regulated in poplar immediately after transfer to short-day (SD) conditions, leading to bud formation and dormancy induction (Ruttink et al., 2007). FIE is a component of the Polycomb repressive complex 2 (PRC2) involved in the trimethylation of H3 lysine 27 (H3K27me3), a chromatin modification associated with stable gene silencing, whereas PKL is an ATP-dependent chromatin-remodeller that regulates embryo identity traits and associates with multiple H3K27me3-enriched loci (Ogas et al., 1999; Zhang et al., 2012). In Arabidopsis, fie mutants show fertilization-independent development of the endosperm (Ohad et al., 1999), precocious flower-like structures (Kinoshita et al., 2001), and seeds with impaired dormancy (Bouyer et al., 2011). According to these data suggesting a role of Polycomb complexes in SD-induction of dormancy, transgenic hybrid aspen lines with RNAi-mediated downregulation of FIE are not able to establish dormancy, even though growth cessation and bud formation processes seem not to be affected (Petterle, 2011).
Other genes up-regulated during the activity–dormancy transition in hybrid aspen encode putative histone deacetylases (HDA14 and HDA08), histone lysine methyltranferase (SUVR3), and HUB2, while several genes belonging to the Trithorax family of factors counteracting the repressive effect of Polycomb complex and putative DEMETER-like DNA glycosylases are down-regulated during the same period (Karlberg et al., 2010). These results prompted the authors to propose a model in which some unknown target genes are repressed by chromatin compaction during dormancy induction due to histone deacetylation and methylation, histone ubiquitination, DNA methylation, and Polycomb activity. Interestingly, dormancy release upon chilling treatment is accompanied by the up-regulation of other histone deacetylase genes (HDA9 and SIN3) and also a DEMETER-like gene, suggesting that chromatin repression mechanisms with distinct target specificity could act on the different stages of the activity–dormancy cycle (Karlberg et al., 2010).
RESEMBLANCES OF BUD DORMANCY AND VERNALIZATION
The similarities of bud dormancy in perennials with the vernalization treatment for flowering in Arabidopsis and cereals reside in their common requirement of a time period of quantitative and cumulative chilling, and the participation of certain MADS-box domain transcription factors with key regulatory tasks (Chouard, 1960; Horvath et al., 2003; Hemming and Trevaskis, 2011). The MADS-box genes FLOWERING LOCUS C (FLC) and VERNALIZATION1 (VRN1) control, respectively, the vernalization response in Arabidopsis and cereals (Sheldon et al., 2000; Trevaskis et al., 2007), whereas DAM genes exert an analogous role in bud dormancy processes. Interestingly, the vernalization and bud dormancy responses are mediated by the regulation of FLC, VRN1, and DAM gene expression through common epigenetic mechanisms. After prolonged cold exposure, concomitantly with gene repression and dormancy release, the chromatin in the promoter of leafy spurge DAM1 shows a decrease in trimethylation of H3 lysine 4 (H3K4me3) and an increase of H3K27me3 (Horvath et al., 2010), two histone modifications found also associated with the vernalization-dependent repression of FLC in Arabidopsis (Bastow et al., 2004; Kim et al., 2005), and in VRN1 in barley before vernalization (Oliver et al., 2009). In peach, DAM6 shows similar epigenetic changes associated with gene repression after dormancy release, in addition to a decrease of H3 acetylation in the chromatin around the ATG region (Leida et al., 2012a; Figure 1). Such changes occurred at different dates in two different cultivars, in close agreement with their particular chilling requirements and dormancy release dates (Leida et al., 2012a).
In addition to histone modifications, the synthesis and action of long non-coding RNAs (lncRNAs) play a crucial role in the epigenetic regulation of FLC gene in Arabidopsis (Turck and Coupland, 2011). Both, a natural antisense transcript called COOLAIR expressed from a promoter located in the 3′ flanking region of FLC, and a sense intronic lncRNA named COLDAIR, participate in different phases of the cold-induced repression and the stable silencing of FLC by PRC2 (Swiezewski et al., 2009; Heo and Sung, 2011).
Other non-coding RNAs acting on the epigenetic regulation of many plant processes are microRNAs (miRNAs). Recently, a genome-wide approach for the isolation of conserved and novel peach miRNAs has allowed the identification of miRNAs differently expressed in chilled vegetative buds (Barakat et al., 2012). Some of these miRNAs colocalize with known QTLs for chilling requirement and blooming date traits, offering new tools and targets for the genetic analysis of dormancy mechanisms. An epigenetic mechanism involving the synthesis of specific miRNA has been also proposed for an adaptive memory of temperature observed in Norway spruce, in which the environmental temperature during embryo development determines the bud phenology and cold acclimation (Yakovlev et al., 2010,2011). An miRNA cascade involving miR156 and miR172 and their respective targets SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL), and APETALA2 (AP2)-like genes modulates flowering induction in Arabidopsis through the regulation of FT and other flowering-related genes (Khan et al., 2014; Spanudakis and Jackson, 2014). Evidence on the differential expression of SPL genes and miR172 during dormancy induction has been obtained from transcriptomic studies in poplar and leafy spurge (Ruttink et al., 2007; Doğramaci et al., 2013), which suggests that this miRNA pathway may also play a regulating role in dormancy processes.
PERSPECTIVES
As new details of the epigenetic regulation of gene expression during bud dormancy processes emerge, its mechanistic similarities with the well-known regulation of Arabidopsis FLC gene are becoming increasingly evident. Based on such analogy, one may reasonably figure out the remaining pieces of the “alarm clock” outlined above. The chromatin of the transcription start site of pre-vernalized FLC is trimethylated at H3K36 and monoubiquitinated at H2B, favored by the interaction of chromatin modification enzymes with the transcriptional machinery (He, 2012). These modifications and proteins could be similarly identified in dormant buds of perennial species where DAM-like genes are transcriptionally active, prior to cold-dependent down-regulation. Also cold-induced COOLAIR-like and COLDAIR-like lncRNAs could be involved, respectively, in the initial down-regulation of DAM genes and the function of PRC2 complexes. In fact, the presence of a large intron shortly after the beginning of the coding region in DAM genes in peach resembles the situation of the first intron of FLC, particularly important in the synthesis of COLDAIR and the nucleation of the PRC2-dependent trimethylation of H3K27 (Song et al., 2012). In spite of the coincident identification of H3K27me3 in the chromatin of DAM-like genes in dormancy-released buds of leafy spurge and peach, the components and the role of PRC2 complexes in the stable silencing of DAM need to be established in temperate perennials, as already performed in Arabidopsis (Jiang et al., 2008). In summary, the rich literature describing vernalization in model plants may radically accelerate the knowledge of the epigenetics of bud dormancy regulation over the next few years, but it will require the implementation of more informative biochemical and functional approaches in perennial species to complement previous genetic and genomic studies.
In addition to these mechanistic commonalities between the regulation of vernalization and bud dormancy, specific regulatory elements are expected to operate in buds. In particular, the seasonality of bud dormancy with undefined repeated cycles of growth–dormancy along the life of an individual, the major role of hormones such as ABA, and the occurrence of dormancy-related processes on preformed flowers located within buds instead of vegetative tissues, among other particularities, suggest the existence of yet unknown exciting pieces in the “clock.”
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA) – FEDER, and the Ministry of Science and Innovation of Spain (grant no. AGL2010-20595).
REFERENCES
- Allona I., Ramos A., Ibáñez C., Contreras A., Casado R., Aragoncillo C. (2008). Molecular control of winter dormancy establishment in trees. Span. J. Agric. Res. 6 201–210 10.5424/sjar/200806S1-389 [DOI] [Google Scholar]
- Arend M., Schnitzler J. P., Ehlting B., Hänsch R., Lange T., Rennenberg H., et al. (2009). Expression of the Arabidopsis mutant abi1 gene alters abscisic acid sensitivity, stomatal development, and growth morphology in gray poplars. Plant Physiol. 151 2110–2119 10.1104/pp.109.144956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R., Rowland L. J., Tanino K. (2003). Induction and release of bud dormancy in woody perennials: a science comes of age. HortScience 38 911–921 [Google Scholar]
- Bai S., Saito T., Sakamoto D., Ito A., Fujii H., Moriguchi T. (2013). Transcriptome analysis of Japanese pear (Pyrus pyrifolia Nakai) flower buds transitioning through endodormancy. Plant Cell Physiol. 54 1132–1151 10.1093/pcp/pct067 [DOI] [PubMed] [Google Scholar]
- Barakat A., Sriram A., Park J., Zhebentyayeva T., Main D., Abbott A. (2012). Genome wide identification of chilling responsive microRNAs in Prunus persica. BMC Genomics 13:481 10.1186/1471-2164-13-481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastow R., Mylne J. S., Lister C., Lippman Z., Martienssen R. A., Dean C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427 164–167 10.1038/nature02269 [DOI] [PubMed] [Google Scholar]
- Bielenberg D. G., Wang Y., Fan S., Reighard G. L., Scorza R., Abbott A. G. (2004). A deletion affecting several gene candidates is present in the evergrowing peach mutant. J. Hered. 95 436–444 10.1093/jhered/esh057 [DOI] [PubMed] [Google Scholar]
- BöBielenberg D. G., Wang Y., Li Z., Zhebentyayeva T., Fan S., Reighard G. L., et al. (2008). Sequencing and annotation of the evergrowing locus in peach (Prunus persica [L.] Batsch) reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genet. Genomes 4 495–507 10.1007/s11295-007-0126-9 [DOI] [Google Scholar]
- Böhlenius H., Huang T., Charbonnel-Campaa L., Brunner A. M., Jansson S., Strauss S. H., et al. (2006). CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312 1040–1043 10.1126/science.1126038 [DOI] [PubMed] [Google Scholar]
- Bouyer D., Roudier F., Heese M., Andersen E. D., Gey D., Nowack M. K., et al. (2011). Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet. 7:e1002014 10.1371/journal.pgen.1002014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouard P. (1960). Vernalization and its relations to dormancy. Annu. Rev. Plant Physiol. 11 191–238 10.1146/annurev.pp.11.060160.001203 [DOI] [Google Scholar]
- Chuine I. (2010). Why does phenology drive species distribution? Philos. Trans. R. Soc. Lond. B Biol. Sci. 365 3149–3160 10.1098/rstb.2010.0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke J. E. K., Eriksson M. E., Junttila O. (2012). The dynamic nature of bud dormancy in trees: environmental control and molecular mechanisms. Plant Cell Environ. 35 1707–1728 10.1111/j.1365-3040.2012.02552.x [DOI] [PubMed] [Google Scholar]
- Couvillon G. A., Erez A. (1985). Influence of prolonged exposure to chilling temperatures on bud break and heat requirement for bloom of several fruit species. J. Am. Soc. Hort. Sci. 110 47–50 [Google Scholar]
- Coville F. V. (1920). The influence of cold in stimulating the growth of plants. Proc. Natl. Acad. Sci. U.S.A. 6 434–435 10.1073/pnas.6.7.434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Riquelme J., Grimplet J., Martínez-Zapater J. M., Carmona M. J. (2012). Transcriptome variation along bud development in grapevine (Vitis vinifera L.). BMC Plant Biol. 12:181 10.1186/1471-2229-12-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doğramaci M., Foley M. E., Chao W. S., Christoffers M. J., Anderson J. V. (2013). Induction of endodormancy in crown buds of leafy spurge (Euphorbia esula L.) implicates a role for ethylene and cross-talk between photoperiod and temperature. Plant Mol. Biol. 81 577–593 10.1007/s11103-013-0026-3 [DOI] [PubMed] [Google Scholar]
- El Kayal W., Allen C. C., Ju C. J., Adams E., King-Jones S., Zaharia L. I., et al. (2011). Molecular events of apical bud formation in white spruce, Picea glauca. Plant Cell Environ. 34 480–500 10.1111/j.1365-3040.2010.02257.x [DOI] [PubMed] [Google Scholar]
- Fan S. (2010). Mapping Quantitative Trait Loci Associated with Chilling Requirement and Bloom Date in Peach. Ph.D. thesis, Clemson University, Clemson; [DOI] [PubMed] [Google Scholar]
- Fan S., Bielenberg D. G., Zhebentyayeva T. N., Reighard G. L., Okie W. R., Holland D., et al. (2010). Mapping quantitative trait loci associated with chilling requirement, heat requirement and bloom date in peach (Prunus persica). New Phytol. 185 917–930 10.1111/j.1469-8137.2009.03119.x [DOI] [PubMed] [Google Scholar]
- Hänninen H., Tanino K. (2011). Tree seasonality in a warming climate. Trends Plant Sci. 16 412–416 10.1016/j.tplants.2011.05.001 [DOI] [PubMed] [Google Scholar]
- He Y. (2012). Chromatin regulation of flowering. Trends Plant Sci. 17 556–562 10.1016/j.tplants.2012.05.001 [DOI] [PubMed] [Google Scholar]
- Hedley P. E., Russell J. R., Jorgensen L., Gordon S., Morris J. A., Hackett C. A., et al. (2010). Candidate genes associated with bud dormancy release in blackcurrant (Ribes nigrum L.). BMC Plant Biol. 10:202 10.1186/1471-2229-10-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming M. N., Trevaskis B. (2011). Make hay when the sun shines: the role of MADS-box genes in temperature-dependent seasonal flowering responses. Plant Sci. 180 447–453 10.1016/j.plantsci.2010.12.001 [DOI] [PubMed] [Google Scholar]
- Heo J. B., Sung S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331 76–79 10.1126/science.1197349 [DOI] [PubMed] [Google Scholar]
- Hoenicka H., Nowitzki O., Hanelt D., Fladung M. (2008). Heterologous overexpression of the birch FRUITFULL-like MADS-box gene BpMADS4 prevents normal senescence and winter dormancy in Populus tremula L. Planta 227 1001–1011 10.1007/s00425-007-0674-0 [DOI] [PubMed] [Google Scholar]
- Horvath D. (2009). Common mechanisms regulate flowering and dormancy. Plant Sci. 177 523–531 10.1016/j.plantsci.2009.09.002 [DOI] [Google Scholar]
- Horvath D. P., Anderson J. V., Chao W. S., Foley M. E. (2003). Knowing when to grow: signals regulating bud dormancy. Trends Plant Sci. 8 534–540 10.1016/j.tplants.2003.09.013 [DOI] [PubMed] [Google Scholar]
- Horvath D. P., Chao W. S., Suttle J. C., Thimmapuram J., Anderson J. V. (2008). Transcriptome analysis identifies novel responses and potential regulatory genes involved in seasonal dormancy transitions of leafy spurge (Euphorbia esula L.). BMC Genomics 9:536 10.1186/1471-2164-9-536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath D. P., Sung S., Kim D., Chao W., Anderson J. (2010). Characterization, expression and function of DORMANCY ASSOCIATED MADS-BOX genes from leafy spurge. Plant Mol. Biol. 73 169–179 10.1007/s11103-009-9596-5 [DOI] [PubMed] [Google Scholar]
- Hsu C.-Y., Adams J. P., Kim H., No K., Ma C., Strauss S. H., et al. (2011). FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc. Natl. Acad. Sci. U.S.A. 108 10756–10761 10.1073/pnas.1104713108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Wang Y., Wang Y., He Y. (2008). Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS ONE 3:e3404 10.1371/journal.pone.0003404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez S., Lawton-Rauh A. L., Reighard G. L., Abbott A. G., Bielenberg D. G. (2009). Phylogenetic analysis and molecular evolution of the dormancy associated MADS-box genes from peach. BMC Plant Biol. 9:81 10.1186/1471-2229-9-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez S., Li Z., Reighard G. L., Bielenberg D. G. (2010a). Identification of genes associated with growth cessation and bud dormancy entrance using a dormancy-incapable tree mutant. BMC Plant Biol. 10:25 10.1186/1471-2229-10-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez S., Reighard G. L., Bielenberg D. G. (2010b). Gene expression of DAM5 and DAM6 is suppressed by chilling temperatures and inversely correlated with bud break rate. Plant Mol. Biol. 73 157–167 10.1007/s11103-010-9608-5 [DOI] [PubMed] [Google Scholar]
- Karlberg A., Englund M., Petterle A., Molnar G., Sjödin A., Bakó L., et al. (2010). Analysis of global changes in gene expression during activity–dormancy cycle in hybrid aspen apex. Plant Biotechnol. 27 1–16 10.5511/plantbiotechnology.27.1 [DOI] [Google Scholar]
- Khan M. R., Ai X. Y., Zhang J. Z. (2014). Genetic regulation of flowering time in annual and perennial plants. Wiley Interdiscip. Rev. RNA 5 347–359 10.1002/wrna.1215 [DOI] [PubMed] [Google Scholar]
- Kim S. Y., He Y., Jacob Y., Noh Y. S., Michaels S., Amasino R. (2005). Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase. Plant Cell 17 3301–3310 10.1105/tpc.105.034645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Harada J. J., Goldberg R. B., Fischer R. L. (2001). Polycomb repression of flowering during early plant development. Proc. Natl. Acad. Sci. U.S.A. 98 14156–14161 10.1073/pnas.241507798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law R. D., Suttle J. C. (2003). Transient decreases in methylation at 5′-CCGG-3′ sequences in potato (Solanum tuberosum L.) meristem DNA during progression of tubers through dormancy precede the resumption of sprout growth. Plant Mol. Biol. 51 437–447 10.1023/A:1022002304479 [DOI] [PubMed] [Google Scholar]
- Leida C., Conesa A., Llácer G., Badenes M. L, Ríos G. (2012a). Histone modifications and expression of DAM6 gene in peach are modulated during bud dormancy release in a cultivar-dependent manner. New Phytol. 193 67–80 10.1111/j.1469-8137.2011.03863.x [DOI] [PubMed] [Google Scholar]
- Leida C., Conejero A., Arbona V., Gómez-Cadenas A., Llácer G., Badenes M. L., et al. (2012b). Chilling-dependent release of seed and bud dormancy in peach associates to common changes in gene expression. PLoS ONE 7:e35777 10.1371/journal.pone.0035777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leida C., Terol J., Martá G., Agustá M., Llácer G., Badenes M. L., et al. (2010). Identification of genes associated with bud dormancy release in Prunus persica by suppression subtractive hybridization. Tree Physiol. 30 655–666 10.1093/treephys/tpq008 [DOI] [PubMed] [Google Scholar]
- Li Z., Reighard G. L., Abbott A. G., Bielenberg D. G. (2009). Dormancy-associated MADS genes from the EVG locus of peach (Prunus persica [L.] Batsch) have distinct seasonal and photoperiodic expression patterns. J. Exp. Bot. 60 3521–3530 10.1093/jxb/erp195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Li W., Zheng P., Xu T., Chen L., Liu D., et al. (2012). Transcriptomic analysis of ‘Suli’ pear (Pyrus pyrifolia white pear group) buds during the dormancy by RNA-Seq. BMC Genomics 13:700 10.1186/1471-2164-13-700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Koornneef M., Soppe W. J. (2007). The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 19 433–444 10.1105/tpc.106.049221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathiason K., He D., Grimplet J., Venkateswari J., Galbraith D. W., Or E., et al. (2009). Transcript profiling in Vitis riparia during chilling requirement fulfillment reveals coordination of gene expression patterns with optimized bud break. Funct. Integr. Genomics 9 81–96 10.1007/s10142-008-0090-y [DOI] [PubMed] [Google Scholar]
- Mazzitelli L., Hancock R. D., Haupt S., Walker P. G., Pont S. D., McNicol J., et al. (2007). Co-ordinated gene expression during phases of dormancy release in raspberry (Rubus idaeus L.) buds. J. Exp. Bot. 58 1035–1045 10.1093/jxb/erl266 [DOI] [PubMed] [Google Scholar]
- Melzer S., Lens F., Gennen J., Vanneste S., Rohde A., Beeckman T. (2008). Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat. Genet. 40 1489–1492 10.1038/ng.253 [DOI] [PubMed] [Google Scholar]
- Ogas J., Kaufmann S., Henderson J., Somerville C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 96 13839–13844 10.1073/pnas.96.24.13839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad N., Yadegari R., Margossian L., Hannon M., Michaeli D., Harada J. J., et al. (1999). Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11 407–416 10.1105/tpc.11.3.407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver S. N., Finnegan E. J., Dennis E. S., Peacock W. J., Trevaskis B. (2009). Vernalization-induced flowering in cereals is associated with changes in histone methylation at the VERNALIZATION1 gene. Proc. Natl. Acad. Sci. U.S.A. 106 8386–8391 10.1073/pnas.0903566106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J. E. (2010). Light and temperature sensing and signaling in induction of bud dormancy in woody plants. Plant Mol. Biol. 73 37–47 10.1007/s11103-010-9620-9 [DOI] [PubMed] [Google Scholar]
- Olukolu B. A., Trainin T., Fan S., Kole C., Bielenberg D. G., Reighard G. L., et al. (2009). Genetic linkage mapping for molecular dissection of chilling requirement and budbreak in apricot (Prunus armeniaca L.). Genome 52 819–828 10.1139/g09-050 [DOI] [PubMed] [Google Scholar]
- Petterle A. (2011). ABA and Chromatin Remodeling Regulate the Activity–Dormancy Cycle in Hybrid Aspen. Ph.D. thesis, Swedish University of Agricultural Sciences, å [Google Scholar]
- Powell L. E. (1987). Hormonal aspects of bud and seed dormancy in temperate zone woody plants. HortScience 22 845–850 [Google Scholar]
- Quilot B., Wu B. H., Kervella J., Génard M., Foulongne M., Moreau K. (2004). QTL analysis of quality traits in an advanced backcross between Prunus persica cultivars and the wild relative species P.davidiana. Theor. Appl. Genet. 109 884–897 10.1007/s00122-004-1703-z [DOI] [PubMed] [Google Scholar]
- Rodriguez J., Sherman W. B., Scorza R., Wisniewski M., Okie W. R. (1994). ‘Evergreen’ peach, its inheritance and dormant behavior. J. Am. Soc. Hort. Sci. 119 789–792 [Google Scholar]
- Rohde A., Bhalerao R. P. (2007). Plant dormancy in the perennial context. Trends Plant Sci. 12 217–223 10.1016/j.tplants.2007.03.012 [DOI] [PubMed] [Google Scholar]
- Rohde A., Prinsen E., De Rycke R., Engler G., Van Montagu M., Boerjan W. (2002). PtABI3 impinges on the growth and differentiation of embryonic leaves during bud set in poplar. Plant Cell 14 1885–1901 10.1105/tpc.003186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde A., Ruttink T., Hostyn V., Sterck L., Driessche K. V., Boerjan W. (2007). Gene expression during the induction, maintenance, and release of dormancy in apical buds of poplar. J. Exp. Bot. 58 4047–4060 10.1093/jxb/erm261 [DOI] [PubMed] [Google Scholar]
- Rohde A., Storme V., Jorge V., Gaudet M., Vitacolonna N., Fabbrini F., et al. (2011). Bud set in poplar–genetic dissection of a complex trait in natural and hybrid populations. New Phytol. 189 106–121 10.1111/j.1469-8137.2010.03469.x [DOI] [PubMed] [Google Scholar]
- Romeu J. F., Monforte A. J., Sánchez G., Granell A., Garcáa-Brunton J., Badenes M. L., et al. (2014). Quantitative trait loci affecting reproductive phenology in peach. BMC Plant Biol. 14:52 10.1186/1471-2229-14-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruttink T., Arend M., Morreel K., Storme V., Rombauts S., Fromm J., et al. (2007). A molecular timetable for apical bud formation and dormancy induction in poplar. Plant Cell 19 2370–2390 10.1105/tpc.107.052811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Bai S., Ito A., Sakamoto D., Saito T., Ubi B. E., et al. (2013). Expression and genomic structure of the dormancy-associated MADS box genes MADS13 in Japanese pears (Pyrus pyrifolia Nakai) that differ in their chilling requirement for endodormancy release. Tree Physiol. 33 654–667 10.1093/treephys/tpt037 [DOI] [PubMed] [Google Scholar]
- Sánchez-Pérez R., Dicenta F, Martánez-Gómez P. (2012). Inheritance of chilling and heat requirements for flowering in almond and QTL analysis. Tree Genet. Genomes 8 379–389 10.1007/s11295-011-0448-5 [DOI] [Google Scholar]
- Santamaría M. E., Hasbún R., Valera M. J., Meijón M., Valledor L., Rodráguez J. L., et al. (2009). Acetylated H4 histone and genomic DNA methylation patterns during bud set and bud burst in Castanea sativa. J. Plant Physiol. 166 1360–1369 10.1016/j.jplph.2009.02.014 [DOI] [PubMed] [Google Scholar]
- Santamaría M. E., Rodráguez R., ñal M. J., Toorop P. E. (2011). Transcriptome analysis of chestnut (Castanea sativa) tree buds suggests a putative role for epigenetic control of bud dormancy. Ann. Bot. 108 485–498 10.1093/aob/mBR185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki R., Yamane H., Ooka T., Jotatsu H., Kitamura Y., Akagi T., et al. (2011). Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot. Plant Physiol. 157 485–497 10.1104/pp.111.181982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon C. C., Rouse D. T., Finnegan E. J., Peacock W. J., Dennis E. S. (2000). The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. U.S.A. 97 3753–3758 10.1073/pnas.97.7.3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Angel A., Howard M., Dean C. (2012). Vernalization–a cold-induced epigenetic switch. J. Cell Sci. 125 3723–3731 10.1242/jcs.084764 [DOI] [PubMed] [Google Scholar]
- Spanudakis E., Jackson S. (2014). The role of microRNAs in the control of flowering time. J. Exp. Bot. 65 365–380 10.1093/jxb/ert453 [DOI] [PubMed] [Google Scholar]
- Srinivasan C., Dardick C., Callahan A., Scorza R. (2012). Plum (Prunus domestica) trees transformed with poplar FT1 result in altered architecture, dormancy requirement, and continuous flowering. PLoS ONE 7:e40715 10.1371/journal.pone.0040715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S., Liu F., Magusin A., Dean C. (2009). Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462 799–802 10.1038/nature08618 [DOI] [PubMed] [Google Scholar]
- Trainin T., Bar-Ya’akov I., Holland D. (2013). ParSOC1, a MADS-box gene closely related to Arabidopsis AGL20/SOC1, is expressed in apricot leaves in a diurnal manner and is linked with chilling requirements for dormancy break. Tree Genet. Genomes 9 753–766 10.1007/s11295-012-0590-8 [DOI] [Google Scholar]
- Trevaskis B., Hemming M. N., Dennis E. S., Peacock W. J. (2007). The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 12 352–357 10.1016/j.tplants.2007.06.010 [DOI] [PubMed] [Google Scholar]
- Turck F., Coupland G. (2011). When vernalization makes sense. Science 331 36–37 10.1126/science.1200786 [DOI] [PubMed] [Google Scholar]
- Ubi B. E., Sakamoto D., Ban Y., Shimada T., Ito A., Nakajima I., et al. (2010). Molecular cloning of dormancy-associated MADS-box gene homologs and their characterization during seasonal endodormancy transitional phases of Japanese pear. J. Am. Soc. Hort. Sci. 135 174–182 [Google Scholar]
- Wu R. M., Walton E. F., Richardson A. C., Wood M., Hellens R. P., Varkonyi-Gasic E. (2012). Conservation and divergence of four kiwifruit SVP-like MADS-box genes suggest distinct roles in kiwifruit bud dormancy and flowering. J. Exp. Bot. 63 797–807 10.1093/jxb/err304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovlev I. A., Asante D. K. A., Fossdal C. G., Junttila O, Johnsen Ø . (2011). Differential gene expression related to an epigenetic memory affecting climatic adaptation in Norway spruce. Plant Sci. 180 132–139 10.1016/j.plantsci.2010.07.004 [DOI] [PubMed] [Google Scholar]
- Yakovlev I. A., Fossdal C. G, Johnsen Ø. (2010). MicroRNAs, the epigenetic memory and climatic adaptation in Norway spruce. New Phytol. 187 1154–1169 10.1111/j.1469-8137.2010.03341.x [DOI] [PubMed] [Google Scholar]
- Yamane H., Kashiwa Y., Ooka T., Tao R., Yonemori K. (2008). Suppression subtractive hybridization and differential screening reveals endodormancy-associated expression of an SVP/AGL24-type MADS-box gene in lateral vegetative buds of Japanese apricot. J. Am. Soc. Hort. Sci. 133 708–716 [Google Scholar]
- Yamane H., Ooka T., Jotatsu H., Hosaka Y., Sasaki R., Tao R. (2011). Expressional regulation of PpDAM5 and PpDAM6, peach (Prunus persica) dormancy-associated MADS-box genes, by low temperature and dormancy-breaking reagent treatment. J. Exp. Bot. 62 3481–3488 10.1093/jxb/err028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S. K., Chung K. S., Kim J., Lee J. H., Hong S. M., Yoo S. J., et al. (2005). CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 139 770–778 10.1104/pp.105.066928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Luedeling E., Xu J. (2010). Winter and spring warming result in delayed spring phenology on the Tibetan Plateau. Proc. Natl. Acad. Sci. U.S.A. 107 22151–22156 10.1073/pnas.1012490107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Bishop B., Ringenberg W., Muir W. M., Ogas J. (2012). The CHD3 remodeler PICKLE associates with genes enriched for trimethylation of histone H3 lysine 27. Plant Physiol. 159 418–432 10.1104/pp.112.194878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhebentyayeva T. N., Fan S., Chandra A., Bielenberg D. G., Reighard G. L., Okie W. R., et al. (2014). Dissection of chilling requirement and bloom date QTLs in peach using a whole genome sequencing of sibling trees from an F2 mapping population. Tree Genet. Genomes 10 35–51 10.1007/s11295-013-0660-6 [DOI] [Google Scholar]
- Zhong W., Gao Z., Zhuang W., Shi T., Zhang Z., Ni Z. (2013). Genome-wide expression profiles of seasonal bud dormancy at four critical stages in Japanese apricot. Plant Mol. Biol. 83 247–264 10.1007/s11103-013-0086-4 [DOI] [PubMed] [Google Scholar]