Abstract
Transcription-coupled repair (TCR) is one of the key of the nucleotide excision repair (NER) pathways required to preserve genome integrity. Although understanding TCR is still a major challenge, recent single-molecule experiments have brought new insights into the initial steps of TCR leading to new perspectives.
Keywords: DNA repair, Mfd, nucleotide excision repair (NER), single-molecule, stalled RNAp, transcription-coupled repair (TCR)
In all kingdoms of life, genetic information is contained in DNA. While genome integrity is usually preserved under normal growth conditions (and therefore contributes to the species specificity), mutations can be determinant in the survival process of the whole organism or species when drastic environmental changes occur. Mutations in the genome arise because of errors during DNA replication or external environmental factors, and several repair systems exist in the cell to maintain genome integrity. Here, we will only discuss one such nucleotide excision repair (NER) pathway associated with damages resulting from UV radiations (which leads to bulky DNA adducts, e.g., thymine dimers). This process seems to be conserved in all living organisms and can be divided in two major sub-pathways: a global genomic repair (GGR) and a transcription-coupled repair (TCR), involving the transcribing RNA polymerase.1,2These two repair mechanisms differ only in how the damage is recognized: TCR involves the RNA polymerase as a marker for the damage while, in GGR, the change in helix rigidity at the lesion3 triggers the recruitment of proteins that facilitate excision, strand removal, DNA synthesis and re-ligation (see below).
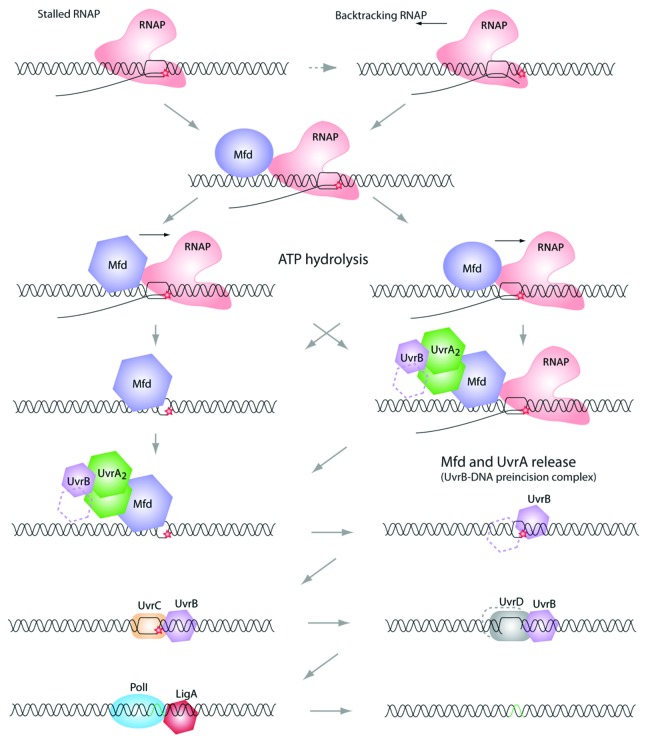
In prokaryotes, the common steps of the NER pathways involve UvrABCD, DNA Pol I and a Ligase (LigA) (Fig. 1).4 As the whole repair pathway is still not fully understood, it is commonly admitted that a dimer of UvrA cannot directly find and bind to the DNA lesion site but needs to be pre-associated to UvrB (one or two monomers).5,6 After this preformed UvrAB complex recognizes and binds the DNA lesion, UvrA is released and the UvrC endonuclease can then interact with UvrB (still loaded on the DNA). UvrC then nicks the DNA on either side of the lesion and the UvrD helicase removes the oligonucleotide, allowing the DNA Polymerase I to “re-synthesize” a new DNA using the complementary strand of DNA as template.7-13 To complete the repair process, the LigA ligase joins the newly synthesized DNA to the adjacent pre-existing strand (Fig. 1).
Figure 1. Transcription coupled repair pathway. When the elongating RNA polymerase reaches a DNA lesion on the template strand, it becomes stalled. At this point, some backtracking events may occur (the 3′-end of the nascent RNA is extruded from the catalytic site). Because a stalled RNA polymerase masks the damage from other proteins (e.g., UvrA and UvrB, see below), it needs to be displaced. This task is achieved by the Mfd (TRCF). Mfd dissociates the RNA polymerase and the nascent transcript, and recruits the UvrAB complex. Then, UvrB-DNA is forms the “pre-incision complex” allowing the recruitment of UvrC, which double-incises the DNA molecule. Finally, UvrD removes the damaged nucleotide sequence and PolI synthesizes a new cDNA, which will be ligated by LigA. Note that the precise mechanism of this repair pathway is still not fully understood and several important questions need to be answered. Among them: does Mfd interact with the RNA polymerase and UvrA at the same time? What is responsible for Mfd conformational changes that allow the “UvrA interacting domain” to be exposed?
Under normal growth conditions, the amount of repair complexes present in the cell is relatively low. Also, the number of DNA lesions present in the genome is usually low (compared with the total genome length) and there is a large variety and abundance of proteins that are naturally bound to the DNA (and therefore might inhibit the loading of repair complexes to damaged DNA). Identifying damage is a difficult task for the cell and so there are different repair mechanisms associated with specific types of damages (e.g., MutH/MutLMlh/Pms1/MutSMsh - eukaryote homologs in superscript, when identified - associated with errors occurring during DNA replication). For the TCR pathway, the cell uses a trick based on a transcribing RNA polymerase. The RNA polymerase is a processive enzyme that uses the DNA as template to produce RNA. This enzyme is then able to “walk” on the DNA at a relatively high speed rate.14,15 When a lesion is present on a transcribed DNA strand, the transcribing RNA polymerase is stopped in its progression with a nascent RNA associated (stalled RNA polymerase on DNA). This stalled RNA polymerase serves as a “marker” for the repair machinery and is therefore a reliable system allowing the cell to monitor lesions.16 In bacteria, the stalled polymerases are recognized by Mfd (Mutation Frequency Decline, also called TRCF-Transciption Repair Coupling Factor).17-21 This TCR process was first observed by Witkin and collaborators,22 who reported that the UV-induced damage might be repaired more efficiently in some part of the genome than in others. In addition, Mellon and Hanawalt23 have shown directly the impact of transcription on the DNA repair using the induced lac operon as a model system and showed that the transcribed strand was more efficiently repaired than the non template one. The role of Mfd is 2-fold. First, it recognizes, binds and displaces the stalled RNA polymerase (and the nascent RNA). This mechanism is important as upstream RNA polymerases, while being able to displace backtracked downstream RNA polymerases,24 are not able to displace a DNA-lesion-dependent stalled-RNA polymerase.25,26 Second, Mfd recruits UvrAB, allowing active DNA-lesion repair.
In eukaryotes, the repair of UV radiation induced damages involves more proteins but follows a mechanism somehow similar to what is found in prokaryotes: for the TCR pathway, RNA polymerase II triggers the recruitment of additional proteins that remodel chromatin (facilitate DNA access27) and repair DNA. Among them, the CSB (Cockayne Syndrome B28) protein plays a central role. CSB belongs to the SWI/SNF (SWItch/Sucrose Non Fermentable) chromatin remodeler protein family and exhibits ATPase activity and a conserved helicase motif. While not being capable of displacing RNA polymerase II, CSB is the first recruited protein, interacts with RNAP and governs the recruitment of repair proteins. As such CSB is believed to be the analog of Mfd. Defects in CSB can result in serious developmental and neurological problems29 whereas, in prokaryotes, inactivating Mfd or UvrA30 has little effect on the survival of E. coli exposed to DNA-damaging agents such as UV. An explanation would be that other pathways for TCR exist in bacteria. For instance, it has been suggested that NusA (a protein associated with a large number of cellular processes and that is known to act as a transcriptional factor) might also play a role in the recruitment of NER proteins.31
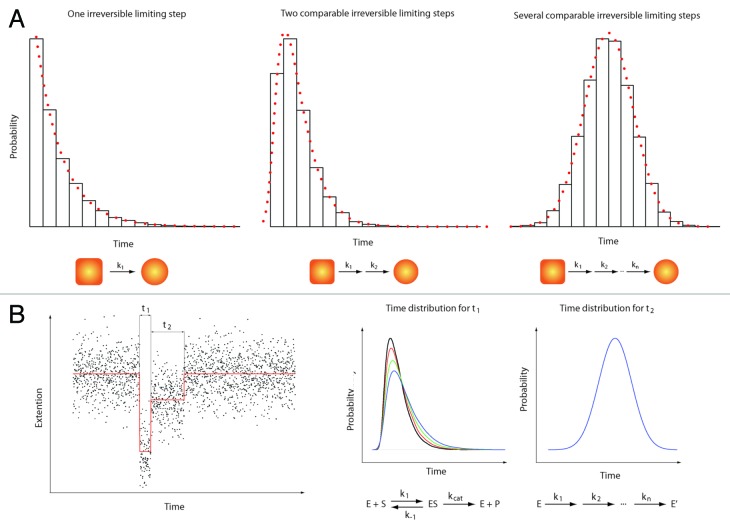
It is becoming clear that many more experiments need to be performed to study initial steps of the NER process. These studies should address the following questions: (i) the precise sequence of events occurring during the repair process (ii) the characterization, (iii) the identification of the limiting steps taking place in this process and (iv) the nature of the different factors and their interactions. To address these questions, new methods like single-molecule approaches might provide important and new insights.32 These techniques (force- or fluorescence-based techniques) not only allow the identification of sub-populations but also probe reaction intermediates. An important point is that the identification of rate-limiting steps in a chemical reaction does not rely on the formation of products or detectable/measurable conformational changes. Indeed, a single chemical reaction (rate-limiting step) is a stochastic process and the probability for this reaction to occur at a certain time is a process that does not dependent on the age and the history of the system (Poissonian process). As such, a histogram showing the number of occurrences vs. time should follow an exponential decay when a single-rate limiting step is present in a chemical reaction. When multiple, substrate-dependent steps are present in a chemical reaction, the shape of the histogram changes accordingly (see Figure 2). It is important to emphasize that such measurements are difficult, if not impossible, to perform in standard (bulk) biochemical assays due to the difficulty of obtaining a fully synchronized population of the species studied.
Figure 2. Single molecule enzymology. The most obvious feature of biochemical processes is their stochastic nature. This implies that any biochemical kinetics usually studied in bulk experiments on a large ensemble of molecules can also be cast in terms of probabilities.39 Here, we give a quick overview of the information contained in such statistical distributions. (A) Three examples of statistical distributions (experimental histograms and corresponding probability density functions) obtained for typical bio-chemical processes. From left to right: 1) Single exponential distribution characteristic of one single limiting step with a rate k1. 2) Two-dimensional exponential characteristic of two rate-limiting steps with rate k1 and k2 respectively. 3) Gaussian-like distribution characteristic of several limiting steps with comparable rates (from k1 to kn with k1 ≈k2 ≈… ≈kn). In a typical experiment, hundreds of events (i.e. hundred of single-molecule traces) are needed to build such histograms and get a reliable description of the bio-chemical reactions. (B) Left panel: A theoretical single-molecule extension vs. time trace (e.g DNA extension vs. time trace). This example shows two different events (different DNA extensions) with two different characteristic times t1 and t2. Right panel: The time distribution for t1 (that would be obtained from different molecules) shows a two-dimensional profile. Changing the substrate concentration (e.g., ATP) dramatically affects the shape of the distribution (different colors corresponding to four different substrate concentrations). This behavior is typical of a Michaelis-Menten kinetic.40 In contrast, the distribution of time t2 shows a Gaussian-like behavior that does not depend on the substrate concentration. Such a distribution could be obtained when the transition from two enzymatic forms (say E and E’) occurs through a large number of irreversible rate-limiting steps.
These new methods are very effective to address specific mechanistic and kinetic questions of biological processes such as repair processes. For instance, Jing Zhou et al.33 employed an optical trapping assay to probe the effect of NusA on transcription pauses; Uphoff et al.34 used single molecule photo-activation, localization and tracking in live bacteria to directly visualized DNA repair processes. In a recent study,35 we used magnetic tweezers to perform single-molecule measurements on the initial steps of the TCR pathway (without the UvrABCD proteins). We have shown that Mfd, when displacing RNA polymerase (stalled at +20 from the transcription start site) acts by catalyzing two irreversible, ATP dependent transitions with different structural, kinetic and mechanistic transitions.
In the absence of UvrAB, Mfd begins by initiating a first catalytic step (mean duration of 20 sec) and then stabilizes a long-lived reaction intermediate (mean duration of 300s). The first catalytic step, which is relatively slow, could be explained by some major conformational changes of Mfd, dependent on ATP, allowing the unmasking of UvrA binding surface at the level of the Mfd D2-D7 domain (consistent with the recent SAXS - Small-angle X-ray scattering - results).36,37 This idea is also supported by the observations that (i) a second RNA polymerase could only bind the promoter when the first RNA polymerase is trapped in this long-lived intermediate and (ii) the size of the transcription bubble is noticeably modified after the first catalytic step.
Interestingly, the distribution of times associated with the second reaction step shows a broaden Gaussian-like distribution, typical of a succession of several irreversible steps with comparable lifetimes. For such a distribution (mean, variance), the probability for Mfd to unbind in a short amount of time (around 100s) is low (in contrast to what would be expected for a single reaction intermediate).This finding has important implications in vivo. Indeed, this will guarantee that UvrA has ample time to find Mfd and initiate the repair machinery.
Understanding the TCR pathway is still a major challenge. While the initial step (Mfd-dependent) of this process is, to some extent, well characterized in bacteria, there are a number of questions that need to be answered. In particular, the nature of the different proteins present at each step of the process remains to be determined. One way to address these questions would be to use a single molecule-based assay, coupling topological signal tracking at the level of the DNA and also fluorescence signal on the protein (using different fluorophores38). This approach (using hybrid instruments combining TIRF - total internal reflection fluorescence- and magnetic Tweezers and/or Curtains) most likely provide new insights into the sequential recruitment of the different actors of the TCR and their role in the changes of the DNA topology. Single-molecule experiments involving eukaryotic proteins might also provide new and important information on the initial steps of the NER process (e.g., studying the activity of CSB and its interaction with RNA polymerase using high-resolution single-molecule experiments).
Acknowledgments
JM is founded by the AVIESAN ITMO Cancer Consortium through Frontières du Vivant (FdV) PhD program. WG is founded by University of Paris Diderot (Paris VII). NJ and TRS are founded by CNRS. The lab is supported by CNRS, University of Paris Diderot (Paris VII), with a core funding by a EURYI grant to TRS. We apologize for all the references that we were not able to cite due to the space limitation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/24934
References
- 1.Kamileri I, Karakasilioti I, Garinis GA. Nucleotide excision repair: new tricks with old bricks. Trends Genet. 2012;28:566–73. doi: 10.1016/j.tig.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Savery N. Prioritizing the repair of DNA damage that is encountered by RNA polymerase. Transcription. 2011;2:168–72. doi: 10.4161/trns.2.4.16146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang W. Structure and mechanism for DNA lesion recognition. Cell Res. 2008;18:184–97. doi: 10.1038/cr.2007.116. [DOI] [PubMed] [Google Scholar]
- 4.Van Houten B, Croteau DL, DellaVecchia MJ, Wang H, Kisker C. ‘Close-fitting sleeves’: DNA damage recognition by the UvrABC nuclease system. Mutat Res. 2005;577:92–117. doi: 10.1016/j.mrfmmm.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 5.Theis K, Chen PJ, Skorvaga M, Van Houten B, Kisker C. Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair. EMBO J. 1999;18:6899–907. doi: 10.1093/emboj/18.24.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pakotiprapha D, Samuels M, Shen K, Hu JH, Jeruzalmi D. Structure and mechanism of the UvrA-UvrB DNA damage sensor. Nat Struct Mol Biol. 2012;19:291–8. doi: 10.1038/nsmb.2240. [DOI] [PubMed] [Google Scholar]
- 7.Savery NJ. The molecular mechanism of transcription-coupled DNA repair. Trends Microbiol. 2007;15:326–33. doi: 10.1016/j.tim.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Kad NM, Van Houten B. Dynamics of lesion processing by bacterial nucleotide excision repair proteins. Prog Mol Biol Transl Sci. 2012;110:1–24. doi: 10.1016/B978-0-12-387665-2.00001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atkinson J, Guy CP, Cadman CJ, Moolenaar GF, Goosen N, McGlynn P. Stimulation of UvrD helicase by UvrAB. J Biol Chem. 2009;284:9612–23. doi: 10.1074/jbc.M808030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grossman L, Yeung AT. The UvrABC endonuclease system of Escherichia coli--a view from Baltimore. Mutat Res. 1990;236:213–21. doi: 10.1016/0921-8777(90)90006-Q. [DOI] [PubMed] [Google Scholar]
- 11.Deaconescu AM, Savery N, Darst SA. The bacterial transcription repair coupling factor. Curr Opin Struct Biol. 2007;17:96–102. doi: 10.1016/j.sbi.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westblade LF, Campbell EA, Pukhrambam C, Padovan JC, Nickels BE, Lamour V, et al. Structural basis for the bacterial transcription-repair coupling factor/RNA polymerase interaction. Nucleic Acids Res. 2010;38:8357–69. doi: 10.1093/nar/gkq692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deaconescu AM, Chambers AL, Smith AJ, Nickels BE, Hochschild A, Savery NJ, et al. Structural basis for bacterial transcription-coupled DNA repair. Cell. 2006;124:507–20. doi: 10.1016/j.cell.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 14.Davenport RJ, Wuite GJ, Landick R, Bustamante C. Single-molecule study of transcriptional pausing and arrest by E. coli RNA polymerase. Science. 2000;287:2497–500. doi: 10.1126/science.287.5462.2497. [DOI] [PubMed] [Google Scholar]
- 15.Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem Soc Trans. 2006;34:1062–6. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- 16.Trautinger BW, Jaktaji RP, Rusakova E, Lloyd RG. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol Cell. 2005;19:247–58. doi: 10.1016/j.molcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Selby CP, Sancar A. Gene- and strand-specific repair in vitro: partial purification of a transcription-repair coupling factor. Proc Natl Acad Sci U S A. 1991;88:8232–6. doi: 10.1073/pnas.88.18.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–8. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 19.Selby CP, Sancar A. Characterization of transcription-repair coupling factors in E. coli and humans. Methods Enzymol. 2003;371:300–24. doi: 10.1016/S0076-6879(03)71023-4. [DOI] [PubMed] [Google Scholar]
- 20.Selby CP, Witkin EM, Sancar A. Escherichia coli mfd mutant deficient in “mutation frequency decline” lacks strand-specific repair: in vitro complementation with purified coupling factor. Proc Natl Acad Sci U S A. 1991;88:11574–8. doi: 10.1073/pnas.88.24.11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park JS, Marr MT, Roberts JWE. E. coli Transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell. 2002;109:757–67. doi: 10.1016/S0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 22.Witkin EM. Time, temperature, and protein synthesis: a study of ultraviolet-induced mutation in bacteria. Cold Spring Harb Symp Quant Biol. 1956;21:123–40. doi: 10.1101/SQB.1956.021.01.011. [DOI] [PubMed] [Google Scholar]
- 23.Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342:95–8. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 24.Nudler E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012;149:1438–45. doi: 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Epshtein V, Nudler E. Cooperation between RNA polymerase molecules in transcription elongation. Science. 2003;300:801–5. doi: 10.1126/science.1083219. [DOI] [PubMed] [Google Scholar]
- 26.Epshtein V, Toulmé F, Rahmouni AR, Borukhov S, Nudler E. Transcription through the roadblocks: the role of RNA polymerase cooperation. EMBO J. 2003;22:4719–27. doi: 10.1093/emboj/cdg452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Czaja W, Mao P, Smerdon MJ. The Emerging Roles of ATP-Dependent Chromatin Remodeling Enzymes in Nucleotide Excision Repair. Int J Mol Sci. 2012;13:11954–73. doi: 10.3390/ijms130911954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lake RJ, Fan HY. Structure, function and regulation of CSB: A multi-talented gymnast. Mech Ageing Dev. 2013 doi: 10.1016/j.mad.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–70. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 30.Schalow BJ, Courcelle CT, Courcelle J. Mfd is required for rapid recovery of transcription following UV-induced DNA damage but not oxidative DNA damage in Escherichia coli. J Bacteriol. 2012;194:2637–45. doi: 10.1128/JB.06725-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen SE, Walker GC. New discoveries linking transcription to DNA repair and damage tolerance pathways. Transcription. 2011;2:37–40. doi: 10.4161/trns.2.1.14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chemla YR, Moffitt JR, Bustamante C. Exact solutions for kinetic models of macromolecular dynamics. J Phys Chem B. 2008;112:6025–44. doi: 10.1021/jp076153r. [DOI] [PubMed] [Google Scholar]
- 33.Zhou J, Ha KS, La Porta A, Landick R, Block SM. Applied force provides insight into transcriptional pausing and its modulation by transcription factor NusA. Mol Cell. 2011;44:635–46. doi: 10.1016/j.molcel.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uphoff S, Reyes-Lamothe R, Garza de Leon F, Sherratt DJ, Kapanidis AN. Single-molecule DNA repair in live bacteria. Proceedings of the National Academy of Sciences of the United States of America 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Howan K, Smith AJ, Westblade LF, Joly N, Grange W, Zorman S, et al. Initiation of transcription-coupled repair characterized at single-molecule resolution. Nature. 2012;490:431–4. doi: 10.1038/nature11430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deaconescu AM, Sevostyanova A, Artsimovitch I, Grigorieff N. Nucleotide excision repair (NER) machinery recruitment by the transcription-repair coupling factor involves unmasking of a conserved intramolecular interface. Proc Natl Acad Sci U S A. 2012;109:3353–8. doi: 10.1073/pnas.1115105109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manelyte L, Kim YI, Smith AJ, Smith RM, Savery NJ. Regulation and rate enhancement during transcription-coupled DNA repair. Mol Cell. 2010;40:714–24. doi: 10.1016/j.molcel.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Long X, Parks JW, Bagshaw CR, Stone MD. Mechanical unfolding of human telomere G-quadruplex DNA probed by integrated fluorescence and magnetic tweezers spectroscopy. Nucleic Acids Res. 2013;41:2746–55. doi: 10.1093/nar/gks1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sunney Xie X. Single-molecule approach to dispersed kinetics and dynamic disorder: Probing conformational fluctuation and enzymatic dynamics. J Chem Phys. 2002;117:11024–32. doi: 10.1063/1.1521159. [DOI] [Google Scholar]
- 40.Kou SC, Cherayil BJ, Min W, English BP, Xie XS. Single-molecule Michaelis-Menten equations. J Phys Chem B. 2005;109:19068–81. doi: 10.1021/jp051490q. [DOI] [PubMed] [Google Scholar]