Abstract
Core histones are the building block of chromatin and among the most highly conserved proteins in eukaryotes. The related “deviant” histones share the histone-fold domain, and serve various roles in DNA metabolism. We provide here a structural and functional outlook of H2A/H2B-like deviant histones in transcription, replication and remodeling.
Keywords: chromatin, transcription factors, nucleosome, histone-fold, posttranslational modifications, H2A/H2B, NF-Y, NC2, TAF4/12, chromatin remodeling complex
The nucleosome is formed by “core” histones (H2A/H2B/H3/H4) assembled in an octameric protein core around which 146 nucleotides of DNA are wrapped.1 Histones are composed of a central histone-fold domain (HFD), directly responsible for the formation of the octamer and DNA contacts, and of N- and C-terminal tails. The HFD is formed by a minimum of 3 helices (α1, α2 and α3) separated by two loops (L1 and L2). Histones undergo extensive post-translational modifications (PTMs), defining chromatin territories that regulate DNA accessibility to DNA-metabolizing enzymes.2 A set of “variant” histones, showing > 90% sequence identity to core histones, mark nucleosomes in specific genomic regions and play important roles in specific DNA metabolisms.
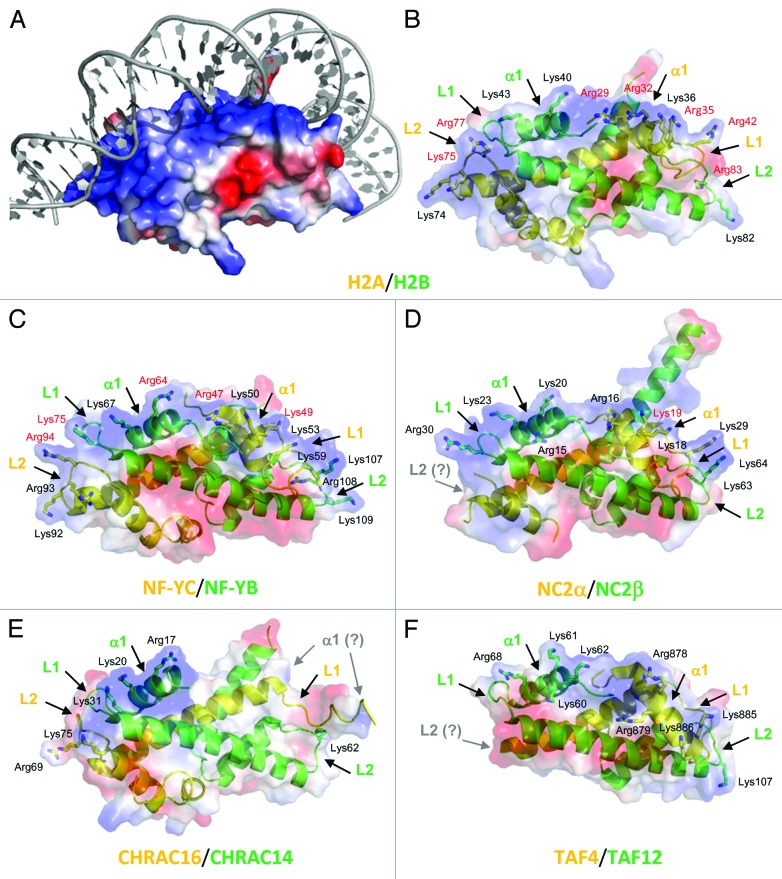
The HFD module can be recognized in several other nuclear proteins. Collectively, these proteins can be viewed as “deviant” histones: typically, only 15–18% sequence identity with core histones is observed along the 65–80 aminoacids of the HFDs. We will focus here on the deviant histones of the H2A-H2B subtype, for which structural studies documented the presence of bona fide HFDs (Fig. 1): (i) the sequence-specific transcription factor NF-Y, (ii) the TBP/TATA-binding NC2, (iii) the small subunits of the chromatin remodeling complex CHRAC and DNA Polymerase ε, and (iv) the TBP-Associated Factors TAF4/TAF12, components of the general transcription factor TFIID.3-6 Of these, NC2α/β, Pole3/Pole4, Pole3/CHRAC15 and NF-YB/NF-YC subunits show sequence identities of 25–35% over the HFD module, extending to an additional α helix (αC) at the HFD C-terminus (Fig. 2).
Figure 1. Structural comparison of HFD proteins. (A) Electrostatic surface of nucleosome H2A/H2B dimer in complex with DNA (PDB-code 1AOI). For clarity, histone tails are not shown. The blue and red colors highlight positively and negatively charged surfaces, respectively. The bound DNA molecule is shown in gray. (B-F) Ribbon representation of the H2A/H2B and deviant histones dimers (H2A-like in yellow, H2B-like in green). The protein electrostatic surface is displayed in semitransparent colors, allowing the view of underlying secondary structure elements. The α1, L1, and L2 regions are indicated, with positively charged residues highlighted as sticks and labeled. For clarity, all HFD proteins are shown in the same orientation. The positively charged (DNA-interacting) surface falls on the top of each protein molecule. In panels (B-D) the DNA present in the crystal structures is omitted for clarity, and Arg/Lys residues whose side chain is in direct contact with DNA are indicated in red color. (B) The Xenopus H2A/H2B dimer (PDB-code 1AOI), with indicated residues numbered accordingly1 (see Fig. 2B, note three residues difference with human H2B). (C) The NF-YB/NF-YC dimer (PDB-code 4AWL).6 (D) NC2α/β (PDB-code 1JFI). The L2 loop is disordered in NC2α (labeled in gray color) due to the absence of DNA contacts.4 (E) CHRAC-14/CHRAC-16 (PDB-code 2BYK; DNA not present in the crystal structure). The α1 region is disordered in CHRAC-16 (gray color).5 (F) TAF4/TAF12 (PDB-code 1H3O; DNA not present in the crystal structure). The C-terminal 26 residues of TAF4, including the L2 loop (gray color), are disordered within the crystal lattice.3
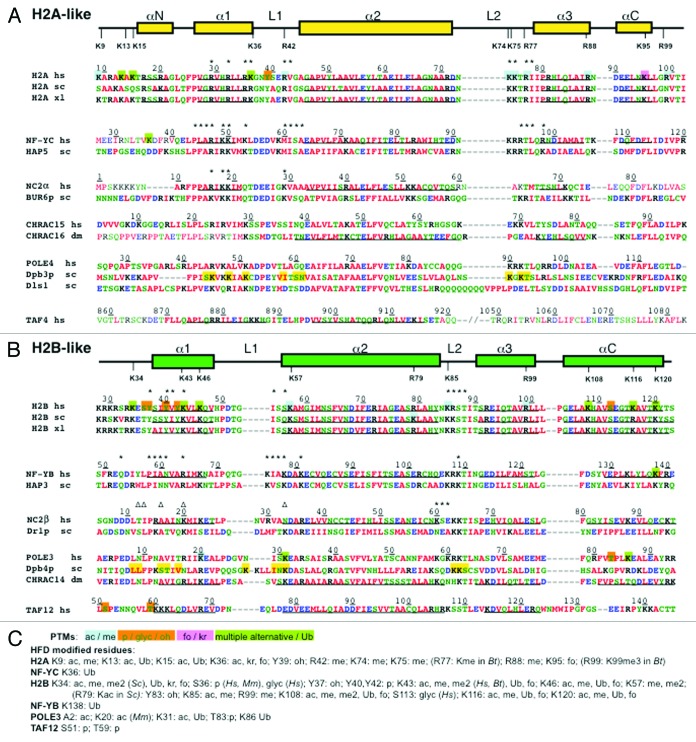
Figure 2. Sequence alignment of the HFD of H2A and H2B with deviant histones proteins The secondary structure arrangement of the histone-fold domain of human H2A or H2B [panels (A) and (B), respectively] is shown above their sequences, with post-translationally modified K/R residues indicated. Sequence alignment of the deviant histone proteins discussed in the text, from human, S. cerevisiae, X. laevis and D. melanogaster (hs, sc, xl, and dm, respectively) is shown below, according to secondary structure determination (helical residues underlined),1,3-6 including gaps to optimize the alignment; in TAF4 the predicted α3 in the CCTD sequence is also aligned, separated by two slashes indicating the ≈100 aa loop.3 Font color indicates aminoacid side chain properties. Residue numbering is shown on top of each human sequence, with asterisks indicating aminoacids involved in DNA contacts as determined in the crystal structures of H2A/H2B dimers, NF-YC/NF-YB and NC2α/NC2β (from PDB codes: 1AOI, 4AWL, 1H3O, respectively). Triangles indicate NC2β DNA contacts with a symmetry related oligonucleotide.4 Regions not present in crystallized proteins, or whose structure was disordered and not determined are in plain font. Identified modified residues in the HFD of histones and histone-like proteins are color shaded for the different PTMs as indicated in (C); in (A) and (B), H2A and H2B modified residues are color shaded only on the human sequences. In Dpb3p and Dpb4p yellow color highlights HFD mutations described by Tsubota et al.19 (C) Color coding and list of post-translational modifications identified in the HFDs (from UniProtKB databank, and Wagner, et al.29): alternative acetylation (ac) or methylation (me) are indicated in blue; phosphorylation (p), hydroxylation (oh), or glycosylation (glyc) in orange; krotonylation (kr) or formylation (fo) in magenta; multiple alternative modifications and/or ubiquitylation (Ub) in green.
NF-Y
NF-Y consists of three subunits: the HFD, dimer NF-YB/NF-YC and NF-YA.7 We recently unraveled the architecture of the NF-Y/DNA assembly by solving the crystal structure of NF-Y in complex with a 25 bp CCAAT box oligonucleotide.6 The NF-Y heterotrimer is based on a head-to-tail assembly of NF-YB and NF-YC, to which the NF-YA subunit associates as an extended structure, hosting two helices. Helix A1 mediates trimerization with the HFD heterodimer; helix A2 and a novel GxGGRF motif provide sequence-specific contacts to the CCAAT box, widening the DNA minor groove. NF-YC and NF-YB interact extensively with the DNA phosphates, in a way that is extremely similar of the H2A/H2B-DNA assembly in the nucleosome (Fig. 1).
The knowledge of genomic binding of NF-Y is derived by ENCODE8: 25% of NF-Y sites are in promoters and a comparable number are located at tissue specific enhancers. In promoters, NF-Y is at -60/-100 from the transcriptional start site (TSS). In general, NF-Y cooperates with neighboring transcription factors (TFs), including growth-controlling and oncogenic ones,8 consistent with the enrichment of CCAAT motifs in the promoters of genes overexpressed in cancer.9 As for binding to other deviant HFDs, NF-Y was reported to bind to TAF11/TAF13, TAF6/TAF9 and TAF4/TAF12; however, the interactions were not mediated by HFDs, but by other domains, or by NF-YA.10
NC2
Originally characterized biochemically as a TBP-binding co-repressor, cloning of NC2 genes (also named Dr1/DRAP1) highlighted the presence of HFDs. In vitro work indicated that NC2 displaces TFIIA and TFIIB interactions with TBP/TATA.11,12 The crystal structure of the TBP/NC2/TATA trimeric complex is illustrative of how NC2 prevents binding of TFIIB to TBP with consequent repression of the TATA-dependent transcription.4 The structure shows that NC2α and NC2β dimerize and interact with DNA in a H2A/H2B-like manner, but the limited length of the bound oligo prevents a complete understanding of the HFD-DNA interactions (Fig. 1). Interestingly, the NF-Y/CCAAT and NC2/TBP/TATA complexes share two common features: first, sequence-specificity relies on minor groove binding of a non-HFD protein (NF-YA and TBP, respectively), and, despite completely different structures, there is a resemblance in the way Phe residues of TBP and NF-YA read the DNA sequence, by inserting deeply between bases.6 The second is the severe bending of DNA imparted by TBP and NF-YA, stabilized on the sides by the HFD subunits which interact with the DNA phosphates backbone. In general, CCAAT promoters tend to be TATA-less,7 so one could imagine that NF-Y and NC2 serve a similar architectural role on non-overlapping sets of promoters, close to the TSS.
NC2 also activates promoters with downstream promoter elements (DPEs). Work in yeast, Drosophila and human cells has shown a positive effect of NC2 on large sets of genes: how this is achieved mechanistically is not clear, but genome-wide experiments further cement the idea that NC2 is largely associated to core promoters of actively transcribed genes.13 One key NC2 associated factor regulating TATA vs DPE transcription is Mot1p, which serves the role of mobilizing TATA-binding complexes, shifting the balance toward TFIID-binding DPEs. Apparently, NC2β is released in the process, and since only HFD heterodimers have been characterized so far, this poses an interesting structural question regarding the (NC2) HFD composition; one possibility is that NC2β is substituted with a different H2B-like partner in the complex with NC2α.
Pole3/Pole4, Pole3/CHRAC15
Small HFD proteins were identified upon purification of at least three different complexes: (i) Pole3/Pole4 (Dpb4/Dpb3 in yeast) are associated to the catalytic subunit of DNA Polymerase ε, one of the major enzymes involved in DNA replication, which is also involved in DNA repair;14 (ii) Pole3/Pole4 were reported to be part of the histone acetylating ATAC complex,15 although in other purification schemes only Pole3 was present,16 or they were absent;17 (iii) Pole3/CHRAC15 dimers are associated to the ISWI chromatin assembly factor ACF, in the remodeling complex CHRAC.5,18
In S. cerevisiae, DPB4 and DPB3 are non essential genes, but dpb4Δdpb3Δ strains show an increased rate of mutations, because of defects in the proofreading capacity of DNA Polymerase δ.14 The mechanism is apparently linked to destabilization of the DNA Polymerase ε/DNA contacts during replication. Indeed, the HFD subunits are important for DNA-binding of the complex: mutations in the α1, L1 and L2 regions of the HFDs—based on the nucleosomal H2A/H2B-DNA contacts—lead to a loss of DNA-binding in EMSAs, and to a decrease in telomeric silencing in vivo.19 In S. Pombe, Dpb4 is not essential,20 but dpb4Δ cells are synthetically lethal with several genes involved in DNA replication (cdc20, cut5/dpb11, sna41 and cdc21/mcm4), signaling DNA replication defects; Dpb3, instead, is essential for cell cycle progression, as dpb3Δ cells accumulate in S-phase. As for the Pole3/CHRAC15 incorporated in CHRAC, they facilitate nucleosome sliding mediated by the ATP-dependent ISWI/Acf1 subunits.5,18
TAF4/TAF12
Originally identified in TBP-containing TFIID, several TAFs were later found as subunits of TBP-less complexes with acetyl-transferase activity.21 The HFD module has been identified in nine TAFs, and implicated in their heterodimerization, with TAF4/TAF12 being the H2A/H2B-likes of the lot. TAFs are present in the SAGA complex as well, which has histone acetylation capacity: specifically, TAF4 is replaced by Ada1 found in partnership with TAF12. TAF4 has an unusual configuration, with an extra domain of unknown structure inserted between the HFD α2 and α3 helices, absent in the other H2A-like.3 Genetic and biochemical experiments indicate that TAF4 plays an important role in transcription of specific sets of genes, specifically enriched in TATA-less Initiator (Inr) units.22 The HFDs are essential for interactions with other TAFs, and indeed required for the formation of a “core” TFIID complex, which consists of two copies of TAF4/TAF12, of H3/H4-like TAF6/TAF9 and of TAF5.23 The presence of a TAF-mediated nucleosome-like structure in TFIID was not confirmed by the recent EM investigations of “core” TFIID complexes containing HFD TAFs.24 DNA-binding of TFIID to DPE elements was detected in Drosophila and ascribed to direct contacts of TAF6/TAF9 with the DPE.25 Interestingly, the TAF4/TAF12 dimer was also shown to be able to bind to DNA in vitro,22 with the TAF4 extra domain being important, in addition to the HFDs. A certain sequence preference for DNA was reported, but this was less important than the length of the DNA (> 70 bps). The current model is that TFIID associates to Inr and DPE elements,26 and TAF4/TAF12, along with TAF6/TAF9, might be in part responsible for these contacts. In this respect, we notice that while the α1 and L2 DNA-binding regions display a high level of similarity with the bona fide DNA-binding relatives, including basic residues contacting the phosphates backbone, the L1 loops of both TAF4 and TAF12 show important differences, notably the presence of acidic residues in TAF12 (Figs. 1, 2). Further biochemical work is required to tackle the daunting task of determining the relative contribution of HFD heterodimers to the TFIID DNA-binding capacity and specificity.
Open Questions
Mixed dimers
Core and variant histones are strict in their heterodimeric partnerships. This is apparently not the case for the deviant ones, as exemplified by Pole3 being incorporated in different complexes by dimerizing with Pole4 or CHRAC15. NC2β is found in ATAC complexes, without a conventional H2A-like partner,16 but together with YEATS2, another histone fold protein;15 indeed, a bona fide heterodimerization of the two recombinant HFDs was reported. The NC2α subunit was shown to have additional functions distinct from NC2β in yeast;27 the tissue distribution of the two subunits in mammals is only partially overlapping, suggesting that they can have roles independent of their established partner. TAF12 also has alternative partners: TAF4 in the “core” TFIID and Ada1 in SAGA. As for NF-Y HFDs they have yet to be found in unorthodox partnerships in mammals, but systematic Y2H analysis of the large families of NF-Y genes in A. thaliana did identify promiscuous interactions between At-NC2α and At-NF-YB2/9, and between At-Pole3 and At-NF-YC8.28 It is difficult to say whether this finding is related to the emergence of “middle-of-the-road” partners in the vast catalog of NF-Y plant genes, or whether this feature is shared in metazoans.
Deviant histones PTMs
Core histones mediate regional organization of chromatin through their PTMs:2 specifically, HFD residues involved in important functional switches are known in H3 (K56 and K79). Thus, a relevant point is to verify how conserved, in structure and function, are modified residues in deviant histones. We showed that NF-YB is ubiquitinated at residue K138 of αC, which corresponds to H2BK120 (K123 in yeast), and that this PTM is important for transcriptional activation.6 The H2BK120Ub is genetically and biochemically upstream of important histone methylations, notably H3K4me3 and H3K79me2/3,2 and indeed NF-Y binding is required for deposition of these PTMs,7 as well as NF-YB-Ub.6 This is the proof of principle that at least one PTM has been conserved in a deviant histone. Hence, it is possible that other PTMs are shared by core, variant, and deviant histones.
Inspection of available data sets of ubiquitinated proteins indicates that the H2B-like Pole3 is also ubiquitinated in the αC K86.29 Alignment of H2B-like proteins highlights two notable similarities (Fig. 2): (i) the DNA-contacting L2 of H2B (KRST) is similar in NF-YB and Pole3 (KRKT), NC2β (EKKT) and TAF12 (KSST), with lysines being involved in DNA-binding. In particular, H2BK85 is also a site of alternative acetylation or methylation. (ii) In helix α1, the H2B K46 is not involved in direct DNA-contacts but is target of different alternative PTMs, including ubiquitylation. Such lysine is conserved in NF-YB and NC2β, where it is also not involved in DNA-contacts. Additional post-translationally modified residues conserved in this subfamily are α1 H2BK43 (present in NC2β, an R in NF-YB and Pole3), α2 H2BK57 (methylated, present in an adjacent position in Pole3K31 and NF-YBK78), α3 H2BR99 (methylated, present in TAF12). The exercise for H2A conveys a similar picture: widespread conservation in L2, with H2AK74 and K75 being methylated; in the α1, H2AK36, targeted by different PTMs and involved in DNA-binding, is conserved in TAF4, as the adjacent K53 in NF-YC, also conserved in CHRAC15 and Pole4. In summary, we propose that conservation of certain residues might be due not only to their local structural importance, but also to their specific modifications.
Specificity for different DNA metabolisms?
So far, NF-Y, NC2, TAF4/TAF12 have been mostly implicated in transcription, but this might merely reflect the historical background of the assays employed in their study. Different examples indicate today that these proteins might also impact other DNA metabolisms, notably DNA repair. The variant histones included in CHRAC/ACF remodeling complexes were shown to physically associate with the KU proteins and were implicated in DNA double-strand break recognition/repair mechanisms.30 TAF12 was shown to recruit proteins of the NER complex on rRNA genes.31 Checkpoint activation in response to replication stress and DNA damage is influenced by chromatin structure, and the cellular response acts through two different mechanisms. The first pathway is modulated by RAD6/BRE1-mediated H2B ubiquitination, and by Dot1-mediated H3K79 methylation; the second is histone independent and requires Dpb11. DNA Polymerase ε biochemically and genetically interacts with Dpb11 (TopBP1 in human cells), sensing replication stress caused by unbalanced dNTP pools due to hydroxyurea treatment.32 The finding of ubiquitylation of Pole3 in the HFD αC helix (Fig. 2) could lead to the hypothesis that it is involved in the both mechanisms, via the H2B-like PTM, and the association to DNA Polymerase ε. Given the requirement of NF-YB-Ub for H2B-Ub and H3K79me2 in active CCAAT genes, one should also wonder whether NF-Y is involved in DNA repair of such transcribed genes following DNA damage. Should a similar modification be identified on NC2β, a similar scenario could also operate in parallel on TATA genes, controlled by NC2. Furthermore, links between NF-Y and TopBP1 recently surfaced:9 TopBP1 is required for the activation of NF-Y-dependent growth promoting genes by mutp53, leading to decreased sensitivity to DNA-damaging agents in mutp53-bearing cancer cells, and tumor formation in vivo: indeed, mutp53, NF-YA and TopBP1 directly interact. Since cells transiently eliminated of NF-YA show S-phase progression defects,9 one can hypothesize that NF-Y might be directly involved in DNA replication or, during such process, in DNA repair.
Conclusions
A large set of genetic and biochemical data indicate that the interpretation of the expansion of HFDs as solely being protein-protein interaction modules is not correct. Rather, the H2A/H2B-likes evolved to maintain features of core histones, notably the capacity to bind DNA and to be modified post-translationally, providing the complexes they are inserted in with a direct link to core chromatin structures. Deviant histone dimers play a role at the interface between their assemblies and chromatin, and they are most likely involved in the complex set of modifications, exploiting and expanding the “histone code” of their distant relatives. Their reciprocal interplay, specialization in terms of protein interactions, modifications, and implication in specific DNA metabolisms, as well as their interplay with core histones, are all areas for future investigations.
Acknowledgments
This work was supported by Regione Lombardia Nepente grant to RM.
Glossary
Abbreviations:
- DPE
downstream promoter element
- HFD
histone-fold domain
- Inr
initiator element
- PTM
post translational modification
- TF
transcription factor
- TSS
transcription start site
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/25002
References
- 1.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Annu Rev Biochem. 2011;80:473–99. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- 3.Werten S, Mitschler A, Romier C, Gangloff YG, Thuault S, Davidson I, et al. Crystal structure of a subcomplex of human transcription factor TFIID formed by TATA binding protein-associated factors hTAF4 (hTAF(II)135) and hTAF12 (hTAF(II)20) J Biol Chem. 2002;277:45502–9. doi: 10.1074/jbc.M206587200. [DOI] [PubMed] [Google Scholar]
- 4.Kamada K, Shu F, Chen H, Malik S, Stelzer G, Roeder RG, et al. Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex. Cell. 2001;106:71–81. doi: 10.1016/S0092-8674(01)00417-2. [DOI] [PubMed] [Google Scholar]
- 5.Hartlepp KF, Fernández-Tornero C, Eberharter A, Grüne T, Müller CW, Becker PB. The histone fold subunits of Drosophila CHRAC facilitate nucleosome sliding through dynamic DNA interactions. Mol Cell Biol. 2005;25:9886–96. doi: 10.1128/MCB.25.22.9886-9896.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nardini M, Gnesutta N, Donati G, Gatta R, Forni C, Fossati A, et al. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell. 2013;152:132–43. doi: 10.1016/j.cell.2012.11.047. [DOI] [PubMed] [Google Scholar]
- 7.Dolfini D, Gatta R, Mantovani R. NF-Y and the transcriptional activation of CCAAT promoters. Crit Rev Biochem Mol Biol. 2012;47:29–49. doi: 10.3109/10409238.2011.628970. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22:1798–812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imbriano C, Gnesutta N, Mantovani R. The NF-Y/p53 liaison: well beyond repression. Biochim Biophys Acta 2011; 1825:131-39. [DOI] [PubMed]
- 10.Frontini M, Imbriano C, diSilvio A, Bell B, Bogni A, Romier C, et al. NF-Y recruitment of TFIID, multiple interactions with histone fold TAF(II)s. J Biol Chem. 2002;277:5841–8. doi: 10.1074/jbc.M103651200. [DOI] [PubMed] [Google Scholar]
- 11.Inostroza JA, Mermelstein FH, Ha I, Lane WS, Reinberg D. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell. 1992;70:477–89. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- 12.Goppelt A, Stelzer G, Lottspeich F, Meisterernst M. A mechanism for repression of class II gene transcription through specific binding of NC2 to TBP-promoter complexes via heterodimeric histone fold domains. EMBO J. 1996;15:3105–16. [PMC free article] [PubMed] [Google Scholar]
- 13.Tora L, Timmers HT. The TATA box regulates TATA-binding protein (TBP) dynamics in vivo. Trends Biochem Sci. 2010;35:309–14. doi: 10.1016/j.tibs.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 14.Aksenova A, Volkov K, Maceluch J, Pursell ZF, Rogozin IB, Kunkel TA, et al. Mismatch repair-independent increase in spontaneous mutagenesis in yeast lacking non-essential subunits of DNA polymerase ε. PLoS Genet. 2010;6:e1001209. doi: 10.1371/journal.pgen.1001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang YL, Faiola F, Xu M, Pan S, Martinez E. Human ATAC Is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J Biol Chem. 2008;283:33808–15. doi: 10.1074/jbc.M806936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suganuma T, Gutiérrez JL, Li B, Florens L, Swanson SK, Washburn MP, et al. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat Struct Mol Biol. 2008;15:364–72. doi: 10.1038/nsmb.1397. [DOI] [PubMed] [Google Scholar]
- 17.Spedale G, Timmers HT, Pijnappel WW. ATAC-king the complexity of SAGA during evolution. Genes Dev. 2012;26:527–41. doi: 10.1101/gad.184705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kukimoto I, Elderkin S, Grimaldi M, Oelgeschläger T, Varga-Weisz PD. The histone-fold protein complex CHRAC-15/17 enhances nucleosome sliding and assembly mediated by ACF. Mol Cell. 2004;13:265–77. doi: 10.1016/S1097-2765(03)00523-9. [DOI] [PubMed] [Google Scholar]
- 19.Tsubota T, Tajima R, Ode K, Kubota H, Fukuhara N, Kawabata T, et al. Double-stranded DNA binding, an unusual property of DNA polymerase epsilon, promotes epigenetic silencing in Saccharomyces cerevisiae. J Biol Chem. 2006;281:32898–908. doi: 10.1074/jbc.M606637200. [DOI] [PubMed] [Google Scholar]
- 20.Spiga MG, D’Urso G. Identification and cloning of two putative subunits of DNA polymerase epsilon in fission yeast. Nucleic Acids Res. 2004;32:4945–53. doi: 10.1093/nar/gkh811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagy Z, Tora L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene. 2007;26:5341–57. doi: 10.1038/sj.onc.1210604. [DOI] [PubMed] [Google Scholar]
- 22.Gazit K, Moshonov S, Elfakess R, Sharon M, Mengus G, Davidson I, et al. TAF4/4b x TAF12 displays a unique mode of DNA binding and is required for core promoter function of a subset of genes. J Biol Chem. 2009;284:26286–96. doi: 10.1074/jbc.M109.011486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright KJ, Marr MT, 2nd, Tjian R. TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc Natl Acad Sci U S A. 2006;103:12347–52. doi: 10.1073/pnas.0605499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bieniossek C, Papai G, Schaffitzel C, Garzoni F, Chaillet M, Scheer E, et al. The architecture of human general transcription factor TFIID core complex. Nature. 2013;493:699–702. doi: 10.1038/nature11791. [DOI] [PubMed] [Google Scholar]
- 25.Burke TW, Kadonaga JT. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 1997;11:3020–31. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cianfrocco MA, Kassavetis GA, Grob P, Fang J, Juven-Gershon T, Kadonaga JT, et al. Human TFIID binds to core promoter DNA in a reorganized structural state. Cell. 2013;152:120–31. doi: 10.1016/j.cell.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Creton S, Svejstrup JQ, Collart MA. The NC2 alpha and beta subunits play different roles in vivo. Genes Dev. 2002;16:3265–76. doi: 10.1101/gad.234002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petroni K, Kumimoto RW, Gnesutta N, Calvenzani V, Fornari M, Tonelli C, et al. The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell. 2012;24:4777–92. doi: 10.1105/tpc.112.105734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics 2011; 10:M111.013284. [DOI] [PMC free article] [PubMed]
- 30.Lan L, Ui A, Nakajima S, Hatakeyama K, Hoshi M, Watanabe R, et al. The ACF1 complex is required for DNA double-strand break repair in human cells. Mol Cell. 2010;40:976–87. doi: 10.1016/j.molcel.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Schmitz KM, Schmitt N, Hoffmann-Rohrer U, Schäfer A, Grummt I, Mayer C. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol Cell. 2009;33:344–53. doi: 10.1016/j.molcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 32.Puddu F, Piergiovanni G, Plevani P, Muzi-Falconi M. Sensing of replication stress and Mec1 activation act through two independent pathways involving the 9-1-1 complex and DNA polymerase ε. PLoS Genet. 2011;7:e1002022. doi: 10.1371/journal.pgen.1002022. [DOI] [PMC free article] [PubMed] [Google Scholar]