Abstract
Recent structural and biochemical studies of human TFIID have significantly increased our understanding of the mechanisms underlying the recruitment of TFIID to promoter DNA and its role in transcription initiation. Structural studies using cryo-EM revealed that modular interactions underlie TFIID’s ability to bind simultaneously multiple promoter motifs and to define a DNA state that will facilitate transcription initiation. Here we propose a general model of promoter binding by TFIID, where co-activators, activators, and histone modifications promote and/or stabilize a conformational state of TFIID that results in core promoter engagement. Within this high affinity conformation, we propose that TFIID’s extensive interaction with promoter DNA leads to topological changes in the DNA that facilitate the eventual loading of RNAP II. While more work is required to dissect the individual contributions of activators and repressors to TFIID’s DNA binding, the recent cryo-EM studies provide a physical framework to guide future structural, biophysical, and biochemical experiments.
Keywords: TFIID, Cryo-EM, core promoter, DNA binding, conformational flexibility, gene regulation
Promoter recognition by TFIID
Gene expression is an essential and complex cellular task that requires the coordinated activities of many proteins and RNAs. At the core of this highly regulated process is transcription initiation by RNA Polymerase II (RNAP II), which synthesizes RNA transcripts from DNA coding sequences. Since RNAP II cannot bind double stranded DNA, nor identify the start of a gene, a host of factors are utilized to guide RNAP II onto promoters in response to environmental cues or cellular signals. This regulatory task requires the combined influence of upstream trans-acting factors in conjunction with promoter cis-acting sequences. Both TFIID and Mediator serve as central players that establish a molecular link between distal activators and RNAP II loading.1 TFIID is a large, multi-subunit complex comprised of TBP and 13–14 TBP-associated factors (TAFs) that, unlike Mediator, contains subunits capable of promoter DNA recognition.2 In addition to TBP-mediated binding of the TATA box,3 other subunits of TFIID make sequence-specific contacts with downstream promoter motifs known as the Initiator (Inr),4 motif ten element (MTE),5 downstream promoter element (DPE),6 and the downstream core element (DCE).7 Recognition of one or more of these promoter motifs, in combination with distal-acting activators and repressors, is integrated by TFIID into a single output, loading of RNAP II at the promoter.
Structural flexibility of human TFIID
Despite decades of biochemical characterization of TFIID’s role in transcription initiation, little is known regarding the structural basis for TFIID’s interaction with promoter DNA. This limitation has been due to the inability to produce large amounts of the holo–TFIID complex from recombinant sources, which has only recently been partially overcome through the reconstitution of a core TFIID subcomplex using the MultiBac system.8 Thus, most structural studies have relied solely on the low yield of endogenous TFIID purified from nuclear extracts, limiting the possibilities for crystallographic study. In contrast to X-ray crystallography, single particle electron microscopy (EM) is ideally suited to visualize the structure of large and scarce protein complexes. Furthermore, recent technical advances in this field are leading to 3D reconstructions of ever improving resolution, as well as an improvement in the unique ability of cryo-EM to detect and describe the structural dynamics exhibited by most macromolecular complexes.9
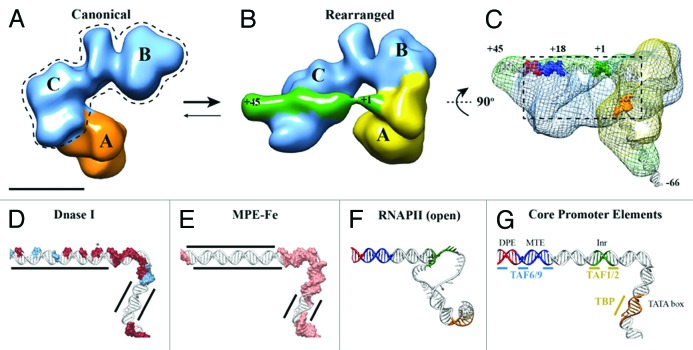
The potential of EM as a structural tool to analyze heterogeneous samples is exemplified by our recent cryo-EM study of human TFIID bound to super core promoter (SCP) DNA.10 In order to determine the 3D structure of promoter-bound human TFIID, it was necessary to overcome a surprising discovery: that lobe A, the major lobe within the horseshoe-shaped human TFIID structure, moves by over 100 Å from a previously characterized “canonical” state into a newly discovered “rearranged” state (Fig. 1A and B). Past TFIID structural studies focused only on a single conformation of the complex, the canonical state,11-14 overlooking the dramatic structural transitions of lobe A as it changes connectivity from lobe C (canonical state) to lobe B (rearranged state) (Fig. 1A and B).
Figure 1. TFIID introduces topological changes in promoter DNA upon formation of the rearranged, DNA-bound conformation. Cryo-EM structures of the canonical conformation of TFIID (A) and the rearranged conformation of TFIID-TFIIA-SCP (B). The BC core is shown in blue, the flexible lobe A in orange (A) and yellow (B), and the SCP DNA in green (B). Promoter DNA positions +1 and +45 are indicated (B). (C) Mesh: Cryo-EM structure of TFIID-TFIIA-SCP(-66) rotated by 90 degrees relative to (B). The structure shows the location of DNA position -66 exiting lobe A. Positions -66, +1, and +45 are indicated and colored according to promoter motifs in (G). (D) DNase I footprint modeled onto the DNA path through TFIID-TFIIA-SCP. Red surfaces: full cleavage; blue surfaces: partial cleavage; (E) MPE-Fe footprinting modeled onto the DNA path through TFIID-TFIIA-SCP. Pink surface: cleavage; MPE-Fe protection by TFIID-IIA indicated by black lines. (F) DNA model from the open complex of TBP/PIC.34 (G) Promoter DNA model from TFIID-TFIIA-SCP colored by promoter motifs and TFIID subunits that make sequence-specific contacts.
The newly characterized structural transitions in TFIID appear highly relevant for its biological function, as indicated by the fact that the presence of TFIIA and/or promoter DNA, natural binding partners of TFIID, affect TFIID’s conformational equilibrium. EM analysis of TFIID in the presence of both TFIIA and DNA showed that the newly discovered rearranged structure binds DNA and becomes the predominant state of the complex. While the functional consequences of the canonical conformation of TFIID remain to be further explored, it may represent a structural state that exhibits differential DNA affinities, whereby downstream DNA interactions by lobe C may be inhibited by the position of lobe A in the canonical state.
Downstream promoter DNA is bound by lobe C
A hallmark of the TFIID-TFIIA-DNA cryo-EM structure is the direct visualization of the downstream DNA bound to TFIID (Fig. 1B, green). The structure shows how TFIID interacts with DNA up to, and including, the MTE (+18 to +27) and DPE (+28 to +32) motifs (Fig. 1C), while density that can account for the last ~15 bps of downstream promoter extends away from the complex. This feature is consistent with DNase I footprinting of TFIID on SCP DNA,10,15 where TFIID only makes contacts up to the DPE motif, thus serving as an internal control for the validity of the DNA path visualized in the TFIID-TFIIA-DNA cryo-EM structure.
The linear path of downstream promoter DNA on the surface of human TFIID also provides a structural explanation for the previously observed pattern of DNase I sensitive sites spaced every 10 bps between positions +3 and +32 (Fig. 1D).5,6,10,15-18 Such a pattern is consistent with the rise of the DNA helix and initially led to the model that the DNA is positioned on the surface of the TFIID structure in a manner analogous to a nucleosome.16 This notion appeared to be supported by the ability of TFIID to induce negative supercoiling on promoter DNA16 and by the presence of a histone-like octamer within TFIID.19-21 However, subsequent studies have shown that TAF6 and TAF9, the two TFIID subunits with a histone fold responsible for binding the MTE and DPE, interact with the DNA using unstructured loops instead of the globular histone fold,22 suggesting that they utilize an atypical DNA binding mode distinct from that of core histones.
The path of downstream promoter DNA observed on the surface of TFIID is most consistent with a non-nucleosomal type of DNA interaction. Unlike nucleosomes, lobe C interacts with the downstream promoter DNA without introducing dramatic distortions (Fig. 1B). This linear mode of interaction may be stabilized by the distributed interaction of TAF6 and TAF9 with the MTE/DPE, which includes three key subregions of contact with the DNA over a 15 bp region (Fig. 1G).5
A novel interaction mode of TFIID on downstream DNA is further suggested by high-resolution chemical footprinting using methidiumpropyl-EDTA-iron(II) (MPE-Fe), which showed TFIID-mediated protection of downstream DNA from MPE-Fe cleavage (Fig. 1E).10,17,18 This result indicates that TFIID either physically prevents MPE-Fe intercalation, induces strain in the helix to prevent the widening necessary for intercalation, or a combination of both. Considering that TAF6 and TAF9 are the only subunits of TFIID providing sequence-specific contacts for the MTE and DPE motifs,5 we propose that the continuous region of MPE-Fe protection from +3 to +18 is likely due to helical compaction caused by the simultaneous binding of TFIID to the Inr, MTE, and DPE, whereas MPE-Fe protection from +19 to +32 is the result of TAF6 and TAF9 interactions. While the underlying importance of this observation remains to be seen, we believe that this extended protection is the result of an important topological change that TFIID imparts on promoter DNA, considering the large MPE-Fe footprint spanning 30 bps of the downstream DNA.
Lobe A and upstream promoter DNA interactions
Across the central channel from lobe C’s interaction with downstream promoter DNA, lobe A interacts with the Inr, TATA box, and upstream DNA sequences. The presence of upstream DNA density extending away from lobe A provided a marker for the -66 position in the promoter DNA, which served to approximate the location of the TATA box based upon its position at -31/30 (Fig. 1C). This localization was corroborated by gold labeling of both the TATA box and TFIIA, which itself demonstrated that, within the context of TFIID, TFIIA interacts with TBP in a manner similar to that observed in crystallographic studies.23,24 While this characterization is an important first step, higher resolution structures will be necessary in order to determine unambiguously the path of DNA through lobe A and the structural changes induced by TFIIA to facilitate DNA binding.
TFIID’s interaction with upstream DNA through the mobile lobe A is supported by the ability of a TFIID subcomplex to direct transcription from promoters containing TATA and Inr motifs.25 Through binding and transcription assays, a subcomplex of TAF1-TAF2-TBP was determined to be sufficient to bind TATA-Inr sequences and to direct transcription initiation in vitro.25 This suggested that TFIID’s sequence recognition of TATA-Inr is contained within a subdomain of the TFIID holo-enzyme. Therefore, given the modular nature of lobe A and its localization by the TATA box and Inr motifs seen in our EM studies, we propose that lobe A minimally comprises TAF1, TAF2, and TBP.
In addition to mediating contacts with TBP in lobe A, TAF1 may also serve as the TFIID subunit responsible for tethering lobe A to the BC core. Analysis of unstructured domains within TFIID’s subunits indicates that TAF1’s C-terminus represents one of the longest regions of predicted unstructured resides (~225 amino acids) based upon thermodynamic calculations of inter-residue interaction energy using algorithms implemented in IUPred (http://iupred.enzim.hu)26 (unpublished observation). This suggests that a large domain of TAF1 is likely unstructured and could serve to tether lobe A to the BC core. This hypothesis is consistent with the observation that TAF1 is sensitive to limited proteolysis in vitro.27 Therefore, we propose a model in which the N-terminus of TAF1 assembles with TBP and TAF2 to form lobe A, while the unstructured C-terminus tethers lobe A to the BC core.
TFIID as a modular transcription factor
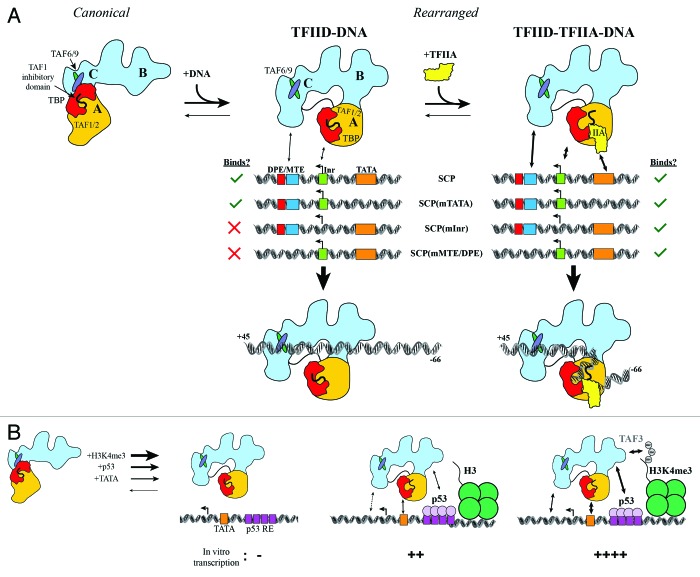
The cryo-EM structure of TFIID-TFIIA-DNA indicates that TFIID utilizes two distinct sites of interaction with promoter DNA. A bipartite model of promoter recognition by TFIID is supported by in vivo and in vitro analysis of TFIID complex integrity. Through a systematic depletion of each subunit of TFIID using RNA interference, a stable core subcomplex of TFIID was identified that comprises subunits TAF4, -5, -6, -9, -12, as knockdown of any of these subunits resulted in the concomitant loss of protein expression for the entire TFIID complex.28 We believe that this core complex corresponds to our structurally defined BC core (Fig. 1A, dotted line), which is capable of interacting with MTE/DPE motifs. As accessory subunits to the core TFIID subcomplex, subunits TAF1, TAF2, TBP, and TAF11 were characterized as components that were dispensable for formation of the core subcomplex. Therefore, as indicated above, we propose this subcomplex to correspond to the flexibly attached lobe A. These lobe assignments are supportive of a model of promoter binding whereby TFIID interacts with upstream and downstream promoter motifs via two distinct structural modules: binding of MTE/DPE by TAF6 and TAF9 within the stable BC core, and upstream DNA binding by the TBP-containing lobe A (Fig. 2A).
Figure 2. Regulatory interplay between co-activators, activators, histone modifications and promoter DNA binding by TFIID. (A) TFIID exists in both canonical and rearranged conformational states, but only the rearranged state interacts efficiently with SCP DNA. Without TFIIA, TFIID interacts only with SCP and SCP(mTATA) (left). TFIIA facilitates TFIID binding to all mutant promoters (right). (B) TATA box DNA, p53, and H3K4me3 likely stabilize TFIID in a rearranged conformation, as these factors cooperatively stimulate transcription initiation by TFIID. Adapted from.43
In further support of this model, a subcomplex of human TFIID was recently reconstituted using MultiBac expression and visualized using cryo-EM.8 This work confirmed the formation of a minimal TFIID subcomplex comprising two copies each of TAF4, 5, 6, 9, 12. The 2-fold symmetry of this complex was broken with the addition of TAF8 and 10, which are present as single copies and induce a conformational change in the complex. The size of this 7-subunit TFIID subcomplex corresponds approximately to the BC core visualized within the intact human TFIID. However, this 7TAF structure cannot be docked within the BC core without further conformational changes of the subcomplex in a manner that would open up the structure (data not shown). This suggests that the 7TAF complex may represent a structural pre-cursor to the BC core seen within holo-TFIID and that the remaining subunits of TFIID cause additional conformational changes that opens the BC core within the holo-TFIID complex. TFIID reconstitution strategies are exciting steps forward toward further structural and biochemical studies aiming to dissect the modular nature of TFIID’s function in transcription initiation.
A modular view of TFIID structure is also consistent with the fact that human TFIID shares a number of subunits with at least two other critical human complexes known as STAGA (Spt3-TAF9-Gcn5-acetyltransferase),29 the homologous human complex for the yeast SAGA complex30,31 and TFTC (TBP-free TAF-containing complex).32 Like SAGA, STAGA acetylates histones via the Gcn5 subunit to create a permissive chromatin environment for transcription.33 The STAGA complex shares subunits TAF5, -6, -9, -10, and -12 with TFIID, all reported to be within the core subcomplex of TFIID.28 These same subunits are found within human TFTC, a protein complex capable of initiating transcription from both TATA-containing and TATA-less promoters that contains additional subunits capable of histone modification.32
We believe that the stable core subcomplex of TFIID observed in vivo, in vitro, and now structurally8,10 (Fig. 1A, dotted outline), serves as a general platform that is directed to specific genes or histone modifications by the ‘plugging-in’ of accessory subunits. In the case of TFIID, the TFIID-specific subunits TAF1, TAF2, and TBP are most likely to form the lobe A subcomplex, which interacts flexibly with the stable BC core, defining either a canonical or a rearranged state of TFIID for binding to promoters with TATA, Inr, MTE, and DPE motifs.
TFIID-induced DNA topology changes around the TSS
While the functional consequences of TFIID’s structural flexibility require further study, we propose that TFIID’s conformational transitions may be necessary to induce specific topological changes in the DNA structure surrounding the TSS. An early suggestion of changes in promoter topology came from footprinting experiments of partially purified TFIID, where TFIID induced a hypersensitive DNase I site at +3 relative to the TSS.17,18 This hypersensitive site has been found consistently for all DNase I footprinting studies performed with highly purified TFIID from Drosophila and human sources.5,10,15 Considering that the strength of this hypersensitive site appears to correlate with levels of in vitro transcription, it has been speculated that TFIID introduces topological changes in DNA necessary for RNAP II loading.
Mapping of DNase I and MPE-Fe footprinting patterns onto the cryo-EM model of DNA bound to TFIID revealed that the DNase I hypersensitive site at +3 is held across the central channel of TFIID and is flanked on either side by protected or exposed DNase I sites (Fig. 1D). Intriguingly, the DNA upstream of the TSS shows MPE-Fe sensitivity, whereas the downstream DNA is protected from MPE-Fe mediated cleavage (Fig. 1E). This result suggests that the DNA may be flexibly bound upstream of the TSS, allowing the MPE-Fe to intercalate between the Inr and TATA box. On the other hand, the protection of DNA immediately downstream of the TSS suggests that the DNA double helix is likely more rigid in this region to prevent MPE-Fe intercalation, since there are no strong protein-DNA contacts between the Inr and MTE motifs. While a higher resolution structure of TFIID-TFIIA-SCP is required to unambiguously model the path of DNA through the electron density, the present cryo-EM structure indicates that the DNA in this critical region may exhibit helical distortions that are exerted by TFIID during promoter binding.
Structural insight into the relevance of these TFIID-induced DNA topology changes may be obtained by comparing the DNA path in the cryo-EM structure of TFIID-TFIIA-DNA with that in the recently determined cryo-EM structure of an open TBP-based pre-initiation complex (TBP/PIC) comprising TBP-TFIIA-TFIIB-RNAP II-TFIIF-TFIIE34 (Fig. 1F). Since DNA melting starts between the TATA and Inr motifs, the same region of the promoter that exhibits changes in MPE-Fe and DNase I sensitivity upon TFIID binding, we propose that the DNA path within the cryo-EM structure of TFIID-TFIIA-DNA is positioned and structurally primed to facilitate the eventual melting of the promoter DNA by the open RNAP II complex. Interestingly, when the path of DNA through the both structures is compared, the position of DNA upstream of the TATA box is rotated approximately 90 degrees (Fig. 1F). It is tempting to speculate that lobe A, containing the TBP-TFIIA, may rotate during the switch into the open PIC complex. This additional transition of lobe A with respect to the BC core of TFIID would thus represent yet another essential step during transcription initiation that relies on the conformational flexibility of TFIID
Interaction of TFIID with diverse promoter architectures
Since TATA, Inr, MTE and DPE motifs are differentially utilized across the genome, there are important regulatory implications from studying TFIID bound to promoters of different architectures. For instance, the TATA box and DPE motifs are differentially utilized within the critical Hox gene locus in Drosophila, where specific genes either contain TATA or DPE motifs. From these studies, TATA and DPE-specific enhancers have been characterized as important determinants of body plan patterning through coordinated gene regulation within the Hox cluster, suggesting that changes in promoter sequences can increase regulatory output.35,36
The diversity of promoters recognized by TFIID and the ability for enhancers to activate selectively genes containing specific promoter motifs (e.g., TATA vs. DPE) has led to the proposal that there may be promoter-specific conformations adopted by TFIID.35 This hypothesis was tested through cryo-EM visualization and footprinting experiments of TFIID bound to promoters of three specific architectures: Inr-MTE/DPE, TATA-Inr, and TATA-MTE/DPE. Since these promoter architectures were derived from mutations in the SCP sequence, we refer to them as SCP(mTATA), SCP(mMTE/DPE), and SCP(mInr), respectively. Mutation of the TATA box within the SCP sequence retained strong DNase I footprinting of TFIID on the downstream DNA sequences from +3 to +32, and slight protection of the mutant TATA box was observed, although only when TFIIA was added (Fig. 2A). The indistinguishable DNase I footprinting pattern on the downstream DNA between SCP and SCP(mTATA) suggested that TFIID adopted a similar structure on both of these promoters. This idea was confirmed by the cryo-EM structure of TFIID bound to SCP(mTATA), where the DNA was found in the same configuration on the rearranged state of TFIID.10
In contrast to SCP(mTATA), mutation of the Inr or MTE/DPE motifs resulted in the abolishment of TFIID-DNA interactions in the absence of TFIIA. This loss of affinity was dramatically overcome by the addition of TFIIA, which resulted in the strong protection of the mutant Inr and MTE/DPE sequences. Importantly, the same pattern of DNA protection from +3 to 32 was observed for these mutant promoters as was observed originally for the SCP.10 Therefore, these results strongly argue that TFIID adopts the same rearranged conformation when bound to promoters of varying architecture (Fig. 2A), indicating that TFIID does not adopt a TATA- or DPE-specific conformation.
Cofactors, activators and histone modification regulate TFIID-DNA binding
The experiments described above showed that, despite TFIID’s low intrinsic affinity for promoters lacking Inr or MTE/DPE motifs, TFIIA-mediated anchoring of TFIID to the TATA box increases the avidity of TFIID for downstream promoter DNA. Within TFIID, TBP is inhibited by the N-terminal domain of TAF1 that binds to the concave underside of TBP, mimicking the TATA box DNA.37 This inhibition is released by the binding of TFIIA to the stirrup of TBP, displacing the N-terminus of TAF1 from TBP’s DNA binding surface.38 Thus, when TFIID binds to the SCP(mInr) and SCP(mMTE/DPE) promoters, TFIIA serves as a GTF by stimulating TFIID binding to TATA box DNA, which, in turn, increases the avidity of TFIID for the downstream DNA, ultimately resulting in extensive DNA binding through the rearranged state of TFIID.
While this is a very specific case, we believe it provides the first structural example of the functional interplay between affinity and avidity during DNA binding by TFIID. The complexity in DNA recognition by TFIID is highlighted by the large proportion of promoters in the human genome that contain minimal consensus sequences and a wide range of TSS,39 making it unclear how DNA binding by TFIID is able to drive transcription initiation. Early insight into this problem came through structural comparisons of TBP bound to 10 naturally occurring TATA box sequences.40 While the nature of TBP-DNA interactions changed in a sequence-dependent manner, the convex side of TBP (opposing face from DNA binding) remained nearly unchanged, leading to the proposal that TBP’s ability to recognize a variety of TATA boxes does not affect subsequent steps during transcription initiation. We believe that the same conceptual framework is useful for understanding TFIID-DNA recognition.
The highly concerted nature of transcription initiation requires that TFIID adopt a specific conformation that is compatible with the binding of the rest of the general transcription factors. We propose that TFIID composition, promoter DNA architecture, nucleosome modifications, and activators have the potential to stimulate transcription through (i) stabilization of TFIID in the rearranged conformation to facilitate DNA binding, (ii) anchoring TFIID near promoter DNA, thus increasing the local concentration TFIID on DNA, or (iii) both (i) and (ii). Given that TFIID subunits exist in multiple isoforms and subcomplexes in vivo,41 it is likely that changes in TFIID composition can further modulate the affinity of TFIID for DNA, changing the promoter-selectivity of the complex. Irrespective of the mechanisms underlying the recruitment of TFIID to DNA, we believe that the formation of a rearranged state is critical for TFIID function by resulting in a high-affinity/avidity DNA binding state that facilitates the loading of RNAP II at the TSS.
In addition to activators serving as a selective anchor to recruit TFIID to specific promoters, histone modification-mediated recruitment of TFIID can also provide affinity and specificity for TFIID recruitment. Previous biochemical and structural work has demonstrated interactions between p53 and TFIID,11 in addition to the ability of the PHD domain within TAF3 to bind H3K4me3 modifications.42 A recent study has now characterized the regulatory interplay between p53, histone H3 lysine 4 trimethylation (H3K4me3), and TFIID-mediated transcription initiation.43 Using promoter DNA with a functional TATA box, the authors studied the transcription output from naked or chromatinized DNA templates in the presence and absence of p53.43 In this reconstituted system, there were no detectable RNA transcripts for TFIID alone (Fig. 2B). However, the addition of p53, alone or in combination with H3K4me3, resulted in the activation of transcription initiation, whereby RNA levels increased dramatically in the combined presence of all these factors (Fig. 2B). While this effect was diminished when the TATA box was mutated, the overall transcription levels in the presence of H3K4me3 and p53 remained higher for the mutant promoter than those for the intact TATA box in the presence of p53 but in the absence of H3K4me3.
These data suggest that p53, which has been shown to interact directly with TFIID in single particle EM reconstructions,11 and histone modifications (H3K4me3) could anchor TFIID to promoter DNA in order to facilitate transcription from promoters that have an intrinsically weak affinity for TFIID. While the authors did not carry out footprinting experiments, we believe that the anchoring of TFIID to promoter DNA through interactions with the TATA box, p53, and H3K4me3 likely correlates with TFIID binding to downstream DNA through the rearranged conformation, in a manner that would parallel what we have previously observed for TFIID-TFIIA binding at promoters lacking functional MTE/DPE motifs (Fig. 2A).10 Therefore, we would like to speculate that transcriptional activation could be due to the stabilization of the rearranged state of TFIID, leading to the optimized loading of RNAP II. Since TFIID’s footprint on promoter DNA co-occupies the binding site for nucleosomes on the core promoter, it is possible that TFIID could also activate transcription by blocking nucleosome access to the core promoter, which would otherwise repress transcription initiation.44,45 We envision that in its rearranged conformation, TFIID’s extensive footprint on upstream and downstream promoter DNA results in the remodeling of DNA just upstream of the TSS, priming it for melting and loading of RNAP II.
Acknowledgments
We would like to acknowledge funding from NIGMS and from the HFSP to E.N. E.N. is a Howard Hughes Medical Institute Investigator.
Glossary
Abbreviations:
- TFIID
transcription factor for RNAP II fraction ‘D’
- RNAP II
RNA Polymerase II
- GTF
general transcription factor
- DPE
downstream promoter element
- MTE
motif ten element
- Inr
initiator element
- TBP
TATA-binding protein
- MPE-Fe
methidiumpropyl-EDTA-iron(II)
- SCP
super core promoter
- TBP/PIC
TBP-based pre-initiation complex
- EM
electron microscopy
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/25291
References
- 1.Näär AM, Lemon BD, Tjian R. Transcriptional coactivator complexes. Annu Rev Biochem. 2001;70:475–501. doi: 10.1146/annurev.biochem.70.1.475. [DOI] [PubMed] [Google Scholar]
- 2.Albright SR, Tjian R. TAFs revisited: more data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. doi: 10.1016/S0378-1119(99)00495-3. [DOI] [PubMed] [Google Scholar]
- 3.Burley SK, Roeder RG. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–99. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 4.Smale ST, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–13. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 5.Theisen JW, Lim CY, Kadonaga JT. Three key subregions contribute to the function of the downstream RNA polymerase II core promoter. Mol Cell Biol. 2010;30:3471–9. doi: 10.1128/MCB.00053-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burke TW, Kadonaga JT. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 1997;11:3020–31. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee DH, Gershenzon N, Gupta M, Ioshikhes IP, Reinberg D, Lewis BA. Functional characterization of core promoter elements: the downstream core element is recognized by TAF1. Mol Cell Biol. 2005;25:9674–86. doi: 10.1128/MCB.25.21.9674-9686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bieniossek C, Papai G, Schaffitzel C, Garzoni F, Chaillet M, Scheer E, et al. The architecture of human general transcription factor TFIID core complex. Nature. 2013;493:699–702. doi: 10.1038/nature11791. [DOI] [PubMed] [Google Scholar]
- 9.Leschziner AE, Nogales E. Visualizing flexibility at molecular resolution: analysis of heterogeneity in single-particle electron microscopy reconstructions. Annu Rev Biophys Biomol Struct. 2007;36:43–62. doi: 10.1146/annurev.biophys.36.040306.132742. [DOI] [PubMed] [Google Scholar]
- 10.Cianfrocco MA, Kassavetis GA, Grob P, Fang J, Juven-Gershon T, Kadonaga JT, et al. Human TFIID binds to core promoter DNA in a reorganized structural state. Cell. 2013;152:120–31. doi: 10.1016/j.cell.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu WL, Coleman RA, Ma E, Grob P, Yang JL, Zhang Y, et al. Structures of three distinct activator-TFIID complexes. Genes Dev. 2009;23:1510–21. doi: 10.1101/gad.1790709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu WL, Coleman RA, Grob P, King DS, Florens L, Washburn MP, et al. Structural changes in TAF4b-TFIID correlate with promoter selectivity. Mol Cell. 2008;29:81–91. doi: 10.1016/j.molcel.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grob P, Cruse MJ, Inouye C, Peris M, Penczek PA, Tjian R, et al. Cryo-electron microscopy studies of human TFIID: conformational breathing in the integration of gene regulatory cues. Structure. 2006;14:511–20. doi: 10.1016/j.str.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 14.Andel F, 3rd, Ladurner AG, Inouye C, Tjian R, Nogales E. Three-dimensional structure of the human TFIID-IIA-IIB complex. Science. 1999;286:2153–6. doi: 10.1126/science.286.5447.2153. [DOI] [PubMed] [Google Scholar]
- 15.Juven-Gershon T, Cheng S, Kadonaga JT. Rational design of a super core promoter that enhances gene expression. Nat Methods. 2006;3:917–22. doi: 10.1038/nmeth937. [DOI] [PubMed] [Google Scholar]
- 16.Oelgeschläger T, Chiang CM, Roeder RG. Topology and reorganization of a human TFIID-promoter complex. Nature. 1996;382:735–8. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- 17.Van Dyke MW, Roeder RG, Sawadogo M. Physical analysis of transcription preinitiation complex assembly on a class II gene promoter. Science. 1988;241:1335–8. doi: 10.1126/science.3413495. [DOI] [PubMed] [Google Scholar]
- 18.Sawadogo M, Roeder RG. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985;43:165–75. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- 19.Hoffmann A, Chiang CM, Oelgeschläger T, Xie X, Burley SK, Nakatani Y, et al. A histone octamer-like structure within TFIID. Nature. 1996;380:356–9. doi: 10.1038/380356a0. [DOI] [PubMed] [Google Scholar]
- 20.Xie X, Kokubo T, Cohen SL, Mirza UA, Hoffmann A, Chait BT, et al. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature. 1996;380:316–22. doi: 10.1038/380316a0. [DOI] [PubMed] [Google Scholar]
- 21.Selleck W, Howley R, Fang Q, Podolny V, Fried MG, Buratowski S, et al. A histone fold TAF octamer within the yeast TFIID transcriptional coactivator. Nat Struct Biol. 2001;8:695–700. doi: 10.1038/90408. [DOI] [PubMed] [Google Scholar]
- 22.Shao H, Revach M, Moshonov S, Tzuman Y, Gazit K, Albeck S, et al. Core promoter binding by histone-like TAF complexes. Mol Cell Biol. 2005;25:206–19. doi: 10.1128/MCB.25.1.206-219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bleichenbacher M, Tan S, Richmond TJ. Novel interactions between the components of human and yeast TFIIA/TBP/DNA complexes. J Mol Biol. 2003;332:783–93. doi: 10.1016/S0022-2836(03)00887-8. [DOI] [PubMed] [Google Scholar]
- 24.Tan S, Hunziker Y, Sargent DF, Richmond TJ. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature. 1996;381:127–51. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- 25.Chalkley GE, Verrijzer CP. DNA binding site selection by RNA polymerase II TAFs: a TAF(II)250-TAF(II)150 complex recognizes the initiator. EMBO J. 1999;18:4835–45. doi: 10.1093/emboj/18.17.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dosztányi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–4. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 27.Ozer J, Mitsouras K, Zerby D, Carey M, Lieberman PM. Transcription factor IIA derepresses TATA-binding protein (TBP)-associated factor inhibition of TBP-DNA binding. J Biol Chem. 1998;273:14293–300. doi: 10.1074/jbc.273.23.14293. [DOI] [PubMed] [Google Scholar]
- 28.Wright KJ, Marr MT, 2nd, Tjian R. TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc Natl Acad Sci U S A. 2006;103:12347–52. doi: 10.1073/pnas.0605499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez E, Kundu TK, Fu J, Roeder RG. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J Biol Chem. 1998;273:23781–5. doi: 10.1074/jbc.273.37.23781. [DOI] [PubMed] [Google Scholar]
- 30.Grant PA, Schieltz D, Pray-Grant MG, Steger DJ, Reese JC, Yates JR, 3rd, et al. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998;94:45–53. doi: 10.1016/S0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- 31.Ogryzko VV, Kotani T, Zhang X, Schiltz RL, Howard T, Yang XJ, et al. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/S0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 32.Wieczorek E, Brand M, Jacq X, Tora L. Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature. 1998;393:187–91. doi: 10.1038/30283. [DOI] [PubMed] [Google Scholar]
- 33.Tse C, Sera T, Wolffe AP, Hansen JC. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol Cell Biol. 1998;18:4629–38. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He Y, Fang J, Taatjes DJ, Nogales E. Structural visualization of key steps in human transcription initiation. Nature. 2013;495:481–6. doi: 10.1038/nature11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohtsuki S, Levine M, Cai HN. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 1998;12:547–56. doi: 10.1101/gad.12.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Juven-Gershon T, Hsu JY, Kadonaga JT. Caudal, a key developmental regulator, is a DPE-specific transcriptional factor. Genes Dev. 2008;22:2823–30. doi: 10.1101/gad.1698108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu D, Ishima R, Tong KI, Bagby S, Kokubo T, Muhandiram DR, et al. Solution structure of a TBP-TAF(II)230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP. Cell. 1998;94:573–83. doi: 10.1016/S0092-8674(00)81599-8. [DOI] [PubMed] [Google Scholar]
- 38.Bagby S, Mal TK, Liu D, Raddatz E, Nakatani Y, Ikura M. TFIIA-TAF regulatory interplay: NMR evidence for overlapping binding sites on TBP. FEBS Lett. 2000;468:149–54. doi: 10.1016/S0014-5793(00)01213-8. [DOI] [PubMed] [Google Scholar]
- 39.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–35. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 40.Patikoglou GA, Kim JL, Sun L, Yang SH, Kodadek T, Burley SK. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 1999;13:3217–30. doi: 10.1101/gad.13.24.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodrich JA, Tjian R. Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation. Nat Rev Genet. 2010;11:549–58. doi: 10.1038/nrg2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 43.Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH, et al. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell. 2013;152:1021–36. doi: 10.1016/j.cell.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lomvardas S, Thanos D. Nucleosome sliding via TBP DNA binding in vivo. Cell. 2001;106:685–96. doi: 10.1016/S0092-8674(01)00490-1. [DOI] [PubMed] [Google Scholar]