In contrast to BRCA1, CtIP has indispensable roles in promoting resection and embryonic development.
Abstract
Homologous recombination (HR) is initiated by DNA end resection, a process in which stretches of single-strand DNA (ssDNA) are generated and used for homology search. Factors implicated in resection include nucleases MRE11, EXO1, and DNA2, which process DNA ends into 3′ ssDNA overhangs; helicases such as BLM, which unwind DNA; and other proteins such as BRCA1 and CtIP whose functions remain unclear. CDK-mediated phosphorylation of CtIP on T847 is required to promote resection, whereas CDK-dependent phosphorylation of CtIP-S327 is required for interaction with BRCA1. Here, we provide evidence that CtIP functions independently of BRCA1 in promoting DSB end resection. First, using mouse models expressing S327A or T847A mutant CtIP as a sole species, and B cells deficient in CtIP, we show that loss of the CtIP-BRCA1 interaction does not detectably affect resection, maintenance of genomic stability or viability, whereas T847 is essential for these functions. Second, although loss of 53BP1 rescues the embryonic lethality and HR defects in BRCA1-deficient mice, it does not restore viability or genome integrity in CtIP−/− mice. Third, the increased resection afforded by loss of 53BP1 and the rescue of BRCA1-deficiency depend on CtIP but not EXO1. Finally, the sensitivity of BRCA1-deficient cells to poly ADP ribose polymerase (PARP) inhibition is partially rescued by the phospho-mimicking mutant CtIP (CtIP-T847E). Thus, in contrast to BRCA1, CtIP has indispensable roles in promoting resection and embryonic development.
Individuals with mutations in BRCA1 have a high risk of developing breast and ovarian cancer. Although BRCA1 is implicated in many cellular processes, its role in HR (Moynahan et al., 1999; Stark et al., 2004) is thought to be critical for tumor suppression and maintenance of genomic stability (Silver and Livingston, 2012). Tumor suppressor functions of BRCA1 are mediated by the BRCA1 carboxyl-terminal (BRCT) domain (Shakya et al., 2011), a motif that binds phosphorylated serine motifs in three different DNA repair proteins: BACH1 (BRIP1), ABRAXAS (CCDC98), and CtIP (Rbbp8; Li and Greenberg, 2012). Among these complexes, BRCA1 association with CtIP has been implicated in nucleolytic resection of DNA double strand breaks (DSBs; Sartori et al., 2007; Chen et al., 2008). Although the BRCA1–CtIP complex is not known to possess nuclease activity itself, it enhances the nuclease activity of the MRE11–RAD50–NBS1 DSB sensor complex, which is required for effective resection (Sartori et al., 2007). According to the current “two-step” model, the BRCA1–CtIP–MRE11 complex initiates end processing, then EXO1 and BLM generate longer stretches of ssDNA, which are stabilized by RPA (Symington and Gautier, 2011). Finally, formation of the RAD51–ssDNA nucleoprotein filament promotes strand invasion and HR. In the absence of BRCA1 or CtIP, RAD51 binding to DSB sites and HR are reduced, resulting in mutagenic DNA repair, genome instability, and tumorigenesis (Bunting and Nussenzweig, 2013).
CtIP is directly phosphorylated by cyclin-dependent kinases (CDKs; Ferretti et al., 2013). CDK phosphorylation of CtIP at T847 promotes ssDNA generation, RPA recruitment and chromosome integrity (Huertas and Jackson, 2009). Whereas changing T847 to alanine impairs resection, mutating T847 to glutamic acid mimics constitutive phosphorylation, which promotes limited resection (Huertas and Jackson, 2009). The BRCA1–CtIP interaction (Wong et al., 1998; Yu et al., 1998) is mediated by CDK phosphorylation of CtIP at S327 (equivalent to S326 in mouse) during G2 phase of the cell cycle (Yu and Chen, 2004). In chicken DT40 cells expressing human CtIP, mutation of S327 into a nonphosphorytable residue inhibits HR repair (Yun and Hiom, 2009). Moreover, in mammalian cells, CtIP–BRCA1 complex formation facilitates removal of 53BP1 binding protein RIF1 from DSB regions, which otherwise blocks resection (Escribano-Díaz et al., 2013). However, the physiological role of S327 phosphorylation has been questioned by the finding that the chicken CtIP-S332A protein can efficiently promote DSB repair by HR, apparently independently of BRCA1 interaction (Nakamura et al., 2010) and by the fact that that knock-in mice homozygous for CtIP-S326A allele are neither tumor prone or HR deficient (Reczek et al., 2013). Thus, the biological importance of the BRCA1–CtIP interaction remains unclear.
Further progress in understanding how BRCA1 promotes HR was made by demonstrating that loss of 53BP1 rescues the lethality, tumorigenesis, and genome instability of BRCA1-deficient mice (Cao et al., 2009; Bouwman et al., 2010; Bunting et al., 2010). It was shown that 53BP1 inhibits resection, but that BRCA1 antagonizes 53BP1, allowing the nucleolytic processing of DNA ends in S phase (Bunting et al., 2010; Chapman et al., 2012). Furthermore PTIP and RIF1, both of which act downstream of 53BP1, inhibit BRCA1-associated DNA metabolism (Callen et al., 2013; Chapman et al., 2013; Escribano-Díaz et al., 2013; Feng et al., 2013; Zimmermann et al., 2013). Nevertheless, the mediators of resection in BRCA1/53BP1, BRCA1/PTIP, and BRCA1/RIF double-deficient cells have not been identified.
Here, we establish that both the increased resection associated with the loss of 53BP1 and the rescue of HR defects in doubly deficient 53BP1/BRCA1 cells are dependent on CtIP but independent of EXO1. Loss of CtIP or its ability to be phosphorylated on T847 leads to embryonic lethality, constitutive DNA damage signaling, genome instability, and defective resection. In contrast, constitutive activation of CtIP by expressing a CtIP phospho-mimicking mutant CtIP (CtIP-T847E) reverts the embryonic lethality, and partially rescues BRCA1-deficient chromosomal instability even in the presence of 53BP1. Thus, whereas BRCA1 is dispensable for HR under some conditions, CtIP-mediated resection is an essential activity that can proceed independently of BRCA1.
RESULTS AND DISCUSSION
Loss of CtIP in B cells is compatible with limited cellular proliferation
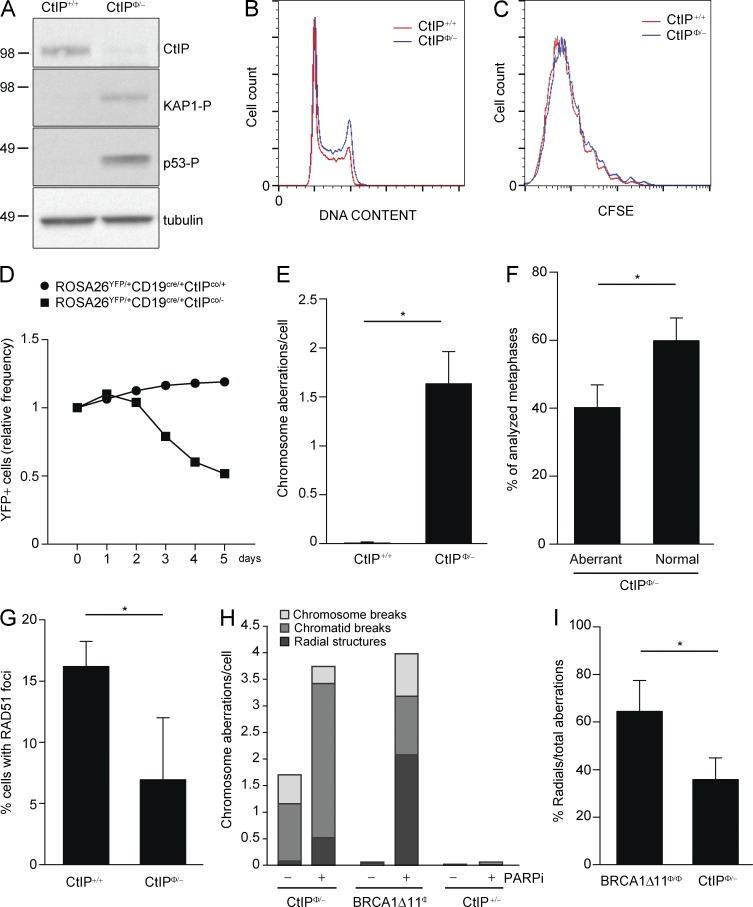
The analysis of CtIP functions in vivo has been hampered by the lethality of CtIP knockout mice, with mutant embryos dying at embryonic day E3.5 (Chen et al., 2005). To circumvent this, we generated a B cell–specific deletion of CtIP by intercrossing CD19cre mice (Rickert et al., 1997) with mice carrying conditional and null alleles (CtIPco/−; Chen et al., 2008; Bothmer et al., 2013). The resulting CtIP-null (CtIPΦ/−) B cells expressed little or no CtIP protein as determined by Western blot analysis (Fig. 1 A). Upon ex vivo stimulation with LPS and IL-4 cells, splenic B cells enter the cell cycle and undergo class switch recombination (CSR). Despite the fact that CtIP-null mice invariably experience early embryonic death (Chen et al., 2005), CtIPΦ/− B cells were able to proliferate, exhibited a normal cell cycle distribution (Fig. 1 B), and underwent a similar number of cellular divisions as WT cells, as determined by CFSE dye dilution (Fig. 1 C). Moreover, CtIP-deficient B cells have similar level of class switching compared with WT (Bothmer et al., 2013).
Figure 1.
Increased spontaneous genomic instability and DNA damage signaling in CtIP-deficient B cells. (A) Expression of CtIP, KAP1 phosphorylation, and p53 phosphorylation as detected by Western blotting in CtIP-deficient and WT B cells. (B) Representative cell cycle analysis of CtIP-deficient (blue line) and WT (red line) B cells. After 2 d in culture, cells were fixed, stained with PI, and analyzed by flow cytometry. (C) Cellular division measured by CFSE dye dilution detected by FACS. One out of three experiments is shown. (D) Frequency of YFP+ cells relative to the percentage at day 0 (the day in which B cells were isolated). (E) Analysis of spontaneous genomic instability in metaphases from B cells isolated from mice of indicated genotypes. Graph shows the mean number of aberrant chromosome structures per cell measured in 3 independent experiments (n = 255 metaphases/genotype; P = 0.0131; two-tailed paired Student’s t test). (F) Percentage of CtIP-deficient B cells with normal chromosomes or with spontaneous chromosomal abnormalities is quantified. Data are derived from 10 independent experiments (n > 600 metaphases; P = 0.0013: two-tailed paired Student’s t test). (G) Percentage of cells with >5 RAD51 foci after 10 Gy irradiation relative to WT. For each experiment more than 600 cells were counted. The mean and SD of 3 independent experiments is shown (P = 0.0258; one-tailed paired Student’s t test). (H) Analysis of genomic instability in metaphase spreads before and after PARPi treatment (1 µM for 16 h) in B cells (n = 50 metaphases). Percentage of radials, chromatid, and chromosome breaks is quantified. (I) Mean percentage of radial structures among total aberrations counted in 4 different experiments (n > 200 metaphases) in BRCA1 or CtIP-deficient B cells upon PARPi treatment (1 µM for 16 h; P = 0.0103; two-tailed paired Student’s t test). For each independent experiment, one mouse per genotype was used.
Although CtIP deletion in B cells did not lead to a marked defect of cellular proliferation in live cells, we detected heightened phosphorylation of KAP-1 and p53 (Fig. 1 A), indicative of constitutive DNA damage signaling. To determine whether there was a selection against CtIP-deficient cells in culture, we compared the efficiency of CD19-CRE–mediated excision in CtIPco/− versus CtIPco/+ B cells by crossing these mice with the reporter line ROSA26-STOP-EYFP, in which YFP expression is induced upon CRE mediated excision. We found that there was a gradual depletion of ROSA26YFP/+CD19cre/+ CtIPco/− but not ROSA26YFP/+CD19cre/+CtIPco/+ B cells after 2 d in culture, indicating that the CtIP deletion confers a growth disadvantage (Fig. 1 D). We therefore restricted our analyses to day 2 stimulated B cells in subsequent experiments to maximize the number of CtIP-deficient and actively cycling cells.
Spontaneous and PARPi-induced genome instability in CtIP-deficient cells
The observed constitutive phosphorylation of KAP-1 and p53 (Fig. 1 A) suggested that loss of CtIP might lead to spontaneous chromosomal damage. To test this, we analyzed metaphase spreads generated from stimulated WT or CtIPΦ/− B cells. We observed that CtIP-deficient B cells showed spontaneous accumulation of chromosome aberrations (Fig. 1, E and F). Indeed, ∼40% of CtIPΦ/− B cells exhibited chromosomal aberrations (Fig. 1 F), and on average, there were 1.5 aberrant chromosomes per mutant metaphase, whereas no instability was observed in controls (Fig. 1 E). To exclude the possibility that the instability we observed is dependent on AID expression induced by activating B cells with IL-4 and LPS, we stimulated CtIP-deficient and WT B cells with RP105. Upon RP105 activation, B cells do not express AID and do not undergo CSR. We found similar levels of spontaneous genomic instability in CtIPΦ/− B cells independent of stimulation conditions (unpublished data). Because CtIP has been implicated in HR, we monitored IR induced Rad51 focus formation in CtIP–deficient B cells. We found that the percentage of RAD51 focus-positive cells was decreased on average to 50% of the WT level (Fig. 1 G), which is consistent with decreased resection activity in CtIP-deficient cells (Sartori et al., 2007). Thus, CtIP loss leads to considerable DNA damage accumulation associated with defective HR.
Cells with impaired HR are hypersensitive to compounds that induce replication-associated breaks. For example, BRCA-deficient cells are hypersensitive to PARPi (Bryant et al., 2005; Farmer et al., 2005; Jackson and Bartek, 2009). Consistent with this, treating cells with the PARPi (olaparib) resulted in a nearly twofold increase in the total amount of chromosomal aberrations in CtIP-null B cells (Fig. 1 H), whereas such PARPi treatment lead to little or no induction of aberrations in control cells. Although the total amount of DNA damage induced by PARPi was equivalent in both CtIP-deficient and BRCA1-deficient cells, radial structures accumulated at a greater frequency in the absence of BRCA1 (Fig. 1, H and I). We conclude that BRCA1- and CtIP-deficient B cells are both hypersensitive to PARPi, but CtIP-deficient cells accumulate considerably more spontaneous damage.
53BP1 or Ku80 deficiency does not rescue embryonic lethality of CtIP knockout mice
Because loss of 53BP1 rescues the viability of BRCA1-deficient mice (Cao et al., 2009), we tested whether deletion of 53BP1 could similarly rescue the embryonic lethality of CtIP−/− mice. However 53BP1−/−CtIP+/− intercrosses did not generate any of the expected 15 CtIP/53BP1 double knockout mice out of 60 live births (Table 1). In Schizosacchoromyces pombe, the hypersensitivity of CtIP (Ctp1)-deficient cells to camptothecin (which yields DSBs during S-phase that rely on resection and HR for their repair) is rescued by loss of KU (Langerak et al., 2011). To see whether a similar relationship exists in mammals, we intercrossed CtIP+/−KU80+/− mice to determine if deletion of KU80 could rescue CtIP−/− embryonic lethality. Again, no CtIP−/− mice were generated among 129 offspring (Table 1). Collectively, these findings revealed that neither 53BP1-deficiency nor KU-deficiency can rescue embryonic lethality in CtIP−/− mice.
Table 1.
Loss of either 53BP1 or KU80 does not rescue CtIP knockout mouse lethality
| CtIP+/-53BP1−/− x CtIP+/−53BP1−/− | CtIP+/−KU80+/− x CtIP+/−Ku80+/− | |||||
| Genotypes | CtIP+/+ 53BP1−/− | CtIP+/− 53BP1−/− | CtIP−/− 53BP−/− | CtIP+/+ KU80−/− | CtIP+/− KU80−/− | CtIP−/− KU80−/− |
| Live-born pups | ||||||
| Observed | 25 | 35 | 0 | 1 | 15 | 0 |
| Expected | 15 | 30 | 15 | 8.0625 | 16.125 | 8.0625 |
| Total mice screened | 60 | 129 | ||||
CtIP+/− 53BP1−/− mice and CtIP+/−Ku80+/− were intercrossed. Genotypes and number of animals generated by the breeding are indicated.
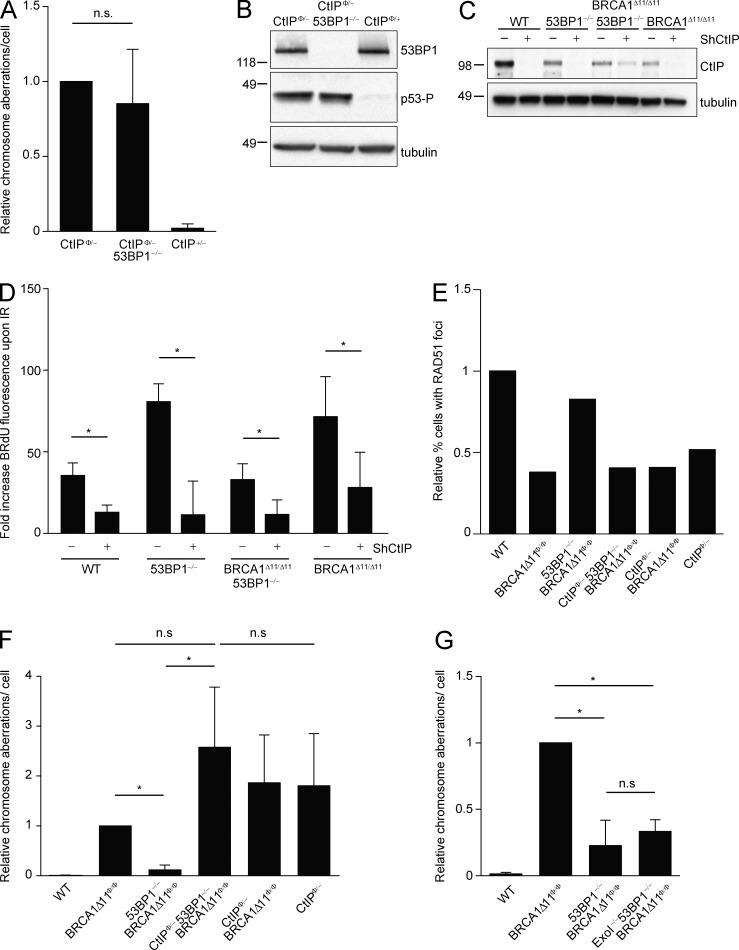
Loss of 53BP1 normalizes the HR defects in BRCA1-deficient cells, at least in part by increasing DSB resection (Bunting et al., 2010). To determine what impact 53BP1 deletion might have on CtIP−/−-associated genomic instability, we generated CtIP/53BP1 double knockout B cells by crossing CD19cre/+CtIPco/− with 53BP1−/− mice (Fig. 2 A). We found that CtIP/53BP1 double-mutant B cells harbored spontaneous chromosomal aberrations and constitutive DNA damage signaling similar to the levels observed in CtIP single-mutant cells (Fig. 2, A and B). Thus, in contrast to its ability to rescue BRCA1-associated phenotypes, loss of 53BP1 does not restore genomic integrity in the absence of CtIP.
Figure 2.
Increased resection of 53BP1−/− cells is CtIP dependent. (A) Analysis of spontaneous genomic instability in metaphases from B cells isolated from mice of the indicated genotypes. Genomic instability measured in five independent experiments is normalized to levels seen in CtIP-deficient B cells (n > 200 metaphases/genotype; P = 0.4179: two-tailed paired Student’s t test). (B) Constitutive levels of p53 phosphorylation in the CtIPΦ/– and CtIPΦ/– 53BP1−/− double mutant B cells. (C) Western blot analysis showing levels of CtIP expression in MEFs infected with CtIP shRNA (shCtIP). (D) Fold increase of mean BrdU fluorescence of irradiated (30 Gy) versus unirradiated MEFs of the indicated genotypes that were either infected or not with ShCtIP. The mean and SD of three independent experiments is shown (significance tests were analyzed with one-tailed paired Student’s t test; *, P < 0.05). (E) Percentage of cells with >5 RAD51 foci after 10 Gy irradiation relative to WT (16.5% of WT cells were positive for RAD51 foci). For each experiment, >600 cells were counted. (F) Analysis of genomic instability in metaphases from B cells treated with PARPi (1 µM; 16 h; n > 200 metaphases/genotype were counted; the mean of 4 independent experiments is reported relative to BRCA1-deficient cells; significance tests were analyzed with two-tailed paired Student’s t test; *, P < 0.05; n.s., not significant). (G) Analysis of genomic instability in B cells after PARPi treatment (1 µM for 16 h). The graph shows the average number of aberrant chromosome structures per cell measured in three independent experiments relative to genome instability in BRCA1-deficient cells (n = 150 metaphases/genotype; significance tests were analyzed with two-tailed paired Student’s t test; *, P < 0.05; n.s., not significant). For each independent experiment, one mouse per genotype was used.
CtIP but not EXO1 mediates the rescue of genomic stability of BRCA1–53BP1-deficient cells
Loss of 53BP1 increases resection in BRCA1-deficient cells (Bunting et al., 2010, 2012). To determine whether and how 53BP1 impacts on resection in CtIP-deficient cells, we used a previously described nondenaturing immunofluorescence protocol, in which MEFs are pulsed with BrdU and ssDNA accumulation after irradiation (IR) is monitored by flow cytometry with an anti-BrdU antibody (Bunting et al., 2012). WT, 53BP1−/−, BRCA1Δ11/Δ1153BP1−/−, and BRCA1Δ11/Δ11 MEFs were infected with a short hairpin RNA (shRNA) specifically targeting the CtIP mRNA (shCtIP; Bunting et al., 2010; Fig. 2 C). The extent of knockdown differed in each cell line, making it difficult to compare the effects of CtIP deletion across the nonisogenic lines (Fig. 2 C). However, in all individual cell lines loss of CtIP expression decreased the extent of IR-induced ssDNA (Fig. 2 D). Thus, DNA damage stimulates an increase in end-resection activity in BRCA1- and 53BP1-deficient cell lines that is CtIP dependent.
To determine the impact of CtIP deficiency on RAD51 foci formation, we crossed CD19cre/+CtIPco/−, BRCA1Δ11f/f and 53BP1−/− mice to generate triple CtIP–BRCA1–53BP1 mutant B cells. Consistent with the decrease of ssDNA in CtIP depleted BRCA1–53BP1 MEFs (Fig. 2 D), we found that CtIP deficiency in B cells led to a decrease in IR induced RAD51 foci formation relative to WT (Fig. 2 E). Moreover, PARPi treatment of CtIP–BRCA1–53BP1 triply deficient cells led to a 13-fold increase in the level of DNA damage relative to BRCA1Δ11Φ/Φ 53BP1−/− (Fig. 2 F). Together with the finding that the IR-induced increase in resection in BRCA1–53BP1 double-knockout cells was CtIP dependent (Fig. 2 D), these results suggest that CtIP-mediated resection is critical for the rescue of HR in BRCA1–53BP1 mutant cells.
In the two-step model for resection, CtIP carries out initial end processing, which is followed by EXO1- or DNA2/BLM-mediated extension of the resected tracks (Symington and Gautier 2011). Because EXO1 recruitment and exonuclease activity are reported to be CtIP dependent (Eid et al., 2010), we tested whether the rescue of HR in BRCA–53BP1-deficient cells was EXO1 dependent. To do this, EXO1+/− mice were intercrossed with CD19cre/+Brca1Δ11f/f53BP1−/− mice, and the resulting CD19cre/+Brca1Δ11f/f53BP1−/−EXO1+/− mice were intercrossed to generate triple BRCA1–53BP1–EXO1 mutant B cells. We found that BRCA1Δ11Φ/Φ 53BP1−/−EXO1−/− cells were as insensitive to PARPi as BRCA1Δ11Φ/Φ 53BP1−/− cells and much less sensitive than BRCA1Δ11Φ/Φ or triple mutant CtIP–BRCA1–53BP1 cells (Fig. 2, F and G). These data thus establish that CtIP but not EXO1 mediates the rescue of genomic stability of BRCA1–53BP1 deficient cells.
CDK mediated phosphorylation of CtIP at T847 but not S327 is essential for viability
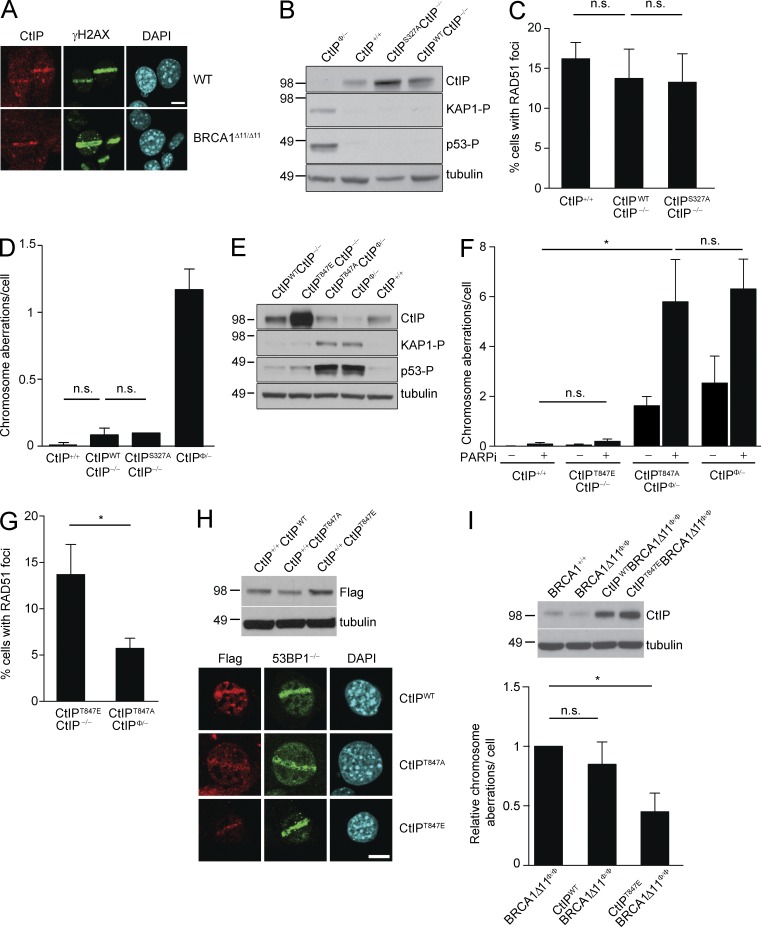
The aforementioned findings suggested that CtIP could act independently of BRCA1 to promote resection. Consistent with this, we found that BRCA1 was not required for recruitment of CtIP to DNA damage sites (Fig. 3 A). To explore the physiological role of the CtIP–BRCA1 interaction mediated by CDK-dependent phosphorylation of CtIP at S327 (Yu and Chen, 2004), we generated BAC transgenic mouse models. By introducing the human gene coding for CtIP (hereafter, referred to as CtIPWT) as a transgenic copy carried by a bacterial artificial chromosome (BAC RP11-104H10), we were able to rescue the lethality caused by homozygous deletion of mouse CtIP (Table 2). CtIPWT was expressed at a similar level compared with endogenous CtIP (Fig. 3 B). Moreover, B cells in CtIPWTCtIP−/− mice were equivalent to CtIP+/+ with respect to RAD51 foci formation and genome stability (Fig. 3, C and D).
Figure 3.
CDK-dependent phosphorylation of CtIPT847 is essential for CtIP function. (A) WT and BRCA1Δ11/Δ11 MEFs were irradiated with a 364-nm laser line derived from a LSM510 microscope. After 10-min recovery, cells were processed for immunofluorescence analysis of CtIP (red) and γH2AX (green). Bar, 15 µm. (B) Western blot showing expression of transgenic CtIPWT and CtIPS327A protein and phosphorylated KAP1 and p53 in B cells. (C) Percentage of cells with >5 RAD51 foci after 10 Gy irradiation. Three independent experiments are reported. For each experiment, >600 cells were counted (significance tests were analyzed with two-tailed paired Student’s t test; *, P < 0.05; n.s., not significant). (D) Analysis of total genomic instability in metaphases from B cells isolated from mice of indicated genotypes after treatment for 16 h with 1 µM PARPi. The mean number of chromosome aberrations per cell measured in 3 independent experiments is shown (n = 250 metaphases; significance tests were analyzed with two-tailed paired Student’s t test; *, P < 0.05; n.s., not significant). (E) Levels of transgenic CtIPWT, CtIPT847E, and CtIPT847A protein expression and KAP1 and p53 phosphorylation in mutant B cells detected by Western blotting. (F) Analysis of total genomic instability in metaphases from B cells with or without PARPi treatment. Mean of three independent experiments is shown (significance tests were analyzed with two-tailed paired Student’s t test; *, P < 0.05; n.s., not significant). (G) Percentage of B cells with >5 RAD51 foci after 10 Gy IR. Three independent experiments are reported. >600 cells were counted for each genotype (P = 0.0251; two-tailed paired t test). (H) Western blot showing the level of CtIP expression in infected cells using anti-FLAG antibody (top). WT MEFs expressing FLAG-CtIPWT, FLAG-CtIPT847A, or FLAG-CtIPT847E were treated with Hoechst 33342 and irradiated with the 364-nm laser line and after 20-min recovery FLAG (red) and 53BP1 (green) were detected by immunofluorescence (bottom). Bar, 15 µm. (I) Western blot showing the levels of CtIP protein in B cells from mice of the indicated genotype (top). Genomic instability in B cells isolated from mice of the indicated genotypes, relative to total instability measured in BRCA1-deficient B cells. Cells were treated with 1 µM PARPi for 16 h (bottom). Three independent experiments are reported (significance tests were analyzed with two-tailed paired Student’s t test; *, P < 0.05; n.s., not significant). For each independent experiment, one mouse per genotype was used.
Table 2.
Effect of CtIP transgenes on CtIP−/− lethality
| CtIPtgCtIP+/− x CtIP+/− | ||||
| Transgenic (CtIPtg) | CtIPWTCtIP−/− | CtIPT847ACtIP−/− | CtIPT847ECtIP−/− | CtIPS327ACtIP−/− |
| Live-born pups | ||||
| Observed | 7 | 0 | 13 | 14 |
| Expected | 12.75 | 11.75 | 10.375 | 13.375 |
| Total mice screened | 102 | 94 | 83 | 107 |
Rescue of CtIP−/− was assessed in transgenic mice generated by intercrossing CtIP heterozygous mice with one parent carrying a copy of CtIP transgene (CtIP-WT, CtIP-T847A, CtIP-T847E, or CtIP-T327A).
We next generated BAC transgenic mice expressing a CtIP mutant in which Ser-327 was substituted for Ala (CtIPS327A) to abrogate phosphorylation by CDK at this site. Like CtIPWT, expression of mutant CtIPS327A rescued the lethality of CtIP knockout mice (Table 2). Indeed, CtIPS327ACtIP−/− mice developed normally, without notable defects in growth, weight, lymphocyte development, or fertility (unpublished data). Moreover, in stark contrast to CtIP deficiency, CtIPS327ACtIP−/− B cells were not markedly hypersensitive to PARPi, did not harbor constitutive DNA damage signaling, and formed normal levels of IR–induced RAD51 foci (Fig. 3, B–D). These results are consistent with a recent characterization of CtIP-S326A knock-in mice, which concluded that CDK-dependent phosphorylation of CtIP–BRCA1 interaction domain is dispensable for HR (Reczek et al., 2013).
In addition to S327, the other CDK-dependent phosphorylation site of CtIP implicated in DSB resection is Thr-847 (Huertas and Jackson, 2009). To assess whether this residue affects CtIP function, we used BAC recombineering to mutate threonine into either alanine (CtIPT847A) or a potentially phospho-mimicking glutamic acid (CtIPT847E), and then generated transgenic mice. We found that the CtIPT847A transgene did not rescue the lethality of CtIP knockout mice, whereas CtIPT847E did (Table 2). By isolating embryos from timed pregnancies, we determined that CtIPT847ACtIP−/− embryos died earlier than E9.5 (Table 3), consistent with the early embryonic lethality of CtIP−/− mice (Chen et al., 2005). Thus, phosphorylation of CtIP at T847, in contrast to S327, is essential for embryonic viability.
Table 3.
Role of CDK-mediated phosphorylation of CtIP at T847 on embryonic viability
| CtIPT847ACtIP+/− x CtIP+/− | ||||||
| Genotype | CtIP−/− | CtIP−/− | CtIP+/+ | CtIPT847A CtIP−/− | CtIPT847A CtIP+/− | CtIPT847A CtIP+/+ |
| E9.5 embryos | ||||||
| Observed | 0 | 2 | 1 | 0 | 14 | 6 |
| Expected | 2.875 | 5.75 | 2.875 | 2.875 | 5.75 | 2.875 |
| Total embryos screened | 23 | |||||
CtIP heterozygous mice were intercrossed with one parent carrying a copy of the transgenic CtIP-T847A mutation, and embryos were genotyped at E9.5.
Phosphorylation of CtIP at T847 is essential for genome stability
To examine the consequence of CtIPT847A protein expression, we generated CtIPT847ACD19cre/+CtIPco/− B cells. Western blot analysis confirmed that CtIPT847A was expressed in these cells at similar levels as CtIPWT but lower than in CtIPT847E (Fig. 3 E). CtIPT847A mutant B cells exhibited high levels of spontaneous genomic instability, persistent DNA damage signaling, and defective IR-induced RAD51 focus formation similar to those seen with CtIP deficiency (Fig. 3, E–G). In contrast, CtIPT847E mutant B cells were normal in all these respects (Fig. 3, E–G). The finding that CtIPT847A phenocopies CtIP deficiency thereby implies that CDK-dependent phosphorylation of T847 is essential for CtIP functions in mice.
Given that phosphorylation of CtIPT847 is essential for efficient resection (Huertas and Jackson, 2009), we wondered whether CDK-dependent phosphorylation favors the recruitment of CtIP to damaged chromatin. To address this, we generated FLAG-tagged CtIPWT, CtIPT847A, and CtIPT847E retroviral constructs, introduced them into WT MEFs, and then induced targeted DNA damage by laser microirradiation. Using an antibody that recognizes exogenously expressed FLAG-tagged proteins (Fig. 3 H) we could readily detect CtIPWT, CtIPT847A, and CtIPT847E at DNA damage sites. Given that CtIPT847A can be recruited to DSB (Fig. 3 H), we conclude that the phosphorylation of CtIP-T847, although critical for resection, is not required for CtIP recruitment to damage regions. Instead, chromatin binding of CtIP appears to require DNA damage inducible phosphorylation at CtIP-T859 by ATR kinase (Peterson et al., 2013).
The sensitivity of BRCA1-deficient cells to PARP inhibition is partially rescued by CtIP-T847E
Because the CtIP-T847E mutant has been reported to be proficient in allowing DSB resection (Huertas and Jackson, 2009), we asked whether this phospho-mimic version of the protein could rescue the sensitivity of BRCA1-deficient cells to PARPi. To do so, we crossed CD19cre/+BRCA1Δ11f/f mice with either CtIPWT or CtIPT847E transgenic mice to generate BRCA1-deficient B cells expressing either CtIPWT or CTIPT847E protein (Fig. 3 I). We found that the genomic instability induced by PARPi treatment was significantly reduced in BRCA1Δ11 mutant B cells expressing CtIPT847E relative to that found in Brca1Δ11Φ/Φ cells (Fig. 3 I). CtIPWT, which expressed slightly lower levels of CtIP than CtIPT847E (Fig. 3 I), also reduced the level of chromosomal aberrations in BRCA1-deficient mice, but this was not significant (Fig. 3 I). These data therefore indicate that overexpression of CtIP can partially overcome 53BP1’s block to resection in BRCA1-deficient cells.
Model for functions of BRCA1 and CtIP in DSB resection
Previous studies have suggested that BRCA1 plays a role in resection and that this is mediated by the CtIP-BRCA1 interaction. These conclusions were based on the findings that BRCA1-deficient cells exhibit a mild decrease in IR-induced RPA focus formation (Chen et al., 2008; Escribano-Díaz et al., 2013), that BRCA1 and CtIP inhibit RIF1 end-blocking activity in S/G2 (Escribano-Díaz et al., 2013; Zimmermann et al., 2013), and that cells expressing CtIP-327A are defective in HR (Yun and Hiom, 2009). However, subsequent studies in chicken (Nakamura et al., 2010), frog (Peterson et al., 2011), and mouse cells (Reczek et al., 2013; analogous to our CtIP-null cells reconstituted with CtIP-S327A), and direct measurements of gene conversion (Chandramouly et al., 2013), argue that the CtIP-BRCA1 interaction is dispensable for HR. Moreover, our finding that restoration of HR in BRCA1–53BP1-deficient cells and increased resection that occurs in the absence of 53BP1 are CtIP dependent, demonstrates that CtIP-mediated resection is BRCA1 independent. These results likely explain why loss of 53BP1 cannot reverse the embryonic lethality or genomic instability of CtIP-deficient mice. Localization of CtIP to damage sites must take place by mechanisms other than association with BRCA1, such as direct association of CtIP with DNA (You et al., 2009) or through phosphorylation by the ATR kinase (Peterson et al., 2013).
If CDK-mediated phosphorylation of CtIP is essential for its function (Table 1 and Fig. 3, E–G), whereas BRCA1 is dispensable for resection, how does BRCA1 promote HR? We have previously suggested that BRCA1’s principal role in DSB repair is to antagonize 53BP1-dependent end protection (Bunting et al., 2010). In this model, in the absence of BRCA1, CtIP activity is limited by 53BP1 and downstream cofactors RIF1 and PTIP, which results in a modest defect in RPA focus formation (Escribano-Diaz et al., 2013). Nevertheless CtIP-mediated resection can partly overcome 53BP1 end blocking activity, evident by our finding that mimicking constitutive CtIP phosphorylation (CtIPT847E) partially alleviated genomic instability of BRCA1-deficient/53BP1+/+ cells. Furthermore, although 53BP1 antagonizes CtIP function, complete absence of 53BP1 allows full access and activity of CtIP, which significantly increases ssDNA formation. Thus, we propose that BRCA1 facilitates CtIP mediated resection in the presence of 53BP1 but is no longer needed in its absence. In addition, BRCA1 plays functions in meiosis and cross-link repair that are 53BP1 independent (Bunting et al., 2012).
The pro-NHEJ (non-homologous end joining) functions that promote class switch recombination, and the antirecombination functions that lead to loss of genome stability in BRCA1-deficient cells are mediated by distinct phospho-dependent interactions with 53BP1 (Callen et al., 2013). PTIP associates with phosphorylation sites at the extreme N terminus of 53BP1 to suppress HR in S phase, whereas RIF1 binds to independent residues to promote NHEJ in G1 (Callen et al., 2013). We have proposed that PTIP and RIF1 could interfere with a distinct set of nucleases (Callen et al., 2013). Consistent with this idea, we have found that the rescue of genome stability in BRCA1–53BP1-deficient cells is dependent on ATM (Bunting et al., 2010) and CtIP (Fig. 2 F), but occurs independently of EXO1 (Fig. 2 G). In contrast, extensive resection during class switching in 53BP1−/− cells is ATM-independent (Yamane et al., 2013), and is mediated by both CtIP and EXO1 (Bothmer et al., 2013). Similarly, unrepaired G1-phase breaks during V(D)J recombination are processed by CtIP (Helmink et al., 2011). These results suggest that, whereas CtIP promotes nucleolytic processing of DSBs in all cell cycle phases, other factors that stimulate or antagonize the extension of resected tracks may be distinct and regulated in a cell cycle–dependent manner.
MATERIALS AND METHODS
Generation of mice.
CtIPco/− (Bothmer et al., 2013), CtIP+/− (Chen et al., 2005), 53BP1−/− (Ward et al., 2004), BRCA1f(Δ11)/f(Δ11) (here reported BRCA1Δ11f/f; NCI mouse repository), Ku80−/− (Nussenzweig et al., 1996), CD19Cre (Rickert et al., 1997), EXO1−/− (Wei et al., 2003), and ROSA26-STOP-EYFP (Srinivas et al., 2001) mice have been described. BAC RP11-104H10 containing human CtIP genomic sequence was used to generate transgenic mice. S327A, T847A, and T847E mutations were targeted using BAC recombineering as previously described (Difilippantonio et al., 2005). Transgenic CtIP was detected by PCR screening of tail DNA using the following primer pairs: hEx1F 5′-AGCACAACCACACTGAATGC-3′ and hEx1R 5′-CACCACAGGTATTCTCACACGG-3′, which amplify a product of 305 bp in the intronic sequence specific for the human gene. All experiments with mice were conducted in agreement with protocols approved by the National Institutes of Health Institutional Animal Care and Use Committee.
B cell culture, flow cytometry, metaphase analysis.
B cells were isolated from WT or mutant spleen with anti-CD43 Microbeads (anti-Ly48; Miltenyi Biotec) and were cultured with LPS (25 µg/ml; Sigma-Aldrich), IL-4 (5 ng/ml; Sigma-Aldrich), and RP105 (0.5 µg/ml; BD). Cell proliferation was analyzed by CFSE (5 µM; Molecular Probes) labeling at 37°C for 10 min. For cell cycle analysis, B cells were fixed in methanol and stained with propidium iodide (PI). BrdU incorporation and detection was performed as described (Bunting et al., 2012). Samples were acquired on a FACSCalibur (BD). For genomic instability analysis, B cells were harvested after 2 d in culture; metaphase spreads were generated and processed for FISH analysis as previously described (Callen et al., 2013). PARP inhibitor (KU58948; Astra Zeneca) was added to cells stimulated ex vivo for one day.
Generation of CtIP retroviral clones.
Coding sequences of the mouse CtIP was cloned into PMX-IRES-GFP plasmid (CtIPWT). A FLAG-tag was cloned by PCR at the N terminus of CtIP coding sequence (FLAG-CtIPWT). Mutant FLAG-CtIPT847E and FLAG-CtIPT847A were generated by targeting Flag-CtIPWT using QuikChange II XL Site-Directed Mutagenesis kit (Agilent Technologies), and mutations were confirmed by sequencing. Infected MEFs were grown and selected in medium containing 2 µg/ml puromycin for 1 wk, and then maintained in 1 µg/ml puromycin.
Immunoblotting, immunofluorescence, and laser microirradiation.
Western blotting was performed with the following primary antibodies: mouse anti-tubulin (Sigma-Aldrich), mouse anti–FLAG-M2 (Sigma-Aldrich), mouse anti-CtIP (mAb 14–1; gift from R. Baer, Columbia University, New York, NY), rabbit anti-CtIP (developed in collaboration with Epitomics), rabbit anti-KAP-1 pS824 (Bethyl Laboratories), rabbit anti–p53 pS15 (Cell Signaling Technology), and rabbit anti-53BP1 (Novus). For RAD51 immunofluorescence, cells were irradiated at 10 Gy allowed to recover for 4 h, and then fixed and processed as previously described (Celeste et al., 2003). For microirradiation, cells were presensitized in DMEM media containing 0.1 µg/ml of Hoechst 33342 for 60 min before replacing with phenol red free media containing 5 mM Hepes, and then irradiated with the 364-nm laser line on a LSM510 confocal microscope (Carl Zeiss, Inc.) equipped with a heated stage. Cells were allowed to recover for the indicated time before processing for immunofluorescence (Celeste et al., 2003). Primary antibodies for immunofluorescence were rabbit anti-CtIP and anti-53BP1, mouse anti-FLAG-M2, mouse anti–γ-H2AX (Upstate Biotechnology), and rabbit anti-RAD51 (Santa Cruz Biotechnology, Inc.).
Acknowledgments
We thank Davide Robbiani for the CtIP shRNA construct, Junjie Chen for 53BP1−/− mice, Winfried Edelmann for EXO1−/− mice, and the members of Nussenzweig laboratory for helpful discussions.
The work was supported by the Intramural Research Program of the National Institutes of Health, the National Cancer Institute, and the Center for Cancer Research and by a Department of Defense grant to A. Nussenzweig (BC102335).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- BAC
- bacterial artificial chromosome
- CSR
- class switch recombination
- DSB
- DNA double strand break
- HR
- homologous recombination
- IR
- ionizing radiation
- NHEJ
- nonhomologous end joining
- PARPi
- poly(ADP-ribose) polymerase inhibitor
- PI
- propidium iodide
- shCtIP
- CtIP shRNA
- shRNA
- short hairpin RNA
- ssDNA
- single-strand DNA
References
- Bothmer A., Rommel P.C., Gazumyan A., Polato F., Reczek C.R., Muellenbeck M.F., Schaetzlein S., Edelmann W., Chen P.L., Brosh R.M., Jr, et al. 2013. Mechanism of DNA resection during intrachromosomal recombination and immunoglobulin class switching. J. Exp. Med. 210:115–123 10.1084/jem.20121975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P., Aly A., Escandell J.M., Pieterse M., Bartkova J., van der Gulden H., Hiddingh S., Thanasoula M., Kulkarni A., Yang Q., et al. 2010. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 17:688–695 10.1038/nsmb.1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., Helleday T. 2005. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 434:913–917 10.1038/nature03443 [DOI] [PubMed] [Google Scholar]
- Bunting S.F., Nussenzweig A. 2013. End-joining, translocations and cancer. Nat. Rev. Cancer. 13:443–454 10.1038/nrc3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting S.F., Callén E., Wong N., Chen H.T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L., et al. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 141:243–254 10.1016/j.cell.2010.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting S.F., Callén E., Kozak M.L., Kim J.M., Wong N., López-Contreras A.J., Ludwig T., Baer R., Faryabi R.B., Malhowski A., et al. 2012. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol. Cell. 46:125–135 10.1016/j.molcel.2012.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E., Di Virgilio M., Kruhlak M.J., Nieto-Soler M., Wong N., Chen H.T., Faryabi R.B., Polato F., Santos M., Starnes L.M., et al. 2013. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell. 153:1266–1280 10.1016/j.cell.2013.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Xu X., Bunting S.F., Liu J., Wang R.H., Cao L.L., Wu J.J., Peng T.N., Chen J., Nussenzweig A., et al. 2009. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol. Cell. 35:534–541 10.1016/j.molcel.2009.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A., Fernandez-Capetillo O., Kruhlak M.J., Pilch D.R., Staudt D.W., Lee A., Bonner R.F., Bonner W.M., Nussenzweig A. 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5:675–679 10.1038/ncb1004 [DOI] [PubMed] [Google Scholar]
- Chandramouly G., Kwok A., Huang B., Willis N.A., Xie A., Scully R. 2013. BRCA1 and CtIP suppress long-tract gene conversion between sister chromatids. Nat Commun. 4:2404 10.1038/ncomms3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J.R., Sossick A.J., Boulton S.J., Jackson S.P. 2012. BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. J. Cell Sci. 125:3529–3534 10.1242/jcs.105353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J.R., Barral P., Vannier J.B., Borel V., Steger M., Tomas-Loba A., Sartori A.A., Adams I.R., Batista F.D., Boulton S.J. 2013. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell. 49:858–871 10.1016/j.molcel.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P.L., Liu F., Cai S., Lin X., Li A., Chen Y., Gu B., Lee E.Y., Lee W.H. 2005. Inactivation of CtIP leads to early embryonic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol. Cell. Biol. 25:3535–3542 10.1128/MCB.25.9.3535-3542.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Nievera C.J., Lee A.Y., Wu X. 2008. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J. Biol. Chem. 283:7713–7720 10.1074/jbc.M710245200 [DOI] [PubMed] [Google Scholar]
- Difilippantonio S., Celeste A., Fernandez-Capetillo O., Chen H.T., Reina San Martin B., Van Laethem F., Yang Y.P., Petukhova G.V., Eckhaus M., Feigenbaum L., et al. 2005. Role of Nbs1 in the activation of the Atm kinase revealed in humanized mouse models. Nat. Cell Biol. 7:675–685 10.1038/ncb1270 [DOI] [PubMed] [Google Scholar]
- Eid W., Steger M., El-Shemerly M., Ferretti L.P., Peña-Diaz J., König C., Valtorta E., Sartori A.A., Ferrari S. 2010. DNA end resection by CtIP and exonuclease 1 prevents genomic instability. EMBO Rep. 11:962–968 10.1038/embor.2010.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano-Díaz C., Orthwein A., Fradet-Turcotte A., Xing M., Young J.T., Tkáč J., Cook M.A., Rosebrock A.P., Munro M., Canny M.D., et al. 2013. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell. 49:872–883 10.1016/j.molcel.2013.01.001 [DOI] [PubMed] [Google Scholar]
- Farmer H., McCabe N., Lord C.J., Tutt A.N., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C., et al. 2005. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 434:917–921 10.1038/nature03445 [DOI] [PubMed] [Google Scholar]
- Feng L., Fong K.W., Wang J., Wang W., Chen J. 2013. RIF1 counteracts BRCA1-mediated end resection during DNA repair. J. Biol. Chem. 288:11135–11143 10.1074/jbc.M113.457440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti L.P., Lafranchi L., Sartori A.A. 2013. Controlling DNA-end resection: a new task for CDKs. Front Genet. 4:99 10.3389/fgene.2013.00099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmink B.A., Tubbs A.T., Dorsett Y., Bednarski J.J., Walker L.M., Feng Z., Sharma G.G., McKinnon P.J., Zhang J., Bassing C.H., Sleckman B.P. 2011. H2AX prevents CtIP-mediated DNA end resection and aberrant repair in G1-phase lymphocytes. Nature. 469:245–249 10.1038/nature09585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P., Jackson S.P. 2009. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 284:9558–9565 10.1074/jbc.M808906200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S.P., Bartek J. 2009. The DNA-damage response in human biology and disease. Nature. 461:1071–1078 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langerak P., Mejia-Ramirez E., Limbo O., Russell P. 2011. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 7:e1002271 10.1371/journal.pgen.1002271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.L., Greenberg R.A. 2012. Links between genome integrity and BRCA1 tumor suppression. Trends Biochem. Sci. 37:418–424 10.1016/j.tibs.2012.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan M.E., Chiu J.W., Koller B.H., Jasin M. 1999. Brca1 controls homology-directed DNA repair. Mol. Cell. 4:511–518 10.1016/S1097-2765(00)80202-6 [DOI] [PubMed] [Google Scholar]
- Nakamura K., Kogame T., Oshiumi H., Shinohara A., Sumitomo Y., Agama K., Pommier Y., Tsutsui K.M., Tsutsui K., Hartsuiker E., et al. 2010. Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genet. 6:e1000828 10.1371/journal.pgen.1000828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig A., Chen C., da Costa Soares V., Sanchez M., Sokol K., Nussenzweig M.C., Li G.C. 1996. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature. 382:551–555 10.1038/382551a0 [DOI] [PubMed] [Google Scholar]
- Peterson S.E., Li Y., Chait B.T., Gottesman M.E., Baer R., Gautier J. 2011. Cdk1 uncouples CtIP-dependent resection and Rad51 filament formation during M-phase double-strand break repair. J. Cell Biol. 194:705–720 10.1083/jcb.201103103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson S.E., Li Y., Wu-Baer F., Chait B.T., Baer R., Yan H., Gottesman M.E., Gautier J. 2013. Activation of DSB processing requires phosphorylation of CtIP by ATR. Mol. Cell. 49:657–667 10.1016/j.molcel.2012.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczek C.R., Szabolcs M., Stark J.M., Ludwig T., Baer R. 2013. The interaction between CtIP and BRCA1 is not essential for resection-mediated DNA repair or tumor suppression. J. Cell Biol. 201:693–707 10.1083/jcb.201302145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickert R.C., Roes J., Rajewsky K. 1997. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 25:1317–1318 10.1093/nar/25.6.1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P. 2007. Human CtIP promotes DNA end resection. Nature. 450:509–514 10.1038/nature06337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakya R., Reid L.J., Reczek C.R., Cole F., Egli D., Lin C.S., deRooij D.G., Hirsch S., Ravi K., Hicks J.B., et al. 2011. BRCA1 tumor suppression depends on BRCT phosphoprotein binding, but not its E3 ligase activity. Science. 334:525–528 10.1126/science.1209909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver D.P., Livingston D.M. 2012. Mechanisms of BRCA1 tumor suppression. Cancer Discov. 2:679–684 10.1158/2159-8290.CD-12-0221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S., Watanabe T., Lin C.S., William C.M., Tanabe Y., Jessell T.M., Costantini F. 2001. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1:4 10.1186/1471-213X-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark J.M., Pierce A.J., Oh J., Pastink A., Jasin M. 2004. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol. Cell. Biol. 24:9305–9316 10.1128/MCB.24.21.9305-9316.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L.S., Gautier J. 2011. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45:247–271 10.1146/annurev-genet-110410-132435 [DOI] [PubMed] [Google Scholar]
- Ward I.M., Reina-San-Martin B., Olaru A., Minn K., Tamada K., Lau J.S., Cascalho M., Chen L., Nussenzweig A., Livak F., et al. 2004. 53BP1 is required for class switch recombination. J. Cell Biol. 165:459–464 10.1083/jcb.200403021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K., Clark A.B., Wong E., Kane M.F., Mazur D.J., Parris T., Kolas N.K., Russell R., Hou H., Jr, Kneitz B., et al. 2003. Inactivation of exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 17:603–614 10.1101/gad.1060603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A.K., Ormonde P.A., Pero R., Chen Y., Lian L., Salada G., Berry S., Lawrence Q., Dayananth P., Ha P., et al. 1998. Characterization of a carboxy-terminal BRCA1 interacting protein. Oncogene. 17:2279–2285 10.1038/sj.onc.1202150 [DOI] [PubMed] [Google Scholar]
- Yamane A., Robbiani D.F., Resch W., Bothmer A., Nakahashi H., Oliveira T., Rommel P.C., Brown E.J., Nussenzweig A., Nussenzweig M.C., Casellas R. 2013. RPA accumulation during class switch recombination represents 5′-3′ DNA-end resection during the S-G2/M phase of the cell cycle. Cell Rep. 3:138–147 10.1016/j.celrep.2012.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z., Shi L.Z., Zhu Q., Wu P., Zhang Y.W., Basilio A., Tonnu N., Verma I.M., Berns M.W., Hunter T. 2009. CtIP links DNA double-strand break sensing to resection. Mol. Cell. 36:954–969 10.1016/j.molcel.2009.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Chen J. 2004. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell. Biol. 24:9478–9486 10.1128/MCB.24.21.9478-9486.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Wu L.C., Bowcock A.M., Aronheim A., Baer R. 1998. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J. Biol. Chem. 273:25388–25392 10.1074/jbc.273.39.25388 [DOI] [PubMed] [Google Scholar]
- Yun M.H., Hiom K. 2009. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 459:460–463 10.1038/nature07955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M., Lottersberger F., Buonomo S.B., Sfeir A., de Lange T. 2013. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science. 339:700–704 10.1126/science.1231573 [DOI] [PMC free article] [PubMed] [Google Scholar]