TNF signaling inactivation sensitizes AML cells to NF-kB inhibition but protects healthy hematopoietic stem progenitor cells from this treatment.
Abstract
Leukemic stem cells (LSCs) isolated from acute myeloid leukemia (AML) patients are more sensitive to nuclear factor κB (NF-κB) inhibition-induced cell death when compared with hematopoietic stem and progenitor cells (HSPCs) in in vitro culture. However, inadequate anti-leukemic activity of NF-κB inhibition in vivo suggests the presence of additional survival/proliferative signals that can compensate for NF-κB inhibition. AML subtypes M3, M4, and M5 cells produce endogenous tumor necrosis factor α (TNF). Although stimulating HSPC with TNF promotes necroptosis and apoptosis, similar treatment with AML cells (leukemic cells, LCs) results in an increase in survival and proliferation. We determined that TNF stimulation drives the JNK–AP1 pathway in a manner parallel to NF-κB, leading to the up-regulation of anti-apoptotic genes in LC. We found that we can significantly sensitize LC to NF-κB inhibitor treatment by blocking the TNF–JNK–AP1 signaling pathway. Our data suggest that co-inhibition of both TNF–JNK–AP1 and NF-κB signals may provide a more comprehensive treatment paradigm for AML patients with TNF-expressing LC.
NF-κB is a major mediator of immunity, inflammation, tissue regeneration, and cancer promotion signaling. It regulates multiple cell behaviors such as proliferation, survival, differentiation, and migration (Naugler and Karin, 2008; DiDonato et al., 2012; Perkins, 2012). Leukemic cells (LCs), including leukemic stem cells (LSCs), demonstrate increased NF-κB activity, which provides a critical survival signal (Kuo et al., 2013). In vitro studies demonstrated that NF-κB inhibition can largely eliminate LSC with minimal effects on normal hematopoietic stem and progenitor cells (HSPCs), suggesting the potential for NF-κB inhibition as an anti-leukemia therapy (Guzman et al., 2001). However, the use of NF-κB inhibitors alone in vivo does not effectively eliminate the acute myeloid leukemia (AML) cells, indicating that additional survival signals might be compensating for the effects of NF-κB inhibition. In addition, the clinical use of NF-κB inhibitors is also limited by potential side effects, including compromised T/B cell immunity, inflammatory tissue damage, and skin/liver cancer development (Chen et al., 2001; Zhang et al., 2004, 2007; Maeda et al., 2005; Sakurai et al., 2006; Luedde et al., 2007; Bettermann et al., 2010; Ke et al., 2010).
TNF, a pro-inflammatory cytokine, has been shown to be a key mediator of inflammatory reactions in tumor tissues and is responsible for elevated NF-κB activity in many tumors. NF-κB levels are significantly increased in most tumor tissues, being produced by tumor-infiltrating hematopoietic cells, tumor cells, and/or tumor stromal cells (Anderson et al., 2004; Balkwill, 2006; Mantovani et al., 2008; Grivennikov and Karin, 2011; Ren et al., 2012). Animal model studies demonstrate that TNF plays an essential role in the pathogenesis of many types of cancer such as skin, liver, and colon cancers by directly stimulating tumor cell proliferation/survival or by inducing a tumor-promoting environment (Moore et al., 1999; Knight et al., 2000; Sethi et al., 2008; Balkwill, 2009; Oguma et al., 2010). Also, supportive care for some cancers includes inhibition of TNF signaling through use of soluble receptors and neutralizing antibodies (Egberts et al., 2008; Popivanova et al., 2008).
Elevated serum TNF levels have been identified in patients with BM failure, including aplastic anemia and myelodysplastic syndromes (MDSs), suggesting that the hematopoietic-repressive activity of TNF might contribute to the cytopenic phenotype of such patients (Molnár et al., 2000; Dybedal et al., 2001; Dufour et al., 2003; Lv et al., 2007). The observed increased levels of TNF during disease progression in MDS patients imply that TNF might also be involved in the leukemic transformation of mutant HSPC (Tsimberidou and Giles, 2002; Stifter et al., 2005; Li et al., 2007; Fleischman et al., 2011). Increased levels of TNF are detected in the peripheral blood (PB) and BM of most human leukemia patients and are correlated to higher white blood cell counts and poorer prognosis (Tsimberidou et al., 2008). In fact, the importance of TNF in leukemogenesis is further documented in Fancc knockout mice and Bcr-Abl–transduced chronic myelogenous leukemia (CML) animal models. In these animals, TNF is required for inducing the leukemic transformation of Fancc mutant cells and promotes the proliferation of CML stem cells (Gallipoli et al., 2013).
TNF can stimulate both survival and death signals within the same type of cells in a context-dependent fashion. TNF-dependent survival signals are mediated primarily by canonical NF-κB signaling (Sakurai et al., 2003; Skaug et al., 2009; Vallabhapurapu and Karin, 2009), whereas the TNF-induced death signal is driven by caspase-8–dependent apoptosis or RIP1/3-dependent necroptosis (Wang et al., 2008; He et al., 2009; Zhang et al., 2009, 2011; Feoktistova et al., 2011; Günther et al., 2011; Kaiser et al., 2011; Oberst et al., 2011; Tenev et al., 2011; Xiao et al., 2011). In addition, TNF can also stimulate the activation of MKK4/7-JNK signaling, although the role of the MKK4/7-JNK signaling pathway is also cell context–dependent (Liu and Lin, 2005; Bode and Dong, 2007; Kim et al., 2007; Chen, 2012). Many studies suggest that TNF-induced MKK4/7-JNK signaling is responsible for most of the side effects associated with NF-κB signal inactivation (Chen et al., 2001; Zhang et al., 2004, 2007; Maeda et al., 2005; Sakurai et al., 2006; Luedde et al., 2007; Ke et al., 2010).
The role of MKK4/7-JNK signaling in the regulation of hematopoiesis is not entirely clear. Sustained JNK activation has been reported in many types of AML cells, coordinating with AKT/FOXO signaling to maintain an undifferentiated state (Sykes et al., 2011). In Bcr/Abl-induced CML, JNK1-AP1 signaling is required for the development of leukemia by mediating key survival signals (Hess et al., 2002). In Fanconi anemia, JNK is required for the TNF-induced leukemic clonal evolution of Fancc mutant HSPC (Li et al., 2007). These studies suggest that the JNK signal promotes the development and progression of leukemia by inducing proliferative and survival activities (Chen et al., 2001; Zhang et al., 2004, 2007; Maeda et al., 2005; Sakurai et al., 2006; Luedde et al., 2007; Bettermann et al., 2010; Ke et al., 2010).
In this study, we searched for the survival signals which compensate for the inhibition of NF-κB signaling in AML stem and progenitor cells. We found that TNF stimulates JNK and NF-κB, which act as parallel survival signals in LC, whereas TNF acts through JNK to induce a death signal in HSPC. Inhibition of TNF-JNK signaling not only significantly sensitizes AML stem and progenitor cells to NF-κB inhibitor treatment but also protects HSPC from the toxicity of such treatment.
RESULTS
Many types of AML cells express and produce TNF
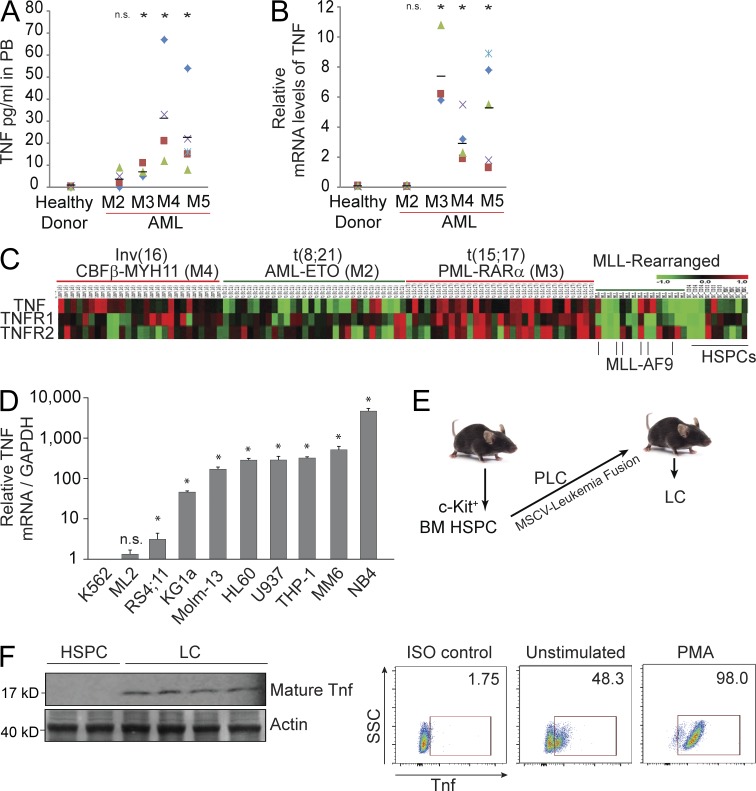
We found that TNF levels are significantly higher in the PB of AML patients having subtypes M3, M4, and M5 as compared with healthy donors (Fig. 1 A). To study whether TNF is produced directly by LC, we first examined LC isolated from these patients for TNF transcription and found that TNF mRNA levels correlate with TNF protein levels in PB (Fig. 1 B). We then examined TNF expression by cDNA array in 106 AML patients with four different leukemic fusions. When compared with hematopoietic cells (CD34+ HSPC, CD33+ myeloid progenitors), many types of LC, especially AML subtypes t(15:17) M3, inv(16) M4, and MLL-AF9 (MA9) M5 cells, express higher levels of TNF (Fig. 1 C). We also found elevated TNF expression in many established human AML cell lines (Fig. 1 D). Furthermore, in our murine AML model, MA9-LC generated from transplanted HSPC expressing the MA9 fusion gene in an MSCV retrovirus (pre-LC [PLC]; Fig. 1 E) produces Tnf, as determined by Western blotting and intracellular staining assays (Fig. 1 F). These data suggest that a significant portion of circulating TNF in many AML patients may be produced directly by tumor cells.
Figure 1.
Many types of AML cells produce TNF. (A) TNF concentrations in PB of AML patients as shown by ELISA from individual patient samples. (B) TNF expression in LC isolated from individual AML patient samples as shown by qRT-PCR assay. Horizontal bars in A and B indicate the mean. (C) Microarray analysis of TNF, TNFR1, and R2 in AML patient samples. t(8:21), t(15:17), and inv(16) are AML subtypes M2, M3, and M4, respectively. MLL leukemias include MLL-AF4 (B-ALL) and MLL-AF9 (AML-M5). CD34+ HSPCs, CD33+ myeloid progenitors, and/or MNC from healthy donors were used as controls. (D) Relative mRNA levels of TNF produced in established human AML cell lines. Values shown are mean ± 1 SD. (E) To generate WT and Tnfr−/− murine LCs, HSPCs isolated from mice were transduced with MSCV containing leukemia fusions creating PLC. PLCs were transplanted into recipient mice. Spleens were harvested for LCs after leukemia developed. (F) Murine MA9-LCs produce TNF as shown by Western blot assay. Each lane represents LC from an individual mouse. TNF protein levels were examined in murine MA9-LC with or without PMA (phorbol 12-myristate 13-acetate) stimulation by intracellular antibody staining and read by flow cytometry. Results shown are representative of three individual experiments. * indicates P < 0.05 compared with healthy donor or other control as determined by Student’s t test two-tailed analysis (A, B, and D).
Tnf promotes the expansion of clonogenic LC but represses the growth of normal HSPC
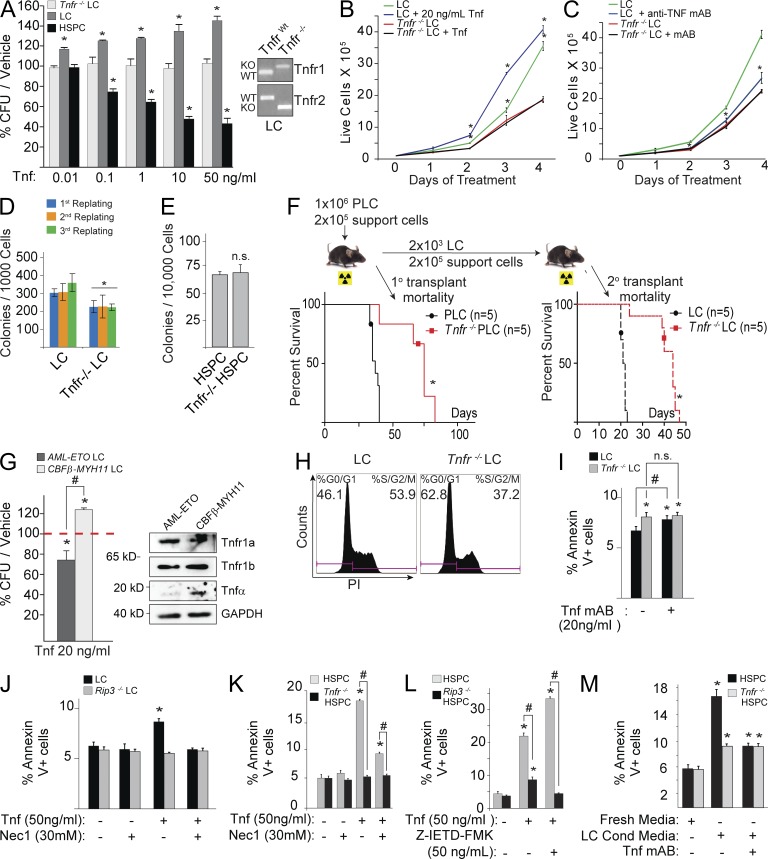
To investigate the role of Tnf in LC and normal HSPC, we generated MA9-transduced Tnfr−/− (Tnf receptors 1 and 2 knockout) and TnfrWT murine LC by infecting HSPC isolated from Tnfr−/− and TnfrWT mice, respectively, with MA9-expressing virus (Fig. 1 E). We found that exogenous Tnf promotes LC growth in methylcellulose colony-forming assays and liquid culture (Fig. 2, A and B) but significantly represses the growth of normal HSPC colonies (Fig. 2 A). Inactivation of Tnf signaling by either Tnfr knockout or a neutralizing monoclonal antibody restricts LC growth but has no obvious effect on normal HSPC (Fig. 2 C-E). Furthermore, transplantation studies demonstrated that Tnfr−/− LC-transplanted mice required a longer latency for leukemia development in vivo than TnfrWT LC (Fig. 2 F). A similar response to exogenous Tnf was also observed in CBFβ-MYH11–transduced murine LC (Tnf-expressing, M4) versus AML-ETO–transduced murine LC (Tnf-nonexpressing, M2), suggesting that such effects are not MA9-LC specific (Fig. 2 G). All these suggest that in these Tnf-producing leukemias, Tnf might function as an enhancer for leukemia development and a repressor for normal hematopoiesis. For simplicity, WT HSPC will be described as HSPC, and murine WT LC will be described as LC. Other genetic deletions, such as Tnfr−/− LC, will be specified.
Figure 2.
Tnf promotes LC growth but represses HSPC growth in vitro. (A) CFU from MA9-LC and HSPC treated with indicated doses of Tnf. The numbers of CFU of each type of cell in Tnf-treated groups were normalized to the CFU number in vehicle-treated group. Genotype results are shown for LC and Tnfr−/− LC. (B and C) LC and Tnfr−/− LC were treated with 20 ng/ml Tnf (B) or 20 µg/ml anti-Tnf mAb (C) in suspension culture. Growth curves of LC were examined by counting the number of live cells daily by trypan blue exclusion. (D) Serial colony-forming ability was compared between LC and Tnfr−/− LC. (E) CFU/20,000 cells shown for Tnfr−/− HSPC and HSPC. (F) Leukemogenic capacity of LC and Tnfr−/− LC was compared by in vivo transplantation. (G) CFUs from AML-ETO and CBFβ-MYH11–transduced murine LCs were treated with Tnf. Expression of Tnf and its receptors are shown by Western blot analysis. (H) Cell cycle was compared between LC and Tnfr−/− LC. (I) Tnfr−/− LC and LC were cultured overnight in the presence of anti-Tnf mAb. Cell death was measured by Annexin V/PI assay. Significance is measured relative to vehicle control in LC, unless otherwise noted. (J) LCs were treated with Tnf with or without Nec-1 overnight, and cell death was measured by Annexin V/PI. Rip3−/− LCs were used as necroptosis-negative controls. (K) HSPC and Tnfr−/− HSPC were cultured in medium with or without exogenous Tnf. Nec1 was used to block necroptosis. (L) HSPCs from WT (Rip3 WT) and Rip3−/− mice were cultured with or without Tnf. Z-IETD-FMK was used to inhibit Caspase 8 activity. Cell death was examined 24 h after Tnf treatment. (M) Tnf secreted by MA9-LC is sufficient to induce cell death in HSPC. WT and Tnfr−/− HSPCs were incubated in fresh medium or 1:5 conditioned/fresh medium from LC culture and treated with or without anti-Tnf mAb. * indicates P < 0.05 when compared with vehicle treated control (A–C and G–M) or WT control (D–F) as determined by Student’s t test two-tailed analysis. # indicates P < 0.05 significant difference when compared with indicated conditions. Values shown are mean ± SD analysis from three independent trials, unless otherwise notated. Western blot in G and readout for H are representative results from three independent trials.
To study the mechanism by which Tnf promotes the growth of LC, we compared the proliferation and survival of Tnfr−/− LC and LC. We found that the proliferation of Tnfr−/− LC is significantly reduced when compared with LC (Fig. 2 H). In addition, we also consistently observed slightly but significantly more Tnfr−/− LC undergoing apoptosis compared with LC in our 4 cytokine culture condition. Apoptosis in LC can be enhanced by culturing in the presence of Tnf-neutralizing antibody (Fig. 2 I). We propose that these effects, although significant, are minimal because Tnf is not the only survival signal for LC. There was a slight but significant increase in cell death in LC upon treatment with high concentrations of exogenous Tnf (50 ng/ml). We believed that such cell death is due to necroptosis because it could be prevented by the Rip1 inhibitor Necrostatin-1 (Nec1) or by Rip3 deletion (Fig. 2 J). This concentration of Tnf still stimulated colony formation by LC (Fig. 2 A), suggesting that Tnf might selectively induce necroptosis in partially differentiated, nonclonogenic LC while promoting the proliferation/survival of clonogenic LC.
Consistent with our previous study, we found that Tnf treatment of normal HSPC kills a portion of cells through inducing RIP1/3-mediated necroptosis, which can be prevented by Nec1treatment (Fig. 2 K) or by Rip3 deletion (Fig. 2 L; Xiao et al., 2011). Finally, we found that Tnf produced by LC is sufficient to induce death in HSPC (Fig. 2 M), suggesting that Tnf produced by LC might contribute to the repressed hematopoiesis observed in leukemia patients. These data suggest that, in contrast to its function in HSPC, Tnf drives a critical survival and proliferative signal in undifferentiated clonogenic LC.
Tnf signal inactivation sensitizes LC to NF-κB inhibition–induced death but protects HSPC from such death
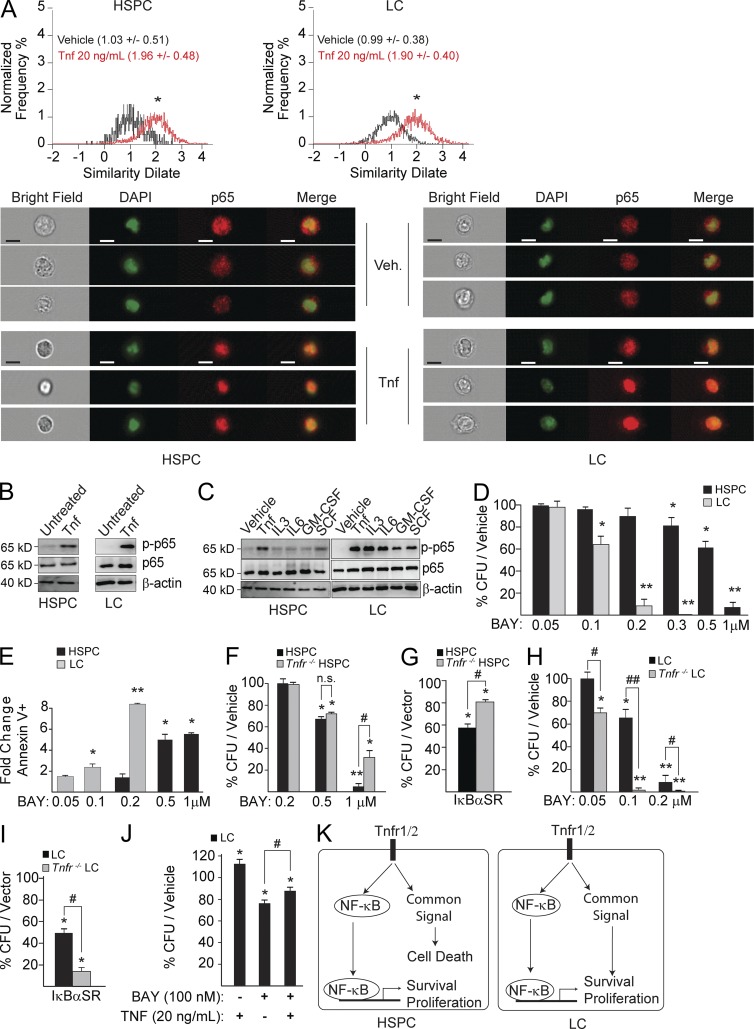
Previous reports showed that NF-κB activity is significantly increased in LC and LSC (Guzman et al., 2001). We confirmed such elevated NF-κB activity in our murine LC compared with HSPC (unpublished data). In addition, we found that NF-κB activity in LC is cytokine-dependent. p65 (NF-κB) activity was turned off when LCs were incubated in a cytokine-free medium and was restored upon Tnf stimulation as measured by both nuclear localization (Fig. 3 A) and p65 phosphorylation (Fig. 3 B). We also found that all hematopoietic cytokines used in our cultures induced NF-κB activity in LC while only Tnf stimulated NF-κB in HSPC, suggesting how NF-κB activity can be elevated in subtypes of LC which do not express TNF (Fig. 3 C). Consistent with a previous report (Guzman et al., 2001), our murine LCs are more sensitive to NF-κB inhibition by BAY11-7085 (a reversible small molecule inhibitor of IκBα phosphorylation, BAY hereafter) in vitro as shown by a reduction in CFU (Fig. 3 D) and an increase in cell death (Fig. 3 E) when compared with HSPC.
Figure 3.
Tnf signal inactivation sensitizes LC to NF-κB inhibitor treatment but protects healthy HSPC from such treatment. (A and B) Tnf stimulation of NF-κB activity in both LC and HSPC as shown by p65 (NF-κB1) nuclear localization (values in parenthesis are mean similarity dilate peak, sample size = 5,000 cells) as determined by ImageStreamX in A or by p65 phosphorylation in B. Bars, 10 µM. (C) HSPC and LC stimulated with individual cytokines show different patterns of p65 phosphorylation. (D and E) HSPC and LC were treated in parallel with indicated doses of BAY11-7085 (BAY) for CFU assay in D or analyzed for cell death in E. (F) HSPC and Tnfr−/− HSPC were treated with increasing doses of BAY and plated for CFU assay. (G) HSPC and Tnfr−/− HSPC were transduced with IκBαSR-GFP. GFP+ cells were sorted and plated for CFU assay. The number of CFUs in each genotype of HSPC was normalized to vector-only-transduced corresponding genotype control. (H) LC and Tnfr−/− LC were treated with increasing doses of BAY and plated for CFU assay. (I) LC and Tnfr−/− LC were transduced with IκBαSR-GFP. GFP+ cells were sorted and plated for CFU assay. The number of CFUs in each genotype of HSPC or LC was normalized to vector-only-transduced corresponding genotype control. (J) LCs were co-treated with BAY and TNF. CFUs were measured 1 wk after treatment. (K) Experiment model. All values shown are mean ± SD normalized to vehicle-treated control, performed on three independent trials. * (P < 0.05) and ** (P < 0.01) indicate significant reduction compared with corresponding vehicle groups or vector-only groups as determined by Student’s t test two-tailed analysis. # indicates P < 0.05 and ## indicates P < 0.01 significant difference when compared with indicated conditions.
Most tissue cells with NF-κB inactivation show sensitivity to Tnf-induced cell death. To study the effects of Tnf signal inactivation on NF-κB inhibition in HSPC and LC, we treated Tnfr−/− LC and LC as well as Tnfr−/− HSPC and HSPC with either BAY or retroviral transduction using a mutant IκBα (IκBαSR: a nonphosphorylatable serine-alanine mutant form of IκBα). We found that NF-κB inhibition-induced cell death by both BAY and IκBαSR is largely protected in Tnfr−/− HSPC compared with HSPC (Fig. 3, F and G). However, both BAY and IκBαSR further significantly decreased the in vitro clonogenic growth of Tnfr−/− LC when compared with LC (Fig. 3 H, I). The addition of exogenous Tnf can also enhance the colony formation of LC, even when the NF-κB signal is inhibited (Fig. 3 J). These data suggest that, as in most other normal tissue cells, NF-κB is required to protect HSPC from Tnf-induced death. However, in LC, the synergistic killing effects of TNF signal inactivation and NF-κB signaling inhibition suggests that Tnf stimulates survival/proliferation signaling in an NF-κB–independent manner. We speculate that Tnf-stimulated death in HSPC and survival in LC might be mediated by the same signaling pathway (Fig. 3 K).
JNK mediates a TNF-stimulated survival signal in LC but a death signal in HSPC
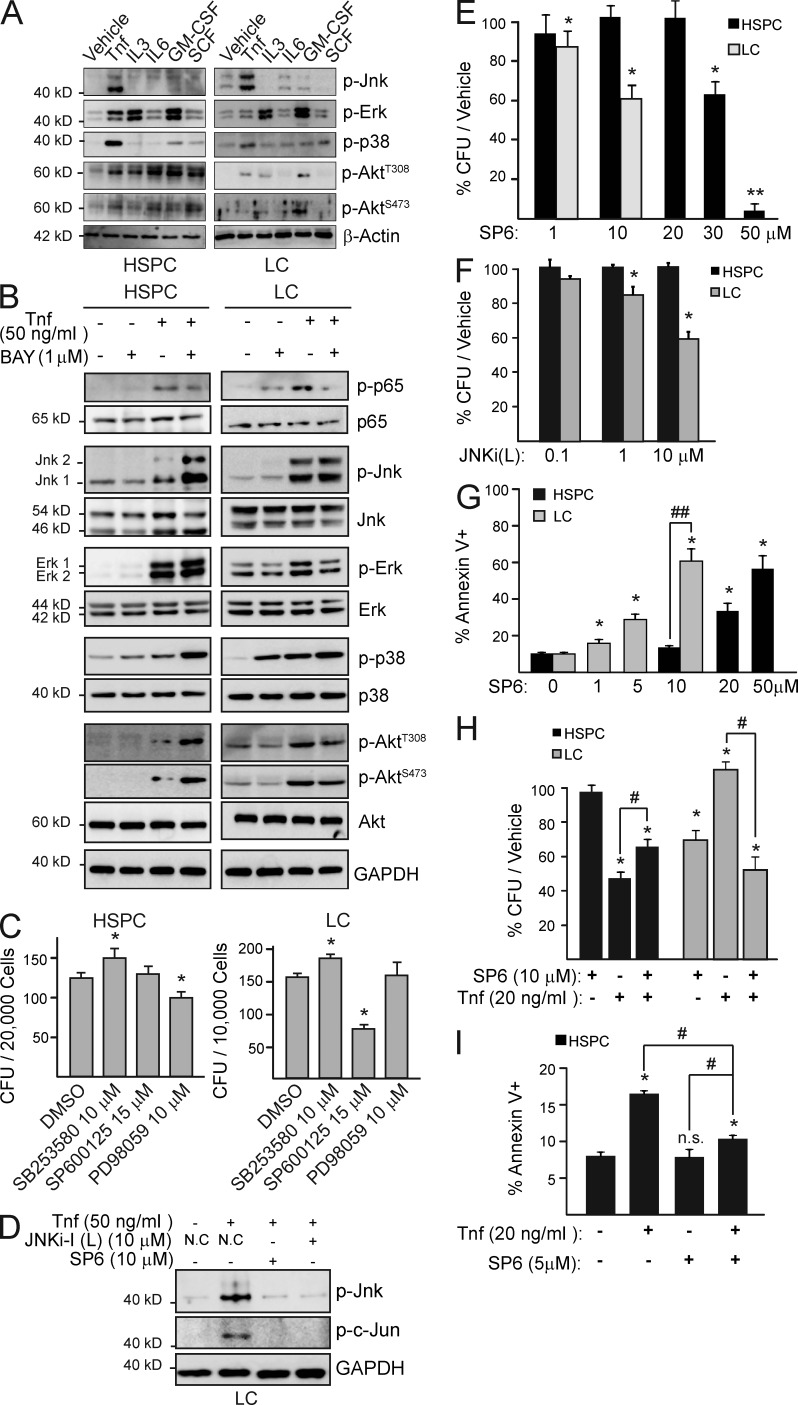
To search for the signaling pathway which mediates the contradictory effects of Tnf stimulation in LC and HSPC, we compared the activities of Jnk1/2, Erk1/2, p38, and Akt, all known Tnf-stimulated signaling mediators, between LC and HSPC after stimulation with either Tnf or the individual hematopoietic cytokines used in our cultures. We found that although all of these signaling mediators can be activated by Tnf and some other cytokines in both LC and HSPC, Jnk is only activated by Tnf but not any of the other cytokines (Fig. 4 A).
Figure 4.
Jnk functions as a Tnf-induced, NF-κB–independent survival signal in LC, while being a death signal in HSPC. (A) HSPC and LC were treated with individual hematopoietic cytokines for 15 min. Jnk, Erk, p38, and Akt activities were analyzed by Western blot. (B) HSPC and LC were pretreated with BAY before stimulation with TNF. p65 and Jnk activities were analyzed by Western blot. (C) HSPC and LC were treated with indicated doses of p38 inhibitor SB253580, Jnk inhibitor SP600125, and Erk inhibitor PD98059 for 12 h and then seeded for CFU assay. DMSO treatment was used as a negative control. (D) LCs pretreated with JNKi-I (L) or SP600125 were analyzed for Jnk and c-Jun activity after 15 min of Tnf stimulation. (E) HSPC and LC were treated with increasing concentrations of SP6 overnight, and then seeded for CFU assay. Numbers of CFU were normalized to the corresponding vehicle-treated group. (F) HSPC and LC were treated in parallel with indicated doses of JNKi-1 (L) overnight, and then plated in methylcellulose. Numbers of CFU were normalized to the corresponding vehicle-treated group. (G) HSPC and LC were treated in suspension culture with increasing concentrations of SP6 overnight. Cell death was analyzed by Annexin-V/7AAD staining. (H) LC and HSPC were treated with Jnk inhibitor SP6 and Tnf individually or in combination overnight, and then seeded for CFU assay. Numbers of CFU were normalized to the corresponding vehicle-treated group. (I) HSPCs were treated overnight with indicated doses of Tnf and SP6. Cell death is shown as Annexin V+ cells. Values shown are mean ± SD normalized to vehicle-treated control from three independent trials. * (P < 0.05) and ** (P < 0.01) indicate significant difference when compared with vehicle-treated control in C, E, F, and H as determined by Student’s t test two-tailed analysis. # indicates P < 0.05 and ## indicates P < 0.01 significant difference when compared with indicated conditions. A, B, and D are representative results of three independent trials.
To study which of these signals accounts for Tnf-induced death in HSPC and NF-κB–independent survival/proliferation in LC, we looked for the signals that are activated by Tnf independently of NF-κB. Jnk and p38 activities were sustained during NF-κB inhibition in both LC and HSPC, suggesting independence from NF-κB signaling (Fig. 4 B). Akt and Erk signals were significantly repressed by NF-κB inhibition in LC but were unchanged or enhanced after BAY treatment in HSPC (Fig. 4 B). To study whether the p38 or the Jnk signal mediates Tnf-induced cell death in HSPC and survival/proliferation in LC, we examined whether their inhibition could prevent Tnf-induced death in HSPC and repress the growth of LC. We found that p38 inhibition promotes the colonial growth of both HSPC and LC (Fig. 4 C). Jnk inhibition by treatment with either SP600125 (an inhibitor targeted to both JNK1 and 2, SP6 hereafter) or JNKi-1(L) (a small peptide of the JNK-binding domain) represses the clonogenic capacity of LC in a dosage-dependent manner but has fewer effects on HSPC. Treatment of LC with 10 µM SP6 and JNKi-1(L) was able to completely block Jnk phosphorylation (Fig. 4 D) and was also able to repress the clonogenic ability of LC (Fig. 4, E and F) and significantly induce cell death with no effects on HSPC (Fig. 4 G). The significant increase in cell death and reduction of clonogenic ability in HSPC were only observed when SP6 was used at >20 µM, suggesting an off-target effect of SP6 at high concentration (Fig. 4, E and G). Blocking Jnk by SP6 partially prevents Tnf-induced cell death and rescues CFU reduction in HSPC but represses Tnf-stimulated LC colony growth as shown by CFU assay and Annexin V staining (Fig. 4, H and I), suggesting that Jnk mediates the Tnf-mediated death signal in HSPC but a survival/proliferation signal in LC.
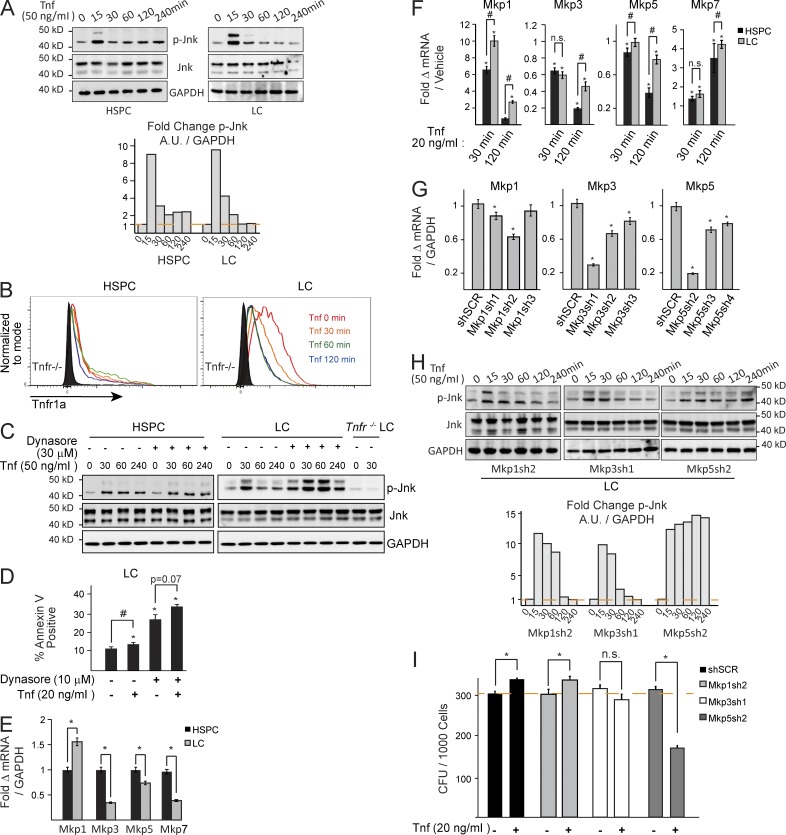
Endocytosis of Tnf receptors and negative regulation of Jnk by Mkp5 determines the response of LC and HSPC to Tnf stimulation by restricting Jnk signal duration
Studies show that the contradictory functions of the Jnk signal stimulated by Tnf are in part determined by the duration of Jnk activity. A short duration (<2 h) promotes survival/proliferation, whereas longer duration (>2 h) promotes cell death (Sakon et al., 2003; Liu and Lin, 2005). We found that Jnk signal duration is limited in LC compared with HSPC in response to TNF stimulation (Fig. 5 A). We predicted that extending the duration of Tnf–induced Jnk activation might convert the survival signal into death signal in LC.
Figure 5.
Tnf receptor endocytosis and Mkps regulate Jnk activity in LC and HSPC. (A) HSPC and LC were stimulated with TNF for the indicated times (in minutes). Jnk activity was examined by Western blot and the normalized ratios of p-Jnk levels were quantified in bar graph to the right. Arbitrary units are normalized to vehicle-treated lane. (B) LCs endocytose Tnfr1 in response to Tnf stimulation as determined by mean fluorescence intensity by flow cytometry. (C) HSPC and LC were pretreated with Dynasore followed by Tnf for the indicated times. Jnk activity was examined by Western blot. (D) LCs were treated with Tnf and/or Dynasore overnight, and assayed the next day for cell death by Annexin V/PI analysis. (E) Basal expression of Mkps was compared between HSPC and LC by qRT-PCR. (F) HSPC and LC were treated with Tnf for the indicated times. Expression of Jnk-specific Mkps were examined by qRT-PCR. Transcript levels were normalized to the corresponding basal levels. (G) shRNA knockdown of Mkps 1, 3, and 5 in LC was evaluated by qRT-PCR. (H) LCs were transduced with shRNA-specific Mkp1, Mkp3, or Mkp5, respectively. The transduced cells were treated with the indicated dosages of Tnf, and Jnk activity was examined by Western blotting of p-Jnk. Quantification of p-Jnk Western blot analysis is shown to the right. Arbitrary units are normalized to vehicle treated lane. (I) The CFU of Mkp1, Mkp3, and Mkp5 shRNA-transduced LCs were examined after Tnf treatment. Results shown are mean values ± 1 SD from three independent trials. * indicates P < 0.05 when compared with vehicle-treated/shSCR control as determined by Student’s t test two-tailed analysis. # indicates P < 0.05 significant difference when compared with the indicated conditions. A, B, C, and H are representative results from three independent trials.
It was known that Tnf-induced Jnk signaling can be turned off by endocytosis of Tnf receptor and/or MAP kinase phosphatase (Mkps)–mediated negative regulation (Kamata et al., 2005). We found that LCs endocytose Tnf receptors in response to Tnf stimulation (Fig. 5 B). Blocking endocytosis of Tnf receptors by Dynasore, a dynamin-specific inhibitor, slightly enhances Jnk signal strength and can also slightly prolong the Jnk signal in LC (Fig. 5 C). Although repression of Tnf receptor endocytosis by Dynasore partially enhances cell death in LC, treatment with additional Tnf does not significantly increase cell death (Fig. 5 D). Because blocking Tnf receptor endocytosis cannot completely prolong the Jnk signal, we next studied the role of Mkps in restricting the Tnf-stimulated Jnk signal in LC.
To investigate which of the Mkps’s restrict Tnf-mediated Jnk signaling in LC, we compared the basal levels of Mkps 1, 3, 5, and 7 (Jnk-specific Mkps) between LC and HSPC (Fig. 5 E) and their expression after Tnf stimulation (Fig. 5 F). We determined that LCs express higher levels of Mkp1 but lower levels of Mkps 3, 5, and 7 than do HSPC (Fig. 5 E). Tnf stimulation induces a significant increase in Mkp 1 and 7 expression in both LC and HSPC; however, Mkp7 levels are comparable between LC and HSPC. Although Mkp3 and Mkp5 did not increase upon Tnf induction, they were sustained in LC but reduced in HSPC 2 h after Tnf treatment (Fig. 5 F). To determine which Mkp was responsible for negatively regulating the Jnk signal, we knocked down Mkps 1, 3, and 5 using retroviral-mediated shRNA transduction (Fig. 5 G). We found that although Mkp1 and 5 knockdown can increase the duration of Jnk activity (Fig. 5 H), only Mkp5 knockdown results in both the complete prevention of Jnk dephosphorylation and the reversal of Tnf-stimulated colony formation (Fig. 5 I). This study suggested that sustained Mkp5 might be responsible for limiting the duration of Tnf-stimulated Jnk signaling in LC.
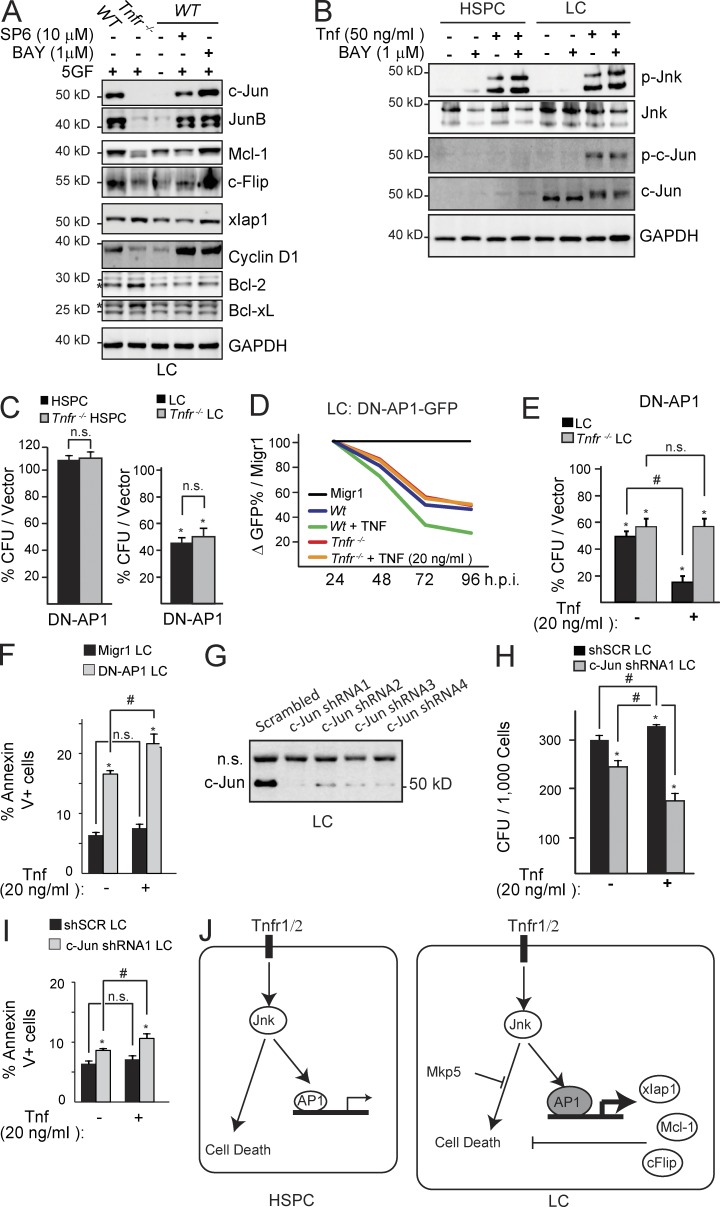
Tnf promotes LC growth via the Jnk–Ap1 signaling pathway, up-regulating survival genes
To determine whether Tnf–Jnk stimulates survival/proliferation in LC by inducing survival/proliferation-related gene expression, we cytokine-starved LC to reset the internal signaling to an unstimulated basal rate and then pretreated the cells with either SP6 or BAY to block Jnk or NF-κB activity, respectively. After SP6 or BAY treatment, we stimulated the cells with Tnf plus four hematopoietic cytokines (five total growth factors, 5GF) to examine the expression of which survival/proliferation-related genes are regulated through Jnk. We found that c-Jun, JunB, c-Flip, Mcl1, and cyclin D1 are down-regulated in Tnfr−/− LC compared with TnfrWT LC, suggesting that these are Tnf target genes. Among them, cytokine-induced c-Jun, Mcl-1, and c-Flip expression were repressed by Jnk inhibitor treatment but not by NF-κB inhibitor, suggesting they are Tnf-Jnk–specific targets. We found that xIap1 was not reduced in Tnfr−/− LC but was reduced by the addition of SP6, suggesting that xIap1 is a Jnk-specific, but Tnf-independent, survival gene. JunB and Cyclin D1 were unaffected by inhibitors of either Jnk or NF-κB. The elevated levels of Bcl-2 and Bcl-xL in Tnfr−/− LC compared with TnfrWT LC likely indicate the presence of a feedback mechanism due to the lack of Mcl-1, c-Flip, and xIap1. (Fig. 6 A).
Figure 6.
c-Jun mediates the Tnf-Jnk survival signal in LC. (A) LC and Tnfr−/− LC, grown in medium containing 10% FBS, were pretreated with BAY or SP6 before addition of growth factors and Tnf. Anti-apoptotic and proliferation-related signals were examined by Western blotting. (B) HSPC and LC were pretreated with BAY followed by Tnf stimulation. p-c-Jun and c-Jun expression were examined by Western blotting. (C) HSPC and Tnfr−/− HSPC as well as LC and Tnfr−/− LC were transduced with DN-AP1 and sorted for CFU assay. (D and E) LC and Tnfr−/− LC were transduced with DN-AP1-GFP and were observed in suspension culture for reduction of GFP+ cell percentage (D) or sorted for CFU with or without 20 ng/ml Tnf treatment (E). (F) DN-AP1–transduced LCs were treated with Tnf or vehicle and assayed for cell death. (G) c-Jun knockdown by specific shRNA was verified by Western blotting. (H) c-Jun shRNA-transduced HSPC and LC were treated with TNF or vehicle and plated in methylcellulose for CFU assay. (I) c-Jun knockdown in LC enhances cell death when treated with Tnf. (J) Experiment model, a mechanism by which Jnk signal promotes survival of LC. Results shown are mean values ± 1 SD from three independent trials. * indicates P < 0.05 significant difference when compared with vehicle in E, F, H, and I, vector-only (Migr1) in C as determined by Student’s t test two-tailed analysis. # indicates P < 0.05 significant difference when compared with indicated conditions. A, B, D, and G are representative results from three independent trials.
The proliferative and survival activities of the Jnk signal are primarily mediated by its nuclear protein substrates. One of the key nuclear substrates of Jnk is c-Jun, which has been identified as a major tumor-promoting gene in many types of cancer (Shaulian, 2010). We found that LCs express higher levels of c-Jun compared with HSPCs, and that Tnf-induced Jnk/c-Jun activation is NF-κB independent (Fig. 6 B). Genetic repression of c-Jun by a dominant-negative AP1 (DN-AP1) has no discernible effects on HSPC but does result in a significant decrease in CFU in LC (Fig. 6 C). Overexpressing DN-AP1 resensitizes LC to Tnf-induced death signaling, as demonstrated by a dynamic reduction in GFP percentage over time with Tnf treatment in suspension culture (Fig. 6 D), a reduction in CFU in the presence of Tnf (Fig. 6 E), and increased cell death (Fig. 6 F). We confirmed the necessity of c-Jun to prevent Tnf-mediated cell death in LC by knocking down c-Jun with a specific shRNA (Fig. 6 G). We found that, similar to overexpression of DN-AP1, c-Jun knockdown resulted in a restored sensitivity of LC to Tnf-induced death signaling as demonstrated by a decrease in CFU (Fig. 6 H) and showed a moderate increase in cell death when stimulated with Tnf (Fig. 6 I). These data suggest that the Jnk signaling pathway is critical for Tnf-stimulated proliferation/survival activities in LC and that Jnk’s survival signal is primarily mediated through c-Jun/AP1 (Fig. 6 J). We proposed then that Tnf, through Jnk, promotes a survival signal that is auxiliary to NF-κB. Therefore, we tested the combined inhibition of both NF-κB and Jnk in LC and HSPC.
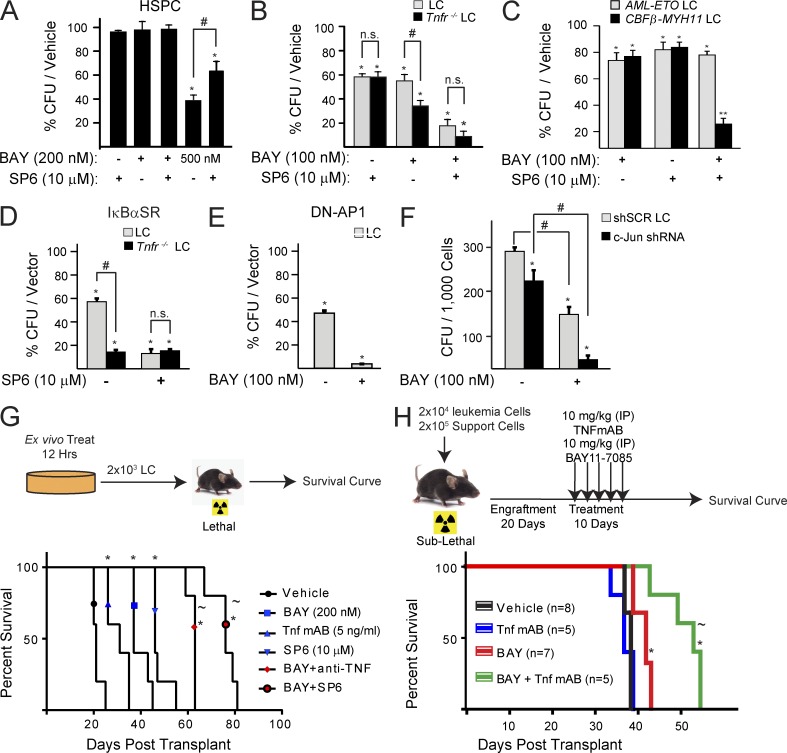
Jnk–AP1 signal inactivation sensitizes LC to NF-κB inhibition-induced cell death but protects HSPC from death
We observed that LCs lacking Tnf receptors are more sensitive to NF-κB inhibition than their WT counterparts (Fig. 3). We then found that Tnf stimulates Jnk as an NF-κB–independent survival signal in LC, but a death signal in HSPC (Fig. 4), through short Jnk signal duration (Fig. 5) and activation of c-Jun/AP1 in LC (Fig. 6). Therefore, we hypothesize that Jnk inhibition should sensitize LC to NF-κB inhibition while protecting HSPC from the effects of NF-κB inhibition. To investigate this, we treated LC and HSPC with BAY and SP6 individually or in combination. We found that SP6 treatment can partially prevent death and rescue the decrease in CFU of HSPC resulting from high concentrations of BAY (Fig. 7 A). However, in LC, addition of SP6 can sensitize LC to BAY-induced cell death at the same rate as in Tnfr−/− LC (Fig. 7 B). We found that inhibition of both signals eliminated more clonogenic LC than inhibiting either individual signal. Such additive inhibitory effects were also observed in the Tnf-expressing CBFβ-MYH11 LC but not in Tnf-nonexpressing AML-ETO LC (Fig. 7 C). To confirm our inhibitor specificity, we genetically inactivated NF-κB or Jnk–AP1 signaling by transducing LC with IκBαSR, DN-AP1, or c-Jun-shRNA. We found that genetic inactivation of NF-κB or Jnk–AP1/c-Jun was able to sensitize LC to SP6 or BAY, respectively, reproducing the conclusion from our inhibitor combination studies (Fig. 7, D–F).
Figure 7.
Jnk inhibition sensitizes LC to NF-κB inhibitor-induced cell death and partially protects HSPC. (A) CFU from HSPC treated with indicated doses of SP6 and BAY. (B) CFU from LC and Tnfr−/− LC treated with indicated doses of SP6 and BAY. (C) CFU from AML-ETO and CBFβ-MYH11-transduced LC treated with Bay, SP6, or combination. (D) LC and Tnfr−/− LC were transduced with IκBαSR-GFP. GFP+ cells were sorted and plated in methylcellulose with or without SP6 treatment. Values are normalized to vector-only-transduced control cells. (E) LCs were transduced with DN-AP1-GFP. GFP+ cells were sorted and plated in methylcellulose with or without BAY treatment. (F) LCs were transduced with c-Jun-shRNA1; GFP+ cells were sorted and plated in methylcellulose with or without BAY treatment. shSCR transduction was used as control. (G) LCs were treated with indicated inhibitors for 12 h in vitro. Surviving cells were transplanted into lethally irradiated recipient mice. Survival of the recipient mice was analyzed by Kaplan-Meier survival graphing. Leukemia was confirmed at the time of death for each transplant mouse. Five mice were used in each treatment group. (H) Sublethally irradiated mice were engrafted with LC and treated with BAY, TNF mAb, or in combination. Survival of the mice was measured over time and analyzed by Kaplan-Meier survival graphing. Leukemia was confirmed at the time of death for each transplanted mouse. Numbers of mice used are indicated in the panel. All values shown are mean values ± 1 SD from three independent trials. In A–F, * (P < 0.05) and ** (P < 0.01) indicate notated significance when compared with vehicle-treated control as determined by Student’s t test two-tailed analysis. In G and H, * indicates P < 0.05 when compared with vehicle treatment. # indicates P < 0.05 significant difference when compared with indicated conditions. ∼ indicates P < 0.05 in G and H when compared with BAY-treated LC/mice as determined by Log-Rank test.
Leukemia stem cells (LSCs) are a small population of LCs which can regenerate leukemia in recipient mice upon transplantation and are proposed to be the key type of cells responsible for cases of leukemia relapse. To study whether inhibition of Tnf–Jnk–AP1 and NF-κB can eliminate LSC, we treated LC in vitro for 12 h with anti-Tnf antibody, SP6, or BAY individually or in combinations, and then evaluated leukemogenicity by in vivo transplantation. Compared with vehicle-only treatment, anti-Tnf, SP6, or BAY treatment significantly delayed the development of leukemia. The combinations of anti-TNF or SP6 plus BAY further increased latency to the development of leukemia (Fig. 7 G). To test the in vivo suitability of a combination of treatments, we engrafted LC into sublethally irradiated recipient mice. After leukemia engraftment (20 d after transplantation), we treated the mice with BAY, anti-TNF antibody, SP6, or JNKi-I(L) individually or in combination. We found that BAY treatment can prolong the lifetimes of the mice. Although anti-TNF antibody treatment failed to extend the lifetime of the mice, it further prolonged the lifetime of the mice when combined with BAY (Fig. 7 H). However, we did not obtain data for co-inhibition of both NF-κB and JNK signaling due to the toxicity of SP6 and JNKi-I(L) in vivo. Nevertheless, we found that overexpression of DN-Ap1 can largely repress the leukemogenic capacity of LC (only 1/5 of recipient mice developed leukemia; unpublished data). These data suggest that both NF-κB and Jnk signals are required for the expansion of LSC in vitro, and that inhibition of both pathways provides better anti-leukemic effects in vivo.
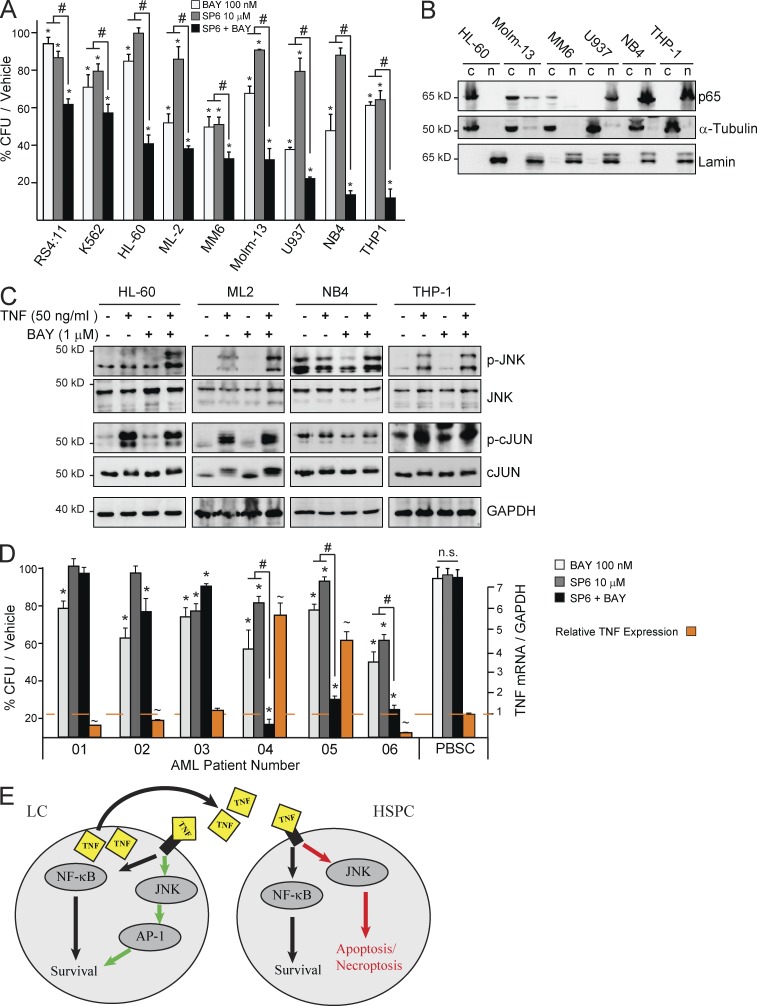
TNF-expressing human LCs are sensitive to a treatment combination of NF-κB and JNK signaling inhibitors
To study whether combination treatment using both NF-κB and JNK signaling inhibitors is also additive in eliminating human LC, we compared the responses of human AML cell lines to the treatments. By incubating the LC with NF-κB and JNK inhibitors individually or in combination for 12 h and then seeding for CFU to examine the reduction of clonogenic LC, we observed additive inhibitory effects of the two inhibitors in almost all AML cell lines. We noted that the sensitivity of AML cell lines to such additive inhibitory effects are correlated to levels of TNF expression (Fig. 1 D). Cell lines RS4:11 (B-ALL), K562 erythroid LC, and ML-2 that had a reduced response also express lower levels of TNF (Fig. 8 A), whereas the TNF-expressing cell lines, including HL-60 (M2/3), ML-2 (M5), Molm-13 (M5), U937 (M5), NB4 (M3), and THP-1 (M5), showed exceptional sensitivity to the combined inhibitor treatment. In addition, we found that, although all the sensitive cell lines showed TNF-stimulated NF-κB–independent JNK–c-JUN activation (Fig. 8 C), the most sensitive AML cell lines (U937, NB4, and THP1) had increased basal levels of NF-κB and JNK-c-JUN activities as shown by elevated nuclear p65 and total p-JNK/p-c-JUN (Fig. 8, B and C). We also confirmed the correlation of additive effects of our combination treatment to TNF expression in primary human LC isolated from newly diagnosed AML patients. TNF expression accurately predicted sensitivity to combined BAY and SP6 inhibitor treatment in each case. Interestingly, one AML patient sample was sensitive to combined inhibitor treatment but did not express TNF (Fig. 8 D). We are currently investigating the role of proinflammatory cytokines other than TNF in promoting NF-κB–independent survival signals in this AML patient sample. Collectively, our studies suggest that the combination treatment of NF-κB and JNK inhibitors might be a useful pharmacological strategy for AML patients whose LC produce TNF.
Figure 8.
Co-inhibition of NF-κB and JNK is synergistic in TNF-expressing human AML. (A) CFU from human LC lines treated with indicated doses of SP, BAY, or combination. (B) Nuclear fractionation analysis showing NF-κB activity in AML cell lines. (C) Several TNF-high–expressing AML cell lines were treated with TNF and BAY individually or in combination. Levels of p-JNK and p-cJUN were examined by Western blotting. (D) Primary AML cells were treated with indicated doses of BAY, SP, or combination, and then assessed for CFU. Right axis indicates relative TNF mRNA expression levels relative to average healthy donor PB stem cells (PBSCs; n = 3). mRNA transcripts were set to GAPDH as control. ∼ indicates increased TNF mRNA level compared with PB stem cell control, P < 0.05. * indicates P < 0.05 when compared with vehicle-treated control, unless otherwise noted, as determined by one-way ANOVA. # indicates P < 0.05 significant difference when compared with indicated conditions. Values shown are mean values ± 1 SD from three independent trials. B and C are representative results from three independent trials. (E) Proposed mechanism for TNF function in AML.
DISCUSSION
Increased NF-κB activity has been detected in a variety of types of human cancers including nearly all types of hematopoietic malignancies (Naugler and Karin, 2008; DiDonato et al., 2012; Perkins, 2012). However, mutations of the key regulators of NF-κB signaling have been detected only in some B cell lymphomas and multiple myeloma. In these B lymphocytic malignancies, abnormal activation of NF-κB signaling is the result of active mutations of upstream regulatory components which are less dependent on cytokine stimulation. Inhibition of NF-κB signaling in these malignancies has been demonstrated to be an effective treatment option, inducing disease remission and significant improvement in patient survival (Hernandez-Ilizaliturri et al., 2011; Lim et al., 2012). In AML, there is generally a lack of identified mutations in the components of NF-κB signaling, suggesting that enhanced NF-κB activity might be induced by microenvironmental factors. Our studies suggest that TNF, produced by LC, promotes growth of these cells through autocrine stimulation of NF-κB and JNK–AP1 as parallel proliferation/survival signals. Therefore, the anti-leukemic effects of NF-κB inhibition in AML in vivo are likely compensated by TNF-activating anti-apoptotic genes operating through JNK. Additionally, we showed that paracrine TNF released from LC represses the growth of normal HSPC, suggesting a link to the hematopoietic repression observed in AML patients. Therefore, we speculate that co-inhibition of both TNF-JNK and NF-κB signals might be a more comprehensive treatment for TNF-expressing AML by synergistically repressing the growth of LC and simultaneously protecting HSPC.
In many solid tumors, inflammation is one of the key components of the tumor environment which promotes tumor development and progression (Naugler and Karin, 2008; DiDonato et al., 2012; Perkins, 2012). In these tumors, TNF promotes tumor cell growth by directly stimulating proliferative/survival signals in tumor cells, inducing the transformation of BM-derived mesenchymal stem cells (MSCs) and myeloid progenitors to tumor-supportive cancer associated fibroblasts and macrophages, and/or inducing a tumor-promoting microenvironment by stimulating the production of tumor-promoting cytokines (Anderson et al., 2004; Balkwill, 2006; Mantovani et al., 2008; Grivennikov and Karin, 2011; Ren et al., 2012). In addition, TNF also causes the inflammation-related damage observed in noncancerous surrounding tumors (Anderson et al., 2004; Balkwill, 2006; Mantovani et al., 2008; Grivennikov and Karin, 2011; Ren et al., 2012). However, the role of inflammation in AML has not been well studied, despite the fact that AML cells develop from the progenitors of inflammatory cells, are normally surrounded by inflammatory cells and MSC, and produce inflammatory cytokines. We found that TNF is significantly increased in the BM and PB of AML patients, especially in the M3, M4, and M5 subtypes. LC in such patients may generate an inflammatory environment by secreting TNF. Our studies support the notion that TNF directly promotes the growth of tumor cells by stimulating proliferative/survival signals. Interestingly, despite the consistent observation of necroptotic cell death in a small proportion of LC upon exogenous TNF treatment, such treatment also increases clonogenic capacity of LC. Therefore, we speculate that TNF promotes the growth of LC primarily by inducing the expansion of the function of leukemic stem and progenitor cells but kills some of the partially differentiated LC. Whether TNF also induces BM MSC and stromal cells to generate a tumor-promoting microenvironment in AML patients requires further study.
In skin and liver tissues, TNF-induced tissue damage is primarily mediated by JNK signaling inducing apoptosis/necroptosis. Normally, TNF-induced damage in these tissues is prevented by NF-κB. NF-κB signal inactivation results in severe inflammation-related tissue damage due to the excessive sensitivity of signal-inactivated cells to TNF-JNK–induced death signaling (Chen et al., 2001; Zhang et al., 2004, 2007; Maeda et al., 2005; Sakurai et al., 2006; Luedde et al., 2007; Bettermann et al., 2010; Ke et al., 2010). The tumor-promoting effects of TNF in these tissues are also mediated by JNK signaling through a “complementary proliferation” and/or senescence-repressing mechanisms (Chen et al., 2001; Zhang et al., 2004, 2007; Maeda et al., 2005; Sakurai et al., 2006; Luedde et al., 2007; Bettermann et al., 2010; Ke et al., 2010). NF-κB functions as a tumor suppressor in these tissues by repressing TNF-JNK activity. Therefore, blocking the TNF-JNK signal can largely repress both tissue damage and tumor generation in NF-κB signal-inactivated animals. These studies provide a strong empirical basis for our combination treatment approach consisting of inhibition of both signals simultaneously. Doing so should not only enhance the anti-leukemic effects but may also reduce the side effects in other tissues usually induced by NF-κB inhibitors.
The two contradictory activities of TNF-JNK signaling, pro-death and pro-survival/proliferation, in normal/benign tissue cells and malignant cells have been reported in other systems. In Fanconi anemia, JNK is required for TNF-induced leukemic clonal evolution of Fancc mutant HSPC by inducing apoptosis in mutant cells and stimulating proliferation/survival activities in LC (Li et al., 2007). In Drosophila, TNF represses tumor growth of scribble (a tumor suppressor) mutant cells by stimulating a JNK-mediated death signal. However, via JNK, TNF promotes tumor progression and metastasis in scribble mutant cells when either oncoprotein RAS or Notch is expressed (Cordero et al., 2010). In skin epidermis, via the Tnfr1–Jnk2–Ap1 signaling pathway, TNF promotes epidermal cell proliferation by up-regulating the cell cycle positive regulator CDK4 and down-regulating cell cycle negative regulators such as P16Ink4a. Such signals which prevent growth restraint and the induction of senescence are required for epidermal neoplasia induced by NF-κB inactivation, chemicals, UV irradiation, or oncogenic Ras (Zhang et al., 2004, 2007; Ke et al., 2010). In hepatocytes, TNF-JNK induces apoptosis in large numbers of cells and stimulates compensatory proliferation in remaining cells, which are required for chemical or genetic lesion (such as Nemo deletion)–induced hepatocellular carcinoma (Maeda et al., 2005; Sakurai et al., 2006; Luedde et al., 2007; Bettermann et al., 2010). These studies suggest that a combination of NF-κB and JNK inhibitors might also be useful in the treatment of these cancers. However, in certain types of cancer cells, the pro-survival activities of JNK might be dependent on NF-κB signaling activity. In such cells, inactivation of the NF-κB signal will convert the pro-survival activity of JNK signaling to pro-death activity (Papa et al., 2004). Therefore, to best use such combination treatment in cancer therapy, it is important to distinguish whether or not the pro-survival/proliferative activity of JNK is NF-κB signal dependent.
Evidence also suggests that whether a cell undergoes death or proliferation/survival fate in response to TNF–JNK signaling is determined by the duration of JNK activity. Elongated JNK activity (>2 h) induces cell death (as shown in HSPC), whereas limited JNK activity (<2 h) promotes cell proliferation/survival (as shown in LC). We found that the duration of JNK in murine LC is primarily limited by MPK5 which is independent of NF-κB signal. Thus, experiments to determine whether expression of MKP5 is elevated in primary human LC will be necessary in future studies. Such studies will allow us to evaluate if inactivation of MKP5 could be a treatment for AML patients.
MATERIALS AND METHODS
Mice and genotyping.
All experiments using mice were performed according to the guidelines of Loyola University Medical Center and were approved by the Loyola University Institutional Animal Care and Use Committee. Tnfr1−/−2−/− mice (B6.129S-Tnfrsf1aTnfrsf1b) were purchased from The Jackson Laboratory. BM hematopoietic cells from Rip3−/− (Rip3-knockout) mice were provided by J. Zhang (Thomas Jefferson University, Philadelphia, PA). Genotypes of all mice were determined by PCR assay using the following primers (5′–3′): Tnfr1-1, GGATTGTCACGGTGCCGTTGAAG (WT = 120 bp); Tnfr1-2, TGACAAGGACACGGTGTGTGGC (Mut = 155 bp); Tnfr1-3, TGCTGATGGGGATACATCCATC; Tnfr1-4, CCGGTGGATGTGGAATGTGTG; Tnfr2-1, CCGGTGGATGTGGAATGTGTG (WT = 257 bp); Tnfr2-2, AGAGCTCCAGGCACAAGGGC (Mut = 160 bp); Tnfr2-3, AACGGGCCAGACCTCGGGT; Rip3-1, GGCTTTCATTGTGGAGGTAAGCTGAGA (WT = 280 bp); and Rip3-2, GAACCCGTGGATAAGTGCACTTGAAT (Floxed = 320 bp).
Generation of murine leukemia cell lines.
CD117+ HSPCs were isolated from WT, Tnfr−/− (knockout of both Tnfr 1 and 2), and Rip3−/− mice and infected with MA9-neoexpressing retrovirus. Infected cells were selected with G418 for 2 wk in four-cytokine medium (10 ng/ml IL-3, 25 ng/ml IL6, 100 ng/ml SCF, and 20 ng/ml GM-SCF) to generate MA9-immortalized cells (PLC in the text) with different genetic mutations. Such PLCs were transplanted into lethally irradiated recipient mice to generate leukemic mice. WT, Tnfr−/−, and Rip3−/− LCs isolated from spleens and BM of the corresponding leukemic mice were used in our studies.
To generate NF-κB signal-inactivated MA9-LC, WT-LC, or Tnfr−/−, LCs generated from the above experiments were infected with IκBαSR-GFP–expressing virus and selected by FACS; these were used in our inhibitor treatment studies. IκBαSR is a mutant form of IκBα with S32A/S36A substitutions. The protein product of IκBαSR is stable and has more effective NF-κB inhibitory ability than does WT IκBα. To generate AP1-repressed MA9-LCs, WT-LCs generated from the above experiments were infected with DN-AP1-GFP–expressing virus. Infected LCs were purified by FACS and subsequently used in our studies.
Reagents.
Rip-1 inhibitor Necrostatin (Nec1) was purchased from Santa Cruz Biotechnology, Inc. Recombinant murine-IL-3 (rm-IL-3), rm-IL-6, rm-SCF, and rm-GM-CSF were purchased from eBioscience. TNF was purchased from BD. BAY11-7085 and SP600125 small molecule inhibitors were purchased from Millipore. LY294002 was purchased from LC Laboratories. JNKi I peptide and negative control were obtained from EMD Millipore. Cell lysis buffer (10×) was obtained from Cell Signaling Technology and supplemented with proteinase inhibitors and phosphatase inhibitors (Roche). Anti-TNF monoclonal antibody used for blocking TNF signaling in culture was obtained from Amgen Inc. Anti–β-actin, GAPDH, Bcl-2, and Bcl-xL antibodies were obtained from Santa Cruz Biotechnology, Inc. Anti-IκBα, p-IκBα, P65, p-P65, Jnk, p-Jnk, Erk, p-Erk, p38, p-p38, Akt, p-AktS473, p-AktT308, Mcl-1, xIAP1, and Cyclin D primary and requisite secondary antibodies were also obtained from Cell Signaling Technology. c-FLIP antibody was obtained from Assay Gate. Tri-reagent used for RNA extraction was purchased from Sigma-Aldrich. TNF antibody was purchased from Novus Biologicals.
Cell culture.
All cells were incubated at 37°C, 100% humidity, and 5% CO2. c-Kit+ HSPC from indicated genotypes of mice were enriched by EasySep mouse CD117 Positive Selection kit (STEMCELL Technologies). HSPCs were cultured in 6-well plates in RPMI-1640 medium supplemented with 10% FBS, 1% penicillin/streptomycin, 100 ng/ml rmSCF, 50 ng/ml rmIL-6, 20 ng/ml rmIL-3, and 20 ng/ml rmGM-CSF. All murine LCs generated in this study were cultured in such medium. Human LC lines were cultured according to instructions from American Type Culture Collection.
Cell counting.
Cell number was determined by trypan blue exclusion using 0.4% trypan blue (Gibco). At each 24-h interval, all cells were harvested from the plate, centrifuged, resuspended in 1 ml of complete medium, and a sample was taken for counting. All cell counting was performed using trypan blue exclusion as visualized in a hemocytometer under 40× magnification. After counting, cells were resuspended in a total of 3 ml of medium and replated. This was repeated every 24 h for four repetitions.
Colony-forming unit assay.
Indicated genotypes of murine LC or BM cells were seeded into MethoCult GF M3434 (STEMCELL Technologies) at 1,000 cells/ml (LC) or 20,000 cells/ml (BM HSPC), incubated at 37°C, 100% humidity, and 5% CO2 for 7 d (LC) or 10 d (HSPC cells). Numbers of colonies were counted according to the manufacturer’s instructions. Triplicate experiments were performed in all of our studies. All data were verified by three individual experiments.
Human AML cell lines were seeded into MethoCult base medium without cytokines at 1,000 cells/ml and incubated and read as murine LC. Primary AML patient samples were seeded into MethoCult 4035 Optimum without EPO and incubated at 37°C, 100% humidity, and 5% CO2. Colonies were read 14 d after seeding.
Cell death assay.
HSPCs or LCs were incubated in medium with or without inhibitors for 24 h. Cells were then stained with alloyphycocyanin-conjugated Annexin V in Annexin binding buffer according to the manufacturer’s instructions (BD). Cell death was determined by the percentages of Annexin V+ cells.
Detection of nuclear localization of NF-κB by Amnis ImageStreamX.
Cells were fixed and permeabilized using the Fix/Perm kit (BD) as described by the manufacturer. Intracellular staining was performed using p65 antibodies (Cell Signaling Technology), Alexa Fluor 647 secondary (eBioscience) and DAPI, and was visualized using Amnis Imagestream X. Nuclear localization of p65 was determined using a similarity dilate algorithm determined by ImageStream IDEAS software (EMD Millipore) and analyzed per 5,000 cells. Significant differences were determined using mean and SD values provided by ImageStream Ideas and compared using Student’s t test.
Retroviral infection.
High titer retrovirus was produced by co-transfecting Phoenix cells with a retroviral vector containing the indicated genes together with packaging vectors using the Calphos Mammalian Transfection kit (Takara Bio Inc.). Retroviral supernatants were harvested 24 and 48 h after transfection. Retroviral plasmids MSCV-GFP and MSCV-Cre-GFP were provided by C. Niu (University of Southern California, Los Angeles, CA), MSCV-MLL-AF9-neo was provided by N. Zeleznik-Le (Loyola University Chicago, Chicago, IL.), and MSCV-IκBαaa-GFP was generated by subcloning the IκBαaa gene from pBabe-IκBαaa-puro plasmids (provided by M. Denning, Loyola University Chicago, Chicago, IL) into the MSCV-IRES-GFP vector. pMieg-DN-AP1 was obtained through Addgene. Viruses were generated using these retroviral vectors. LCs or c-kit+ HSPCs were transduced with such virus-expressing genes of interest by spinoculation at 32°C., 2,000 rpm for 4 h. Cells were then incubated using standard culture conditions as described above, and GFP percentage of representative samples were read by flow cytometry every 24 h. This assay allowed us to evaluate the functions of genes on cell growth with both internal control (GFP− nontransduced cells) and external control (MSCV-GFP infection). Transduced cells were also purified by FACS for CFU assay.
Western blot analysis.
106 LCs/HSPCs were plated in 6-well plates for 5 h in RPMI-1640 medium supplemented with 10% FBS. FBS was present at all times during experiments and did not show any effect on the measured signals. Growth factors were removed during this phase to reset the cell signaling to an unstimulated, basal rate. After 5 h of growth factor withdrawal, inhibitors were added if required for 1.5 h. After inhibition, cells were stimulated if required for 10 min and removed from the plates and then lysed using cell lysis buffer (Cell Signaling Technology) supplemented with protease and phosphatase inhibitor cocktails (Roche). Contents were resolved on 10% SDS-PAGE and visualized after transfer to nitrocellulose membranes.
Ex vivo transplantation.
104 LCs (CD45.2+) were plated in each well in a suspension culture and treated with indicated doses of BAY11-7085, SP600125, and anti-TNF antibody (Amgen) in indicated combinations for 12 h. All cells in each well were harvested and mixed with 106 support BM cells (CD45.1+) and then equally transplanted into five lethally irradiated recipient mice (CD45.1+). Mice were monitored for leukemia development by observing for symptoms: hunched body, significant weight loss, or hind-limb paralysis. Leukemia was confirmed by examining CD45.2+ LC in PB, spleen, and BM, as well as liver and kidney infiltration.
In vivo transplantation and treatment.
2 × 104 LCs were transplanted into sublethally irradiated C57BL6/J mice with 2 × 105 support BM cells via tail vein injection. 20 d after transplantation, mice were treated with 10 mg/kg InVivomAb anti–m-TNF (Bio X Cell) and 10 mg/kg BAY11-7085 individually or in combinations every other day for 10 d. Mice were monitored for leukemia development by lethargy, paralysis, significant weight loss, and/or enlarged abdomen. Leukemia was verified after the mice were sacrificed by examining infiltration of LC in livers, lungs, and spleens.
shRNA knockdown.
LCs were transduced with retrovirus-expressing shRNAs (OriGene) specifically targeted to c-Jun (TG501139), or Mkps (1:TG514083; 3:TG514782; 5:TG513100). The transduced cells were selected for 1 wk using puromycin to obtain stably transduced cells. Knockdown efficiency was examined by Western blot (c-Jun) or RT-PCR (Mkps). Scrambled shRNAs were transduced and studied in parallel as controls.
Primary human samples.
PB samples from AML patients were obtained from the clinic at Loyola University Medical Center in accordance with the IRB protocol. Leukemic blasts in PB of all patients were 30–90% when samples were collected. Samples were processed for mononuclear cells (MNCs) by Ficoll-Paque gradient centrifugation. A portion of MNC was used for RNA extraction and TNF expression analysis; another fraction of MNC was plated in StemSpan serum-free medium (STEMCELL Technologies) supplemented with recombinant human SCF (100 ng/ml), Flt-3L (100 ng/ml), TPO (20 ng/ml), IL-6 (20 ng/ml), and IL-3 (20 ng/ml). Cytokines were obtained from Humanzyme. After overnight culture, 3 × 105 cells from each sample were harvested and treated with the indicated doses of BAY11-7085, SP600125, or TNF, and plated in methylcellulose (STEMCELL Technologies) for CFU assay. Colonies were read after 14 d. In addition, serum was collected from the same patients for examination of TNF and other cytokines.
Statistical analysis.
Significant differences were determined by Student’s t test unless otherwise noted.
Acknowledgments
The authors would like to dedicate this work to the memory of Bernard J. Enders (1929–2012).
IκBα SR construct was provided by Dr. Mitchell Denning, Loyola University Chicago, Division of Health Sciences. The authors also thank the staff of the Department of Comparative Medicine at Loyola University Medical Center for excellent animal care services, and Drs. Manuel Diaz, Nancy Zeleznik-Le, and Andrew Dingwall for ongoing professional collaboration and scientific suggestions and discussions that improved the present studies.
This work was supported by the US Department of Defense (grant PR080447 through Loyola University Chicago), the National Natural Science Foundation of China (project 81071774), a grant from the Science and Technology Commission of Shanghai Municipality (10540503400), National Basic Research Program of China (project 2013CB966800), and the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning through Shanghai Normal University. P. Breslin is supported in part by the Jimmy Burns Foundation. A. Volk was supported by an Experimental Immunology T32 Training grant (administered through Loyola University Chicago Department of Microbiology and Immunology) and is currently supported with an F31 pre-doctoral fellowship from the National Cancer Institute (F31CA174147-01).
The authors declare no competing financial interests.
Author contributions: A. Volk, J. Li, J.P. Xin, D. You, X.L. Liu, Y.C. Xiao, P. Breslin, Z.J. Li, W. Wei, R. Schmidt, X.Y. Li, Jun Zhang, Z. Zhang, S. Nand, J.K. Zhang, and J.J. Chen performed the research and analyzed the data. A. Volk and Jiwang Zhang designed and performed the research and analyzed the data. A. Volk, P. Breslin, and Jiwang Zhang wrote the manuscript.
Footnotes
Abbreviations used:
- AML
- acute myeloid leukemia
- CML
- chronic myelogenous leukemia
- HSPC
- hematopoietic stem and progenitor cell
- LC
- leukemic cell
- LSC
- leukemic stem cell
- Mkps
- MAP kinase phosphatase
- MNC
- mononuclear cell
- MSC
- mesenchymal stem cell
- PB
- peripheral blood
- PLC
- pre-LC
References
- Anderson G.M., Nakada M.T., DeWitte M. 2004. Tumor necrosis factor-α in the pathogenesis and treatment of cancer. Curr. Opin. Pharmacol. 4:314–320 10.1016/j.coph.2004.04.004 [DOI] [PubMed] [Google Scholar]
- Balkwill F. 2006. TNF-α in promotion and progression of cancer. Cancer Metastasis Rev. 25:409–416 10.1007/s10555-006-9005-3 [DOI] [PubMed] [Google Scholar]
- Balkwill F. 2009. Tumour necrosis factor and cancer. Nat. Rev. Cancer. 9:361–371 10.1038/nrc2628 [DOI] [PubMed] [Google Scholar]
- Bettermann K., Vucur M., Haybaeck J., Koppe C., Janssen J., Heymann F., Weber A., Weiskirchen R., Liedtke C., Gassler N., et al. 2010. TAK1 suppresses a NEMO-dependent but NF-κB-independent pathway to liver cancer. Cancer Cell. 17:481–496 10.1016/j.ccr.2010.03.021 [DOI] [PubMed] [Google Scholar]
- Bode A.M., Dong Z. 2007. The functional contrariety of JNK. Mol. Carcinog. 46:591–598 10.1002/mc.20348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F. 2012. JNK-induced apoptosis, compensatory growth, and cancer stem cells. Cancer Res. 72:379–386 10.1158/0008-5472.CAN-11-1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Nomura M., She Q.B., Ma W.Y., Bode A.M., Wang L., Flavell R.A., Dong Z. 2001. Suppression of skin tumorigenesis in c-Jun NH(2)-terminal kinase-2-deficient mice. Cancer Res. 61:3908–3912 [PubMed] [Google Scholar]
- Cordero J.B., Macagno J.P., Stefanatos R.K., Strathdee K.E., Cagan R.L., Vidal M. 2010. Oncogenic Ras diverts a host TNF tumor suppressor activity into tumor promoter. Dev. Cell. 18:999–1011 10.1016/j.devcel.2010.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDonato J.A., Mercurio F., Karin M. 2012. NF-κB and the link between inflammation and cancer. Immunol. Rev. 246:379–400 10.1111/j.1600-065X.2012.01099.x [DOI] [PubMed] [Google Scholar]
- Dufour C., Corcione A., Svahn J., Haupt R., Poggi V., Béka’ssy A.N., Scimè R., Pistorio A., Pistoia V. 2003. TNF-α and IFN-γ are overexpressed in the bone marrow of Fanconi anemia patients and TNF-α suppresses erythropoiesis in vitro. Blood. 102:2053–2059 10.1182/blood-2003-01-0114 [DOI] [PubMed] [Google Scholar]
- Dybedal I., Bryder D., Fossum A., Rusten L.S., Jacobsen S.E. 2001. Tumor necrosis factor (TNF)–mediated activation of the p55 TNF receptor negatively regulates maintenance of cycling reconstituting human hematopoietic stem cells. Blood. 98:1782–1791 10.1182/blood.V98.6.1782 [DOI] [PubMed] [Google Scholar]
- Egberts J.H., Cloosters V., Noack A., Schniewind B., Thon L., Klose S., Kettler B., von Forstner C., Kneitz C., Tepel J., et al. 2008. Anti–tumor necrosis factor therapy inhibits pancreatic tumor growth and metastasis. Cancer Res. 68:1443–1450 10.1158/0008-5472.CAN-07-5704 [DOI] [PubMed] [Google Scholar]
- Feoktistova M., Geserick P., Kellert B., Dimitrova D.P., Langlais C., Hupe M., Cain K., MacFarlane M., Häcker G., Leverkus M. 2011. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol. Cell. 43:449–463 10.1016/j.molcel.2011.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischman A.G., Aichberger K.J., Luty S.B., Bumm T.G., Petersen C.L., Doratotaj S., Vasudevan K.B., LaTocha D.H., Yang F., Press R.D., et al. 2011. TNFα facilitates clonal expansion of JAK2V617F positive cells in myeloproliferative neoplasms. Blood. 118:6392–6398 10.1182/blood-2011-04-348144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallipoli P., Pellicano F., Morrison H., Laidlaw K., Allan E.K., Bhatia R., Copland M., Jørgensen H.G., Holyoake T.L. 2013. Autocrine TNF-α production supports CML stem and progenitor cell survival and enhances their proliferation. Blood. 122:3335–3339 10.1182/blood-2013-02-485607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S.I., Karin M. 2011. Inflammatory cytokines in cancer: tumour necrosis factor and interleukin 6 take the stage. Ann. Rheum. Dis. 70:i104–i108 10.1136/ard.2010.140145 [DOI] [PubMed] [Google Scholar]
- Günther C., Martini E., Wittkopf N., Amann K., Weigmann B., Neumann H., Waldner M.J., Hedrick S.M., Tenzer S., Neurath M.F., Becker C. 2011. Caspase-8 regulates TNF-α-induced epithelial necroptosis and terminal ileitis. Nature. 477:335–339 10.1038/nature10400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman M.L., Neering S.J., Upchurch D., Grimes B., Howard D.S., Rizzieri D.A., Luger S.M., Jordan C.T. 2001. Nuclear factor-κB is constitutively activated in primitive human acute myelogenous leukemia cells. Blood. 98:2301–2307 10.1182/blood.V98.8.2301 [DOI] [PubMed] [Google Scholar]
- He S., Wang L., Miao L., Wang T., Du F., Zhao L., Wang X. 2009. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-α. Cell. 137:1100–1111 10.1016/j.cell.2009.05.021 [DOI] [PubMed] [Google Scholar]
- Hernandez-Ilizaliturri F.J., Deeb G., Zinzani P.L., Pileri S.A., Malik F., Macon W.R., Goy A., Witzig T.E., Czuczman M.S. 2011. Higher response to lenalidomide in relapsed/refractory diffuse large B-cell lymphoma in nongerminal center B-cell-like than in germinal center B-cell-like phenotype. Cancer. 117:5058–5066 10.1002/cncr.26135 [DOI] [PubMed] [Google Scholar]
- Hess P., Pihan G., Sawyers C.L., Flavell R.A., Davis R.J. 2002. Survival signaling mediated by c-Jun NH(2)-terminal kinase in transformed B lymphoblasts. Nat. Genet. 32:201–205 10.1038/ng946 [DOI] [PubMed] [Google Scholar]
- Kaiser W.J., Upton J.W., Long A.B., Livingston-Rosanoff D., Daley-Bauer L.P., Hakem R., Caspary T., Mocarski E.S. 2011. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature. 471:368–372 10.1038/nature09857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamata H., Honda S., Maeda S., Chang L., Hirata H., Karin M. 2005. Reactive oxygen species promote TNFα-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell. 120:649–661 10.1016/j.cell.2004.12.041 [DOI] [PubMed] [Google Scholar]
- Ke H., Harris R., Coloff J.L., Jin J.Y., Leshin B., Miliani de Marval P., Tao S., Rathmell J.C., Hall R.P., Zhang J.Y. 2010. The c-Jun NH2-terminal kinase 2 plays a dominant role in human epidermal neoplasia. Cancer Res. 70:3080–3088 10.1158/0008-5472.CAN-09-2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.S., Morgan M.J., Choksi S., Liu Z.G. 2007. TNF-induced activation of the Nox1 NADPH oxidase and its role in the induction of necrotic cell death. Mol. Cell. 26:675–687 10.1016/j.molcel.2007.04.021 [DOI] [PubMed] [Google Scholar]
- Knight B., Yeoh G.C., Husk K.L., Ly T., Abraham L.J., Yu C., Rhim J.A., Fausto N. 2000. Impaired preneoplastic changes and liver tumor formation in tumor necrosis factor receptor type 1 knockout mice. J. Exp. Med. 192:1809–1818 10.1084/jem.192.12.1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo H.P., Wang Z., Lee D.F., Iwasaki M., Duque-Afonso J., Wong S.H., Lin C.H., Figueroa M.E., Su J., Lemischka I.R., Cleary M.L. 2013. Epigenetic roles of MLL oncoproteins are dependent on NF-κB. Cancer Cell. 24:423–437 10.1016/j.ccr.2013.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Sejas D.P., Zhang X., Qiu Y., Nattamai K.J., Rani R., Rathbun K.R., Geiger H., Williams D.A., Bagby G.C., Pang Q. 2007. TNF-α induces leukemic clonal evolution ex vivo in Fanconi anemia group C murine stem cells. J. Clin. Invest. 117:3283–3295 10.1172/JCI31772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.H., Yang Y., Staudt L.M. 2012. Pathogenetic importance and therapeutic implications of NF-κB in lymphoid malignancies. Immunol. Rev. 246:359–378 10.1111/j.1600-065X.2012.01105.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Lin A. 2005. Role of JNK activation in apoptosis: a double-edged sword. Cell Res. 15:36–42 10.1038/sj.cr.7290262 [DOI] [PubMed] [Google Scholar]
- Luedde T., Beraza N., Kotsikoris V., van Loo G., Nenci A., De Vos R., Roskams T., Trautwein C., Pasparakis M. 2007. Deletion of NEMO/IKKγ in liver parenchymal cells causes steatohepatitis and hepatocellular carcinoma. Cancer Cell. 11:119–132 10.1016/j.ccr.2006.12.016 [DOI] [PubMed] [Google Scholar]
- Lv L., Kerzic P., Lin G., Schnatter A.R., Bao L., Yang Y., Zou H., Fu H., Ye X., Gross S.A., et al. 2007. The TNF-α 238A polymorphism is associated with susceptibility to persistent bone marrow dysplasia following chronic exposure to benzene. Leuk. Res. 31:1479–1485 10.1016/j.leukres.2007.01.014 [DOI] [PubMed] [Google Scholar]
- Maeda S., Kamata H., Luo J.L., Leffert H., Karin M. 2005. IKKβ couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 121:977–990 10.1016/j.cell.2005.04.014 [DOI] [PubMed] [Google Scholar]
- Mantovani A., Allavena P., Sica A., Balkwill F. 2008. Cancer-related inflammation. Nature. 454:436–444 10.1038/nature07205 [DOI] [PubMed] [Google Scholar]
- Molnár L., Berki T., Hussain A., Németh P., Losonczy H. 2000. Detection of TNFα expression in the bone marrow and determination of TNFα production of peripheral blood mononuclear cells in myelodysplastic syndrome. Pathol. Oncol. Res. 6:18–23 10.1007/BF03032653 [DOI] [PubMed] [Google Scholar]
- Moore R.J., Owens D.M., Stamp G., Arnott C., Burke F., East N., Holdsworth H., Turner L., Rollins B., Pasparakis M., et al. 1999. Mice deficient in tumor necrosis factor-alpha are resistant to skin carcinogenesis. Nat. Med. 5:828–831 10.1038/10552 [DOI] [PubMed] [Google Scholar]
- Naugler W.E., Karin M. 2008. NF-κB and cancer-identifying targets and mechanisms. Curr. Opin. Genet. Dev. 18:19–26 10.1016/j.gde.2008.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberst A., Dillon C.P., Weinlich R., McCormick L.L., Fitzgerald P., Pop C., Hakem R., Salvesen G.S., Green D.R. 2011. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature. 471:363–367 10.1038/nature09852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguma K., Oshima H., Oshima M. 2010. Inflammation, tumor necrosis factor and Wnt promotion in gastric cancer development. Future Oncol. 6:515–526 10.2217/fon.10.13 [DOI] [PubMed] [Google Scholar]
- Papa S., Zazzeroni F., Bubici C., Jayawardena S., Alvarez K., Matsuda S., Nguyen D.U., Pham C.G., Nelsbach A.H., Melis T., et al. 2004. Gadd45 β mediates the NF-κB suppression of JNK signalling by targeting MKK7/JNKK2. Nat. Cell Biol. 6:146–153 10.1038/ncb1093 [DOI] [PubMed] [Google Scholar]
- Perkins N.D. 2012. The diverse and complex roles of NF-κB subunits in cancer. Nat. Rev. Cancer. 12:121–132 [DOI] [PubMed] [Google Scholar]
- Popivanova B.K., Kitamura K., Wu Y., Kondo T., Kagaya T., Kaneko S., Oshima M., Fujii C., Mukaida N. 2008. Blocking TNF-α in mice reduces colorectal carcinogenesis associated with chronic colitis. J. Clin. Invest. 118:560–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G., Zhao X., Wang Y., Zhang X., Chen X., Xu C., Yuan Z.R., Roberts A.I., Zhang L., Zheng B., et al. 2012. CCR2-dependent recruitment of macrophages by tumor-educated mesenchymal stromal cells promotes tumor development and is mimicked by TNFα. Cell Stem Cell. 11:812–824 10.1016/j.stem.2012.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakon S., Xue X., Takekawa M., Sasazuki T., Okazaki T., Kojima Y., Piao J.H., Yagita H., Okumura K., Doi T., Nakano H. 2003. NF-κB inhibits TNF-induced accumulation of ROS that mediate prolonged MAPK activation and necrotic cell death. EMBO J. 22:3898–3909 10.1093/emboj/cdg379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai H., Suzuki S., Kawasaki N., Nakano H., Okazaki T., Chino A., Doi T., Saiki I. 2003. Tumor necrosis factor-α-induced IKK phosphorylation of NF-κB p65 on serine 536 is mediated through the TRAF2, TRAF5, and TAK1 signaling pathway. J. Biol. Chem. 278:36916–36923 10.1074/jbc.M301598200 [DOI] [PubMed] [Google Scholar]
- Sakurai T., Maeda S., Chang L., Karin M. 2006. Loss of hepatic NF-κB activity enhances chemical hepatocarcinogenesis through sustained c-Jun N-terminal kinase 1 activation. Proc. Natl. Acad. Sci. USA. 103:10544–10551 10.1073/pnas.0603499103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi G., Sung B., Aggarwal B.B. 2008. TNF: a master switch for inflammation to cancer. Front. Biosci. 13:5094–5107 10.2741/3066 [DOI] [PubMed] [Google Scholar]
- Shaulian E. 2010. AP-1—The Jun proteins: Oncogenes or tumor suppressors in disguise? Cell. Signal. 22:894–899 10.1016/j.cellsig.2009.12.008 [DOI] [PubMed] [Google Scholar]
- Skaug B., Jiang X., Chen Z.J. 2009. The role of ubiquitin in NF-κB regulatory pathways. Annu. Rev. Biochem. 78:769–796 10.1146/annurev.biochem.78.070907.102750 [DOI] [PubMed] [Google Scholar]
- Stifter G., Heiss S., Gastl G., Tzankov A., Stauder R. 2005. Over-expression of tumor necrosis factor-alpha in bone marrow biopsies from patients with myelodysplastic syndromes: relationship to anemia and prognosis. Eur. J. Haematol. 75:485–491 10.1111/j.1600-0609.2005.00551.x [DOI] [PubMed] [Google Scholar]
- Sykes S.M., Lane S.W., Bullinger L., Kalaitzidis D., Yusuf R., Saez B., Ferraro F., Mercier F., Singh H., Brumme K.M., et al. 2011. AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Cell. 146:697–708 10.1016/j.cell.2011.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenev T., Bianchi K., Darding M., Broemer M., Langlais C., Wallberg F., Zachariou A., Lopez J., MacFarlane M., Cain K., Meier P. 2011. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol. Cell. 43:432–448 10.1016/j.molcel.2011.06.006 [DOI] [PubMed] [Google Scholar]
- Tsimberidou A.M., Giles F.J. 2002. TNF-α targeted therapeutic approaches in patients with hematologic malignancies. Expert Rev. Anticancer Ther. 2:277–286 10.1586/14737140.2.3.277 [DOI] [PubMed] [Google Scholar]
- Tsimberidou A.M., Estey E., Wen S., Pierce S., Kantarjian H., Albitar M., Kurzrock R. 2008. The prognostic significance of cytokine levels in newly diagnosed acute myeloid leukemia and high-risk myelodysplastic syndromes. Cancer. 113:1605–1613 10.1002/cncr.23785 [DOI] [PubMed] [Google Scholar]
- Vallabhapurapu S., Karin M. 2009. Regulation and function of NF-κB transcription factors in the immune system. Annu. Rev. Immunol. 27:693–733 10.1146/annurev.immunol.021908.132641 [DOI] [PubMed] [Google Scholar]
- Wang L., Du F., Wang X. 2008. TNF-α induces two distinct caspase-8 activation pathways. Cell. 133:693–703 10.1016/j.cell.2008.03.036 [DOI] [PubMed] [Google Scholar]
- Xiao Y., Li H., Zhang J., Volk A., Zhang S., Wei W., Zhang S., Breslin P., Zhang J. 2011. TNF-α/Fas-RIP-1–induced cell death signaling separates murine hematopoietic stem cells/progenitors into 2 distinct populations. Blood. 118:6057–6067 10.1182/blood-2011-06-359448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.Y., Green C.L., Tao S., Khavari P.A. 2004. NF-κB RelA opposes epidermal proliferation driven by TNFR1 and JNK. Genes Dev. 18:17–22 10.1101/gad.1160904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.Y., Adams A.E., Ridky T.W., Tao S., Khavari P.A. 2007. Tumor necrosis factor receptor 1/c-Jun-NH2-kinase signaling promotes human neoplasia. Cancer Res. 67:3827–3834 10.1158/0008-5472.CAN-06-4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.W., Shao J., Lin J., Zhang N., Lu B.J., Lin S.C., Dong M.Q., Han J. 2009. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 325:332–336 10.1126/science.1172308 [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhou X., McQuade T., Li J., Chan F.K., Zhang J. 2011. Functional complementation between FADD and RIP1 in embryos and lymphocytes. Nature. 471:373–376 10.1038/nature09878 [DOI] [PMC free article] [PubMed] [Google Scholar]