Abstract
Using Exserohilum rostratum-specific and panfungal real-time PCR, we studied 24 blood samples and 2 synovial fluid specimens from 20 patients with persistent or worsening pain following injections of contaminated methylprednisolone. Seven blood specimens from 6 patients were significantly positive for fungal DNA by panfungal PCR, with multiple fungal species identified.
TEXT
To date, the tragic outbreak in the United States of fungal meningitis and other infections associated with injections of contaminated methylprednisolone has reached 20 states, causing 751 fungal infections with 64 deaths. Exserohilum rostratum has been identified as the principal causative agent of most of infections, although other fungi also have been implicated (1). With progression of the outbreak, there are many patients experiencing persistent or worsening symptoms yet without positive magnetic resonance imaging (MRI) or fungal culture results. Earlier during this outbreak, we developed a rapid E. rostratum-specific real-time PCR assay, which allows the rapid primary detection of the infecting pathogen in patients with suspected infection (2). Therefore, these patients requested that they be tested by our novel real-time PCR assay for the signature of E. rostratum or other fungal DNA in their blood. Through a research protocol approved by the Rutgers University institutional review board (IRB), blood samples from these patients were drawn at their primary care physicians' local clinics or hospitals, deidentified, blind coded, and sent to the Public Health Research Institute (PHRI), New Jersey Medical School, Rutgers, The State University of New Jersey, for real-time PCR testing following IRB approval. Herein, we summarize the molecular testing results for these patients.
Between 22 February and 11 December 2013, we received 24 blood samples and 2 synovial fluid samples from 20 patients in 8 states (Indiana, 4; Kentucky, 3; West Virginia, 3; Tennessee, 3; Florida, 3; New Jersey, 2; Texas, 1; Pennsylvania, 1) (Table 1). Basic demographic information was available for only 11 patients due to the limited accessibility. All patients received contaminated methylprednisolone solution by injection once or multiple times during August and September in 2012 and developed a variety of clinical manifestations, including meningitis, arachnoiditis, back/neck pain or other persistent pain around the injection site, and fatigue. Blood samples from 6 healthy adult volunteers were also collected and tested as normal controls. All healthy volunteers gave written informed consent, and the study protocol and amendments were approved by the Rutgers IRB.
TABLE 1.
Summary of characteristics of patients and panfungal detection results
| Patient | Gender/agea | Location | Sample |
Panfungal real-time PCR |
Sequencing IDe | ||
|---|---|---|---|---|---|---|---|
| Typeb | Collection datec | Resultd | CT | ||||
| 1 | Male/59 | Evansville, IN | Blood | 2/20/13 | + | 33.4 | E. rostratum |
| Blood | 5/28/13 | ± | 34.6 | Alternaria sp. | |||
| Blood | 11/6/13 | − | 35.6 | NT | |||
| 2 | NA/NA | Webster, TX | Blood | 3/11/13 | + | 33.5 | Pichia sp.; Aureobasidium sp. |
| 3 | Male/NA | Paducah, KY | Blood | 5/20/13 | ± | 34.8 | Chaetothyriales sp. |
| 4 | Female/41 | Evansville, IN | Blood | 7/14/13 | − | 36.2 | NT |
| 5 | Male/46 | Evansville, IN | Blood | 7/23/13 | ± | 34.5 | A. fumigatus |
| 6 | NA/NA | Parkersburg, WV | Blood | 8/5/13 | + | 28.5 | A. fumigatus; Capnodium sp. |
| Blood | 10/1/13 | + | 33.3 | A. fumigatus | |||
| 7 | Female/48 | Mullica Hill, NJ | Blood | 8/8/13 | − | 35.6 | NT |
| Blood | 11/14/13 | − | 36.9 | NT | |||
| SF | 11/14/13 | − | 36.7 | NT | |||
| SF | 11/14/13 | − | 35.1 | NT | |||
| 8 | NA/NA | Paducah, KY | Blood | 9/17/13 | − | 36.1 | NT |
| 9 | Female/51 | Elkhart, IN | Blood | 9/23/13 | + | 33.5 | Cryptococcus neoformans |
| 10 | NA/NA | Parkersburg, WV | Blood | 9/6/13 | − | 36.6 | NT |
| 11 | NA/NA | Parkersburg, WV | Blood | 9/10/13 | − | 36.2 | NT |
| 12 | Female/NA | Woodstown, NJ | Blood | 10/8/13 | + | 33.4 | A. fumigatus |
| 13 | Male/54 | Pennsylvania, PA | Blood | 10/15/13 | − | 35.7 | NT |
| 14 | NA/NA | Crossville, TN | Blood | 10/22/13 | − | 36.4 | NT |
| 15 | NA/NA | Crossville, TN | Blood | 10/28/13 | + | 33.3 | Hortaea werneckii |
| 16 | Male/NA | Russellville, KY | Blood | 10/28/13 | − | 36.0 | NT |
| 17 | Female/41 | Ocala, FL | Blood | 10/30/13 | − | 35.7 | NT |
| 18 | Female/52 | Ocala, FL | Blood | 11/3/13 | − | 36.3 | NT |
| 19 | NA/NA | Nashville, TN | Blood | 11/14/13 | + | 34.3 | Filobasidium uniguttulatum; Phleibia sp. |
| 20 | NA/NA | Murdock, FL | Blood | 12/10/13 | ± | 34.0 | A. fumigatus |
NA, not available.
SF, synovial fluid.
Month/day/year.
The ± sign indicates that the samples was positive when the CT cutoff was set at 35 but negative when the CT cutoff was set at 34.
NT, not tested.
EDTA blood (3 to 5 ml) was collected at local clinic or hospital, and shipped immediately on ice to the PHRI. Blood was processed for DNA extraction using a MolYsis Basic5 kit (Molzym GmbH & Co.) in combination with a MasterPure yeast DNA purification kit (Epicentre Biotechnologies, Madison, WI). A 100-μl cell suspension from the last step of MolYsis treatment was mixed with 500 μl of yeast lysis solution from the MasterPure kit and processed as previously described (2). One nanogram of E. rostratum DNA used to spike 5 ml of defibrinated sheep blood (Remel) was extracted as a positive extraction control. Five milliliters of blood from each healthy volunteer was treated in parallel as a negative control.
E. rostratum PCR was performed as previously described (2). Primers and molecular beacon probe for panfungal detection were adapted from a previously established nucleic acid sequence-based amplification assay (3), with the small modification of clipping the T7 promoter sequence off the reverse primer. E. rostratum DNA was used as a positive control. Human genomic DNA was tested as a negative control in parallel. Nuclease-free water was used as a no-template control (NTC). PCR testing was repeated three times on three separate days. Comparison of cycle threshold (CT) values from three different sample groups was performed by one-way analysis of variance. The level of statistical significance was set at a P value of <0.05.
The E. rostratum-positive PCR product was purified using a QIAquick PCR purification kit (Qiagen) and sequenced (Macrogen). For panfungal PCR-positive but E. rostratum-negative samples, the primers ITS1F (5′-GAACCTGCGGAAGGATCATT-3′) and ITS1R (5′-GGAACCAAGAGATCCGTTGT-3′) were used to amplify internal transcribed spacer 1 (ITS1) region, and the redundant sets of primers ITS2F (5′-CATGCCTGTCCGAGCGTCAT-3′) and ITS2R (5′-GTAACCCTACCTGATCCGA-3′) and primers ACITS2F (5′-GATGAAGAACGCAGCGAAAT-3′) and ITS4A (5′-ATGCTTAAGTTCAGCGGGTA-3′) were used to amplify the ITS2 region. PCR was performed on a Bio-Rad iCycler (Bio-Rad) in a 50-μl PCR mixture containing 2 μl of DNA, a 0.2 μM concentration of each primer, and 25 μl of EmeraldAmp master mix (TaKaRa Bio Inc.). The thermal cycling conditions included an initial denaturation step at 98°C for 2 min, 45 cycles of 98°C for 20 s (denaturation), 55°C for 30 s (annealing), and 72°C for 30 s (extension), followed by an extra extension step at 72°C for 2 min. PCR product purification and sequencing were conducted as described above. Finally, the BLAST program was run for molecular identification of the sequenced sample (4). We used a 100% cutoff for species identity.
Negative PCR results were observed for all 6 healthy volunteers, with no CT values in E. rostratum detection and panfungal CT (threshold cycle) values averaging 35.6 (range, 34.9 to 37.0). While no CT cutoff was needed for E. rostratum PCR, we did establish a CT cutoff of 35 for the panfungal detection, which correctly recognized 95% (17/18 replicates) of negative samples from healthy volunteers. Although only one patient blood sample was E. rostratum real-time PCR positive, with a CT of 37.7, 12 samples from 10 patients (50%) were panfungal PCR positive, with a mean CT of 33.4, which was significantly lower than that of healthy controls (P < 0.0001) (Fig. 1). Even when a more rigid CT cutoff of 34 was applied, 7 samples from 6 patients (30% by patient) were still fungal-DNA positive.
FIG 1.
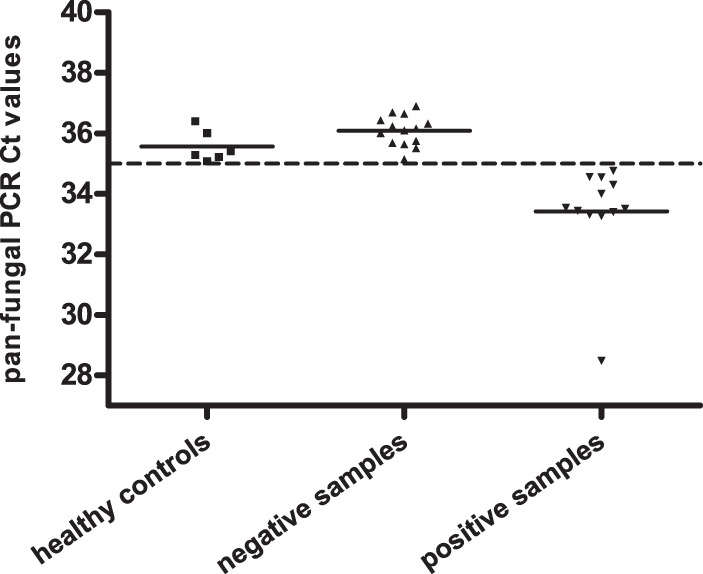
Panfungal real-time PCR CT value distributions in samples from healthy controls, negative samples, and positive samples. The dashed line represents the CT cutoff of 35, and the solid lines denote the mean CT values of healthy controls and patients. All healthy controls were negative, with a mean CT of 35.6, and the mean CT was 36.1 for negative samples tested in this study. In contrast, 11 samples from 10 patients were panfungal PCR positive, with a mean CT of 33.4, significantly lower than that of healthy controls (P < 0.0001). The number of positive samples decreased to 7 when a more rigorous CT cutoff of 34 was applied.
ITS sequencing was performed for all panfungal PCR-positive samples (CT < 35) (Table 1). The E. rostratum PCR-positive sample was confirmed by sequencing. Multiple fungal DNA signatures were identified from other panfungal PCR-positive samples, including Alternaria sp., Aspergillus fumigatus, and other plant- and soil-based fungi (5–8) (Table 1). Mixed fungal signatures (fungal identity of ITS1 different from that of ITS2) were found in three blood samples.
To our knowledge, this is the first report with positive detection of fungal DNA in blood since the outbreak began >18 months ago. Surprisingly, as much as 29% of blood samples were significantly fungal-DNA positive even when a rigid CT cutoff of 34 was applied, comparable to 24% fungal-DNA positivity in cerebrospinal fluid (CSF) and 33% in tissue reported previously (9).
Two intriguing observations surfaced in this study. First, a wide fungal spectrum (E. rostratum and 10 non-Exserohilum fungal species) was identified. Most species were reported to be involved in opportunistic fungal infections in humans (5–8), and of these, A. fumigatus and an Alternaria sp. were detected in clinical samples from a few patients in this outbreak (9). Mixed fungal organisms identified from three blood samples indicate the possibility that at least a subset of the affected patient population has experienced mixed fungal infections with non-Exserohilum species. Nevertheless, it remains difficult to interpret these results due to the absence of blood cultures and the very limited access to detailed clinical information or diagnostic testing results. Second, A. fumigatus may have played a role in some prolonged infections in this outbreak. Although A. fumigatus was isolated and believed to be the cause of the index case of this outbreak, it had not been detected in any of the subsequent 700-plus cases before our study started. In contrast, our test detected this pathogen independently in 5 blood samples from 4 patients residing in Indiana, West Virginia, New Jersey, and Florida. As A. fumigatus is notorious for its low culturability from infection sites, especially blood (10–12), it is reasonable to deduce that this organism may have eluded culture isolation while causing chronically progressive symptoms.
We are not yet able to estimate the value of finding fungal DNA from patient blood in this outbreak, given the small sample size and the fact that there is little knowledge of the natural history of the infection, the fungal pathogenesis and innate host defense, or the therapeutic response and prognosis. However, detecting fungal DNA from blood is valuable in assisting rapid and accurate diagnosis for some types of invasive fungal infections, such as invasive candidiasis and invasive aspergillosis (13–16). Regardless of sample size, 29 to 50% (depending on the cutoff) of tested blood being fungus positive indicated that having an insidious fungal infection may not be just an incidental event in this vulnerable population.
Our study has several limitations. First, the patient group was small in proportion to the large number of patients exposed and suffered from selection referral bias. These patients requested blood tests because they were having severe or worsening symptoms that remained unaccounted for. However, the contamination of the methylprednisolone vials with non-Exserohilum fungi provides a foundation for concern that these signals in patients with localized symptoms reflect a more complex pattern than heretofore appreciated. Second, due to its broad coverage, the panfungal real-time PCR is vulnerable to trace contamination at any step from sample collection to PCR amplification (17, 18). However, bearing in mind the possibility of contamination amid blood testing, we carefully applied extreme precautions to prevent airborne and carryover contaminations, used sufficient negative controls, and applied a rigorous CT cutoff to the analysis of panfungal real-time PCR results. Such efforts sufficiently justified the reliability of our novel finding.
In summary, our finding of fungal DNA in blood from patients exposed to contaminated steroids provides another dimension to our understanding of this outbreak. We further demonstrate the applicability of our recently developed diagnostic PCR assays on clinical samples and provide a proactive generalized screening tool to assist in the early diagnosis of infections associated with this tragic event.
ACKNOWLEDGMENTS
This project was funded by resources provided by the Public Health Research Institute. David S. Perlin receives research support from Merck, Astellas, and Pfizer and has a patent application pending for assays for drug-resistant fungal infections.
We declare that there are no conflicts of interest.
Footnotes
Published ahead of print 9 April 2014
REFERENCES
- 1.Kauffman CA, Pappas PG, Patterson TF. 2013. Fungal infections associated with contaminated methylprednisolone injections. N. Engl. J. Med. 368:2495–2500. 10.1056/NEJMra1212617 [DOI] [PubMed] [Google Scholar]
- 2.Zhao Y, Petraitiene R, Walsh TJ, Perlin DS. 2013. A real-time PCR assay for rapid detection and quantification of Exserohilum rostratum, a causative pathogen of fungal meningitis associated with injection of contaminated methylprednisolone. J. Clin. Microbiol. 51:1034–1036. 10.1128/JCM.03369-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao Y, Park S, Kreiswirth BN, Ginocchio CC, Veyret R, Laayoun A, Troesch A, Perlin DS. 2009. Rapid real-time nucleic acid sequence-based amplification-molecular beacon platform to detect fungal and bacterial bloodstream infections. J. Clin. Microbiol. 47:2067–2078. 10.1128/JCM.02230-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2008. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing: approved guideline, CLSI document MM18-A. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 5.Crous PW, Schubert K, Braun U, de Hoog GS, Hocking AD, Shin HD, Groenewald JZ. 2007. Opportunistic, human-pathogenic species in the Herpotrichiellaceae are phenotypically similar to saprobic or phytopathogenic species in the Venturiaceae. Stud. Mycol. 58:185–217. 10.3114/sim.2007.58.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vicente VA, Attili-Angelis D, Pie MR, Queiroz-Telles F, Cruz LM, Najafzadeh MJ, de Hoog GS, Zhao J, Pizzirani-Kleiner A. 2008. Environmental isolation of black yeast-like fungi involved in human infection. Stud. Mycol. 61:137–144. 10.3114/sim.2008.61.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan W, Liao W, Hagen F, Theelen B, Shi W, Meis JF, Boekhout T. 2012. Meningitis caused by Filobasidium uniguttulatum: case report and overview of the literature. Mycoses 55:105–109. 10.1111/j.1439-0507.2011.02054.x [DOI] [PubMed] [Google Scholar]
- 8.Hawkes M, Rennie R, Sand C, Vaudry W. 2005. Aureobasidium pullulans infection: fungemia in an infant and a review of human cases. Diagn. Microbiol. Infect. Dis. 51:209–213. 10.1016/j.diagmicrobio.2004.10.007 [DOI] [PubMed] [Google Scholar]
- 9.Lockhart SR, Pham CD, Gade L, Iqbal N, Scheel CM, Cleveland AA, Whitney AM, Noble-Wang J, Chiller TM, Park BJ, Litvintseva AP, Brandt ME. 2013. Preliminary laboratory report of fungal infections associated with contaminated methylprednisolone injections. J. Clin. Microbiol. 51:2654–2661. 10.1128/JCM.01000-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barton RC. 2013. Laboratory diagnosis of invasive aspergillosis: from diagnosis to prediction of outcome. Scientifica 2013:29. 10.1155/2013/459405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horvath JA, Dummer S. 1996. The use of respiratory-tract cultures in the diagnosis of invasive pulmonary aspergillosis. Am. J. Med. 100:171–178. 10.1016/S0002-9343(97)89455-7 [DOI] [PubMed] [Google Scholar]
- 12.Kontoyiannis DP, Sumoza D, Tarrand J, Bodey GP, Storey R, Raad II. 2000. Significance of aspergillemia in patients with cancer: a 10-year study. Clin. Infect. Dis. 31:188–189. 10.1086/313918 [DOI] [PubMed] [Google Scholar]
- 13.White PL, Archer AE, Barnes RA. 2005. Comparison of non-culture-based methods for detection of systemic fungal infections, with an emphasis on invasive Candida infections. J. Clin. Microbiol. 43:2181–2187. 10.1128/JCM.43.5.2181-2187.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White PL, Linton CJ, Perry MD, Johnson EM, Barnes RA. 2006. The evolution and evaluation of a whole blood polymerase chain reaction assay for the detection of invasive aspergillosis in hematology patients in a routine clinical setting. Clin. Infect. Dis. 42:479–486. 10.1086/499949 [DOI] [PubMed] [Google Scholar]
- 15.Wellinghausen N, Siegel D, Winter J, Gebert S. 2009. Rapid diagnosis of candidaemia by real-time PCR detection of Candida DNA in blood samples. J. Med. Microbiol. 58:1106–1111. 10.1099/jmm.0.007906-0 [DOI] [PubMed] [Google Scholar]
- 16.Luong ML, Clancy CJ, Vadnerkar A, Kwak EJ, Silveira FP, Wissel MC, Grantham KJ, Shields RK, Crespo M, Pilewski J, Toyoda Y, Kleiboeker SB, Pakstis D, Reddy SK, Walsh TJ, Nguyen MH. 2011. Comparison of an Aspergillus real-time polymerase chain reaction assay with galactomannan testing of bronchoalveolar lavage fluid for the diagnosis of invasive pulmonary aspergillosis in lung transplant recipients. Clin. Infect. Dis. 52:1218–1226. 10.1093/cid/cir185 [DOI] [PubMed] [Google Scholar]
- 17.Loeffler J, Hebart H, Bialek R, Hagmeyer L, Schmidt D, Serey FP, Hartmann M, Eucker J, Einsele H. 1999. Contaminations occurring in fungal PCR assays. J. Clin. Microbiol. 37:1200–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison E, Stalhberger T, Whelan R, Sugrue M, Wingard JR, Alexander BD, Follett SA, Bowyer P, Denning DW. 2010. Aspergillus DNA contamination in blood collection tubes. Diagn. Microbiol. Infect. Dis. 67:392–394. 10.1016/j.diagmicrobio.2010.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]


