Abstract
Hepatitis E virus (HEV) infection is recognized as an emerging and often undiagnosed disease in industrialized countries, with asymptomatic infections actually occurring in blood donors. Sensitive detection of HEV-RNA is crucial for diagnosis and monitoring of disease progression. We evaluated the analytical sensitivity and performance of three HEV RT-PCR assays (RealStar HEV reverse transcription-PCR [RT-PCR], hepatitis@ceeramTools, and ampliCube HEV RT-PCR) for screening of individuals for HEV infections (ID-nucleic acid amplification technology [ID-NAT]) and for blood donor pool screening (minipool-NAT [MP-NAT]). RNA was extracted using NucliSens easyMAG (ID-NAT) and a high-volume extraction protocol (4.8 ml, chemagic Viral 5K, MP-NAT). Three NAT assays were evaluated for ID-NAT but only two assays for MP-NAT due to inhibition of the ampliCube HEV RT-PCR kit using the corresponding RNA extract. Assays provided good analytical sensitivity, ranging from 37.8 to 180.1 IU/ml (ID-NAT) and from 4.7 to 91.2 IU/ml (MP-NAT). The applicability of HEV antigen (HEV-Ag) screening was compared to that of RT-PCR screening and detection of HEV-IgM antibodies using seroconversion panels of 10 HEV genotype 3-infected individuals. Four individuals revealed a positive HEV-Ag detection result, with corresponding viremias ranging from 1.92E + 03 to 2.19E + 05 IU/ml, while the progression of HEV-Ag followed that of HEV viremia. The other six individuals showed no presence of HEV-Ag although the corresponding viremias were also in the range of >1.0E + 03. Anti-HEV-IgM antibodies were detectable in seven donors; one donor presented parallel positivities of HEV-Ag and anti-HEV IgM. The evaluated NAT methods present powerful tools providing sensitive HEV detection. Application of HEV-Ag or anti-HEV IgM screening is currently inferior for the early detection of HEV infection due to the decreased sensitivity compared to NAT methods.
INTRODUCTION
Non-travel-associated hepatitis E virus (HEV) infections are increasingly recognized as an emerging disease in industrialized countries (1, 2). HEV is a single-stranded RNA virus belonging to the family of Hepeviridae, with differences regarding the geographical distribution of the four currently described major genotypes (genotypes 1 to 4) (3). In developing countries, HEV genotype 1 and 2 infections are hyperendemic, with transmission by the fecal-oral route, and are restricted to humans (4–6), whereas genotype 3 (Europe, United States, Japan, New Zealand, and Argentina) and 4 (Japan and China [7]) infections are observed in industrialized countries. Genotypes 3 and 4 have been isolated from humans and other mammalian species (e.g., domestic pigs, wild boars, deers, and rabbits [8–11]), and the occurrence of genetically similar HEV isolates suggests a zoonotic or food-borne route of transmission of HEV (12, 13). However, HEV transmission by solid-organ transplantation has also been described (14), and chronic HEV infections were found in transplant patients (15, 16). The transmission of hepatitis E infections by contaminated blood products has already been reported in Europe, Japan, and the United Kingdom (2, 17–21). The occurrence of autochthonous and asymptomatic infections in blood donors, as well as the detection of widespread distribution of HEV in plasma fractionation pools from North America, Europe, and southeast Asia, makes HEV a potential new hazardous blood-borne pathogen with respect to blood safety (2, 22–25).
Anti-HEV-IgM is currently the most prominent and sensitive serological marker for the diagnosis of acute, recent, or ongoing HEV infection, but the somewhat lower specificity of different IgM assays has often been discussed in the past (26, 27). Additionally, the diagnostic window period of HEV viremia prior to the occurrence of HEV-specific antibodies limits the applicability of serological tests (25). To date, detection of HEV-RNA by molecular genetic methods has been considered the “gold standard” (28). Recently, detection of hepatitis E virus antigen (HEV-Ag) was introduced as an additional early diagnostic marker (28, 29). The HEV-Ag is a viral capsid protein detectable within the window period or acute phase of infection with a persistence of 3 to 4 weeks after infection (30).
Nevertheless, a sensitive screening method for blood donors is required to avoid transfusion of HEV-contaminated blood products. The sensitive detection of HEV-RNA is also crucial to assess the progression of HEV infection, especially in transplant patients or other recipients of blood products, in terms of clearance or persistence of viral particles (31). The aim of the present study was comparison of the sensitivities and performances of different commercial HEV-RNA amplification systems for (i) HEV blood donor pool screening and (ii) HEV-RNA detection in individuals with acute or chronic infections. Furthermore, the applicability of HEV-Ag in comparison to molecular genetic screening, as well as the occurrence of HEV-IgM-specific antibodies, was evaluated in consecutive samples of 10 virologically confirmed HEV genotype 3-infected individuals.
MATERIALS AND METHODS
Blood donors.
From July to September 2011, a total of 16,125 individual German blood donors were routinely screened for the presence of HEV-RNA by the Uni.Blutspendedienst OWL, revealing 13 HEV-RNA-positive donations (25). Samples donated in continuous intervals after the initial HEV-RNA-positive donation (day 0) were available for 10 blood donors (sex, male; geographic origin, North Rhine-Westphalia [n = 4], Lower Saxony [n = 1], or Hesse [n = 5]; mean age, 28 years [± 10; range, 20 to 53 years]). All donors underwent a predonation medical examination, denied current diseases or any known risk factors for viral infections, and presented with an asymptomatic hepatitis E virus infection. The study protocol conformed to the ethical guidelines and was approved by the institutional review board of the Ruhr University of Bochum. Informed consent was obtained from each donor.
RNA extraction.
For donor pool screening, high-volume extraction of 4.8 ml of plasma was performed using the chemagic viral DNA/RNA reagent kit (Viral 5k; PerkinElmer chemagen Technologie GmbH, Baesweiler, Germany) combined with the automated chemagic MSMI magnetic separation module (PerkinElmer chemagen Technologie GmbH). Briefly, 4.8 ml of plasma was mixed with 4.8 ml of lysis buffer, 30 μl of protease, and 7 μl of poly(A). Samples were incubated at 55°C for 10 min. Subsequently, lysates were mixed with 15 ml of binding buffer containing 100 μl of magnetic beads. The MSMI module automatically performed the nucleic acid extraction process, including binding, two washes, and elution in a final volume of 100 μl of elution buffer.
For single-sample screening, extraction of total RNA from 500 μl of plasma was performed using the NucliSens easyMAG (bioMérieux, Nürtingen, Germany) automated RNA/DNA extraction system. RNA was eluted in 55 μl of elution buffer.
Real-time RT-PCR.
Three different commercial assays, the RealStar HEV reverse transcription-PCR (RT-PCR) assay (Altona Diagnostic Technologies [ADT], Hamburg, Germany), the hepatitis@ceeramTools kit (Ceeram; S.A.S., La Chapelle sur Erdre, France), and the ampliCube HEV RT-PCR kit (Mikrogen, Neuried, Germany), were compared. Amplification using the Real-Star HEV RT-PCR kit was performed according to the manufacturer's instructions on a Rotor-Gene 3000 system (Corbett Life Sciences, Sydney, Australia). Amplification using the hepatitis@ceeramTools kit and the ampliCube HEV RT-PCR kit was carried out according to the manufacturer's instructions using a LightCycler 480 system (Roche, Mannheim, Germany).
Analytical sensitivity and comparison of different amplification methods.
The analytical sensitivity and the precision of the three different assays for blood donor pool screening or individual patient/donor sample screening were determined using a 2-fold dilution series of plasma samples inoculated with the first WHO international standard for hepatitis E virus RNA for nucleic acid amplification technology (NAT)-based assays (WHO-NAT standard, Paul-Ehrlich Institute, Langen, Germany [32]). Nucleic acids were extracted using the two different extraction methods. The 95% detection limit was calculated by probit analysis with 6 dilution steps and 24 replicates using SPSS software (SPSS GmbH Software, version 14.0; SPSS, Munich, Germany). The HEV concentration in positive plasma samples of different donors was quantified using the WHO-NAT standard.
In order to compare the applicabilities of the different PCR methods for HEV blood donor pool screening, subsequent plasma samples of HEV-RNA-positive donors spanning the originally positive donation detected (25) were diluted with negative human plasma to simulate master pools of 48 or 96 donations mimicking a routine pool screening procedure with different pool sizes. Simulation of master pools was set up by combining 200 μl of EDTA-plasma of the initial HEV-positive donation with negative human plasma to achieve a level of 9.4 ml (pool of 48 samples) or 19 ml (pool of 96 samples); samples were analyzed with the RealStar HEV RT-PCR kit and the hepatitis@ceeramTools kit.
Serological testing.
Screening for HEV-Ag in HEV-RNA-positive donors was performed using the Wantai HEV-Ag enzyme-linked immunosorbent assay (ELISA) (distributed by Axiom Diagnostics, Worms, Germany) according to the manufacturer's instructions. In order to elucidate a potential donor-dependent influence on the detection of HEV-Ag, 2-fold serial dilutions of antigen-positive plasma samples to levels of approximately 1 × 10E + 03 IU/ml were prepared with human plasma negative for anti-HEV immunoglobulins, HEV-RNA, and HEV-Ag; samples were analyzed in duplicate.
Plasma samples of HEV-RNA-positive donors were further screened for the presence of HEV-specific IgM antibodies using a recomWell HEV-IgM immunoassay (version autumn 2012; Mikrogen GmbH, Neuried, Germany). Samples were analyzed according to the manufacturer's instructions.
RESULTS
Analytical sensitivity and specificity and comparison of NAT assays.
For blood donor pool screening, the RealStar HEV RT-PCR assay showed the highest sensitivity, with a 95% detection limit of 4.7 IU/ml (95% confidence interval [CI], 3.6 to 7.6), followed by the hepatitis@ceeramTools kit with 91.2 IU/ml (95% CI, 64.9 to 205.9) (Table 1). The ampliCube HEV RT-PCR kit is not applicable for nucleic acids extracted with the Viral 5k kit; the internal assay control showed complete inhibition. For individual sample screening, the RealStar HEV RT-PCR assay consistently had the highest sensitivity of 37.8 IU/ml (95% CI, 22.2 to 671.2), followed by the hepatitis@ceeramTools kit with 86.8 IU/ml (95% CI, 68.9 to 124.7) and the ampliCube HEV RT-PCR kit with 180.4 IU/ml (95% CI, 128.5 to 355.2). The reproducibility of the assays was demonstrated by analyzing the intra-assay and interassay variations for the CT (crossing threshold) values. The intra-assay variability was calculated from eight replicates; the interassay variability was determined from three independent PCR runs with eight replicates per run. Values are given as means ± standard deviations (SD) and were calculated using GraphPad Prism 5.0 software (GraphPad Software, San Diego, CA). Intra-assay and interassay variations were in acceptable ranges for all evaluated assays and revealed variation coefficients < 5% (Table 2).
TABLE 1.
Analytical sensitivity of different HEV RT-PCR assaysa
| Sample concn (IU/ml) | MP-NAT screening: chemagen viral 5K |
ID-NAT screening: Nuclisens EasyMAG |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Real-Star HEV RT-PCR kit |
Ceeram |
Real-Star HEV RT-PCR kit |
Ceeram |
ampliCube HEV RT-PCR kit |
||||||
| No. of positive results/total no. of results | % of positive results | No. of positive results/total no. of results | % of positive results | No. of positive results/total no. of results | % of positive results | No. of positive results/total no. of results | % of positive results | No. of positive results/total no. of results | % of positive results | |
| 400 | 24/24* | 100.0 | ||||||||
| 200 | 24/24* | 100.0 | 24/24* | 100.0 | 24/24* | 100.0 | 22/24 | 91.7 | ||
| 100 | 24/24 | 100.0 | 24/24 | 100.0 | 24/24 | 100.0 | 19/24 | 79.2 | ||
| 50 | 8/8 | 100.0 | 10/24 | 41.7 | 22/24 | 91.7 | 15/24 | 62.5 | 10/24 | 41.7 |
| 25 | 24/24* | 100.0 | 10/24 | 41.7 | 23/24 | 95.8 | 11/24 | 45.8 | 7/24 | 29.2 |
| 12.5 | 24/24 | 100.0 | 3/24 | 12.5 | 18/24 | 75.0 | 6/24 | 25.0 | 4/24 | 16.7 |
| 6.25 | 24/24 | 100.0 | 1/24 | 4.2 | 13/24 | 54.2 | 6/24 | 25.0 | 0/24 | 0 |
| 3.13 | 19/24 | 79.2 | 5/24 | 20.8 | ||||||
| 1.56 | 11/24 | 45.8 | 3/24 | 12.5 | ||||||
| 0.75 | 10/24 | 41.7 | ||||||||
| 95% detection limit, IU/ml (95% CI) | 4.7 (3.6–7.6) | 91.2 (64.9–205.9) | 37.8 (22.2–671.2) | 86.8 (68.9–124.7) | 180.4 (128.5–355.2) | |||||
*, the corresponding concentration was used to calculate the CT values for intra- and interassay variations for HEV and the internal control for the same assay in Table 2.
TABLE 2.
Precision testing of different HEV RT-PCR assays
| Parametera | MP-NAT screening: chemagen viral 5K |
ID-NAT screening: Nuclisens EasyMAG |
|||
|---|---|---|---|---|---|
| Real-Star HEV RT-PCR kit | Ceeram | Real-Star HEV RT-PCR kit | Ceeram | ampliCube HEV RT-PCR kit | |
| Intra-assay HEV* [mean CT (± SD)/VC] | 34.26 (± 0.43)/1.26 | 31.48 (± 0.17)/0.54 | 30.8 (± 0.57)/1.84 | 35.15 (± 0.60)/1.71 | 35.79 (± 0.67)/1.86 |
| Intra-assay IC* [mean CT (± SD)/VC] | 26.27 (± 0.24)/0.89 | 28.85 (± 0.16)/0.54 | 30.89 (± 0.76)/2.45 | 30.67 (± 0.39)/1.26 | 29.81 (± 0.32)/1.07 |
| Interassay HEV* [mean CT (± SD)/VC] | 32.85 (± 1.19)/3.62 | 31.55 (± 0.37)/1.18 | 31.74 (± 1.36)/4.29 | 35.15 (± 1.02)/2.90 | 36.39 (± 0.83)/2.29 |
| Interassay IC* [mean CT (± SD)/VC] | 27.89 (± 2.56)/3.19 | 28.56 (± 0.26)/0.92 | 32.47 (± 1.58)/4.87 | 29.80 (± 1.00)/3.34 | 30.16 (± 0.66)/2.20 |
IC, internal control; VC, variation coefficient; *, calculated using the concentration corresponding to the entry shown with an asterisk for the same assay in Table 1.
Pool simulation.
The detection frequencies of the RealStar HEV RT-PCR kit and the hepatitis@ceeramTools kit were different due to the determined 95% detection limits (see Table S1 in the supplemental material). The HEV-RNA concentrations of individual samples were converted to the respective pool sizes of 48 and 96 samples. Both assays detected all samples, with calculated pool concentrations above the stated detection limits of 4.7 IU/ml (RealStar HEV RT-PCR assay) and 91.2 IU/ml (hepatitis@ceeramTools kit) (see the gray-shaded data in Table S1 in the supplemental material). Due to the higher sensitivity of the Real-Star HEV RT-PCR kit, 21 samples were detected in a pool size of 48 and 17 samples in a pool size of 96 compared to 13 samples (48-sample pool) or 6 samples (96-sample pool) detected by hepatitis@ceeramTools. Furthermore, the RealStar HEV RT-PCR assay detected 12 additional samples below the determined detection limit compared to the 11 samples additionally detected by the hepatitis@ceeramTools kit.
Comparison of HEV antigen detection and RNA concentration data.
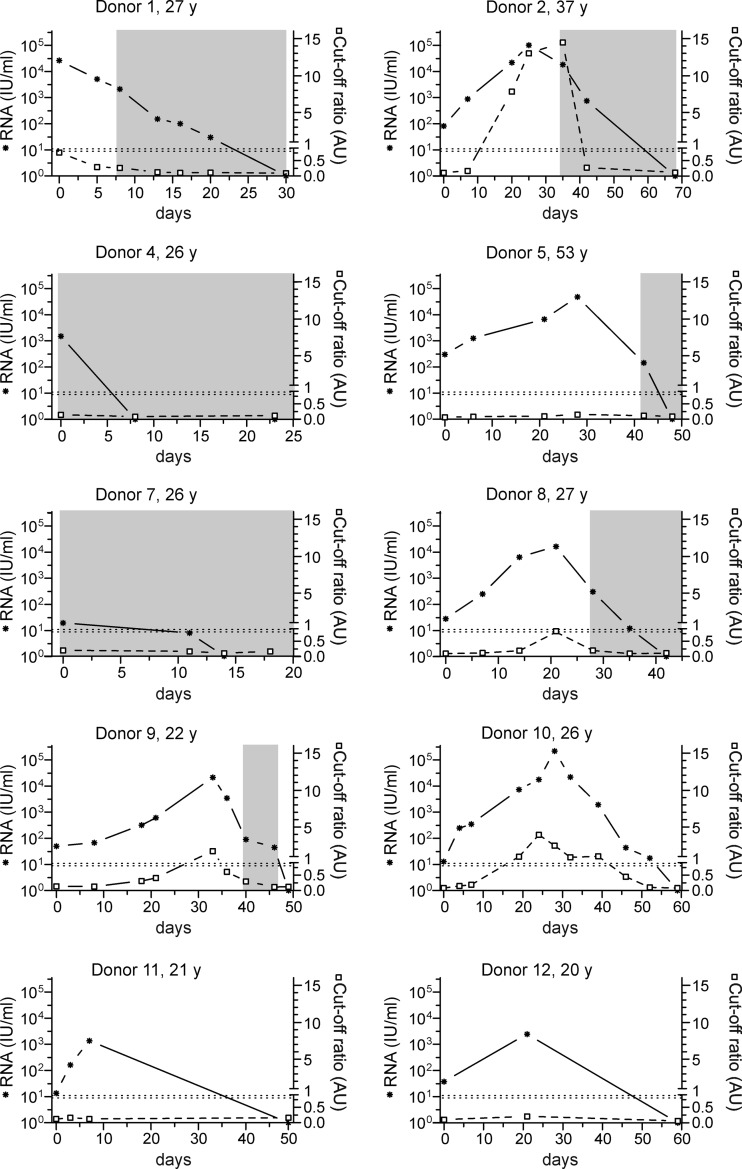
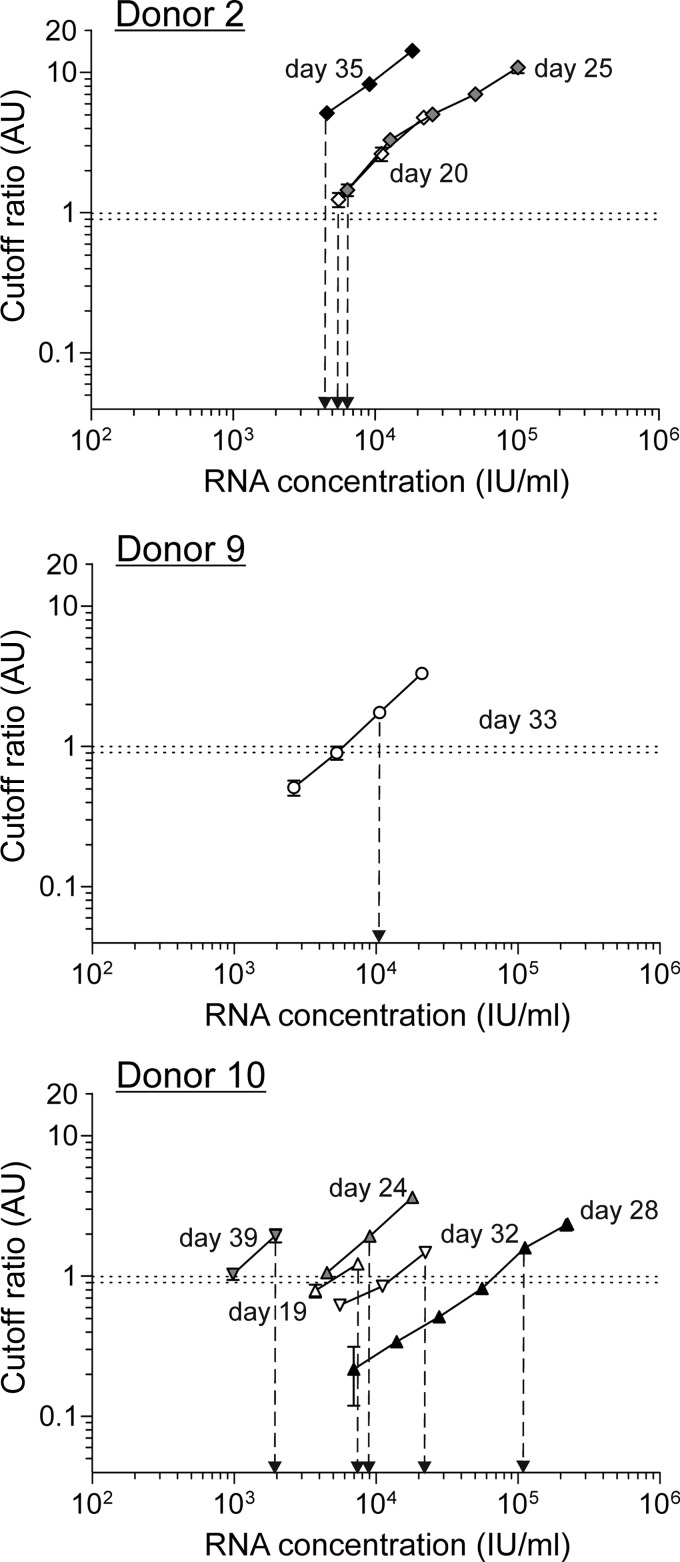
The progression of RNA concentrations and HEV-Ag detection results is shown in Fig. 1. The day of the first detection of hepatitis E virus RNA by PCR screening was defined as day 0 (25). HEV-Ag was detectable only in samples from donors 2, 8 (only borderline results), 9, and 10. In samples from donor 2, HEV-Ag was first detectable at a corresponding HEV concentration of 2.21E + 04 IU/ml on day 20. The antigen detection period continued to day 35 (HEV concentration, 1.83E + 04 IU/ml), culminating in a maximum viremia value of 1.02E + 05 IU/ml on day 25. HEV-Ag was detectable in only one sample from donors 8 (day 21, borderline result) and 9 (day 33) during the progression of HEV viremia, with corresponding HEV concentrations of 1.64 + 04 IU/ml and 2.13E + 04 IU/ml, respectively. For these three donors, HEV-Ag was detectable only at HEV-RNA concentrations of >1.0 × 10E + 04 IU/ml. HEV-Ag was detectable with corresponding HEV concentrations ranging between 7.31E + 03 and 1.92E + 03 IU/m only in samples from donor 10. Antigens were detected in parallel to HEV viremia from day 20 to day 40, with a maximum viremia value of 2.19E + 05 IU/ml. The occurrence of HEV-Ag followed the progression of HEV viremia for these four donors. In samples from all other donors, HEV-Ag was not detectable at any time, although the maximum RNA concentration exceeded the previously determined detection limit of approximately 1 × 10E + 04 IU/ml (e.g., for donor 1, 2.63E + 04 IU/ml, and for donor 5, 4.74E + 04 IU/ml). Analysis of a 2-fold dilution series of positive samples from donors 2, 9, and 10 showed that the detection of HEV-Ag and the lowest corresponding HEV-RNA concentration differed for each donor (Fig. 2), suggesting a potential donor-dependent sensitivity. In samples from donor 9, HEV-Ag was detectable at corresponding RNA concentrations of 5 × 10E + 03 and 1 × 10E + 04 IU/ml, whereas donor 2 still showed clear HEV-Ag positivity at a corresponding RNA concentration of approximately 3 × 10E + 03 IU/ml. Samples from donor 10 demonstrated further differences within the same donor. For example, samples taken on day 28 showed the detection of HEV-Ag at corresponding RNA concentrations > 1 × 10E + 05 IU/ml, whereas samples recovered on day 39 demonstrated HEV-Ag detection at considerably lower RNA concentrations of approximately 1 × 10E + 03 IU/ml. We also tested a 2-fold-dilution series of the WHO-NAT standard due to the absence of a HEV-Ag standard to analyze a potential correlation, but although the WHO-NAT standard contained intact virus particles, HEV-Ag was not detectable in this reference material.
FIG 1.
Comparison of HEV antigen and RNA levels. The course of HEV-RNA concentrations (*) and occurrences of HEV-Ag (□) are displayed. The day of detection of HEV-RNA by PCR screening was defined as day 0. The dotted horizontal line represents the borderline cutoff ratio of the HEV-Ag assay (0.9 to 1.0 AU [arbitrary units]). The time period of HEV-IgM antibody occurrence is indicated with gray shading. y, years of age.
FIG 2.
Correlation of antigen and RNA values. A 2-fold dilution series of the WHO-NAT standard and HEV-Ag-positive samples from donor 2 (three samples, days 20, 25, and 35), donor 9 (one sample, day 33), and donor 10 (five samples, days 19, 24, 28, 32, and 39) were analyzed using the Wantai HEV-Ag test. The dotted horizontal line represents the borderline cutoff ratio of the HEV-Ag assay (0.9 to 1.0 AU [arbitrary units]).
HEV-specific IgM antibodies were detectable in samples from seven donors. However, only donor 2 presented a parallel positivity of HEV-Ag and anti-HEV IgM antibodies. In the other three donors, IgM antibodies were not detectable. Nevertheless, samples were not available within the time frame where IgM seroconversion most likely occurred for donors 11 and 12.
DISCUSSION
The published reports of HEV infections by contaminated blood products (2, 17–20) and the detection of HEV in plasma fractionation pools (23, 24) and samples from blood donors (22, 25, 33, 34) suggest that transfusion transmission of HEV is probably not uncommon. Many of the infections are subclinical (22, 34), with infections being incorrectly diagnosed or undiagnosed due to the applied test regimen. The rates of individual HEV-RNA-positive donors range (21) from 0.012% in Sweden (22) and 0.014% in the United Kingdom (34) to 0.022% to 0.08% in Germany (22, 25) and 0.07% in China (33). The possible clinical courses (asymptomatic, mild hepatitis, acute liver failure) and severities of HEV infections in transfusion recipients are variable, depending on predisposition or immune status. Most likely, the vast majority of HEV infections result in an asymptomatic course (35), but the observed severe courses of HEV infections in major groups of transfusion recipients—e.g., patients with preexisting liver disease (36, 37), pregnant women (38, 39), transplant patients (15, 16), and immunocompromised patients (40)—unalterably raise the issue of blood safety for those patients (2). Currently, the consensus of several authors implies that only NAT testing of blood donors has the potential to effectively prevent viral transmission (22, 25, 41). In this context, the performances of different HEV RT-PCR assays have to be evaluated for the development of screening strategies to estimate the risk of transfusion-transmitted HEV infections. The molecular detection of HEV-RNA is also essential for the diagnosis of acute hepatitis E and the assessment of disease progression (virus clearance versus persistence) (31, 42), at least to detect or monitor HEV infection in major groups of transfusion recipients. Furthermore, the detection of HEV-RNA facilitates the diagnosis of HEV infection in immunocompromised patients without an adequate antibody response (15). The prior assessment of different RT-PCR assays, mainly including in-house tests, revealed a 100- to 1,000-fold difference in their sensitivities (43). It has been further shown that genotype 3 diversity influences RNA quantification (44). Therefore, the first WHO international standard for hepatitis E virus RNA for NAT-based assays was recently developed to allow harmonization of molecular-genetic assays for the detection of HEV-RNA (32).
So far, only one commercially available RT-PCR assay and one in-house RT-PCR assay have been evaluated for HEV blood donor screening, demonstrating a high sensitivity of 4.6 IU/ml (22, 25). Recently, Abravanel et al. compared the RealStar HEV RT-PCR kit and the hepatitis@ceeramTools kit for individual sample screening using the RNAeasy extraction kit, with their results also indicating that the RealStar HEV RT-PCR kit has a higher sensitivity than the hepatitis@ceeramTools kit (42). That study further demonstrated that the usage of an RT protocol based on the ORF3 region is essential to accurately quantify all HEV genotype 3 subtypes because this region is better conserved than most (44). The single-stranded HEV genome has a size of approximately 7.2 kb and contains a short 5′ untranslated region (UTR), three open reading frames (ORF1 to -3), and a short 3′ UTR terminated by a poly(A) tail (45). Abravanel et al. observed that the Ceeram assay is potentially less sensitive for genotype 3e due to a high difference in CT values. Although ORF3 is highly conserved among the four different HEV genotypes, there is still a lack of performance studies referring to commercially available assays, including HEV genotypes 1, 2, and 4 (44), most likely due to the absence of genotype-specific panels. Therefore, the establishment of genotype-specific panels is essential to allow further validation of different assays.
The differences observed for the analytical sensitivities of the evaluated assays in this study are possibly related to differences in the recommended RNA input volumes, starting from 5 μl for the hepatitis@ceeramTools kit, 10 μl for the ampliCube HEV RT-PCR kit, and 25 μl for the RealStar HEV RT-PCR kit. Interestingly, the hepatitis@ceeramTools kit showed similar sensitivities for the two extraction methods, whereas the RealStar HEV RT-PCR kit had approximately 10-fold-lower sensitivity with the NucliSens easyMAG extraction than with the viral 5K extraction. Possible explanations for this observation were so-far-unknown effects caused by the composition of the elution buffers. This statement is in line with the observed inhibition of the ampliCube HEV RT-PCR kit in combination with nucleic acids extracted with the Viral 5k kit. Nevertheless, the manufacturer had no explanation for the observed inhibition (personal communication). The RealStar HEV RT-PCR kit seemed to be most suitable for ID-NAT and MP-NAT, even with a minipool size of 96 samples. The Ceeram assay is rather suitable for minipool screening in a size of 48 samples or lower due to the reduced sensitivity. However, only a little is known about transfusion-associated HEV infections. More data are required regarding the duration of viremia, the infective dose, the role of anti-HEV in the recipient, and the frequency of clinically apparent transfusion-transmitted HEV infections to consider the implementation of HEV NAT blood donor screening (41). Therefore, the required minimum sensitivity has remained undetermined so far.
Studies by Gupta et al. and Majumdar et al. also using the Wantai HEV-Ag assay led to the conclusion that HEV-Ag can be an alternative early diagnostic marker for the detection of active viral replication in acute HEV infection (28, 29). The occurrence of HEV-Ag showed a good concordance with HEV-RNA with similar levels of HEV-Ag and HEV-RNA, whereas the occurrence of anti-HEV IgM did not show any concordance with HEV-RNA (28). In contrast, in the present study conducted with asymptomatic individuals, we observed a significant diagnostic gap between the presence of high viral loads and HEV-Ag occurrence. HEV antigens were detectable in only 4 of 10 individuals, with a diagnostic window ranging from 20 days (donors 2, 8, and 10) to 33 days (donor 9). HEV-Ag was not detectable in the other six donors, and antigen screening alone would have inevitably missed those HEV infections. Although HEV-specific IgM antibodies were detectable in 7 of 10 donors, three donors showed no presence of IgM antibodies at any time. Furthermore, the observed variations in the performances of different anti-HEV IgM assays demand the detection of HEV-RNA for proper diagnosis of infection (42).
In developing countries, a simple and nonlaborious test such as HEV-Ag, enabling early detection of viral infection prior to the occurrence of IgM antibodies, would enhance the management of HEV infection. It has been shown in previous studies that HEV-Ag appears earlier than anti-HEV IgM (HEV-Ag in-house assay [30]) and that the combination of anti-HEV IgM and HEV-Ag compensates for the delay in the detection of anti-HEV IgM (Wantai HEV-Ag assay [46]). The observed differences for the detection of HEV-Ag depending on the corresponding HEV-RNA concentration, including different isolates as well as different samples from the same donor, suggest that the ratio of HEV-Ag to HEV-RNA is most likely not 1/1. This aspect is well known for patients with HBV infections, presenting with an excess (typically 1,000- to 100,000-fold) of empty subviral particles (SVPs) in addition to infectious particles. These SVPs are composed solely of HBV envelope proteins in the form of relatively smaller spheres and filaments of various lengths (47). A related mechanism is unknown for hepatitis E so far, but it is conceivable due to the results of our study. Nevertheless, investigations with a larger cohort are required to establish broad cutoffs. Other explanations might be a shift in the HEV-Ag/HEV-RNA ratio, a shift of antigen conformation, or any other so-far-unknown biological events resulting in release of viral proteins but not nucleic acids during disease progression.
In summary, all of the RT-PCR assays evaluated in this study provided good analytical sensitivity with high reproducibility for both ID-NAT and MP-NAT. Comparison of the anti-HEV IgM assay and HEV-Ag assay with NAT assays tested in this study showed that the identification of HEV-positive blood donors was secured only by molecular genetic screening. The sensitivities of the methods compared in this study allowed a pool screening strategy. However, the pool size needs to be adjusted depending on the respective lower detection limits of the assays used.
Supplementary Material
ACKNOWLEDGMENT
We thank Sarah Kirkby for her linguistic advice.
Footnotes
Published ahead of print 16 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03578-13.
REFERENCES
- 1.Borgen K, Herremans T, Duizer E, Vennema H, Rutjes S, Bosman A, de Roda Husman AM, Koopmans M. 2008. Non-travel related hepatitis E virus genotype 3 infections in the Netherlands; a case series 2004–2006. BMC Infect. Dis. 8:61. 10.1186/1471-2334-8-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adlhoch C, Kaiser M, Pauli G, Koch J, Meisel H. 2009. Indigenous hepatitis E virus infection of a plasma donor in Germany. Vox Sang. 97:303–308. 10.1111/j.1423-0410.2009.01211.x [DOI] [PubMed] [Google Scholar]
- 3.Lu L, Li C, Hagedorn CH. 2006. Phylogenetic analysis of global hepatitis E virus sequences: genetic diversity, subtypes and zoonosis. Rev. Med. Virol. 16:5–36. 10.1002/rmv.482 [DOI] [PubMed] [Google Scholar]
- 4.Mansuy JM, Peron JM, Abravanel F, Poirson H, Dubois M, Miedouge M, Vischi F, Alric L, Vinel JP, Izopet J. 2004. Hepatitis E in the south west of France in individuals who have never visited an endemic area. J. Med. Virol. 74:419–424. 10.1002/jmv.20206 [DOI] [PubMed] [Google Scholar]
- 5.Purcell RH, Emerson SU. 2008. Hepatitis E: an emerging awareness of an old disease. J. Hepatol. 48:494–503. 10.1016/j.jhep.2007.12.008 [DOI] [PubMed] [Google Scholar]
- 6.Guthmann JP, Klovstad H, Boccia D, Hamid N, Pinoges L, Nizou JY, Tatay M, Diaz F, Moren A, Grais RF, Ciglenecki I, Nicand E, Guerin PJ. 2006. A large outbreak of hepatitis E among a displaced population in Darfur, Sudan, 2004: the role of water treatment methods. Clin. Infect. Dis. 42:1685–1691. 10.1086/504321 [DOI] [PubMed] [Google Scholar]
- 7.Rutjes SA, Lodder WJ, Lodder-Verschoor F, van den Berg HH, Vennema H, Duizer E, Koopmans M, de Roda Husman AM. 2009. Sources of hepatitis E virus genotype 3 in The Netherlands. Emerg. Infect. Dis. 15:381–387. 10.3201/eid1503.071472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamar N, Izopet J, Rostaing L. 2013. Hepatitis E virus infection. Curr. Opin. Gastrenterol. 29:271–278. 10.1097/MOG.0b013e32835ff238 [DOI] [PubMed] [Google Scholar]
- 9.Clemente-Casares P, Pina S, Buti M, Jardi R, Martin M, Bofill-Mas S, Girones R. 2003. Hepatitis E virus epidemiology in industrialized countries. Emerg. Infect. Dis. 9:448–454. 10.3201/eid0904.020351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsieh SY, Meng XJ, Wu YH, Liu ST, Tam AW, Lin DY, Liaw YF. 1999. Identity of a novel swine hepatitis E virus in Taiwan forming a monophyletic group with Taiwan isolates of human hepatitis E virus. J. Clin. Microbiol. 37:3828–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krumbholz A, Mohn U, Lange J, Motz M, Wenzel JJ, Jilg W, Walther M, Straube E, Wutzler P, Zell R. 2012. Prevalence of hepatitis E virus-specific antibodies in humans with occupational exposure to pigs. Med. Microbiol. Immunol. 201:239–244. 10.1007/s00430-011-0210-5 [DOI] [PubMed] [Google Scholar]
- 12.Matsuda H, Okada K, Takahashi K, Mishiro S. 2003. Severe hepatitis E virus infection after ingestion of uncooked liver from a wild boar. J. Infect. Dis. 188:944. 10.1086/378074 [DOI] [PubMed] [Google Scholar]
- 13.Yazaki Y, Mizuo H, Takahashi M, Nishizawa T, Sasaki N, Gotanda Y, Okamoto H. 2003. Sporadic acute or fulminant hepatitis E in Hokkaido, Japan, may be food-borne, as suggested by the presence of hepatitis E virus in pig liver as food. J. Gen. Virol. 84:2351–2357. 10.1099/vir.0.19242-0 [DOI] [PubMed] [Google Scholar]
- 14.Schlosser B, Stein A, Neuhaus R, Pahl S, Ramez B, Kruger DH, Berg T, Hofmann J. 2012. Liver transplant from a donor with occult HEV infection induced chronic hepatitis and cirrhosis in the recipient. J. Hepatol. 56:500–502. 10.1016/j.jhep.2011.06.021 [DOI] [PubMed] [Google Scholar]
- 15.Kamar N, Selves J, Mansuy JM, Ouezzani L, Peron JM, Guitard J, Cointault O, Esposito L, Abravanel F, Danjoux M, Durand D, Vinel JP, Izopet J, Rostaing L. 2008. Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N. Engl. J. Med. 358:811–817. 10.1056/NEJMoa0706992 [DOI] [PubMed] [Google Scholar]
- 16.Legrand-Abravanel F, Kamar N, Sandres-Saune K, Garrouste C, Dubois M, Mansuy JM, Muscari F, Sallusto F, Rostaing L, Izopet J. 2010. Characteristics of autochthonous hepatitis E virus infection in solid-organ transplant recipients in France. J. Infect. Dis. 202:835–844. 10.1086/655899 [DOI] [PubMed] [Google Scholar]
- 17.Boxall E, Herborn A, Kochethu G, Pratt G, Adams D, Ijaz S, Teo CG. 2006. Transfusion-transmitted hepatitis E in a ‘nonhyperendemic' country. Transfus. Med. 16:79–83. 10.1111/j.1365-3148.2006.00652.x [DOI] [PubMed] [Google Scholar]
- 18.Colson P, Coze C, Gallian P, Henry M, De Micco P, Tamalet C. 2007. Transfusion-associated hepatitis E, France. Emerg. Infect. Dis. 13:648–649. 10.3201/eid1304.061387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsubayashi K, Kang JH, Sakata H, Takahashi K, Shindo M, Kato M, Sato S, Kato T, Nishimori H, Tsuji K, Maguchi H, Yoshida J, Maekubo H, Mishiro S, Ikeda H. 2008. A case of transfusion-transmitted hepatitis E caused by blood from a donor infected with hepatitis E virus via zoonotic food-borne route. Transfusion 48:1368–1375. 10.1111/j.1537-2995.2008.01722.x [DOI] [PubMed] [Google Scholar]
- 20.Matsubayashi K, Nagaoka Y, Sakata H, Sato S, Fukai K, Kato T, Takahashi K, Mishiro S, Imai M, Takeda N, Ikeda H. 2004. Transfusion-transmitted hepatitis E caused by apparently indigenous hepatitis E virus strain in Hokkaido, Japan. Transfusion 44:934–940. 10.1111/j.1537-2995.2004.03300.x [DOI] [PubMed] [Google Scholar]
- 21.Dreier J, Juhl D. 2014. Autochthonous hepatitis E virus infections: a new transfusion-associated risk? Transfus. Med. Hemother. 41:29–39. 10.1159/000357098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baylis SA, Gartner T, Nick S, Ovemyr J, Blumel J. 2012. Occurrence of hepatitis E virus RNA in plasma donations from Sweden, Germany and the United States. Vox Sang. 103:89–90. 10.1111/j.1423-0410.2011.01583.x [DOI] [PubMed] [Google Scholar]
- 23.Baylis SA, Koc O, Nick S, Blumel J. 2012. Widespread distribution of hepatitis E virus in plasma fractionation pools. Vox Sang. 102:182–183. 10.1111/j.1423-0410.2011.01527.x [DOI] [PubMed] [Google Scholar]
- 24.Corman VM, Drexler JF, Eckerle I, Roth WK, Drosten C, Eis-Hubinger AM. 2013. Zoonotic hepatitis E virus strains in German blood donors. Vox Sang. 104:179–180. 10.1111/j.1423-0410.2012.01638.x [DOI] [PubMed] [Google Scholar]
- 25.Vollmer T, Diekmann J, Johne R, Eberhardt M, Knabbe C, Dreier J. 2012. Novel approach for the detection of hepatitis E virus infection in German blood donors. J. Clin. Microbiol. 50:2708–2713. 10.1128/JCM.01119-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi M, Kusakai S, Mizuo H, Suzuki K, Fujimura K, Masuko K, Sugai Y, Aikawa T, Nishizawa T, Okamoto H. 2005. Simultaneous detection of immunoglobulin A (IgA) and IgM antibodies against hepatitis E virus (HEV) is highly specific for diagnosis of acute HEV infection. J. Clin. Microbiol. 43:49–56. 10.1128/JCM.43.1.49-56.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian DY, Chen Y, Xia NS. 2006. Significance of serum IgA in patients with acute hepatitis E virus infection. World J. Gastroenterol. 12:3919–3923 http://www.wjgnet.com/1007-9327/12/3919.asp [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta E, Pandey P, Pandey S, Sharma MK, Sarin SK. 2013. Role of hepatitis E virus antigen in confirming active viral replication in patients with acute viral hepatitis E infection. J. Clin. Virol. 58:374–377. 10.1016/j.jcv.2013.07.019 [DOI] [PubMed] [Google Scholar]
- 29.Majumdar M, Singh MP, Pujhari SK, Bhatia D, Chawla Y, Ratho RK. 2013. Hepatitis E virus antigen detection as an early diagnostic marker: report from India. J. Med. Virol. 85:823–827. 10.1002/jmv.23529 [DOI] [PubMed] [Google Scholar]
- 30.Zhang F, Li X, Li Z, Harrison TJ, Chong H, Qiao S, Huang W, Zhang H, Zhuang H, Wang Y. 2006. Detection of HEV antigen as a novel marker for the diagnosis of hepatitis E. J. Med. Virol. 78:1441–1448. 10.1002/jmv.20717 [DOI] [PubMed] [Google Scholar]
- 31.Kamar N, Rostaing L, Legrand-Abravanel F, Izopet J. 2013. How should hepatitis E virus infection be defined in organ-transplant recipients? Am. J. Transplant. 13:1935–1936. 10.1111/ajt.12253 [DOI] [PubMed] [Google Scholar]
- 32.Baylis SA, Blümel J, Mizusawa S, Matsubayashi K, Sakata H, Okada Y, Nübling CM, Hanschmann KM HEV Collaborative Study Group. 2013. World Health Organization International Standard to harmonize assays for detection of hepatitis E virus RNA. Emerg. Infect. Dis. 19:729–735. 10.3201/eid1905.121845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo QS, Yan Q, Xiong JH, Ge SX, Shih JW, Ng MH, Zhang J, Xia NS. 2010. Prevalence of hepatitis E virus in Chinese blood donors. J. Clin. Microbiol. 48:317–318. 10.1128/JCM.01466-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ijaz S, Szypulska R, Tettmar KI, Kitchen A, Tedder RS. 2012. Detection of hepatitis E virus RNA in plasma mini-pools from blood donors in England. Vox Sang. 102:272. 10.1111/j.1423-0410.2011.01554.x [DOI] [PubMed] [Google Scholar]
- 35.Beale MA, Tettmar K, Szypulska R, Tedder RS, Ijaz S. 2011. Is there evidence of recent hepatitis E virus infection in English and North Welsh blood donors? Vox Sang. 100:340–342. 10.1111/j.1423-0410.2010.01412.x [DOI] [PubMed] [Google Scholar]
- 36.Dalton HR, Hazeldine S, Banks M, Ijaz S, Bendall R. 2007. Locally acquired hepatitis E in chronic liver disease. Lancet 369:1260. 10.1016/S0140-6736(07)60595-9 [DOI] [PubMed] [Google Scholar]
- 37.Péron JM, Bureau C, Poirson H, Mansuy JM, Alric L, Selves J, Dupuis E, Izopet J, Vinel JP. 2007. Fulminant liver failure from acute autochthonous hepatitis E in France: description of seven patients with acute hepatitis E and encephalopathy. J. Viral Hepat. 14:298–303. 10.1111/j.1365-2893.2007.00858.x [DOI] [PubMed] [Google Scholar]
- 38.Boccia D, Guthmann JP, Klovstad H, Hamid N, Tatay M, Ciglenecki I, Nizou JY, Nicand E, Guerin PJ. 2006. High mortality associated with an outbreak of hepatitis E among displaced persons in Darfur, Sudan. Clin. Infect. Dis. 42:1679–1684. 10.1086/504322 [DOI] [PubMed] [Google Scholar]
- 39.Christensen PB, Engle RE, Hjort C, Homburg KM, Vach W, Georgsen J, Purcell RH. 2008. Time trend of the prevalence of hepatitis E antibodies among farmers and blood donors: a potential zoonosis in Denmark. Clin. Infect. Dis. 47:1026–1031. 10.1086/591970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gérolami R, Moal V, Picard C, Colson P. 2009. Hepatitis E virus as an emerging cause of chronic liver disease in organ transplant recipients. J. Hepatol. 50:622–624. 10.1016/j.jhep.2008.12.008 [DOI] [PubMed] [Google Scholar]
- 41.Juhl D, Baylis SA, Blumel J, Gorg S, Hennig H. 2014. Seroprevalence and incidence of hepatitis E virus infection in German blood donors. Transfusion 54:49–56. 10.1111/trf.12121 [DOI] [PubMed] [Google Scholar]
- 42.Abravanel F, Chapuy-Regaud S, Lhomme S, Dubois M, Peron JM, Alric L, Rostaing L, Kamar N, Izopet J. 2013. Performance of two commercial assays for detecting hepatitis E virus RNA in acute or chronic infections. J. Clin. Microbiol. 51:1913–1916. 10.1128/JCM.00661-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baylis SA, Hanschmann KM, Blumel J, Nubling CM. 2011. Standardization of hepatitis E virus (HEV) nucleic acid amplification technique-based assays: an initial study to evaluate a panel of HEV strains and investigate laboratory performance. J. Clin. Microbiol. 49:1234–1239. 10.1128/JCM.02578-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abravanel F, Sandres-Saune K, Lhomme S, Dubois M, Mansuy JM, Izopet J. 2012. Genotype 3 diversity and quantification of hepatitis E virus RNA. J. Clin. Microbiol. 50:897–902. 10.1128/JCM.05942-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tam AW, Smith MM, Guerra ME, Huang CC, Bradley DW, Fry KE, Reyes GR. 1991. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology 185:120–131. 10.1016/0042-6822(91)90760-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao C, Li L, Harrison TJ, Wang Q, Song A, Fan J, Ma H, Zhang C, Wang Y. 2009. Relationships among viral diagnostic markers and markers of liver function in acute hepatitis E. J. Gastroenterol. 44:139–145. 10.1007/s00535-008-2281-7 [DOI] [PubMed] [Google Scholar]
- 47.Chai N, Chang HE, Nicolas E, Han Z, Jarnik M, Taylor J. 2008. Properties of subviral particles of hepatitis B virus. J. Virol. 82:7812–7817. 10.1128/JVI.00561-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.