Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) is the most prevalent cause of bloodstream infections (BSIs) and is recognized as a major nosocomial pathogen. This study aimed to evaluate a newly designed multiplex real-time PCR assay capable of the simultaneous detection of mecA, S. aureus, and coagulase-negative staphylococci (CoNS) in blood culture specimens. The Real-MRSA and Real-MRCoNS multiplex real-time PCR assays (M&D, Republic of Korea) use the TaqMan probes 16S rRNA for Staphylococcus spp., the nuc gene for S. aureus, and the mecA gene for methicillin resistance. The detection limit of the multiplex real-time PCR assay was 103 CFU/ml per PCR for each gene target. The multiplex real-time PCR assay was evaluated using 118 clinical isolates from various specimen types and a total of 350 positive blood cultures from a continuous monitoring blood culture system. The results obtained with the multiplex real-time PCR assay for the three targets were in agreement with those of conventional identification and susceptibility testing methods except for one organism. Of 350 positive bottle cultures, the sensitivities of the multiplex real-time PCR kit were 100% (166/166 cultures), 97.2% (35/36 cultures), and 99.2% (117/118 cultures) for the 16S rRNA, nuc, and mecA genes, respectively, and the specificities for all three targets were 100%. The Real-MRSA and Real-MRCoNS multiplex real-time PCR assays are very useful for the rapid accurate diagnosis of staphylococcal BSIs. In addition, the Real-MRSA and Real-MRCoNS multiplex real-time PCR assays could have an important impact on the choice of appropriate antimicrobial therapy, based on detection of the mecA gene.
INTRODUCTION
The rapid diagnosis and treatment of bacterial sepsis are requisite to decrease mortality and morbidity rates, because it is a rapidly progressing, life-threatening condition that can cause shock and organ failure (1). Staphylococci are the most commonly isolated organisms, accounting for almost 30% of all hospital-acquired infections and 50% of bloodstream infections (BSIs) (2). Staphylococcus aureus is a leading cause of health care-associated infections ranging from mild conditions such as skin and soft-tissue infections to life-threatening sepsis. Methicillin-resistant S. aureus (MRSA) is recognized as a major nosocomial pathogen associated with intrahospital and interhospital transmission (3). Staphylococcal infections have resulted in significant morbidity, deaths, and longer hospital stays if not treated early with effective antibiotics. The prevalence of MRSA infections exceeds 49% in U.S. hospitals and continues to increase (4). Coagulase-negative staphylococci (CoNS) are also known to be the most common isolates from blood cultures, although many are contaminants.
Fast accurate discrimination of MRSA from methicillin-susceptible staphylococci directly from patient blood samples provides data for proper medical decisions regarding antimicrobial therapy, which plays an important role in the reduction of deaths resulting from sepsis. Currently, bacterial culture is required as a standard method for diagnosis of the presence of bacterial pathogens in clinical samples. However, this technique has some disadvantages with regard to desired detection speed and sensitivity (5). Generally, blood culture samples are incubated for 5 days until they show positive signals in continuous monitoring blood culture systems (CMBCSs). Moreover, blood cultures may lead to false-negative results when fastidious or slowly growing bacteria are involved or when samples are obtained after antimicrobial therapy has been started (6, 7). The early diagnosis and adequate treatment of bacterial infections have great impacts on the outcomes for patients with systemic infections.
Real-time PCR is significantly faster than conventional PCR and other detection methods. The combination of excellent sensitivity and specificity, low contamination risk, ease of performance, and speed has made real-time PCR technology appealing to clinical microbiology laboratories (8). In this study, a multiplex real-time PCR assay using probes specific for S. aureus and a methicillin resistance gene was tested for the rapid detection and identification of MRSA, methicillin-susceptible S. aureus (MSSA), methicillin-resistant CoNS (MRCoNS), and methicillin-susceptible CoNS (MSCoNS) directly from positive blood cultures.
MATERIALS AND METHODS
Bacterial and fungal strains.
A total of 67 bacterial and 28 fungal reference strains and 118 clinical isolates from various specimen types were used to determine the specificity of the multiplex real-time PCR assay (see Tables 2 and 3). A total of 350 positive blood culture samples, including 166 Staphylococcus spp. and 184 nonstaphylococcal strains, were collected to evaluate the diagnostic performance of the multiplex real-time PCR assay. All clinical isolates and positive blood culture samples were collected between January 2013 and May 2013 at Yonsei University Wonju Severance Christian Hospital (Wonju, Republic of Korea). The identification of organisms was conducted with the MicroScan system (Siemens Healthcare Diagnostics, Sacramento, CA) and the Vitek 2 system (bioMérieux, Durham, NC).
TABLE 2.
Specificity of the multiplex real-time PCR assay for detecting the 16S rRNA, nuc, and mecA genes in 95 bacterial and fungal reference strains
| Organism type and genus and species or serovar | Standard straina |
CT for: |
||
|---|---|---|---|---|
| 16S rRNA | nuc | mecA | ||
| Gram-positive bacteria | ||||
| Staphylococcus | ||||
| S. aureus | ATCC 29213 | 28.33 | 29.94 | UDb |
| ATCC 25923 | 13.39 | 17.13 | UD | |
| ATCC 13565 | 14.09 | 14.26 | UD | |
| ATCC 33586 | 16.07 | 15.86 | UD | |
| ATCC BAA-2312 | 22 | 15.76 | 15.24 | |
| S. xylosus | ATCC 29971 | 22.30 | UD | UD |
| S. epidermidis | ATCC 12228 | 12.32 | UD | UD |
| Enterococcus | ||||
| E. hirae | ATCC 9790 | UD | UD | UD |
| E. raffinosus | ATCC 49427 | UD | UD | UD |
| E. sulfureus | ATCC 49903 | UD | UD | UD |
| E. durans | ATCC 19432 | UD | UD | UD |
| E. casseliflavus | ATCC 700327 | UD | UD | UD |
| ATCC 49997 | UD | UD | UD | |
| E. faecium | ATCC 19434 | UD | UD | UD |
| E. faecalis | ATCC 29212 | UD | UD | UD |
| E. mundtii | ATCC 43186 | UD | UD | UD |
| E. cecorum | ATCC 43198 | UD | UD | UD |
| E. gallinarum | ATCC 49573 | UD | UD | UD |
| E. faecalis | ATCC 51299 | UD | UD | UD |
| E. solitarius | ATCC 49428 | UD | UD | UD |
| E. faecium | ATCC 35667 | UD | UD | UD |
| E. malodoratus | ATCC 43197 | UD | UD | UD |
| E. saccharolyticus | ATCC 43076 | UD | UD | UD |
| E. casseliflavus | ATCC 25788 | UD | UD | UD |
| Streptococcus | ||||
| S. pneumoniae | ATCC 49619 | UD | UD | UD |
| S. agalactiae | ATCC 13813 | UD | UD | UD |
| Micrococcus luteus | ATCC 49732 | UD | UD | UD |
| Mycobacterium | ||||
| M. avium | ATCC 25291 | UD | UD | UD |
| M. chelonae | ATCC 35749 | UD | UD | UD |
| M. gastri | ATCC 15754 | UD | UD | UD |
| M. kansasii | ATCC 12478 | UD | UD | UD |
| M. nonchromogenicum | ATCC 19530 | UD | UD | UD |
| M. phlei | ATCC 11758 | UD | UD | UD |
| M. smegmatis | ATCC 19420 | UD | UD | UD |
| M. triviale | ATCC 23292 | UD | UD | UD |
| M. aurum | ATCC 23366 | UD | UD | UD |
| M. farcinogenes | ATCC 35753 | UD | UD | UD |
| M. gilvum | ATCC 43909 | UD | UD | UD |
| M. neoaurum | ATCC 25795 | UD | UD | UD |
| M. parafortuitum | ATCC 19686 | UD | UD | UD |
| M. peregrinum | ATCC 14467 | UD | UD | UD |
| M. septicum | ATCC 700731 | UD | UD | UD |
| M. abscessus | ATCC 19977 | UD | UD | UD |
| Corynebacterium diphtheriae | ATCC 11913 | UD | UD | UD |
| Gram-negative bacteria | ||||
| Escherichia | ||||
| E. coli | ATCC 25922 | UD | UD | UD |
| ATCC 35218 | UD | UD | UD | |
| Enterobacter aerogenes | ATCC 1304 | UD | UD | UD |
| Citrobacter freundii | ATCC 6750 | UD | UD | UD |
| Shigella | ||||
| S. boydii | DML 399 | UD | UD | UD |
| S. dysenteriae | DML 400 | UD | UD | UD |
| S. flexneri | ATCC 9199 | UD | UD | UD |
| Serratia liquefaciens | ATCC 27952 | UD | UD | UD |
| Salmonella | ||||
| S. Typhi | ATCC 19430 | UD | UD | UD |
| S. Enteritidis | ATCC 13076 | UD | UD | UD |
| S. Paratyphi | ATCC 11511 | UD | UD | UD |
| S. Typhimurium | ATCC 13311 | UD | UD | UD |
| S. Newport | ATCC 6962 | UD | UD | UD |
| Klebsiella | ||||
| K. pneumoniae | ATCC 13883 | UD | UD | UD |
| K. oxytoca | ATCC 700324 | UD | UD | UD |
| Proteus | ||||
| P. alcalifaciens | ATCC 51902 | UD | UD | UD |
| P. vulgaris | ATCC 49132 | UD | UD | UD |
| P. mirabilis | ATCC 49132 | UD | UD | UD |
| Pseudomonas | ||||
| P. cepacia | ATCC 25608 | UD | UD | UD |
| P. aeruginosa | ATCC 27853 | UD | UD | UD |
| Haemophilus influenzae | ATCC 49247 | UD | UD | UD |
| Leclercia adecarboxylata | ATCC 23216 | UD | UD | UD |
| Bordetella bronchiseptica | ATCC 10580 | UD | UD | UD |
| Fungi | ||||
| Candida | ||||
| C. albicans | ATCC 36802 | UD | UD | UD |
| ATCC 10231 | UD | UD | UD | |
| C. tropicalis | ATCC 14506 | UD | UD | UD |
| C. glabrata | ATCC 38326 | UD | UD | UD |
| C. parapsilosis | ATCC 7330 | UD | UD | UD |
| C. lusitaniae | ATCC 34449 | UD | UD | UD |
| C. guilliermondii | ATCC 56822 | UD | UD | UD |
| C. krusei | ATCC 2159 | UD | UD | UD |
| Penicillium | ||||
| P. camemberti | ATCC 58608 | UD | UD | UD |
| P. paneum | KACC 44823 | UD | UD | UD |
| Aspergillus | ||||
| A. oryzae var. oryzae | KACC 44847 | UD | UD | UD |
| A. oryzae var. effusus | ATCC 1010 | UD | UD | UD |
| A. clavatus | ATCC 66443 | UD | UD | UD |
| A. sydowii | KACC 41869 | UD | UD | UD |
| A. fumigatus | KCMF 10773 | UD | UD | UD |
| A. flavus | KCMF 10777 | UD | UD | UD |
| A. tamari | ATCC 20054 | UD | UD | UD |
| Fusarium acuminatum | ATCC 10466 | UD | UD | UD |
| Aureobasidium pullulans | KACC 41291 | UD | UD | UD |
| Bipolaris sorokiniana | KACC 44841 | UD | UD | UD |
| Cryptococcus neoformans | KCMF 20047 | UD | UD | UD |
| Kodamaea ohmeri | KCMF 20430 | UD | UD | UD |
| Saccharomyces cerevisiae | KCMF 50427 | UD | UD | UD |
| Trichophyton | ||||
| T. rubrum | KCMF 10444 | UD | UD | UD |
| T. mentagrophytes | KCMF 10515 | UD | UD | UD |
| Microsporum canis | KCMF 10531 | UD | UD | UD |
| Epidermophyton floccosum | ATCC 52063 | UD | UD | UD |
| Malassezia furfur | KCMF 20409 | UD | UD | UD |
ATCC, American Type Culture Collection; DML, Diagnostic Microbiology Laboratory, Biomedical Laboratory Science, Yonsei University; KACC, Korean Agricultural Culture Collection; KCMF, Korea Culture Collection Medical Fungi.
UD, undetermined.
TABLE 3.
Results of the multiplex real-time PCR assay for the discrimination of MRSA, MSSA, MRCoNS, and MSCoNS among 118 clinical isolates
| Culture identification | Total no. of samples | No. of isolates positive for: |
CT values |
|||
|---|---|---|---|---|---|---|
| 16S rRNA | nuc | mecA | Range | Average | ||
| Staphylococcus aureus | 12 | 12 | 12 | 9 | 21.12–30.85 | 24.98 |
| Staphylococcus epidermidis | 4 | 4 | 0 | 3 | 24.18–31.03 | 27.98 |
| Staphylococcus haemolyticus | 3 | 3 | 0 | 3 | 23.55–25.34 | 24.29 |
| Staphylococcus capitis | 1 | 1 | 0 | 1 | 31.66 | 31.66 |
| Streptococcus spp. | 5 | 0 | 0 | 0 | UDa | UD |
| Enterococcus faecalis | 4 | 0 | 0 | 0 | UD | UD |
| Enterococcus faecium | 10 | 0 | 0 | 0 | UD | UD |
| Enterococcus mundtii | 1 | 0 | 0 | 0 | UD | UD |
| Corynebacterium spp. | 1 | 0 | 0 | 0 | UD | UD |
| Escherichia coli | 16 | 0 | 0 | 0 | UD | UD |
| Klebsiella pneumoniae | 13 | 0 | 0 | 0 | UD | UD |
| Pseudomonas aeruginosa | 13 | 0 | 0 | 0 | UD | UD |
| Acinetobacter baumannii | 11 | 0 | 0 | 0 | UD | UD |
| Enterobacter asburiae | 1 | 0 | 0 | 0 | UD | UD |
| Enterobacter cloacae | 1 | 0 | 0 | 0 | UD | UD |
| Enterobacter asburiae | 1 | 0 | 0 | 0 | UD | UD |
| Moraxella catarrhalis | 1 | 0 | 0 | 0 | UD | UD |
| Serratia marcescens | 1 | 0 | 0 | 0 | UD | UD |
| Providencia rettgeri | 1 | 0 | 0 | 0 | UD | UD |
| Morganella morganii | 1 | 0 | 0 | 0 | UD | UD |
| Proteus mirabilis | 1 | 0 | 0 | 0 | UD | UD |
| Aeromonas spp. | 1 | 0 | 0 | 0 | UD | UD |
| Citrobacter freundii | 2 | 0 | 0 | 0 | UD | UD |
| Candida albicans | 5 | 0 | 0 | 0 | UD | UD |
| Candida glabrata | 1 | 0 | 0 | 0 | UD | UD |
| Candida parapsilosis | 3 | 0 | 0 | 0 | UD | UD |
| Candida tropicalis | 2 | 0 | 0 | 0 | UD | UD |
| Saccharomyces cerevisiae | 2 | 0 | 0 | 0 | UD | UD |
| Total | 118 | |||||
UD, undetermined.
Blood culture and collection of blood culture bottle samples.
Two or three pairs of culture bottles for aerobes or anaerobes were incubated in a BacT/Alert 3D (bioMérieux), Bactec 9240 (Becton, Dickinson Diagnostic System, Spark, MD), or Bactec FX (Becton, Dickinson) blood culture system for 5 days after inoculation with blood drawn from the patient at the bedside. If bacterial growth was not detected within 5 days, then the blood culture result was considered negative. When bacterial growth was noted, the culture sample was inoculated onto both blood agar and MacConkey agar plates, which were then cultured overnight at 35°C in a 5% CO2 incubator. Isolates were identified based on colony morphology, Gram staining, and biochemical test results and were further verified with a Vitek 2 (bioMérieux), MicroScan (Siemens), or Vitek 2 yeast identification card (bioMérieux) system. Antimicrobial susceptibility tests were performed with the Clinical and Laboratory Standards Institute (CLSI) recommended disk diffusion test, the Vitek 2 (bioMérieux) system, and the MicroScan (Siemens) system.
DNA preparation.
To prepare DNA templates for the multiplex real-time PCR assay, one colony of each type strain and clinical isolate was suspended in 100 μl DNA extraction solution (M&D, Wonju, Republic of Korea). The suspended bacterial solution was boiled for 10 min. After centrifugation at 13,000 × g for 10 min, the supernatant was used as the DNA template. For preparation of DNA templates from positive blood culture bottle samples, 0.5 ml of blood suspension was taken directly from the blood culture bottle and 1.0 ml phosphate-buffered saline (PBS) (pH 8.0) was added. After centrifugation at 13,000 × g for 1 min, the supernatant was removed and the pellet was resuspended in 1.0 ml of sterile red blood cell (RBC) lysis buffer, for RBC lysis, and centrifuged at 13,000 × g for 1 min. This washing step was repeated twice, and the pellet was resuspended in DNA extraction solution as described above.
Multiplex real-time PCR TaqMan assay.
The multiplex real-time PCR TaqMan assay was carried out with the Real-MRSA and Real-MRCoNS multiplex real-time PCR assay kits (M&D, Wonju, Republic of Korea) using the CFX-96 real-time PCR system (Bio-Rad, Hercules, CA), which was used for thermocycling and fluorescence detection. The real-time PCR amplification was performed in a total volume of 20 μl containing 10 μl of 2× Thunderbird probe quantitative PCR mixture (Toyobo, Osaka, Japan), 5.0 μl of primer and TaqMan probe mixture, and 5 μl of template DNA; distilled water (DW) was added for a final volume of 20 μl. The specific primers and probes used for the identification of Staphylococcus species and S. aureus and for detection of antibiotic resistance are listed in Table 1. The multiplex real-time PCR assay detected the 16S rRNA, nuc, and mecA genes simultaneously, in a single tube, by incorporating three target-specific TaqMan probes labeled with different fluorophores (Cy5, HEX, and FAM).
TABLE 1.
Primers and probes used in this study
| Target and primer/probe name | Nucleotide sequence (5′ to 3′) | Modificationsa |
|---|---|---|
| 16S rRNA | ||
| 16S-472F | AGTGATGAAGGTCTTCGGATCGTAAA | |
| 16S-575R | CGTGGCTTTCTGATTAGGTACCGTC | |
| 16S-513P | GGAAGAACAWAYGTGTAAGTAACTRTGCACRT | Cy5 and BHQ2 |
| nuc | ||
| nuc263-F | AAAGCGATTGATGGTGATACGGTT | |
| nuc355-R | TGCTTTGTTTCAGGTGTATCAACCA | |
| nuc294-P | ATGTACAAAGGTCAACCAATGACATTYAGA | HEX and BHQ1 |
| mecA | ||
| mecA-1501F | GCTCAAATTTCAAACAAAAATTTAGATAATG | |
| mecA-1598R | TGAAAGGATCTGTACTGGGTTAATCAGT | |
| mecA-1542P | AGCTGATTCAGGTTACGGACAAGGTGA | FAM and BHQ1 |
Cy5, cyanine fluorescein; BHQ, black hole quencher; HEX, 4,4,7,2′,4′,5′,7′-hexachloro-6-carboxyfluorescein; FAM, 6-carboxyfluorescein.
Positive and negative controls were included throughout the procedure. No-template controls with sterile DW instead of template DNA were incorporated in each run under the following conditions: 95°C for 3 min and 35 cycles of 95°C for 20 s and 60°C for 40 s in a single real-time PCR assay. The bacterial load was quantified by determining the cycle threshold (CT), which is the number of PCR cycles required for the fluorescence to exceed a value significantly higher than the background fluorescence level.
Spiking experiment to determine the assay detection limit.
To determine the detection limit of the multiplex real-time PCR assay, the cultured S. aureus (ATCC 25923) bacterial reference strain was suspended in 1× (PBS), and the density of the bacterial suspension was adjusted to 0.5 McFarland units (approximately 1.5 × 108 cells/ml). A series of 10-fold dilutions, which ranged from 108 to 100 CFU/ml, were prepared from the bacterial suspensions, and 100 μl from each dilution was spread on a blood agar plate. Plates were incubated at 37°C for 16 h to count CFU. Each dilution was spiked into 1.0 ml of a negative blood culture sample for 5 days, and each spiked sample was used for genomic DNA extraction with DNA extraction solution (M&D, Wonju, Republic of Korea). The eluted DNA (5.0 μl) was used as the template for the multiplex real-time PCR assay.
PCR-reverse blot hybridization assay.
To confirm the results of the multiplex real-time PCR assay for the identification of Staphylococcus spp. and the determination of methicillin resistance with positive blood culture samples, the PCR-reverse blot hybridization assay (REBA), the REBA Sepsis-ID test (M&D), was performed according to the manufacturer's instructions. PCR was performed using a 50-μl reaction mixture (GeNet Bio, Daejeon, Republic of Korea) containing 25 μl of 2× master mix, 10 μl of 1× primer mixture, 5.0 μl of sample DNA, and sterile DW for a final volume of 50 μl. The first 10 PCR cycles consisted of denaturation at 95°C for 30 s followed by annealing and extension at 60°C for 30 s. These 10 cycles were followed by 40 cycles of denaturation at 95°C for 30 s followed by annealing and extension at 54°C for 30 s. After the final cycle, samples were maintained at 72°C for 10 min to complete the synthesis of all strands. The amplified target was visualized as a single band corresponding to a length of 100 to 200 bp, using the ChemiDoc system (Vilber Lourmat, Germany).
For REBA Sepsis-ID, the hybridization and washing processes were performed according to the manufacturer's instructions. In brief, biotinylated PCR products were denatured at 25°C for 5 min in denaturation solution and diluted in 970 μl hybridization solution on the REBA membrane strip, in the provided blotting tray. Denatured single-stranded PCR products were used to hybridize with the probes on the strip at 55°C for 30 min. The strips were then washed twice in 1.0 ml washing solution at 55°C for 10 min with gentle shaking, incubated at 25°C for 30 min with streptavidin-alkaline phosphatase conjugate (Roche Diagnostics, Mannheim, Germany) diluted 1:2,000 in conjugate diluent solution (CDS), and washed twice with 1.0 ml CDS at room temperature for 1 min. Colorimetric hybridization signals were visualized with the addition of alkaline phosphatase-mediated staining solution, i.e., nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate (NBT-BCIP) (Roche Diagnostics), diluted 1:50, and strips were incubated until the color was detected. Finally, the band patterns were read and interpreted.
RESULTS
Multiplex real-time PCR assay sensitivity and specificity with reference strains.
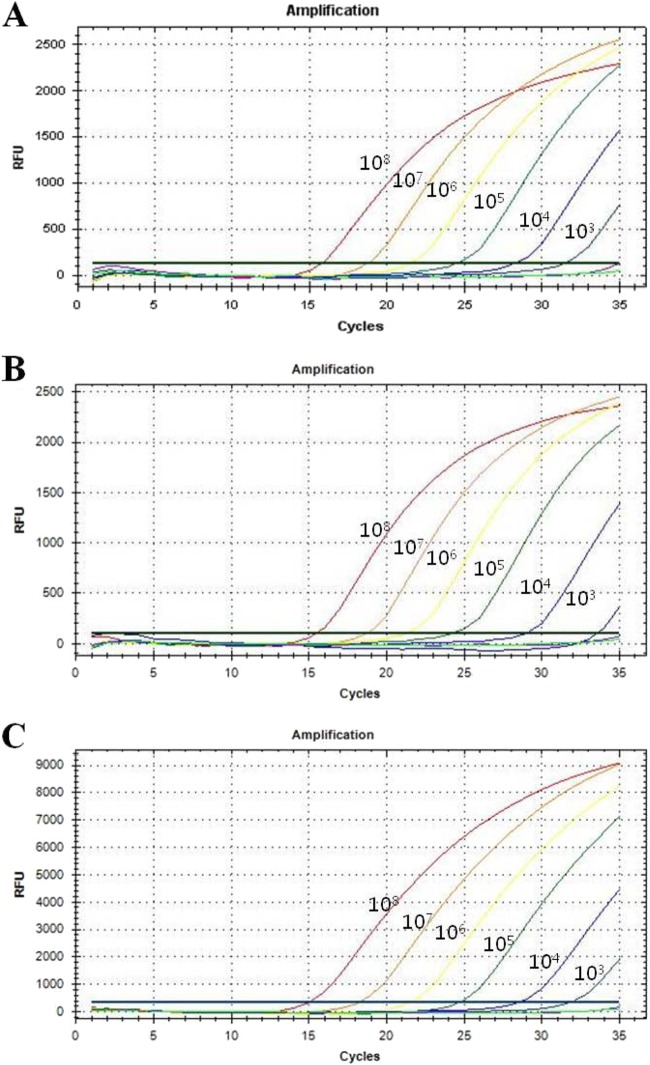
Data from a spiking experiment showed that the detection limit of the multiplex real-time PCR assay for the 16S rRNA, nuc, and mecA genes was 103 CFU/ml (Fig. 1). The CT values for the 16S rRNA, nuc, and mecA genes with each cell concentrate (108 to 103 CFU/ml) ranged from 15 to 31.77, from 15.76 to 31.36, and from 15.24 to 31.15, respectively.
FIG 1.
Detection limits for the three target probes from 10-fold serial dilutions of spiked samples. Serially diluted S. aureus DNA ranging from 108 to 100 CFU/ml was used to determine the detection limit of the multiplex real-time PCR assay. (A) Amplification curve for the 16S rRNA probe for detecting Staphylococcus spp. (B) Amplification curve for the nuc probe for detecting S. aureus. (C) Amplification curve for the mecA probe for detecting methicillin resistance. The overall detection limits of this assay for the 16S rRNA, nuc, and mecA genes were approximately 103 CFU/ml. RFU, relative fluorescence units.
To determine the assay specificity, primers and probes for detecting the 16S rRNA, nuc, and mecA genes were tested with 88 nonstaphylococcal reference strains, including 37 Gram-positive and 23 Gram-negative bacteria and 28 fungi. The multiplex real-time PCR assay for detecting the 16S rRNA, nuc, and mecA genes yielded negative results with all nonstaphylococcal reference strains; thus, no cross-reactivity was detected (Table 2).
All seven staphylococcal reference strains, including five S. aureus strains, one Staphylococcus xylosus strain, and one Staphylococcus epidermidis strain, showed positive fluorescence signals for 16S rRNA in the multiplex real-time PCR assay. Five S. aureus reference strains showed positive results and two non-S. aureus reference strains were negative for the S. aureus-specific nuc gene in the multiplex real-time PCR assay (Table 2). Therefore, the specificity of the multiplex real-time PCR assay for detecting 16S rRNA, nuc, and mecA genes in reference strains was 100%.
Results of the multiplex real-time PCR assay with clinical isolates.
The results of conventional methods and the real-time PCR assay with 118 clinical isolates, including 41 Gram-positive and 64 Gram-negative bacteria and 13 fungi, were completely concordant (100%). All 20 staphylococcal clinical isolates, including 12 S. aureus strains, four S. epidermidis strains, three Staphylococcus haemolyticus strains, and one Staphylococcus capitis strain, showed positive fluorescence signals for detecting 16S rRNA with the multiplex real-time PCR assay. All of the S. aureus and CoNS isolates showed positive and negative signals for the nuc gene, respectively, in the multiplex real-time PCR assay (Table 3).
Antibiotic susceptibility test results revealed that nine of 12 S. aureus (75%) and seven of eight CoNS (87.5%) clinical isolates with known oxacillin resistance (≥4.0 μg/ml) were all positive for the mecA gene, based on results of the multiplex real-time PCR assay (Table 3). These data reveal that the sensitivity and specificity of the multiplex real-time PCR assay for detecting the 16S rRNA, nuc, and mecA genes were 100% with clinical isolates.
Results of the multiplex real-time PCR assay with positive blood culture samples.
To evaluate the performance of the multiplex real-time PCR assay with positive blood culture samples, a total of 350 positive blood culture bottle samples were used. Among 350 positive blood culture samples, 166 cases were identified as Staphylococcus spp. by the standard culture method. They were all positive and 184 non-Staphylococcus cases were negative for 16S rRNA in the multiplex real-time PCR assay (Table 4). These results show that the results of the multiplex real-time PCR assay in targeting 16S rRNA to differentiate Staphylococcus spp. from non-Staphylococcus spp. in positive blood culture samples were totally concordant with standard culture results.
TABLE 4.
Comparison of the multiplex real-time PCR assay, conventional culture identification, and antimicrobial susceptibility test with 166 Staphylococcus-positive blood culture samples
| Culture identification(s) | No. of samples (n = 166) | Multiplex real-time PCR assay results (n [%]) withb: |
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
Staphylococcus-specific probe (n = 166) |
S. aureus-specific probe (n = 166) |
Methicillin-resistant staphylococci probe (n = 118) |
|||||||
| MRSA (n = 23) |
MRCoNS (n = 95) |
||||||||
| 16S rRNA+ | 16S rRNA− | nuc+ | nuc− | mecA+ | mecA− | mecA+ | mecA− | ||
| Staphylococcus | |||||||||
| S. aureus | 36 | 36 (100) | 0 (0) | 35 (97.2) | 1 (2.8) | 22 (95.7) | 1 (4.3) | ||
| CoNS | |||||||||
| S. epidermidis | 63 | 63 (100) | 0 (0) | 0 (0) | 63 (100) | 48 (100) | 0 (0) | ||
| S. hominis | 26 | 26 (100) | 0 (0) | 0 (0) | 26 (100) | 18 (100) | 0 (0) | ||
| S. capitis | 23 | 23 (100) | 0 (0) | 0 (0) | 23 (100) | 15 (100) | 0 (0) | ||
| S. haemolyticus | 12 | 12 (100) | 0 (0) | 0 (0) | 12 (100) | 10 (100) | 0 (0) | ||
| S. saprophyticus | 2 | 2 (100) | 0 (0) | 0 (0) | 2 (100) | 2 (100) | 0 (0) | ||
| S. xylosus | 1 | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | 0 (0) | ||
| S. warneri | 1 | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | ||
| S. intermedius | 1 | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 1 (100) | 0 (0) | ||
| S. epidermidis, S. warneria | 1 | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | ||
| Total | 166 | 166 (100) | 0 (0) | 35 (21.1) | 131 (78.9) | 22 (95.7) | 1 (4.3) | 95 (100) | 0 (0) |
Polymicrobial infection.
Superscript plus symbols indicate the presence of the gene, and superscript minus symbols indicate its absence.
Among 166 Staphylococcus cases, 36 were identified as S. aureus by bacterial culture methods, and all except one case were positive for the S. aureus-specific nuc gene in the multiplex real-time PCR assay. A total of 130 cases, including 63 S. epidermidis cases (48.5%), 26 Staphylococcus hominis cases (20%), 23 S. capitis cases (17.7%), 12 S. haemolyticus cases (9.2%), two Staphylococcus saprophyticus cases (1.5%), one S. xylosus case (0.8%), one Staphylococcus warneri case (0.8%), one Staphylococcus intermedius case (0.8%), and one polymicrobial infection (S. epidermidis and S. warneri) case (0.8%), were identified as CoNS by bacterial culture and all were negative for the nuc gene by multiplex real-time PCR assay testing (Table 4). These results show that, except for one case, the results of the multiplex real-time PCR assay in targeting the nuc gene to differentiate S. aureus from CoNS in positive blood culture samples were highly concordant with standard culture results.
Among 36 S. aureus cases, 23 were resistant to oxacillin and 22 were positive for the methicillin resistance mecA gene (Table 4). Among the 130 CoNS cases, 95 cases, including 48 S. epidermidis cases, 18 S. capitis cases, 15 S. hominis cases, 10 S. haemolyticus cases, two S. saprophyticus cases, and one case each of S. xylosus and S. intermedius, were resistant to oxacillin and all were positive for the mecA gene in the multiplex real-time PCR assay (Table 4). These results show that, except for one case, the results of the multiplex real-time PCR assay in targeting the mecA gene to detect methicillin resistance in S. aureus and CoNS in positive blood culture samples were highly concordant with the results of standard antimicrobial susceptibility tests.
Comparison of bacterial identification and methicillin susceptibility test results with the multiplex real-time PCR assay and the PCR-REBA.
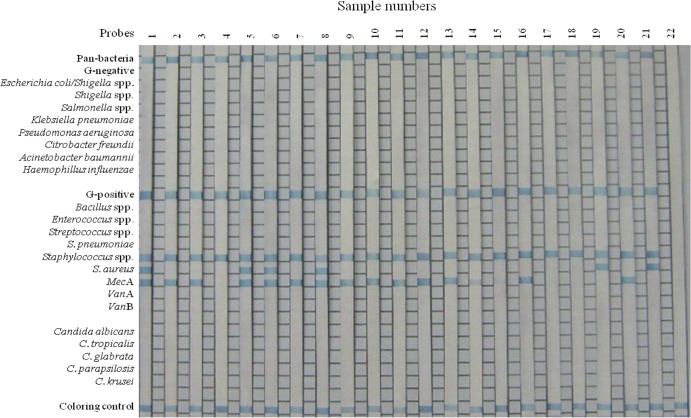
Another molecular assay, the PCR-REBA, was performed with the same DNA samples from positive blood cultures in order to confirm false positivity of the multiplex real-time PCR assay. The results were compared with standard culture method results (Fig. 2). All 166 staphylococcal positive samples from the multiplex real-time PCR assay showed positive signals with Gram-positive strain- and Staphylococcus genus-specific probes in the REBA Sepsis-ID test. Results from the multiplex real-time PCR assay and the PCR-REBA for detecting methicillin resistance were all concordant except for two cases. One case was identified as MRSA by the standard culture method and the PCR-REBA (positive for S. aureus and the mecA gene); however, the multiplex real-time PCR assay results were discordant with the results of those two assays (negative with the nuc and mecA gene-specific probes) (Table 5). The other sample was identified as MRCoNS by the standard culture method and the multiplex real-time PCR assay (positive for the nuc and mecA genes), but the PCR-REBA results were discordant with the results of those two assays (negative with the mecA gene-specific probe) (Table 5).
FIG 2.
Typical results of the PCR-REBA with blood culture bottle samples positive for Staphylococcus spp., as a confirmative assay for the multiplex real-time PCR assay. Lanes 1, 5, 6, and 8, MRSA; lanes 2, 3, 7, 9, 10, 11, 12, 13, 14, 15, 16, and 20, MRCoNS; lanes 4, 17, and 18, MSCoNS; lanes 19 and 21, MSSA; lane 22, negative control. G, Gram.
TABLE 5.
Comparison of results between the multiplex real-time PCR assay and the PCR-REBA
| Culture identification | No. (%) identified | Multiplex real-time PCR assay results (no. [%] of isolates)a |
PCR-REBA results (no. [%] of isolates)a |
||||||
|---|---|---|---|---|---|---|---|---|---|
| 16S rRNA+, nuc+, mecA+ | 16S rRNA+, nuc+, mecA− | 16S rRNA+, nuc−, mecA+ | 16S rRNA+, nuc−, mecA− | 16S rRNA+, nuc+, mecA+ | 16S rRNA+, nuc+, mecA− | 16S rRNA+, nuc−, mecA+ | 16S rRNA+, nuc−, mecA− | ||
| MRSA | 23 (100) | 22 (95.7) | 0 (0) | 0 (0) | 1 (4.3) | 23 (100) | 0 (0) | 0 (0) | 0 (0) |
| MSSA | 13 (100) | 0 (0) | 13 (100) | 0 (0) | 0 (0) | 0 (0) | 13 (100) | 0 (0) | 0 (0) |
| MRCoNS | 95 (100) | 0 (0) | 0 (0) | 95 (100) | 0 (0) | 0 (0) | 0 (0) | 94 (98.9) | 1 (1.1) |
| MSCoNS | 35 (100) | 0 (0) | 0 (0) | 0 (0) | 35 (100) | 0 (0) | 0 (0) | 0 (0) | 35 (100) |
| Total | 166 | 22 | 13 | 95 | 36 | 23 | 13 | 94 | 36 |
Superscript plus symbols indicate the presence of the gene, and superscript minus symbols indicate its absence.
DISCUSSION
Until recently, initial treatment with antimicrobials for bacteremia often followed the identification of bacterial species by Gram staining of positive blood cultures. However, this process requires 48 to 72 h (2 to 3 days) for the identification of pathogens such as MRSA, MSSA, MRCoNS, and MSCoNS, in addition to the accurate determination of antimicrobial sensitivities for prompt optimal patient therapy and infection-control initiatives (9). Molecular diagnostic methods such as real-time PCR are now becoming established in clinical laboratories, and coupling this technology with traditional methods has provided rapid, specific, sensitive detection of microbial pathogens from blood cultures within 2 to 4 h (10, 11).
In this study, a multiplex real-time PCR assay that targets bacterial 16S rRNA to distinguish Staphylococcus spp., the nuc gene to distinguish S. aureus from other Staphylococcus spp., and the mecA gene for the detection of methicillin resistance was evaluated with reference strains and clinical samples. Results of the present study showed that the multiplex real-time PCR assay was rapid, with a turnaround time of usually no more than 4 h, which included 1 h for DNA preparation and 1.5 h for target DNA amplification. It allowed for the rapid identification of S. aureus and CoNS and their methicillin resistance status without a post-PCR process (e.g., agarose gel electrophoresis), and amplicon recognition was achieved by monitoring the accumulation of specific products during each cycle, in comparison with other PCR assays. Another advantage of this assay over standard PCR assays is that the entire process from amplification to analysis is performed in the same tube. This differs from standard PCR assays, in which the PCR product is moved and manipulated into other formats. As a result, there is a decreased possibility of contamination of the product with real-time PCR methods. Furthermore, the assay was very specific and sensitive, because the results were highly correlated with standard bacterial identification and antimicrobial susceptibility test results (Table 6). A possible limitation of the assay is the occurrence of false-negative results due to the presence of PCR inhibitors or polymicrobial infections in direct blood samples.
TABLE 6.
Overall sensitivity and specificity of the multiplex real-time PCR assay in comparison with the standard blood culture method with positive blood culture samples
| Multiplex real-time PCR assay result | No. (%) |
Sensitivity (%) | Specificity (%) | ||||
|---|---|---|---|---|---|---|---|
|
Staphylococcus spp. (n = 166) |
Methicillin-resistant staphylococci (n = 118) |
Nonstaphylococcal strains (n = 184) | |||||
| S. aureus (n = 36) | CoNS (n = 130) | MRSA (n = 23) | MRCoNS (n = 95) | ||||
| 16S rRNA | |||||||
| Positive | 36 (100) | 130 (100) | 0 (0) | 100 | 100 | ||
| Negative | 0 (0) | 0 (0) | 184 (100) | ||||
| nuc | |||||||
| Positive | 35 (97.2) | 0 (0) | 0 (0) | 97.2 | 100 | ||
| Negative | 1 (2.8) | 0 (0) | 184 (100) | ||||
| mecA | |||||||
| Positive | 22 (95.7) | 95 (100) | 0 (0) | 99.2 | 100 | ||
| Negative | 1 (4.3) | 0 (0) | 184 (100) | ||||
In previous reports from other study groups, CoNS was reported to be the major causative microorganism in sepsis (12, 13). In this study, S. epidermidis, which is a CoNS member, was isolated most often from positive blood culture samples, with a total of 76 cases (58.5% of CoNS cases). This was identical to the results from other studies (14).
Among 166 Staphylococcus cases, 118 (71.1%) were resistant to methicillin by the conventional antimicrobial susceptibility tests, and their drug resistance was also tested by the multiplex real-time PCR assay. These results were confirmed with the PCR-REBA molecular diagnostic test. The results of the two different molecular diagnostic assays in detecting methicillin resistance among 118 Staphylococcus positive blood culture samples were highly concordant each other, except for two cases (Table 5); one of the cases was a polymicrobial infection case in which the patient was infected with both Enterococcus faecalis and S. aureus but the case was negative for both the nuc and mecA genes in the multiplex real-time PCR assay. The negative results were attributable to the fact that the signal strength for E. faecalis was much stronger than that for S. aureus in the REBA Sepsis-ID assay, because the E. faecalis genomic DNA concentration may have been much greater than that of S. aureus for target amplification, and the fluorescence signal for E. faecalis could not be detected in the multiplex real-time PCR assay because the 16S rRNA gene of the assay was specifically detected in Staphylococcus spp. The other case was identified as S. saprophyticus that was resistant to oxacillin. However, the multiplex real-time PCR assay results were concordant with the conventional assay results, while the PCR-REBA results were not concordant with the results of the two assays. Therefore, the overall concordance rates between the two molecular diagnostic assays for detecting the 16S rRNA, nuc, and mecA genes were 100%, 98.6%, and 99.5%, respectively.
Additionally, the multiplex real-time PCR assay results for detecting the three target genes were negative for all 184 nonstaphylococcal positive blood culture samples; thus, no cross-reactivity was demonstrated, and the overall specificity of this assay was 100% for each of the target genes. The overall sensitivities of the multiplex real-time PCR assay were 100% (166/166 positive blood culture samples), 97.2% (35/36 positive blood culture samples), and 99.2% (117/118 positive blood culture samples) for detecting the 16S rRNA, nuc, and mecA genes, respectively, in Staphylococcus positive blood culture samples (Table 6).
In conclusion, the use of the multiplex real-time PCR assay showed rapid, highly sensitive, specific results for detecting MRSA, MSSA, MSCoNS, and MRCoNS species directly from positive blood culture bottle samples. Although the use of molecular diagnostic technology is more expensive than conventional methods, the clinical and economic benefits of saving time in treatment remains to be elucidated. Therefore, the Real-MRSA and Real-MRCoNS multiplex real-time PCR assays may provide the essential information to accelerate therapeutic decisions for earlier and adequate antibiotic treatment in the acute phase of sepsis.
ACKNOWLEDGMENT
This study was supported by the Korea Health Technology R&D Project, Ministry of Health and Welfare, Republic of Korea (grant A121030 to H.L.).
Footnotes
Published ahead of print 19 March 2014
REFERENCES
- 1.Engel C, Brunkhorst FM, Bone HG, Brunkhorst R, Gerlach H, Grond S. 2007. Epidemiology of sepsis in Germany: results from a national prospective multicenter study. Intensive Care Med. 33:606–618. 10.1007/s00134-006-0517-7 [DOI] [PubMed] [Google Scholar]
- 2.Gradelski E, Valera L, Aleksunes L, Bonner D, Fung-Tomc J. 2001. Correlation between genotype and phenotypic categorization of staphylococci based on methicillin susceptibility and resistance. J. Clin. Microbiol. 39:2961–2963. 10.1128/JCM.39.8.2961-2963.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huletsky A, Giroux R, Rossbach V, Gagnon M, Vaillancourt M, Bernier M, Gagnon F, Truchon K, Bastien M, Picard FJ, Belkum A, Ouellette M, Roy P, Bergeron M. 2004. New real time PCR assay for rapid identification of MRSA directly from specimens containing a mixture of staphylococci. J. Clin. Microbiol. 42:1875–1884. 10.1128/JCM.42.5.1875-1884.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lodise TP, McKinnon PS, Swiderski L, Rybak MJ. 2003. Outcomes analysis of delayed antibiotic treatment for hospital-acquired Staphylococcus aureus bacteremia. Clin. Infect. Dis. 36:1418–1423. 10.1086/375057 [DOI] [PubMed] [Google Scholar]
- 5.Schuurman T, de Boer RF, Kooistra-Smid AM, van Zwet AA. 2004. Prospective study of use of PCR amplification and sequencing of 16S ribosomal DNA from cerebrospinal fluid for diagnosis of bacterial meningitis in a clinical setting. J. Clin. Microbiol. 42:734–740. 10.1128/JCM.42.2.734-740.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fenollar F, Raoult D. 2007. Molecular diagnosis of bloodstream infections caused by non-cultivable bacteria. Int. J. Antimicrob. Agents 30(Suppl 1):S7–S15 [DOI] [PubMed] [Google Scholar]
- 7.Horz HP, Vianna ME, Gomes BP, Conrads G. 2005. Evaluation of universal probes and primer sets for assessing total bacterial load in clinical samples: general implications and practical use in endodontic antimicrobial therapy. J. Clin. Microbiol. 43:5332–5337. 10.1128/JCM.43.10.5332-5337.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espy MJ, Uhl JR, Sloan LM, Buckwalter SP, Jones MF, Vetter EA, Yao JD, Wengenack NL, Rosenblatt JE, Cockerill FR, III, Smith TF. 2006. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin. Microbiol. Rev. 19:165–256. 10.1128/CMR.19.1.165-256.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown DFJ, Edwards DI, Hawkey PM, Morrison D, Ridgway GL, Towner KJ, Wren WD. 2005. Guidelines for the laboratory diagnosis and susceptibility testing of methicillin-resistant Staphylococcus aureus (MRSA). J. Antimicrob. Chemother. 56:1000–1018. 10.1093/jac/dki372 [DOI] [PubMed] [Google Scholar]
- 10.Klaschik S, Lehmann LE, Raadts A, Book M, Gebel J, Hoeft A, Stuber F. 2004. Detection and differentiation of in vitro-spiked bacteria by real-time PCR and melting-curve analysis. J. Clin. Microbiol. 42:512–517. 10.1128/JCM.42.2.512-517.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohammadi T, Reesink HW, Vandenbroucke-Grauls CM, Savelkoul PH. 2003. Optimization of real-time PCR assay for rapid and sensitive detection of eubacterial 16S ribosomal DNA in platelet concentrates. J. Clin. Microbiol. 41:4796–4798. 10.1128/JCM.41.10.4796-4798.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krediet TG, Mascini EM, van Rooij E, Vlooswijk J, Paauw A, Gerards LJ, Fleer A. 2004. Molecular epidemiology of coagulase-negative staphylococci causing sepsis in a neonatal intensive care unit over an 11-year period. J. Clin. Microbiol. 42:992–995. 10.1128/JCM.42.3.992-995.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoll BJ, Hansen N, Fanaroff AA, Wright LL, Carlo WA, Ehrenkranz RA, Lemons JA, Donovan EF, Stark AR, Tyson JE, Oh W, Bauer CR, Korones SB, Shankaran S, Laptook AR, Stevenson DK, Papile LA, Poole WK. 2002. Late-onset sepsis in very low birth weight neonates: the experience of the NICHD Neonatal Research Network. Pediatrics 110:285–291. 10.1542/peds.110.2.285 [DOI] [PubMed] [Google Scholar]
- 14.Mack D, Horstkotte MA, Rohde H, Knobloch JKM. 2006. Coagulase-negative staphylococci, p 109–153 In Pace JL, Rupp ME, Finch RG. (ed), Biofilms, infection, and antimicrobial therapy. CRC Press, Boca Raton, FL [Google Scholar]