Abstract
Mycobacterium canariasense is a recently described late-pigmenting, rapidly growing mycobacterium linked to bacteremia in patients with underlying malignant diseases. We report a case of M. canariasense infection in a patient from Massachusetts with underlying diffuse B cell lymphoma, which was identified both by multilocus sequence typing and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). To our knowledge, this is the first description after its original identification in Spain and the first report of this opportunistic pathogen in the Americas.
CASE REPORT
We report a case of catheter-related bacteremia by Mycobacterium canariasense in a 42-year-old patient from Massachusetts with a history of intravenous drug abuse, hepatitis C, and diffuse B cell lymphoma.
The patient presented complaining of fever (101.2°F), chills, vomiting, and nausea since 2 days prior to his admission. Six weeks earlier, he had been admitted and diagnosed with presumptive osteomyelitis of the sacroiliac joint for which he received empirical treatment with intravenous vancomycin for 6 weeks, completed a week prior to admission. Multiple sets of blood cultures were negative during this admission. At that time, a bone marrow biopsy was also performed for pancytopenia, and the patient was diagnosed with diffuse large B cell lymphoma with plasmacytoid features. He received two cycles of antineoplastic chemotherapy with cyclophosphamide, doxorubicin hydrochloride, vincristine, and prednisolone (R-CHOP), with the second cycle administered 2 days prior to admission and coinciding with the onset of fever.
He did not report rashes, headache, cough, or gastrointestinal complaints, and his prior medical history was significant for bipolar and major depressive disorder as well as essential hypertension. Liver function tests were monitored for hepatitis C exacerbation but were within reference limits. A complete blood count was notable for total leukocytes of 5,150/mm3, with 50.5% neutrophils and 16.8% lymphocytes, decreased hemoglobin of 8.6 g/dl, hematocrit of 31.2%, and a normal platelet count of 381,000/mm3. Blood urea nitrogen, serum creatinine, and glucose were within reference ranges.
Shortly after admission during a febrile episode, blood cultures were drawn and the peripherally inserted central catheter was removed and sent for culture using the roll-plate semiquantitative technique described by Maki et al. (1). The blood cultures in the aerobic bottles grew Candida dubliniensis, Candida albicans, Pseudomonas aeruginosa, and numerous Gram-variable rods. Candida species were determined with both the API 20C AUX and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) systems. The catheter tip grew numerous Gram-variable rods as well as a group B beta-streptococcus.
The Gram-variable rods (which grew from 4/4 bottles collected in two sets 3 days apart) grew on Lowenstein-Jensen (LJ) medium after 4 days (at 37°C) and revealed small, smooth, nonpigmented colonies that later evolved into larger yellowish shiny colonies (Fig. 1A). Smears confirmed the presence of variably acid-fast organisms (Fig. 1B).
FIG 1.
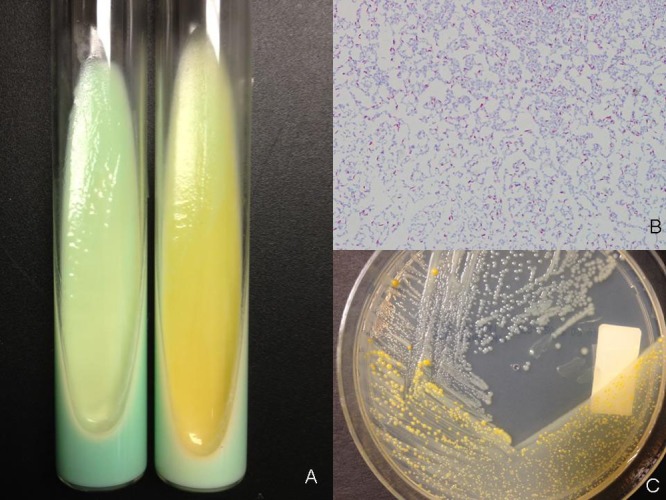
(A) Lowenstein-Jensen medium with young nonpigmented colonies (left) and late colonies (right) characteristically turning yellowish with a moist, shiny appearance; (B) partially acid-fast staining rods; (C) characteristic change in color of the colonies at different stages of growth on Middlebrook medium.
Specimens grown on Lowenstein-Jensen medium were prepared through cell harvesting, saponification, extraction, and clarification to isolate mycolic acids. High-performance liquid chromatography (HPLC) analysis was performed on an Agilent 1100 HPLC system (Wilmington, DE) coupled to MIDI software and using MIDI software for evaluation of total mycolic acid peaks; however, the HPLC pattern obtained did not match any species-specific standard but was closest to the Mycobacterium fortuitum/peregrinum cluster (with a Sim index of 0.348, below the cutoff for a reliable identification). Subsequently, whole-cell fatty acid analysis performed by gas-liquid chromatography (GLC) revealed a profile typical of mycobacterial fatty acids of which the majority were hexadecanoate C16:0 (39.20%) and octadecenoate C18:1 (23.24%), as previously described for this species by Jiménez et al. (2).
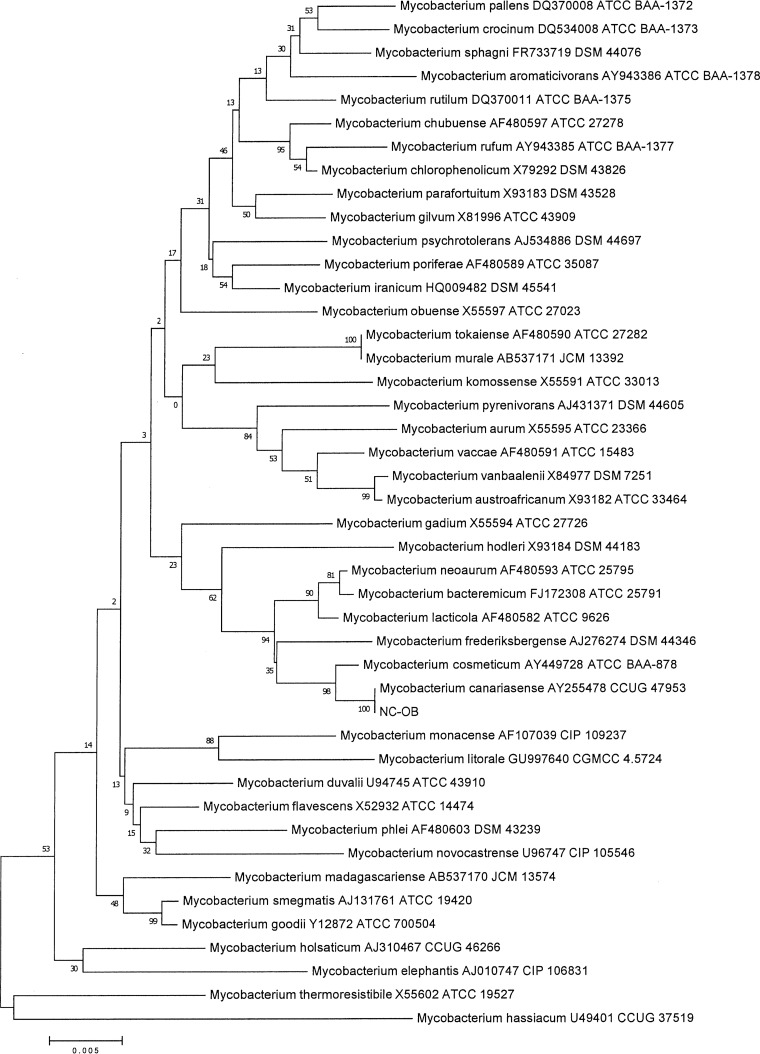
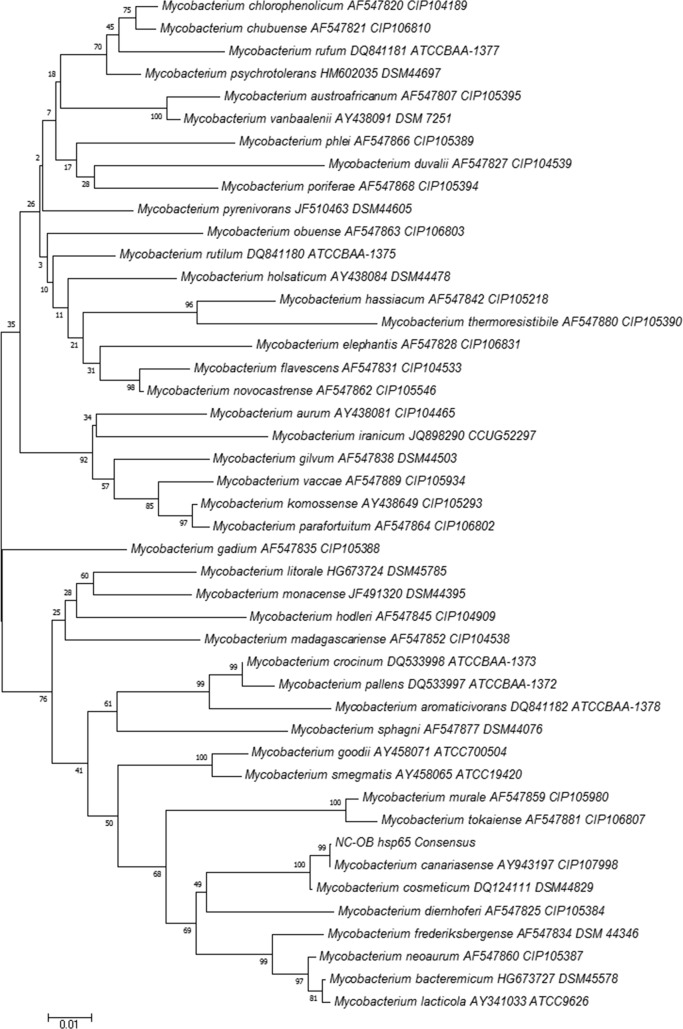
Multilocus sequence typing was performed on the isolate (NC-OB) using the full 16S rRNA gene, partial hsp65 gene, and partial rpoβ (region V) gene, as previously described (3, 4, 14). Sequencing analysis was done using the Ripseq Web application (iSentio, Bergen, Norway). The patient's isolate was a 100% match to the Mycobacterium canariasense type strain by full 16S rRNA and hsp65 gene sequencing, with a detailed sequence comparison suggesting a close relation of this species to Mycobacterium cosmeticum and Mycobacterium diernhoferi (Fig. 2–3). Because there is no region V of rpoβ gene sequencing for the type strain of M. canariasense in GenBank, we were unable to compare the sequence match of this strain to the type strain of M. canariasense. Sequence alignment and phylogenetic trees were constructed using the neighbor-joining method with Kimura's two-parameter distance correction model and 1,000 bootstrap replications in the MEGA (version 6) software (Fig. 2 and 3) (5).
FIG 2.
Neighbor-joining tree of an alignment of the 16S rRNA gene of pigmented rapidly growing mycobacteria and isolate NC-OB (1,431 bp). The scientific name, GenBank accession number, and culture collection number are included on the branch labels for the reference sequences. Bootstrap values are displayed on branch nodes.
FIG 3.
Neighbor-joining tree of an alignment of the partial hsp65 gene of pigmented rapidly growing mycobacteria and isolate NC-OB (421 bp). The scientific name, GenBank accession number, and culture collection number are included on the branch labels for the reference sequences. Bootstrap values are displayed on branch nodes.
Subsequently, both colony morphotypes were grown on Middlebrook agar (Fig. 1C) and extracted following the inactivated mycobacterium bead preparation method, as suggested by the manufacturer. Briefly, a 1-μl loopful of mycobacterial biomass was collected in a 1.5-ml Eppendorf tube containing 300 μl of deionized water and 900 μl of absolute ethanol followed by vortexing and a 10-min (100°C) incubation step. The sample was then centrifuged at maximum speed for 2 min followed by the addition of 500 μl of HPLC-grade water, vortexing, and recentrifugation. Afterward, the supernatant was removed using a pipette, followed by the addition of 50 μl of deionized water and vortexing to resuspend the pellet. Following a 60-min heat inactivation at 100°C, the tube was allowed to cool down and 1,200 μl of precooled absolute ethanol was added. The specimen was centrifuged at maximum speed for 4 min, and ethanol was carefully removed by pipetting, allowing the pellet to air dry for 5 min. Next, 200 μl of 1-mm silica beads was added followed by resuspension of the pellet by vortexing with 30 μl of pure acetonitrile and 30 μl of 70% formic acid. Following mixing by vortexing for 5 s and subsequent centrifugation at maximum speed for 2 min, 1 μl of the supernatant was placed on the MALDI target, allowed to dry, and followed by overlaying of 1 μl of matrix solution for posterior analysis by MALDI-TOF MS using the MicroFlex LT mass spectrometer (Bruker Daltonics Inc., Billerica, MA), which, based on the spectral fingerprint of the isolate, confirmed the identification (identification score, 1.966) as Mycobacterium canariasense, followed by Mycobacterium cosmeticum (1.495) and Microbacterium dextranolyticum (1.346) as second and third best matches.
Biochemical traits for the isolate revealed positive Tween 80 hydrolysis (strong), iron uptake, arylsulfatase, and rapid growth on MacConkey agar without crystal violet and Middlebrook 7H11 (Fig. 1C). Niacin accumulation, nitrate reductase, and growth on 5% NaCl were negative.
Overall, the phenotypic/biochemical characteristics, mycolic and fatty acid patterns, as well as the genotypic and MALDI-MS profiles of the organism were consistent with the published description of M. canariasense: smooth, pale-yellow colonies appearing in 4 days on subculture (Fig. 1C). Susceptibility testing to ten antimycobacterial drugs (amikacin, clarithromycin, cefoxitin, ciprofloxacin, moxifloxacin, trimethoprim sulfamethoxazole, imipenem, doxycycline, minocycline, and linezolid) by broth microdilution was performed following CLSI-approved guidelines, proving that strain NC-OB was highly susceptible to all drugs except clarithromycin, which was reported as intermediate.
The patient was treated with 7 days of fluconazole for the Candida dubliniensis and Candida albicans fungemia and with 5 days of vancomycin and piperacillin-tazobactam for a presumed mixed Gram-positive rod and Pseudomonas infection. Subsequent to catheter removal and therapy, the bacteremia has not recurred.
Mycobacterium canariasense was described by Jimenez et al. for the first time in a suspected nosocomial outbreak that affected 17 patients over a 2-year period (2000 to 2002) at a tertiary care hospital in the Canary Islands in Spain (2).
Microscopically, M. canariasense is characterized by staining partially acid fast. Initially, the colonies which are nonpigmented and smooth grew rapidly (2 to 3 days) to later develop a yellowish and shinier appearance (2, 6). The organism is aerobic, and growth occurs at 30°C to 37°C on Lowenstein-Jensen and MacConkey agars without crystal violet but not on 5% NaCl LJ medium. Isolates exhibit arylsulfatase activity at 3 days, low level of heat-stable catalase, and hydrolysis in Tween 80 but no nitrate reductase activity (2, 6).
Its close relationship to other rapidly growing mycobacteria (RGM) (2, 6) is the reason why conventional methods of identification may fail to recognize this agent, thus prompting confirmation by other methods, such as gene sequencing or mass spectrometry. M. canariasense is closely related to M. diernhoferi, Mycobacterium neoaurum, and Mycobacterium mucogenicum (1, 2) and can be distinguished from these three by its inability to reduce nitrates (2, 6). On the other hand, thin-layer chromatography (TLC) of M. canariasense reveals a pattern identical to those of Mycobacterium abscessus and Mycobacterium chelonae with the presence of α and α′ mycolates, also allowing its differentiation from the aforementioned related species (2). In this case, HPLC failed to accurately identify the isolate, most closely matching it to M. fortuitum, another related member of the group (2). Even though much more discriminative than TLC, HPLC has its limitations, particularly in differentiating species with shared patterns, and especially among the rapid growers, as seen in this case.
To date, M. canariasense has been isolated only from human blood and contaminated central venous catheters (7); no environmental source has been identified, nor has close contact (human-to-human) transmission been reported. Similar to other rapidly growing nontuberculous mycobacteria (NTM), M. canariasense is presumed to be an environmental agent that may act as an opportunistic pathogen. From the original case series published in 2004, 12 of 17 patients had an underlying malignancy such as lymphoma, leukemia, or multiple myeloma as well as other soft tissue tumors (7). Furthermore, 11 of these patients had undergone antineoplastic chemotherapy and had received parenteral antibiotic therapy via a central venous catheter (7). Like the majority of these cases, our patient had recently been diagnosed with a malignancy (diffuse large B cell lymphoma) and was undergoing his second cycle of chemotherapy when he first developed fever.
The facts that no other source of infection was identified based on clinical assessment, that this same agent grew in a blood culture and subsequent catheter tip, and that the patient recovered shortly after removal of the catheter strongly suggest that M. canariasense bacteremia in our patient was likely secondary to an intravascular catheter. Although no specific therapy for this organism was given, the patient has not had a recurrence of mycobacteremia. Of interest, the patient was observed crushing and injecting hydromorphone tablets into his PICC line. This may also explain the coisolation of 2 different Candida species and Pseudomonas aeruginosa in his blood cultures. His blood cultures were negative in a previous episode of presumed osteomyelitis. Though rare, NTM have been reported to cause transient bacteremia and osteomyelitis in intravenous drug users (8).
Though uncommon, bloodstream infections by NTM have been described in immunocompromised patients (9), with increasing reports in catheter-related bloodstream infections (10) being caused most commonly by M. fortuitum and M. mucogenicum (10). The diagnosis of such infections may be challenging, and accurate identification may be hampered by inadequate incubation times of conventional blood cultures to detect mycobacteria, the limited availability of commercial nucleic acid probes for RGM, and potential misidentification with other related organisms, such as Nocardia spp., Rhodococcus, Gordonia spp., and Tsukamurella spp. (10, 11). An example of this is the genus Tsukamurella, species of which are actinomycetales that stain weakly acid fast and have been shown to share the same α and α′ mycolate pattern as those of M. canariasense, M. chelonae, and M. abscessus (12, 13). Prompt and accurate identification is pivotal, since catheter removal and specific antibiotic therapy improve outcomes in catheter-related mycobacteremia.
In conclusion, our case highlights the importance of using a combinatorial approach to identification in the routine mycobacteriology laboratory practice as well as the implementation of novel identification systems such as MALDI-TOF MS for accurate identification of species. In addition, this case confirms the potential role of M. canariasense as an opportunistic pathogen in cancer patients and broadens its geographical spectrum of occurrence to the Americas, suggesting a more cosmopolitan distribution for this agent.
Nucleotide sequence accession numbers.
The nucleotide sequences determined for Mycobacterium canariasense (NC-OB) in this study have been deposited in GenBank under the following accession numbers: for 16S rRNA, KJ720533; for hsp65, KJ720534; and for rpoB, KJ720535.
ACKNOWLEDGMENTS
We thank Myron Sasser and Gary Jackoway (MIDI, Inc., Newark, DE) for their kind assistance with the GLC analyses, Nicholas Parodi, Anita Strong, Martha Gee, and Shannon O'Neil for their skillful help on the susceptibility testing (University of Texas Health Science Center at Tyler, Mycobacteria/Nocardia Laboratory, Tyler, TX), and Keith E. Simmon (Department of Biomedical Informatics, University of Utah, Salt Lake City, UT) for his assistance in the phylogenetic analyses.
Footnotes
Published ahead of print 16 April 2014
REFERENCES
- 1.Maki DG, Weise CE, Sarafin HW. 1977. A semiquantitative culture method for identifying intravenous-catheter-related infection. N. Engl. J. Med. 296:1305–1309. 10.1056/NEJM197706092962301 [DOI] [PubMed] [Google Scholar]
- 2.Jiménez MS, Campos-Herrero MI, García D, Luquin M, Herrera L, García MJ. 2004. Mycobacterium canariasense sp. nov. Int. J. Syst. Evol. Microbiol. 54:1729–1734. 10.1099/ijs.0.02999-0 [DOI] [PubMed] [Google Scholar]
- 3.Simmon KE, Low YY, Brown-Elliott BA, Wallace RJ, Jr, Petti CA. 2009. Phylogenetic analysis of Mycobacterium aurum and Mycobacterium neoaurum with re-description of M. aurum culture collection strains. Int. J. Syst. Evol. Microbiol. 59:1371–1375. 10.1099/ijs.0.007799-0 [DOI] [PubMed] [Google Scholar]
- 4.Turenne CY, Tschetter L, Wolfe J, Kabani A. 2001. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J. Clin. Microbiol. 39:3637–3648. 10.1128/JCM.39.10.3638-3648.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30:2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tortoli E. 2006. The new mycobacteria: an update. FEMS Immunol. Med. Microbiol. 48:159–178. 10.1111/j.1574-695X.2006.00123.x [DOI] [PubMed] [Google Scholar]
- 7.Campos-Herrero MI, García D, Figuerola A, Suárez P, Campo C, García MJ. 2006. Bacteremia caused by the novel species Mycobacterium canariasense. Eur. J. Clin. Microbiol. Infect. Dis. 25:58–60. 10.1007/s10096-005-0079-6 [DOI] [PubMed] [Google Scholar]
- 8.Shimizu H, Mizuno Y, Nakamura I, Fukushima S, Endo K, Matsumoto T. 2013. Vertebral osteomyelitis caused by nontuberculous mycobacteria: case reports and review. J. Infect. Chemother. 19:972–977. 10.1007/s10156-013-0550-8 [DOI] [PubMed] [Google Scholar]
- 9.El Helou G, Viola GM, Hachem R, Han XY, Raad II. 2013. Rapidly growing mycobacterial bloodstream infections. Lancet Infect. Dis. 13:166–174. 10.1016/S1473-3099(12)70316-X [DOI] [PubMed] [Google Scholar]
- 10.Hawkins C, Qi C, Warren J, Stosor V. 2008. Catheter-related bloodstream infections caused by rapidly growing nontuberculous mycobacteria: a case series, including rare species. Diagn. Microbiol. Infect. Dis. 61:187–191. 10.1016/j.diagmicrobio.2008.01.004 [DOI] [PubMed] [Google Scholar]
- 11.Karunakaran R, Halim HA, Ng KP, Hanifah YA, Chin E, Jaafar FL, Abubakar S. 2011. Tsukamurella tyrosinosolvens intravascular catheter-related bacteremia in a haematology patient: a case report. Eur. Rev. Med. Pharmacol. Sci. 15:1343–1346 [PubMed] [Google Scholar]
- 12.Hamid ME, Minnikin DE, Goodfellow M. 1993. A simple chemical test to distinguish mycobacteria from other mycolic-acid-containing actinomycetes. J. Gen. Microbiol. 139:2203–2313. 10.1099/00221287-139-9-2203 [DOI] [PubMed] [Google Scholar]
- 13.Hinrikson HP, Pfyffer GE. 1994. Mycobacterial mycolic acids. Med. Microbiol. Lett. 3:49–57 [Google Scholar]
- 14.Adékambi T, Colson P, Drancourt M. 2003. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J. Clin. Microbiol. 41:5699–5708. 10.1128/JCM.41.12.5699-5708.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]