Abstract
The prevalence of serogroup 6 among 1,206 Streptococcus pneumoniae clinical isolates collected from Korean hospitals over three periods (1996 to 2001, 2004 to 2006, and 2008 to 2009) was investigated. The number of serogroup 6 isolates increased from 9.7 to 17.5% over the three periods. While the proportion of serotype 6A and 6D isolates increased significantly, that of serotype 6B isolates decreased. Twenty-four isolates (2.0%) were typed as the recently identified putative serotype 6E or genetic variants of serotype 6B. The results suggest that the lack of change in frequency of serotype 6B, in spite of the introduction of the PCV7 vaccine as seen in previous studies in South Korea, might be due mainly to the improper inclusion of putative serotype 6E in serotype 6B. All but three serotype 6E isolates belonged to CC90, indicating their clonal expansion.
INTRODUCTION
It is estimated that Streptococcus pneumoniae is responsible for the deaths of approximately 1.6 million persons each year, of which nearly 1 million are children <5 years old. The majority of these deaths occur in developing countries. To prevent pneumococcal disease, the seven-valent pneumococcal conjugate vaccine (PCV7) was introduced in the United States in 2000 and in South Korea in 2003. Although a significant decrease in invasive pneumococcal disease has been reported in many studies since then, an increase in disease caused by serotypes not covered by the PCV7 vaccine is a growing concern (1). Recently, the PCV13 vaccine, which covers the additional 1, 3, 5, 6A, 7F, and 19A serotypes, has been introduced; however, data regarding its effect on pneumococcal serotype prevalence are still limited in most countries.
Traditionally, serogroup 6 was considered to consist of two serotypes, 6A and 6B. However, the 6C and 6D serotypes were characterized in 2007 and 2009, respectively (2, 3), and an additional putative serotype 6E or genetic variants of serotype 6B have also been recently proposed (4). Although putative serotype 6E has not been serologically and biochemically characterized, it showed great sequence divergence in capsular locus genes relative to the other serogroup 6 serotypes (4). In our previous paper, we postulated that putative serotype 6E or genetic variants of serotype 6B may not be rare and may not be restricted to a specific geographic locality (4). In this study, we developed a PCR method to detect putative serotype 6E isolates on the basis of putative structural differences in the capsular polysaccharide genes and traced the changes in serogroup 6 serotypes in South Korea on the basis of data from three periods (1996 to 2001, 2004 to 2006, and 2008 to 2009).
MATERIALS AND METHODS
S. pneumoniae isolates.
We identified 147 S. pneumoniae serogroup 6 isolates among a total of 1,206 nonduplicate S. pneumoniae clinical isolates over three periods; 226 isolates were obtained from May 1996 to December 2001, 649 isolates were obtained from April 2004 to November 2006, and 331 isolates were obtained from May 2008 to May 2009. The isolates were collected from patients of diverse ages in 12 hospitals in various regions of South Korea.
Serotyping and detection of putative serotype 6E.
Serotyping was conducted by using the capsular Quellung reaction with commercial antisera (Statens Serum Institut, Copenhagen, Denmark) as recommended by the manufacturer. For isolates serotyped as serogroup 6, a previously described serotype-specific PCR method was used to identify serotypes 6C and 6D (5). The primer sets used to detect putative serotype 6E or genetic variants of serotype 6B were designed on the basis of an additional four open reading frame (ORFs) (transposons) between the dexB and wzy genes, insertion sequences between the wciN and wciO genes, and the 9-nucleotide (nt) deletion within the wze gene (Table 1) (4). To amplify ORFs between the dexB and wzy genes, PCR was performed as follows: 96°C for 15 min; 35 cycles of 96°C for 30 s, 55°C for 1 min, and 72°C for 3 min; 72°C for 10 min; and holding at 15°C. The insertion sequences between the wciN and wciO genes and the 9-nt deletion within the wze gene were detected simultaneously under the conditions described previously (3).
TABLE 1.
Primers used to detect pneumococcal serogroup 6 serotypes
| Target and primer | Sequence (5′–3′) | PCR product size |
Reference(s) | |
|---|---|---|---|---|
| 6C + 6D | 6E | |||
| Transposon between dexB and wzy | ||||
| dexBF2 | TCCATGGGATGCTTTCTGTGTGG | 2.1 kbp | 4.8 kbp | 4 |
| 3120R | CGAAGTGAAGTTCAATCGCAC | |||
| Insertion sequences between wciN and wciO | ||||
| 5106F | TACCATGCAGGGTGGAATGT | 1.8 kbp | 2.3 kbp, 1,267 bp | 3, 10 |
| 5101F | ATTTGGTGTACTTCCTCC | |||
| 3101R | CCATCCTTCGAGTATTGC | |||
| 9-nt deletion in wze | ||||
| 5113F2 | CGAATACGGTTCTTATGGAAGTTAT | None | 500 bp | This study |
| 3122R3 | CTGATTCTATTAGTCTATCCAATACC | |||
Antimicrobial susceptibility testing.
MICs were determined for 147 isolates of serogroup 6 by the broth microdilution method according to the Clinical and Laboratory Standards Institute guidelines (6). In vitro susceptibilities to 13 antimicrobial agents, penicillin, amoxicillin, amoxicillin-clavulanate, ceftriaxone, cefuroxime, erythromycin, azithromycin, clarithromycin, levofloxacin, moxifloxacin, gatifloxacin, clindamycin, and trimethoprim-sulfamethoxazole, were tested. S. pneumoniae ATCC 49619, Staphylococcus aureus ATCC 29213, and Escherichia coli ATCC 25922 were used as control strains.
Statistical analysis.
Fisher's exact t test was used to determine significant differences in resistance with SPSS version 11.5 for Windows (SPSS, Chicago, IL).
RESULTS AND DISCUSSION
In our previous study, we proposed genetic variants of serotype 6B putatively as a novel pneumococcal serotype, serotype 6E (4). Putative serotype 6E isolates showed high amino acid sequence divergence from the other serotypes of serogroup 6 within the encoded cps locus. In addition to sequence polymorphisms, the cps locus of putative serotype 6E is unique in that it includes four ORFs between the dexB and wzy genes, a deletion of three amino acid codons in the wze gene, and an approximately 300-bp insertion between the wciN and wciO genes. On the basis of these unique regions, we designed specific primer sets to detect putative serotype 6E and investigated the prevalence of serogroup 6 serotypes among 1,206 S. pneumoniae clinical isolates from South Korea.
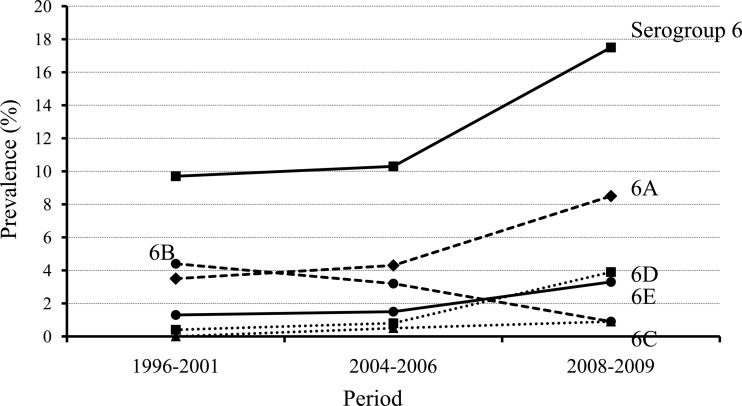
The prevalence of serogroup 6 serotypes is presented in Fig. 1, which shows the change in serotypes over three different periods. In total, serogroup 6 isolates increased from 9.7% in 1996 to 2001 to 17.5% in 2008 to 2009 (P = 0.012). The most prevalent serotype among serogroup 6 isolates was 6A (5.3%), followed by 6B (2.8%) and putative serotype 6E (2.0%). On the other hand, serotype 6C was found in only six isolates. While the proportion of 6A and 6D serotype isolates increased (P = 0.036 and 0.001, respectively), that of serotype 6B decreased over the three periods (P = 0.016). The decrease in serotype 6B can be explained by the introduction of the PCV7 vaccine in South Korea in November, 2003. The increase in serotype 6A might be due to the fact that this serotype is not included in the PCV7 vaccine.
FIG 1.
Serotype changes in S. pneumoniae serogroup 6 isolates from South Korea during three periods, 1996 to 2001, 2004 to 2006, and 2008 to 2009. During these periods, serogroup 6 increased from 9.7 to 17.5% (P = 0.012). While serotype 6B decreased from 4.4 to 0.9% (P = 0.016), serotypes 6A and 6D increased significantly. Putative serotype 6E isolates, most of which were classified as serotype 6B on the basis of the Quellung reaction, also increased from 1.3 to 3.3%, but the increase was not statistically significant (P = 0.146).
In several previous studies performed in South Korea, the decrease in isolates of serotype 6B, which is unlike serotype 19A, since the introduction of PCV7 was not obvious (7–9). Our study suggests that such phenomena might be due mainly to the identification of putative serotype 6E isolates as serotype 6B. All putative serotype 6E isolates but one were identified as serotype 6B by the classical Quellung reaction. In reality, serotype 6B isolates excluding putative serotype 6E decreased in 2008 to 2009. Although serological analysis did not show complete separation between serotype 6B and putative serotype 6E, the PCV7 vaccine does not seem to protect against putative serotype 6E or genetic variants of serotype 6B on the basis of epidemiological evidence. Therefore, it is necessary to classify the serotypes with more specificity and to investigate the epidemiology of S. pneumoniae to better evaluate vaccine efficacy.
Most of the putative serotype 6E isolates showed very high rates of nonsusceptibility to the antimicrobials cefuroxime, azithromycin, clarithromycin, and trimethoprim-sulfamethoxazole (>90%) (Table 2). Erythromycin and clindamycin nonsusceptibility rates were also high among the putative serotype 6E isolates (83.3 and 70.8%, respectively). One putative serotype 6E isolate was resistant to levofloxacin and gatifloxacin. Although antimicrobial nonsusceptibility rates were relatively low in serotype 6C isolates, a significant difference among the serotypes could not be identified. Thus, the increase in serotypes 6A and 6D, as well as putative serotype 6E, might not be due to the selective advantage of antimicrobial resistance.
TABLE 2.
Rates of antimicrobial nonsusceptibility of serogroup 6 isolates
| Antimicrobial agent | No. (%) of nonsusceptible isolatesa |
|||||
|---|---|---|---|---|---|---|
| Serogroup 6 (n = 147) | Serotype 6A (n = 64) | Serotype 6B (n = 34) | Serotype 6C (n = 6) | Serotype 6D (n = 19) | Putative serotype 6E (n = 24) | |
| Penicillin | 3 (2.0) | 2 (3.1) | 1 (2.9) | 0 | 0 | 0 |
| Amoxicillin | 4 (2.7) | 0 | 2 (5.9) | 0 | 0 | 2 (8.3) |
| Amoxicillin-clavulanate | 4 (2.7) | 0 | 2 (5.9) | 0 | 0 | 2 (8.3) |
| Ceftriaxone | 3 (2.0) | 2 (3.1) | 1 (2.9) | 0 | 0 | 0 |
| Cefuroxime | 129 (87.8) | 60 (93.8) | 28 (82.4) | 2 (33.3) | 16 (84.2) | 23 (95.8) |
| Erythromycin | 137 (93.2) | 63 (98.4) | 34 (100) | 3 (50.0) | 17 (89.5) | 20 (83.3) |
| Azithromycin | 129 (87.8) | 57 (89.1) | 34 (100) | 3 (50.0) | 13 (68.4) | 22 (91.7) |
| Clarithromycin | 135 (91.8) | 60 (93.8) | 34 (100) | 3 (50.0) | 15 (78.9) | 23 (95.8) |
| Levofloxacin | 8 (5.4) | 1 (1.6) | 1 (2.9) | 0 | 5 (26.4) | 1 (4.2) |
| Moxifloxacin | 2 (1.4) | 1 (1.6) | 1 (2.9) | 0 | 0 | 0 |
| Gatifloxacin | 8 (5.4) | 1 (1.6) | 1 (2.9) | 0 | 5 (26.4) | 1 (4.2) |
| Clindamycin | 101 (68.7) | 44 (68.8) | 32 (94.1) | 3 (50.0) | 5 (26.4) | 17 (70.8) |
| Trimethoprim-sulfamethoxazole | 124 (84.4) | 55 (85.9) | 34 (100) | 3 (50.0) | 8 (42.1) | 24 (100) |
The MIC nonsusceptibility breakpoints of the antimicrobial agents are as follows: penicillin, 4 mg/liter; amoxicillin, 4 mg/liter; amoxicillin-clavulanate, 4/2 mg/liter; ceftriaxone, 2 mg/liter; cefuroxime, 1 mg/liter; erythromycin, 0.5 mg/liter; azithromycin, 1 mg/liter; clarithromycin, 0.5 mg/liter; levofloxacin, 4 mg/liter; moxifloxacin, 2 mg/liter; gatifloxacin, 2 mg/liter; clindamycin; 0.5 mg/liter; trimethoprim-sulfamethoxazole, 1/19 mg/liter.
The putative serotype 6E isolate genotypes are presented in Table 3. A total of 13 different STs, including a new one (ST9932), were identified among the 24 putative serotype 6E isolates. The most prevalent ST was ST90 (10 isolates, 41.7%), and 10 STs were either single- or double-locus variants. Thus, most putative serotype 6E isolates, except for three (Kor55, K01-04-126, and K08-92), are included in clonal complex 90 (CC90). Putative serotype 6E isolates from other Asian countries belonged mostly to CC90 (4). Thus, it is suggested that most putative serotype 6E isolates derived from the same ancestor, ST90. Very high sequence similarities within the cps locus support the clonal expansion of putative serotype 6E isolates (4). Emergence of putative serotype 6E due to recombination within the cps locus was also expected, but it might not be frequent.
TABLE 3.
Sources of putative serotype 6E isolates and STs determined by multilocus sequence typing
| Period and isolate | Source | ST (allelic profile)a |
|---|---|---|
| 1996-2001 | ||
| K10-5 | Eye discharge | 90 (5-6-1-2-6-3-4) |
| Kor364 | Unknown | 90 (5-6-1-2-6-3-4) |
| Kor55 | Blood | 877 (10-13-4-16-6-1-17)b |
| 2004-2006 | ||
| K01-04-126 | Nasal/nasopharynx | 2842 (4-4-2-4-156-1-1)b |
| K01-05-205 | Nasal/nasopharynx | 90 (5-6-1-2-6-3-4) |
| K01-05-227 | Nasal/nasopharynx | 3387 (5-6-1-2-6-3-26) |
| K01-05-235 | Pus | 90 (5-6-1-2-6-3-4) |
| K01-05-327 | Unknown | 95 (5-6-33-2-6-3-4) |
| K01-05-399 | Sputum | 2156 (5-6-1-2-6-3-14) |
| K01-05-407 | Pus | 90 (5-6-1-2-6-3-4) |
| K01-06-071 | Sputum | 90 (5-6-1-2-6-3-4) |
| K01-04-14 | Pus | 9332 (5-6-19-2-6-3-4) |
| K01-04-19 | Transtracheal aspirate | 3387 (5-6-1-2-6-3-26) |
| 2008-2009 | ||
| K13-21 | Sputum | 1624 (5-6-1-2-6-3-1) |
| K07-6 | Sputum | 273 (5-6-1-2-6-1-14) |
| K08-75 | Sputum | 1624 (5-6-1-2-6-3-1) |
| K08-92 | Sputum | 3418 (7-11-19-1-6-1-50)b |
| K13-2 | Sputum | 4759 (5-4-1-2-6-3-4) |
| K13-10 | Sputum | 90 (5-6-1-2-6-3-4) |
| K13-47 | Sputum | 90 (5-6-1-2-6-3-4) |
| K13-86 | Sputum | 90 (5-6-1-2-6-3-4) |
| K13-105 | Sputum | 4764 (5-6-1-2-6-3-53) |
| K13-123 | Sputum | 90 (5-6-1-2-6-3-4) |
| K16-185 | Blood | 4763 (5-6-1-2-4-3-26) |
aroE-gdh-gki-recP-spi-xpt-ddl.
ST not belonging to CC90.
In this study, we investigated the prevalence of serogroup 6 serotypes, especially the newly identified putative serotype 6E or genetic variants of serotype 6B, in South Korea. Although this study did not analyze the data by separating invasive and carried isolates, our results indicate that the apparent lack of change in the frequency of serotype 6B, in spite of the introduction of the PCV7 vaccine, that was reported in previous studies may have been due mainly to the misidentification of putative serotype 6E as serotype 6B. Most of the putative serotype 6E isolates belonged to CC90, suggesting that they derived from the same ancestor. Continuous investigation of serotypes should be performed to monitor the efficacy of vaccines and to establish a strategy to prevent pneumococcal infections.
ACKNOWLEDGMENTS
The S. pneumoniae isolates used in this study were obtained from the Asian Bacterial Bank (ABB) of the Asia Pacific Foundation for Infectious Diseases (APFID, Seoul, South Korea).
This study was supported by a grant from the Korea Healthcare Technology R&D Project, Ministry of Health, Welfare, and Family Affairs, Seoul, Republic of Korea (A111251).
Footnotes
Published ahead of print 9 April 2014
REFERENCES
- 1.Muñoz-Almagro C, Jordan I, Gene A, Latorre C, Garcia-Garcia JJ, Pallares R. 2008. Emergence of invasive pneumococcal disease caused by nonvaccine serotypes in the era of 7-valent conjugate vaccine. Clin. Infect. Dis. 46:174–182. 10.1086/524660 [DOI] [PubMed] [Google Scholar]
- 2.Jin P, Kong F, Xiao M, Oftadeh S, Zhou F, Liu C, Russell F, Gilbert GL. 2009. First report of putative Streptococcus pneumoniae serotype 6D among nasopharyngeal isolates from Fijian children. J. Infect. Dis. 200:1375–1480. 10.1086/606118 [DOI] [PubMed] [Google Scholar]
- 3.Park IH, Pritchard DG, Cartee R, Brandao A, Brandileone MC, Nahm MH. 2007. Discovery of a new capsular serotype (6C) within serogroup 6 of Streptococcus pneumoniae. J. Clin. Microbiol. 45:1225–1233. 10.1128/JCM.02199-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ko KS, Baek JY, Song JH. 2013. Capsular gene sequences and genotypes of “serotype 6E” Streptococcus pneumoniae isolates. J. Clin. Microbiol. 51:3395–3399. 10.1128/JCM.01645-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobs MR, Bajaksouzian S, Bonomo RA, Good CE, Windau AR, Hujer AM, Massire C, Melton R, Blyn LB, Ecker DJ, Sampath R. 2009. Occurrence, distribution, and origins of Streptococcus pneumoniae serotype 6C, a recently recognized serotype. J. Clin. Microbiol. 47:64–72. 10.1128/JCM.01524-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2011. Performance standards for antimicrobial susceptibility testing, 21st informational supplement, M100–S21. CLSI, Wayne, PA [Google Scholar]
- 7.Baek JY, Ko KS, Kim SH, Kang CI, Chung DR, Peck KR, Song JH. 2011. Comparison of genotypes of Streptococcus pneumoniae serotypes 6A and 6B before and after the introduction of PCV7 vaccination in Korea. Diagn. Microbiol. Infect. Dis. 69:370–375. 10.1016/j.diagmicrobio.2010.10.019 [DOI] [PubMed] [Google Scholar]
- 8.Choi EH, Kim SH, Eun BW, Kim SJ, Kim NH, Lee J, Lee JH. 2008. Streptococcus pneumoniae serotype 19A in children, South Korea. Emerg. Infect. Dis. 14:275–281. 10.3201/eid1402.070807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SH, Song JH, Chung DR, Thamlikitkul V, Yang Y, Wang H, Lu M, So TM, Hsueh PR, Yasin RM, Carlos CC, Pham HV, Lalitha MK, Shimono N, Perera J, Shibl AM, Baek JY, Kang CI, Ko KS, Peck KR, Study Group ANSORP 2012. Changing trends in antimicrobial resistance and serotypes of Streptococcus pneumoniae isolates in Asian countries: an Asian Network for Surveillance of Resistance Pathogens (ANSORP) study. Antimicrob. Agents Chemother. 56:1418–1426. 10.1128/AAC.05658-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mavroidi A, Godoy D, Aanensen DM, Robinson DA, Hollingshead SK, Spratt BG. 2004. Evolutionary genetics of the capsular locus of serotype 6 pneumococci. J. Bacteriol. 186:8181–8192. 10.1128/JB.186.24.8181-8192.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]