Abstract
The Viper HSV-Qx assay was evaluated for the detection of herpes simplex virus 1 (HSV-1) and HSV-2 in specimens from oral, anogenital, and other miscellaneous sites. The HSV-Qx assay was found to be highly sensitive and accurate; however, a gray zone may be required for specimens with values falling between 50 and 800 maximum relative fluorescence units.
TEXT
Herpes simplex virus 1 (HSV-1) and HSV-2 cause a spectrum of diseases that often present as lesions at oral or anogenital sites (1–4). Accurate HSV detection and typing are important for management, and molecular methods are considered the methods of choice (5–10). Recently, the HSV-1 and -2 Qx amplified DNA assay (HSV-Qx) for use on the Viper instrument (Becton Dickinson) was released, but it was licensed for anogenital specimens only. In this study, swabs collected from anogenital, oral, and other sites were used to compare the performance of the HSV-Qx to that of a real-time HSV PCR on the LightCycler 2.0 platform (HSV-LC) (Roche Diagnostics).
For HSV-LC, 200 μl of specimen was subjected to total nucleic acid extraction on a MagNA Pure LC, and 5 μl of eluate was used as the template in PCRs using the HSV-1/-2 detection kit (Roche Diagnostics), as recommended by the manufacturer (9–12). Crossing-point (Cp) and melting-temperature (Tm) analyses were determined by the manufacturer's software. The Tm values for HSV-1 and HSV-2 are 54°C and 68°C (±2.5°C), respectively. For HSV-Qx, 500 μl of specimen was placed into 2 ml Probetec Qx diluent, and processing conditions followed the manufacturer's instructions. The peak fluorescence intensity was expressed as the maximum relative fluorescence units (MaxRFU).
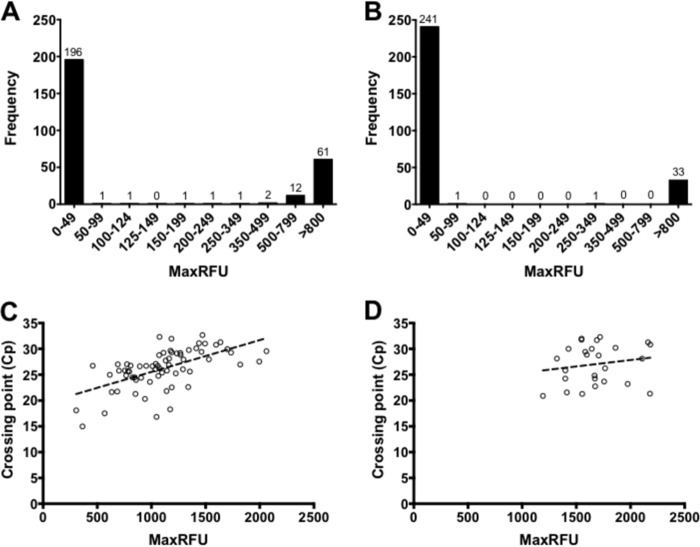
To evaluate analytical specificity, high-titer suspensions of various organisms were used (see Table S1 in the supplemental material), but no cross-reactions were observed for either assay. For analytical sensitivity, cultured HSV-1 and HSV-2 stocks were diluted 10-fold in universal transport medium (UTM) (Copan Diagnostics), and triplicate values were obtained from three independent experiments. Virus stocks were quantified using a standard curve generated with plasmids harboring the HSV target (8, 9). For HSV-LC, inverse linear relationships were observed for HSV-1 (y = −3.354x + 37.5; R2 = 1.000) and HSV-2 (y = −3.597x + 39.93; R2 = 1.000) when the Cp values were plotted against virus concentrations (log copies/ml) (see Fig. S1 in the supplemental material). For HSV-LC, the interexperimental coefficients of variation (%CV) ranged from 0.39 to 0.57% for HSV-1 and from 0.33 to 2.24% for HSV-2, whereas for HSV-Qx, the %CV ranged from 24.61 to 173.21% and from 6.90 to 117.28% for HSV-1 and HSV-2, respectively. Unlike the Cp values obtained with HSV-LC, the MaxRFU values obtained with HSV-Qx were highly variable and did not correlate with HSV viral loads or Cp values (Fig. 1; see also Fig. S1). Overall, both methods were highly sensitive and specific for HSV detection, with HSV-Qx 20-fold more sensitive at ∼10 copies/ml for both targets.
FIG 1.
Distribution of MaxRFU values for HSV-1 and HSV-2 using HSV-Qx. (A and B) Distributions of MaxRFU values are depicted for HSV-1 and HSV-2 results following the categorization provided by the manufacturer. (C and D) Lack of correlation is shown between MaxRFU and Cp values obtained using the LightCycler for HSV-1 (R2 = 0.3644) and HSV-2 (R2 = 0.0726), respectively.
Next, 276 swabs (115 anogenital, 91 oral, and 70 from other anatomical sites) that were submitted to the microbiology laboratory at CDHA between 31 January and 26 April 2013 were tested in parallel using HSV-Qx and HSV-LC. Each method was compared to a modified gold standard, defined as concordant results (positive or negative) between the two methods. Thirteen discrepant results (Table 1) were resolved at Mt. Sinai Hospital (Toronto, ON) following extraction on a NucliSENS easyMAG instrument and amplification with a RealStar alpha herpesvirus PCR kit (αHV-PCR), which can differentiate among HSV-1, HSV-2, and varicella-zoster virus (VZV). HSV-Qx was more sensitive than HSV-LC, regardless of the anatomical site or the HSV target (Table 2). HSV-LC missed four HSV-1 and four HSV-2 results. A single false-negative result that had a MaxRFU value of 124 (near the recommended cutoff value for positivity of ≥125) was obtained with HSV-Qx (Table 1). Overall, the clinical sensitivities for HSV-1 and HSV-2 were 94.6% and 97.1% for HSV-LC and 98.6% and 100% for HSV-Qx, respectively.
TABLE 1.
Summary of discrepant analyses
| Anatomical site | HSV-LC |
HSV-Qx |
αHV-PCR | Final result | Commenta | ||||
|---|---|---|---|---|---|---|---|---|---|
| Cp | Tm | Result | HSV-1 (MaxRFU) | HSV-2 (MaxRFU) | Result | ||||
| Lip | 26.2 | 54.2 | HSV-1 | 124 | 0 | Neg | HSV-1 | HSV-1 | FN HSV-1 (HSV-Qx) |
| Lip | Negb | 1,731 | 14 | HSV-1 | HSV-1 | HSV-1 | FN HSV-1 (HSV-LC) | ||
| Lip | Neg | 1,062 | 3 | HSV-1 | HSV-1 | HSV-1 | |||
| Mouth | Neg | 1,207 | 4 | HSV-1 | HSV-1 | HSV-1 | |||
| Bucca | Neg | 1,982 | 12 | HSV-1 | HSV-1 | HSV-1 | |||
| Lip | Neg | 0 | 340 | HSV-2 | HSV-2 | HSV-2 | FN HSV-2 (HSV-LC) | ||
| Left thigh | Neg | 0 | 2,088 | HSV-2 | HSV-2 | HSV-2 | |||
| Vulva | Neg | 0 | 1,238 | HSV-2 | HSV-2 | HSV-2 | |||
| Mouth | Neg | 192 | 0 | HSV-1 | Neg | Neg | FP HSV-1 (HSV-Qx) | ||
| Mouth | Neg | 513 | 0 | HSV-1 | Neg | Neg | |||
| Vagina | Neg | 202 | 20 | HSV-1 | Neg | Neg | |||
| Miscellaneous | 24.4 | 67.8 | HSV-2 | 1,733 | 1,530 | HSV-1, HSV-2 | HSV-2 | HSV-2 | |
| Labia | Neg | 1,350 | 0 | HSV-1 | HSV Neg, VZV Posc | Neg | FP HSV-1 (HSV-Qx), VZV Pos | ||
FN, false negative; FP, false positive.
Neg, negative.
Pos, positive.
TABLE 2.
Clinical performance of HSV-LC and HSV-Qx
| Anatomical site | Detection of HSV-1 (% [95% CIa]) with: |
Detection of HSV-2 (% [95% CI]) with: |
||||||
|---|---|---|---|---|---|---|---|---|
| HSV-LC |
HSV-Qx |
HSV-LC |
HSV-Qx |
|||||
| Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | |
| Overallb | 94.6 (86.7–98.5) | 100.0 (98.2–100.0) | 98.7 (92.7–100.0) | 97.5 (94.3–99.2) | 97.1 (84.7–99.9) | 100.0 (98.5–100.0) | 100.0 (89.7–100.0) | 100.0 (98.5–100.0) |
| Anogenitalc | 100.0 (87.2–100.0) | 95.9 (95.9–100.0) | 100.0 (87.2–100.0) | 97.7 (92.0–99.7) | 91.3 (72.0–98.9) | 100.0 (96.1–100.0) | 100.0 (85.2–100.0) | 100.0 (96.1–100.0) |
| Orald | 88.6 (73.3–96.8) | 100.0 (93.6–100.0) | 97.1 (85.1–99.9) | 96.4 (87.7–99.6) | 75.0 (19.3–99.4) | 100.0 (95.9–100.0) | 100.0 (39.8–100.0) | 100.0 (95.9–100.0) |
| Miscellaneouse | 100.0 (73.5–100.0) | 100.0 (93.8–100.0) | 100.0 (73.6–100.0) | 98.3 (90.8–99.8) | 87.5 (47.4–99.7) | 100.0 (94.2–100.0) | 100.0 (63.1–100.0) | 100.0 (94.2–100.0) |
CI, confidence interval.
n = 276 swabs; 74 HSV-1, 35 HSV-2.
n = 115 swabs; 27 HSV-1, 23 HSV-2.
n = 91 swabs; 35 HSV-1, 4 HSV-2.
n = 70 swabs; 12 HSV-1, 8 HSV-2.
For HSV-LC, the clinical specificities for HSV-1 and HSV-2 were 100%; however, a genotype was not assigned for six specimens using Tm analysis (Table 3). These were accurately detected and differentiated by HSV-Qx and αHV-PCR (Table 1). For HSV-Qx, a specificity of 100% was observed for HSV-2, but five false positives contributed to a reduced specificity of 98.6% for HSV-1 (Tables 1 and 2). The first false-positive HSV-1 result was seen in a specimen that was confirmed as positive for HSV-2. While coinfection is possible (13), the HSV-1 result was not reproduced by HSV-Qx or confirmed with the other molecular methods (Table 1). The second false-positive result was in a specimen confirmed as positive for VZV by αHV-PCR and a second real-time VZV PCR (11). Interestingly, no cross-reactions were observed with VZV in the specificity panel (see Table S1 in the supplemental material). The last three false-positive HSV-1 results obtained with HSV-Qx had low MaxRFU values (192, 202, and 513) (Table 1).
TABLE 3.
HSV-Qx resolves genotypes in specimens that were problematic for HSV-LC
| Specimen type | HSV-LC |
HSV-Qx |
Discrepant analysis result (αHV-PCR) | ||||
|---|---|---|---|---|---|---|---|
| Cp | Tm | Result | HSV-1 (MaxRFU) | HSV-2 (MaxRFU) | Result | ||
| Throat | 22.01 | 60.48 | HSV | 1,078 | 10 | HSV-1 | HSV-1 |
| Vulva | 28.05 | 60.22 | HSV | 1,151 | 10 | HSV-1 | HSV-1 |
| Buttock | 23.53 | 60.92 | HSV | 0 | 1,710 | HSV-2 | HSV-2 |
| Buttock | 23.44 | 62.98 | HSV | 0 | 1,484 | HSV-2 | HSV-2 |
| Vagina | 20.67 | 60.79 | HSV | 0 | 1,086 | HSV-2 | HSV-2 |
| Unknown | 28.88 | 60.73 | HSV | 15 | 1,552 | HSV-2 | HSV-2 |
With three of five false-positive results for HSV-Qx displaying low MaxRFU values, and a false-negative result near the recommended cutoff for positivity, the distributions of MaxRFU values were plotted for each HSV target (Fig. 1A and B). For HSV-2, 99.3% of the results were classified as either negative or positive, with MaxRFU values of ≤49 and ≥800, respectively (Fig. 1B). For HSV-1, a larger number of results (n = 19; 6.9%) fell between these two categories of MaxRFU values (Fig. 1A). As such, a “gray zone” was implemented where any specimen falling between 50 and 799 MaxRFU would be retested by HSV-Qx and submitted for confirmation using αHV-PCR. Following the implementation of HSV-Qx and the processing of 1,043 specimens, 633 results were negative, 278 were HSV-1 positive, and 125 were HSV-2 positive. Four specimens (0.4%) had MaxRFU values falling into the gray zone (three HSV-1 with MaxRFU values of 158, 234, and 489 and one HSV-2 with a MaxRFU value of 382). αHV-PCR confirmed the HSV-2 and one of the HSV-1 results (MaxRFU of 489). These two had repeat HSV-Qx values of ≥800 and were considered positive. The remaining two results could not be resolved by repeat processing or confirmed by αHV-PCR and therefore were considered indeterminate.
In summary, HSV-Qx is a relatively accurate method for the detection and differentiation of HSV from swabs obtained from anogenital, oral, and other anatomical sites. Swabs in UTM can be processed rapidly using this fully automated system, and HSV-Qx has a lower cost per specimen ($22) compared to that of HSV-LC ($34). However, until an accurate assessment of the cutoff value for positivity can be established, testing of specimens with MaxRFU values falling between 50 and 799 should be repeated. A specimen with a repeat MaxRFU value of ≥800 can be considered positive, but a repeat result of <800 yields an indeterminate result unless confirmed by another method.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Division of Microbiology, Department of Pathology and Laboratory Medicine, at the CDHA (Halifax, NS) for their ongoing support. We also thank Wenda Greer (Division of Hematopathology, Department of Pathology and Laboratory Medicine, CDHA, Halifax, NS) for the human herpesvirus 4 (HHV-4) isolates, Raymond Tellier and Salleen Wong from the Provincial Laboratory for Public Health (Calgary, AB) for the HHV-6a, -6b, and -7 isolates, and Craig MacCormick (Dalhousie University, Halifax, NS) for the HHV-8 isolates. We are indebted to Nathalie Bastien at the National Microbiology Laboratory (Winnipeg, MB) for the VZV strain OKA.
We have no financial conflicts of interest to declare and agree with the final content of the article.
Footnotes
Published ahead of print 2 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03636-13.
REFERENCES
- 1.Whitley RJ, Kimberlin DW, Roizman B. 1998. Herpes simplex viruses. Clin. Infect. Dis. 26:541–553; quiz 554–555. 10.1086/514600 [DOI] [PubMed] [Google Scholar]
- 2.Whitley RJ. 1990. Viral encephalitis. N. Engl. J. Med. 323:242–250. 10.1056/NEJM199007263230406 [DOI] [PubMed] [Google Scholar]
- 3.Sucato G, Wald A, Wakabayashi E, Vieira J, Corey L. 1998. Evidence of latency and reactivation of both herpes simplex virus (HSV)-1 and HSV-2 in the genital region. J. Infect. Dis. 177:1069–1072. 10.1086/515261 [DOI] [PubMed] [Google Scholar]
- 4.Stevens JG. 1989. Human herpesviruses: a consideration of the latent state. Microbiol. Rev. 53:318–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanson KE, Alexander BD, Woods C, Petti C, Reller LB. 2007. Validation of laboratory screening criteria for herpes simplex virus testing of cerebrospinal fluid. J. Clin. Microbiol. 45:721–724. 10.1128/JCM.01950-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burrows J, Nitsche A, Bayly B, Walker E, Higgins G, Kok T. 2002. Detection and subtyping of herpes simplex virus in clinical samples by LightCycler PCR, enzyme immunoassay and cell culture. BMC Microbiol. 2:12. 10.1186/1471-2180-2-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Espy MJ, Uhl JR, Mitchell PS, Thorvilson JN, Svien KA, Wold AD, Smith TF. 2000. Diagnosis of herpes simplex virus infections in the clinical laboratory by LightCycler PCR. J. Clin. Microbiol. 38:795–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LeBlanc JJ, Campbell S, Pettipas J, Hatchette TF, Davidson RJ. 2008. Herpes simplex virus type 2 displays atypical melting-temperature profiles at low viral titers. J. Clin. Microbiol. 46:2786–2789. 10.1128/JCM.02177-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LeBlanc JJ, Heinstein CR, Hatchette TF. 2012. Homogenization with heat treatment: a cost effective alternative to nucleic acid extraction for herpes simplex virus real-time PCR from viral swabs. J. Virol. Methods 179:261–264. 10.1016/j.jviromet.2011.09.018 [DOI] [PubMed] [Google Scholar]
- 10.LeBlanc JJ, Pettipas J, Campbell SJ, Davidson RJ, Hatchette TF. 2008. Uracil-DNA glycosylase (UNG) influences the melting temperature (T(m)) of herpes simplex virus (HSV) hybridization probes. J. Virol. Methods 151:158–160. 10.1016/j.jviromet.2008.03.024 [DOI] [PubMed] [Google Scholar]
- 11.Binkhamis K, Al-Siyabi T, Heinstein CR, Hatchette TF, LeBlanc JJ. 2014. Molecular detection of varicella zoster virus while keeping an eye on the budget. J. Virol. Methods 202:24–27. 10.1016/j.jviromet.2014.02.009 [DOI] [PubMed] [Google Scholar]
- 12.Al-Siyabi T, Binkhamis K, Wilcox M, Wong S, Pabbaraju K, Tellier R, Hatchette TF, LeBlanc JJ. 2013. A cost effective real-time PCR for the detection of adenovirus from viral swabs. Virol. J. 10:184. 10.1186/1743-422X-10-184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perkins D, Chong H, Irvine B, Domagalski J. 2007. Genital coinfection with herpes simplex viruses type 1 and 2: comparison of real-time PCR assay and traditional viral isolation methods. J. Cell. Mol. Med. 11:581–584. 10.1111/j.1582-4934.2007.00045.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.