Abstract
Saprochaete clavata and Magnusiomyces capitatus are human pathogens that are frequently mistaken for each other due to their similar phenotypes and erroneous or limited databases. Based on internal transcribed spacer (ITS) sequences, we propose species-specific carbon assimilation patterns and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) fingerprints that enable the identification of S. clavata, M. capitatus, and Galactomyces candidus to the species level.
TEXT
Saprochaete clavata (de Hoog, Smith, and Guého) de Hoog and Smith 2004 (synonym Geotrichum clavatum de Hoog, Smith, and Guého 1986) is an ascomycetous fungus (1, 2). Colonies are white farinose and dry and consist of true hyphae that branch at acute angles and disarticulate into arthroconidia. This species has rarely been isolated from human samples. Its ecology, reservoir, and importance in agriculture and food are unknown (3). This species is closely related to the known human pathogen Magnusiomyces capitatus (de Hoog, Smith, and Guého) de Hoog and Smith 2004 (previously known as Geotrichum capitatum) (4). Magnusiomyces capitatus is reported to have caused invasive infections, especially in patients with hematological malignancies (5, 6), and it has even been involved in several outbreaks, often associated with contaminated dairy products (7–9). Initially, de Hoog et al. described the new species G. clavatum to distinguish strains identified as G. capitatum that show distinct growth on cellobiose, salicin, and arbutin (2). However, commercially available strips lack salicin and arbutin and are thus useless for obtaining an accurate identification. Furthermore, the databases that come with the automated platforms for microbial identification based on sugar assimilation patterns or mass spectrometry profiles lack this species (10) or show low discrimination for M. capitatus (11, 12).
From analysis of D1/D2 sequence divergence, Phaff et al. mentioned the apparent conspecificity of G. clavatum, Dipodascus spicifer, and G. capitatum (13). In fact, the D1/D2 sequences of the three species have 99% similarity. But, Kurtzman and Robnett considered only G. clavatum and D. spicifer to be synonymous (14). Based on internal transcribed spacer (ITS) sequence analysis, it was demonstrated that G. clavatum differed from G. capitatum (96% similarity) and that it belongs to the Saprochaete/Magnusiomyces clade; therefore, G. clavatum was transferred to the anamorph genus Saprochaete (1). Despite the use of ITS region sequencing, the majority of clinical isolates of S. clavata are identified as M. capitatus because nucleotide sequences of S. clavata available in public databases, such as GenBank, are either misidentified or too short (163 bp). In order to provide clues for species identification without using ITS sequencing, we analyzed phenotypic characteristics of clinical isolates identified as Geotrichum spp. based on profiles generated by routinely available techniques (sugar assimilation pattern [ID32C; bioMérieux] or matrix-assisted laser desorption ionization–time of flight mass spectrometry [MALDI-TOF MS]).
For the 101 clinical isolates received as Geotrichum species since 2003 at the National Reference Center for Mycoses and Antifungals (NRCMA), purity was checked by using a chromogenic medium (BBL CHROMagar; Becton, Dickinson, USA). The urease activity (urea-indole medium; bioMérieux, Marcy l'Etoile, France) and carbon assimilation patterns (ID32C and 50CH; bioMérieux) were determined. The D1/D2 and ITS1–5.8S-ITS2 regions of the ribosomal DNA were sequenced by using universal primers (NL1/NL4 [15] and V9D/LS266 [16, 17], respectively). The sequences of the ITS1–5.8S-ITS2 regions were delimited by the sequences of the primers ITS1 and ITS4 (TCCGTAGGTGAACCTGCGG and GCATATCAATAAGCGGAGGA, respectively), and the sequences were compared with the nucleotide sequences of the S. clavata CBS 425.71 type strain (GenBank accession number KF984489), the M. capitatus CBS 162.80 type strain (accession number KF984490), and the Galactomyces candidus (teleomorph of Geotrichum candidum) CBS 178.71 type strain (accession number KF984491). For each clinical isolate and type strain, a PCR fingerprinting technique was performed with the core sequence of phage M13 (5′-GAGGGTGGCGGTTCT-3′) and OPE4 (5′-GTGACATGCC-3′) as a single primer (7, 18). For 19 clinical isolates (15 S. clavata, 4 M. capitatus) and the 3 type strains, MALDI-TOF fingerprints were obtained using mass spectrometry technology on the Vitek MS automate (bioMérieux) after 24 h and after 48 h of growth on malt extract agar plates with gentamicin and chloramphenicol (Merck) and on Sabouraud agar slants with gentamicin and chloramphenicol (Bio-Rad), and they were analyzed using the Vitek MS version 2.0 reference strain database.
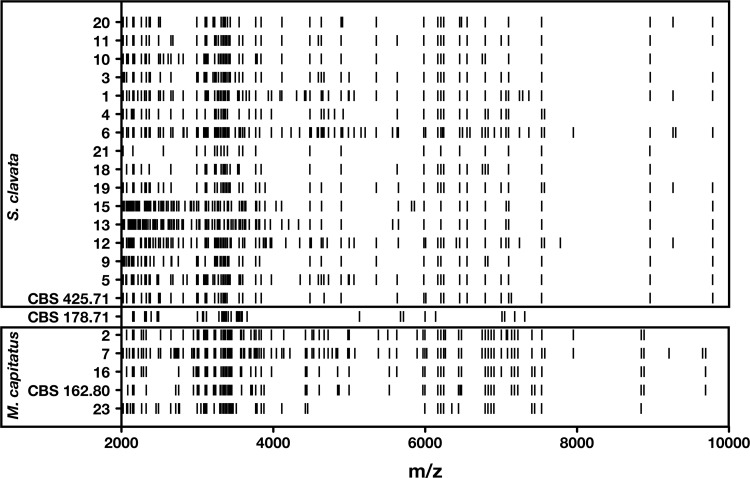
Based on ITS region sequencing, 59/101 isolates were eventually identified as S. clavata, 27 as M. capitatus, and 15 as G. candidus. The delimited sequences of ITS and D1/D2 regions between S. clavata (464 bp) and M. capitatus (454 bp) have 96% and 99% similarity, respectively. Random amplified polymorphic DNA (RAPD) analysis suggests that species-specific fingerprints may be obtained (Fig. 1). This technique, however, cannot be envisioned as a routine means for species identification. Comparison of ID32C profiles allowed for the differentiation of species-specific carbon assimilation profiles (Table 1), with 10 codes specific for M. capitatus, 8 for S. clavata, and 4 for G. candidus. Using the Vitek MS and its corresponding database, the identifications were confirmed for the 4 clinical isolates and the type strain of M. capitatus, but no identifications were obtained for the other 17 isolates in the v2.0 database. However, when analyzing MALDI-TOF MS profiles of the 22 isolates, three groups corresponding to the three species can be delineated based on specific peaks (Fig. 2).
FIG 1.
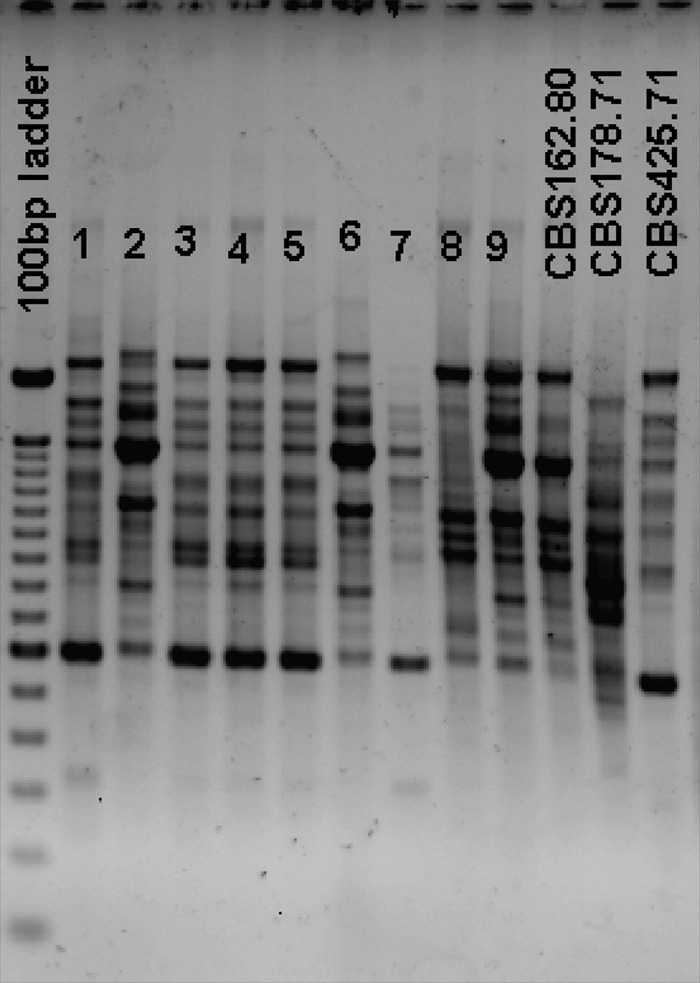
Examples of PCR fingerprints obtained by using M13 primer for clinical isolates of Saprochaete clavata (lanes 1, 3, 4, 5, and 7) and Magnusiomyces capitatus (lanes 2, 6, 8, and 9) and type strains of S. clavata (CBS 425.71), M. capitatus (CBS 162.80), and Galactomyces candidus (CBS 178.71).
TABLE 1.
ID32C profiles for the 101 Geotrichum sp. clinical isolates identified by ITS region sequencing
| Species (no. of isolates) | No. (%) of isolates with the indicated profile | ID32C profile |
|---|---|---|
| Magnusiomyces capitatus (27) | 6 (22.2) | 20000100010 |
| 6 (22.2) | 32000100030 | |
| 3 (11.1) | 22000100010 | |
| 3 (11.1) | 22000100030 | |
| 3 (11.1) | 32000100010 | |
| 2 (7.4) | 30000100010 | |
| 1 (3.7) | 20000100020 | |
| 1 (3.7) | 22000100011 | |
| 1 (3.7) | 30400100030 | |
| 1 (3.7) | 32001100031 | |
| Saprochaete clavata (59) | 23 (38.9) | 30100100031 |
| 16 (27.1) | 30100100011 | |
| 9 (15.2) | 32100100031 | |
| 3 (5) | 30000100030 | |
| 3 (5) | 30000100031 | |
| 2 (3.4) | 30000100011 | |
| 2 (3.4) | 30100100010 | |
| 1 (1.6) | 32100100011 | |
| Galactomyces candidus (15) | 6 (40) | 32003100130 |
| 6 (40) | 32003100131 | |
| 2 (13.3) | 32003100031 | |
| 1 (6.6) | 32003100150 |
FIG 2.
Raw MALDI-TOF MS spectra for 19 clinical isolates and the type strains CBS 425.71 (Saprochaete clavata), CBS 178.71 (Galactomyces candidus), and CBS 162.80 (Magnusiomyces capitatus) after 24-h subculture on 2% malt dextrose agar plates plus gentamicin and chloramphenicol.
These results underline the importance of using specialized databases, such as that of the Centraalbureau voor Schimmelcultures (CBS) (see http://www.cbs.knaw.nl/collections/BioloMICSSequences.aspx?file=all), where the taxonomy is more reliable than that in public repositories, as pointed out by Nilsson and colleagues (19). It also shows the importance of incrementing databases according to the latest developments of fungal taxonomy.
Nucleotide sequence accession numbers.
Newly determined sequence data from this study have been deposited in GenBank under accession numbers KF984489, KF984490, and KF984491.
ACKNOWLEDGMENTS
The technical help of the sequencing facility and specifically that of Laure Diancourt, Anne-Sophie Delannoy, and Jean-Michel Thiberge (Genotyping of Pathogens and Public Health, Institut Pasteur) is gratefully acknowledged. We thank members of the French Mycoses Study Group, who provided the isolates used in the present study (in alphabetical order of the cities), Nathalie Brieu and Evelyne Lagier (Aix-en-Provence), Jean-Philippe Bouchara and Marc Pihet (Angers), Cécile Jensen (Avignon), Frédéric Grenouillet (Besançon), Christian Chochillon (Hôpital Bichat, Paris), Isabelle Accoceberry and Olivier Albert (Bordeaux), Julie Bonhomme (Caen), Nathalie Fauchet (Créteil), Philippe Poirier and Monique Cambon (Clermont-Ferrand), Pierre Cahen (Foch), André Paugam and Marie-Thérèse Baixench (Hôpital Cochin, Paris), Dominique De Briel (Colmar), Frédéric Dalle (Dijon), Bernadette Lebeau (Grenoble), Françoise Botterel (Hôpital Henri Mondor, Paris), Muriel Cornet (Hôpital de l'Hôtel Dieu, Paris), Odile Eloy (Le Chesnay), Boualem Sendid (Lille), Stéphane Ranque (Marseille), Nathalie Bourgeois and Philippe Rispail (Montpellier), Malik Al Nakib (Montsouris, Paris), Marie Machouart (Nancy), Florent Morio (Nantes), Marie-Elisabeth Bougnoux (Hôpital Necker Enfants Malades, Paris), Martine Garri-Toussaint (Nice), Didier Poisson (Orléans), Marie-Francoise David and Najiby Kassis-Chikhani Liliana Mihaila (Villejuif), Charles Soler (Percy, Clamart), Anne Gaschet and Philippe Geudet (Perpignan), Annick Datry and Sophie Brun (Hôpital de la Pitié-Salpêtrière, Paris), Christine Chaumeil (Quinze-Vingt, Paris), Dominique Toubas (Reims), Jean-Pierre Gangneux (Rennes), Stéphane Bonacorsi (Hôpital Robert Debré, Paris), Loïc Favennec and Gilles Gargala (Rouen), Hélène Raberin (St Etienne), Stéphane Bretagne (Hôpital Saint-Louis, Paris), Valérie Letscher-Bru (Strasbourg), and Sophie Cassaing (Toulouse) and our European colleagues Konrad Mühlethaler, Stefan Zimmerli (Institute for Infectious Diseases, University of Bern, Bern, Switzerland), Polona Zalar (Biology Department, Biotechnical Faculty, University of Ljubljana, Ljubljana, Slovenia), and Ferran Sánchez-Reus and Merce Gurgui (Hospital de la Santa Creu i Sant Pau, Barcelona, Spain).
This work was supported by the Institut Pasteur and the Institut de Veille Sanitaire.
Footnotes
Published ahead of print 2 April 2014
REFERENCES
- 1.De Hoog GS, Smith MT. 2004. Ribosomal gene phylogeny and species delimitation in Geotrichum and its teleomorphs. Stud. Mycol. 50:489–515 [Google Scholar]
- 2.De Hoog GS, Smith MT, Guého E. 1986. A revision of the genus Geotrichum and its teleomorphs. Stud. Mycol. 29:1–131 [Google Scholar]
- 3.de Hoog GS, Smith MT. 2011. Saprochaete Coker & Shanor ex D.T.S. Wagner & Dawes (1970), p 1317–1330 In Kurtzman CP, Fell JW, Boekhout T. (ed), The yeasts: a taxonomic study. Elsevier, Amsterdam, the Netherlands [Google Scholar]
- 4.de Hoog GS, Smith MT. 2011. Magnusiomyces Zender (1977), p 565–574 In Kurtzman CP, Fell JW, Boekhout T. (ed), The yeasts: a taxonomic study. Elsevier, Amsterdam, the Netherlands [Google Scholar]
- 5.Garcia-Ruiz JC, Lopez-Soria L, Olazabal I, Amutio E, Arrieta-Aguirre I, Velasco-Benito V, Ponton J, Moragues MD. 2013. Invasive infections caused by Saprochaete capitata in patients with haematological malignancies: report of five cases and review of the antifungal therapy. Rev. Iberoam. Micol. 30:248–255. 10.1016/j.riam.2013.02.004 [DOI] [PubMed] [Google Scholar]
- 6.Girmenia C, Pagano L, Martino B, D'Antonio D, Fanci R, Specchia G, Melillo L, Buelli M, Pizzarelli G, Venditti M, Martino P. 2005. Invasive infections caused by Trichosporon species and Geotrichum capitatum in patients with hematological malignancies: a retrospective multicenter study from Italy and review of the literature. J. Clin. Microbiol. 43:1818–1828. 10.1128/JCM.43.4.1818-1828.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ersoz G, Otag F, Erturan Z, Aslan G, Kaya A, Emekdas G, Sugita T. 2004. An outbreak of Dipodascus capitatus infection in the ICU: three case reports and review of the literature. Jpn. J. Infect. Dis. 57:248–252 [PubMed] [Google Scholar]
- 8.Gurgui M, Sanchez F, March F, Lopez-Contreras J, Martino R, Cotura A, Galvez ML, Roig C, Coll P. 2011. Nosocomial outbreak of Blastoschizomyces capitatus associated with contaminated milk in a haematological unit. J. Hosp. Infect. 78:274–278. 10.1016/j.jhin.2011.01.027 [DOI] [PubMed] [Google Scholar]
- 9.Martino P, Venditti M, Micozzi A, Morace G, Polonelli L, Mantovani MP, Petti MC, Burgio VL, Santini C, Serra P, Mandelli F. 1990. Blastoschizomyces capitatus: an emerging cause of invasive fungal disease in leukemia patients. Rev. Infect. Dis. 12:570–582. 10.1093/clinids/12.4.570 [DOI] [PubMed] [Google Scholar]
- 10.Bader O, Weig M, Taverne-Ghadwal L, Lugert R, Gross U, Kuhns M. 2011. Improved clinical laboratory identification of human pathogenic yeasts by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 17:1359–1365. 10.1111/j.1469-0691.2010.03398.x [DOI] [PubMed] [Google Scholar]
- 11.Lohmann C, Sabou M, Moussaoui W, Prevost G, Delarbre JM, Candolfi E, Gravet A, Letscher-Bru V. 2013. Comparison between the Biflex III-Biotyper and the Axima-SARAMIS systems for yeast identification by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 51:1231–1236. 10.1128/JCM.03268-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giacchino M, Chiapello N, Bezzio S, Fagioli F, Saracco P, Alfarano A, Martini V, Cimino G, Martino P, Girmenia C. 2006. Aspergillus galactomannan enzyme-linked immunosorbent assay cross-reactivity caused by invasive Geotrichum capitatum. J. Clin. Microbiol. 44:3432–3434. 10.1128/JCM.00856-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phaff HJ, Blue J, Hagler AN, Kurtzman CP. 1997. Dipodascus starmeri sp. nov., a new species of yeast occurring in cactus necroses. Int. J. Syst. Bacteriol. 47:307–312. 10.1099/00207713-47-2-307 [DOI] [PubMed] [Google Scholar]
- 14.Kurtzman CP, Robnett CJ. 1998. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73:331–371. 10.1023/A:1001761008817 [DOI] [PubMed] [Google Scholar]
- 15.O'Donnell K. 1993. Fusarium and its near relatives, p 225–233 In Reynolds DR, Taylor JW. (ed), The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. CAB, Wallingford, United Kingdom [Google Scholar]
- 16.de Hoog GS, Gerrits van den Ende AH. 1998. Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses 41:183–189. 10.1111/j.1439-0507.1998.tb00321.x [DOI] [PubMed] [Google Scholar]
- 17.Masclaux F, Gueho E, de Hoog GS, Christen R. 1995. Phylogenetic relationships of human-pathogenic Cladosporium (Xylohypha) species inferred from partial LS rRNA sequences. J. Med. Vet. Mycol. 33:327–338. 10.1080/02681219580000651 [DOI] [PubMed] [Google Scholar]
- 18.Gadea I, Cuenca-Estrella M, Prieto E, Diaz-Guerra TM, Garcia-Cia JI, Mellado E, Tomas JF, Rodriguez-Tudela JL. 2004. Genotyping and antifungal susceptibility profile of Dipodascus capitatus isolates causing disseminated infection in seven hematological patients of a tertiary hospital. J. Clin. Microbiol. 42:1832–1836. 10.1128/JCM.42.4.1832-1836.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nilsson RH, Ryberg M, Kristiansson E, Abarenkov K, Larsson KH, Köljalg U. 2006. Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective. PLoS One 1:e59. 10.1371/journal.pone.0000059 [DOI] [PMC free article] [PubMed] [Google Scholar]