Abstract
Infections due to Pseudomonas fulva remain a rare but emerging concern. A case of ventriculitis due to Enterobacter cloacae and Pseudomonas fulva following placement of an external ventricular drain is described. Similar to other reports, the organism was initially misidentified as Pseudomonas putida. The infection was successfully treated with levofloxacin.
CASE REPORT
A 55-year-old woman with a history of chronic migraines and hypertension was transferred to Emory University Hospital for management of a spontaneous subarachnoid hemorrhage. The patient complained of 1 week of worsening migraine headaches but denied nausea, vomiting, and neurologic disturbances until the day of admission, when she developed generalized paresthesia and progressive decline in the level of consciousness. On initial examination, she was lethargic and confused but her motor, sensory, and cranial nerves were intact. A computed tomography (CT) scan of the head showed a large subarachnoid hemorrhage in the basilar cisterns, bilateral sylvian fissures, and the interhemispheric fissure with hydrocephalus. Subsequently, an external ventricular drain (EVD) was placed. A saccular anterior communicating aneurysm was identified on a CT angiogram of the head, and the patient underwent coil embolization. On day 3 of hospitalization, the patient developed a temperature of 38.1°C and was diagnosed with an Escherichia coli urinary tract infection for which she was given levofloxacin (750 mg daily) during hospital days 4 to 8. Fevers persisted, however, and on day 5 of hospitalization, a grossly bloody cerebrospinal fluid (CSF) specimen from the EVD showed a white blood cell (WBC) count of 350 × 106 cells/liter. CSF Gram stain and culture at that time were negative. Over the next several days, the CSF cell count progressively increased and on the day 9 of hospitalization was 14,000 × 106 cells/liter with 80% neutrophils. During the same time period, the CSF glucose level ranged between 59 and 68 mg/dl (with corresponding plasma glucose levels of 112 to 131 mg/dl) and protein levels ranged between 159 and 270 mg/dl. Three CSF cultures obtained with each EVD aspiration during concurrent levofloxacin treatment for urinary tract infection were negative.
Given worsening fever, three CSF specimens were sent for culture on hospital days 12 to 14. Each culture was positive for two different Gram-negative rods, which were detected on blood agar, chocolate agar, and MacConkey agar and in thioglycolate broth (Remel, Lenexa, KS) after overnight incubation at 35°C in 5% CO2. The first organism was present in a lower quantity than the second (2+ [moderate] in the first two cultures and 1+ [few] in the third) and was identified as Enterobacter cloacae. The other organism was present in high quantity (4+ [many]) in all three cultures, was a lactose nonfermenter, and formed smooth, wet colonies with a yellow nondiffusible pigment (Fig. 1). The oxidase reaction was inconclusive but was ultimately determined to be weakly positive after repeat testing. Biochemical identification and antimicrobial susceptibility were determined on the MicroScan WalkAway Plus (Siemens Healthcare Diagnostics Inc., West Sacramento, CA) instrument using MicroScan Breakpoint Combo Panel Type 31 and the PROMPT (3M Company, St. Paul, MN) inoculation system. The isolate was identified as Pseudomonas putida (probability 99.2%). However, since the yellow pigment of the colonies was not consistent with the identification of P. putida, identification using other systems was attempted.
FIG 1.
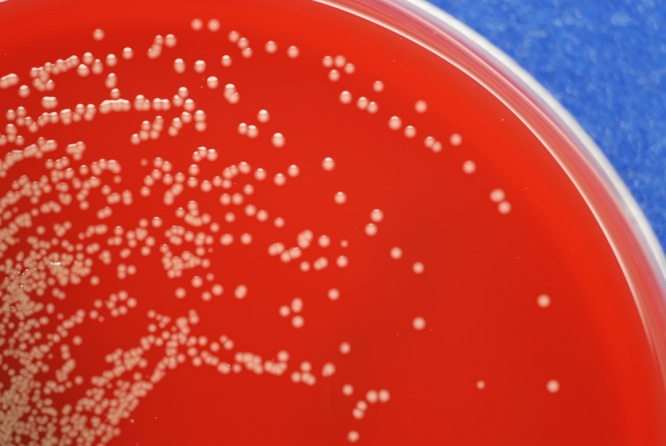
Smooth, wet colonies with yellow nondiffusible pigment growing on a blood agar plate after overnight incubation.
The isolate was identified by the API 20 NE (bioMérieux, Durham, NC) identification strip as P. putida (98.2% probability, biocode 0142455). Biochemical and cell wall fatty acid analysis performed at the Georgia Public Health Laboratory also resulted in an identification of P. putida. Identification using MALDI-TOF mass spectrometry was performed using both the Vitek MS RUO (bioMérieux, Durham, NC) and the Microflex LT (Bruker Daltonics, Billerica, Massachusetts) mass spectrometers. On the Vitek MS RUO instrument, the isolate was tested in duplicate and analyzed using the SARAMIS SuperSpectra database version 4.09. The identification was returned as P. putida but with a low confidence value of 81.9. On the Microflex LT instrument, the isolate was tested in duplicate, with and without formic acid pretreatment, using flexControl and flexAnalysis (Compass 1.4) and BioTyper (version 3.1) software. Reliable identification as Pseudomonas fulva was obtained under both conditions with scores of 1.970 and 1.870 using matrix only and scores of 2.212 and 2.118 with formic acid pretreatment. To further resolve the identification, PCR amplification and sequencing of the 16S rRNA gene was performed by the Georgia Public Health Laboratory using the ABI 3130xl genetic analyzer (Life Technologies, Grand Island, NY). The sequencer internal library generated an identification of Pseudomonas fulva (99.92% sequence homology with P. fulva GenBank accession no. FJ972539). The next closest match was to Pseudomonas straminea (98.63%). Because the species match to P. fulva was >99% and the separation from the next most similar species was greater than 0.8%, the identification as P. fulva was considered definitive (1). An identification of P. putida was not presented as an option by the sequencer for this isolate. When the obtained sequence was applied to a GenBank search, the first identification was P. fulva, with a 99% similarity match to type strain P. fulva NRIC 0180 with 3 mismatches in 482 bp. The next nearest species match was to Pseudomonas fluorescens. Even though the percentages for both identifications were 99%, the total score for P. fulva was 8 points greater than for P. fluorescens. The nearest match to any reference or type strain of P. putida using BLAST was 97% to P. putida strain KT2440.
Biochemical characterization using the Biolog OmniLog identification system (Biolog, Inc., Haywood CA) and GEN III MicroPlate was done in triplicate and resulted in a metabolic fingerprint consistent with an identification of P. fulva with probabilities of 77% (excellent identification), 56%, and 55% (good identification). No other species met the 50% probability required for acceptable species identification. Additional tests showed that the isolate failed to grow at 42°C and did not fluoresce when examined under UV light from a Wood's lamp.
The P. fulva isolate was susceptible to all antibiotics tested: amikacin, aztreonam, cefepime, ceftazidime, gentamicin, levofloxacin, meropenem, piperacillin-tazobactam, and tobramycin. Of the antibiotics tested, the E. cloacae isolate was resistant only to ampicillin. After initial positive CSF cultures, the patient was started on intravenous (i.v.) levofloxacin (750 mg i.v. daily) and rifampin (600 mg i.v. daily) while awaiting final species identification. The fevers resolved, the CSF WBC count improved, the EVD was removed, and rifampin was discontinued on the third day of this regimen. The patient completed an additional 16 days of oral levofloxacin (750 mg per day). The patient remained afebrile and was without evidence of recurrence in follow-up approximately 1 month after completion of antibiotic therapy.
P. fulva is a Gram-negative rod that was initially grouped with other plant-inhabiting Pseudomonas species that also produce a yellow pigment. The classification was revised based on 16S rRNA gene sequences, and P. fulva is now phylogenetically located in the P. putida group, which also includes Pseudomonas monteilii, Pseudomonas mosselii, Pseudomonas oryzihabitans, Pseudomonas plecoglossicida, and P. putida (2). P. fulva cells measure 0.6 to 0.8 by 1.4 to 1 μm, have rounded ends, and are motile with polar flagella. Growth occurs at temperatures between 4°C and 37°C, but P. fulva does not grow at 41°C. Typically, P. fulva colonies are smooth and wet. Although catalase and oxidase are produced, the oxidase reaction has been described as weakly positive (3, 4). As with other Pseudomonas species, metabolism is strictly aerobic. Key biochemical characteristics include production of arginine dihydrolase and lack of reduction of nitrate to nitrite. In contrast to its closely related species, P. fulva produces a water-insoluble yellow pigment and does not assimilate malonate or m-hydroxybenzoate while P. putida produces a water-soluble fluorescent pigment and does assimilate the two compounds.
While P. fulva has been identified mainly in natural environments, including rice and petroleum fields from which it was initially isolated (5), few cases of P. fulva infection in human have been documented. Thus far, two previous cases have highlighted findings of P. fulva in human CSF and bloodstream. Comparing these two reports with our own finding revealed important similarities that support our conclusion and nuances in the presenting characteristics that highlight the complexity of P. fulva as an emerging infection.
Similar to the two previous reported human cases of P. fulva, the initial identification efforts misidentified the organism as P. putida. Almuzara et al. relied on the Vitek 2 system and API 20 NE, both of which indicated the presence of P. putida (4). Similarly, Seok et al. also originally identified their isolate as P. putida by using the Vitek 2 and ID 32 systems (3). Our specimen was subjected to the MicroScan WalkAway Plus Neg Breakpoint Combo Panel Type 41, API 20 NE, and Vitek MS, and all three systems led to P. putida identification. Consistent with the initial identifications in the CSF and bloodstream cases, which had relatively high probabilities of identification of P. putida ranging from 99.0% on the Vitek2 system in the report by Seok et al. to 99.9% from the ID 32 GN system, biochemical testing of our isolate on the MicroScan (99.2%) and API 20 NE (98.2%) also produced high probabilities. We postulate that the initial identifications of our isolate may have suggested P. putida due to limited distinguishing biochemicals on the panels and a database content not inclusive of P. fulva. The API 20 NE strip does not contain any biochemicals that distinguish P. putida from P. fulva, and the MicroScan panel contains three distinguishing biochemicals, inositol, sucrose, and malonate, none of which are assimilated by P. fulva, and the panel gave the expected negative reactions. The Vitek MS does not have P. fulva mass spectra in the reference database and generated an identification of P. putida, but with a probability of only 81.9%.
Similar to the other two cases, we questioned the identification of P. putida due to the inconsistent phenotypic and biochemical characterization of the isolates with the typical P. putida features. Colonies from all three reports were brownish yellow in color, due to production of a water-insoluble pigment, which more closely resembled P. fulva. The isolates were arginine dihydrolase positive, and further substrate utilization profiles showed that the isolates were malonate negative. Similar to our observation of no growth of our isolate at elevated temperatures, the isolates reported by both Seok et al. and Almuzara et al. also did not grow at 41°C (2, 3). These characteristics were consistent with the identification by Uchino et al. of P. fulva (2). In considering oxidase tests, the CSF specimen isolated by Seok et al. (3) was oxidase negative while the bloodstream isolate in the study by Almuzara et al. (4) was found to be weakly oxidase positive, similar to our isolate.
As these characterizations further suggest greater consistency with identification of P. fulva, subsequent verification using the more reliable 16S rRNA gene sequencing method in all three cases corroborated the findings by showing that all three isolates had the highest identity with P. fulva. Further comparisons of antibiotic profiles of the three isolated samples revealed subtle differences, which highlighted the potential variability in P. fulva susceptibility. In the Seok et al. case report (3), the organism was susceptible to piperacillin-tazobactam, ceftazidime, cefotaxime, aztreonam, cefepime, imipenem, meropenem, gentamicin, tobramycin, amikacin, and levofloxacin. However, it demonstrated resistance to chloramphenicol and trimethoprim-sulfamethoxazole when tested by the Etest (3). Our own strain showed a susceptibility profile similar to that of Seok et al. In contrast, the isolate from the report of Almuzara et al. showed susceptibility to cefepime, cefepime plus lithium clavulanate, amikacin, ciprofloxacin, and colistin but resistance to ceftazidime, ceftazidime plus lithium clavulanate, piperacillin-tazobactam, imipenem, meropenem, and gentamicin. Further assessment with EDTA-sodium mercaptoacetic acid (EDTA-SMA) double-disk assays was performed with imipenem and meropenem, to which the P. fulva strain tested susceptible, thus indicating the possible presence of metallo-β-lactamase (MBL). Additional PCR amplification studies showed that the P. fulva isolate did indeed contain the blaVIM gene (4). Integration of the MBL cassette into this isolate raises the concern of possible transmission of these resistance-inducing cassettes across different Pseudomonas species, including P. fulva.
Although P. fulva has traditionally been described within the context of rice and petroleum fields, studies have shown a broad range of environments in which the organism can survive. In a study designed to collect samples from 80 “habitat types” that represented a diversity of microenvironments ranging from vertebrate skin to bathroom counter surfaces in 20 households, P. fulva was among the four most commonly encountered Pseudomonas species (6). Since P. fulva is commonly found in the environment, there is potential for it to be identified from clinical specimens. As the P. fulva in the present study was isolated concomitantly with Enterobacter cloacae, there is strong reason to believe that the P. fulva was hospital acquired. Our case further highlights the possible emergence of P. fulva in the hospital setting and the need for circumspect diagnosis in the setting of phenotypic and biochemical inconsistencies with automated identification.
ACKNOWLEDGMENTS
We thank Theresa Stanley at Children's Healthcare of Atlanta for performing identification of our isolate using the Microflex LT MALDI-TOF mass spectrometer and John Varga at Emory University School of Medicine Rollins Research Center for performing the Biolog identification.
Footnotes
Published ahead of print 19 March 2014
REFERENCES
- 1.CLSI. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing; approved guideline, CLSI document MM18-A. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 2.Uchino M, Shida O, Uchimura T, Komagata K. 2001. Recharacterization of Pseudomonas fulva Iizuka and Komagata 1963, and proposals of Pseudomonas parafulva sp. nov. and Pseudomonas cremoricolorata sp. nov. J. Gen. Appl. Microbiol. 47:247–261. 10.2323/jgam.47.247 [DOI] [PubMed] [Google Scholar]
- 3.Seok Y, Shin H, Lee Y, Cho I, Na S, Yong D, Jeong SH, Lee K. 2010. First report of bloodstream infection caused by Pseudomonas fulva. J. Clin. Microbiol. 48:2656–2657. 10.1128/JCM.01609-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Almuzara MN, Vazquez M, Tanaka N, Turco M, Ramirez MS, Lopez EL, Pasteran F, Rapoport M, Procopio A, Vay CA. 2010. First case of human infection due to Pseudomonas fulva, an environmental bacterium isolated from cerebrospinal fluid. J. Clin. Microbiol. 48:660–664. 10.1128/JCM.01849-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iizuka H, Komagata K. 1963. New species of Pseudomonas belonging to fluorescent group. (Studies on the microorganisms of cereal grains. Part V). Nippon Nogeikagaku Kaishi 37:137–141. 10.1271/nogeikagaku1924.37.137 [DOI] [Google Scholar]
- 6.Remold SK, Brown CK, Farris JE, Hundley TC, Perpich JA, Purdy ME. 2011. Differential habitat use and niche partitioning by Pseudomonas species in human homes. Microb. Ecol. 62:505–517. 10.1007/s00248-011-9844-5 [DOI] [PubMed] [Google Scholar]


