Abstract
Simple, low-cost PCR/PCR-restriction fragment length polymorphism (RFLP) assays targeting cyp51A promoter and codon 98 regions were developed for the detection of triazole-resistant Aspergillus fumigatus strains carrying TR34/L98H mutations. The assays were evaluated using 40 itraconazole-susceptible isolates and 35 itraconazole-resistant isolates. The prevalence of TR34/L98H mutations in clinical/environmental A. fumigatus isolates may now be determined easily from resource-poor settings.
TEXT
Aspergillus fumigatus is the principal etiological agent of invasive aspergillosis (IA), a life-threatening infection in immunocompromised patients. Treatment options are limited due to the low efficacies of some antifungal drugs against clinical A. fumigatus isolates. Oral triazoles (itraconazole, voriconazole, and posaconazole) exhibit excellent activities against A. fumigatus isolates in vitro and are currently being used as first-line therapies in the management and prophylaxis of IA (1). Ten years ago, acquired triazole resistance among clinical A. fumigatus isolates was rare. Clinical failures are now reported frequently, and the frequency of isolation of triazole-resistant clinical A. fumigatus isolates has increased in several countries (2–6). Rapid emergence of triazole-resistant A. fumigatus isolates has been attributed to the exposure of environmental fungi to 14α-demethylase inhibitors (DMIs) which are structurally and functionally related to clinically licensed triazoles. The DMIs are widely used to control fungal growth for crop plant/ornamental flower protection (7). The occurrence of triazole-resistant A. fumigatus strains has been documented in environmental samples from some countries, with isolation frequencies ranging from 5% to 12% (7, 8).
The molecular basis of resistance to triazoles in clinical A. fumigatus isolates involves point mutations at several codons in the cyp51A gene, which encodes 14α-sterol demethylase. However, a dominant mechanism involving a 34-bp tandem repeat (TR34) in the promoter region together with an L98H substitution (TR34/L98H) in cyp51A has been observed in triazole-resistant isolates recovered from environmental sources, treatment-naive subjects, and patients under treatment (6, 9, 10). These studies have largely been carried out at few specialized centers, possibly because these mutations have been detected by sophisticated techniques and expensive instruments typically involving PCR or real-time PCR together with specific probes/molecular beacons or DNA sequencing (6, 10–13). In this report, we describe simple PCR/PCR-restriction fragment length polymorphism (PCR-RFLP) assays for rapid detection of TR34/L98H mutations in the cyp51A gene.
The study was approved by the ethical committee of the Faculty of Medicine, Kuwait University.
Reference A. fumigatus strains CBS 113.26 (carrying wild-type sequences in the promoter region and codon 98 in cyp51A [cyp51A98]) and VPCI1042/09 (carrying mutant [TR34/L98H] sequences in the promoter region and codon 98) were used for the establishment of PCR assays. A total of 75 clinical and environmental A. fumigatus isolates were used for the evaluation of the developed methods. The background information regarding country of origin, source of isolation, susceptibility to itraconazole (including MICs), and presence or absence of TR34/L98H mutations in the cyp51A gene of A. fumigatus isolates tested for the performance of the molecular assays is presented in Table 1. The details of clinical/environmental A. fumigatus isolates from France, The Netherlands, and India that were used in this study have also been published previously (4, 14, 15). Drug susceptibility testing (DST) of A. fumigatus isolates with itraconazole was carried out by Etest as described elsewhere (16). Isolates with reduced susceptibility to itraconazole (MIC of ≥2 μg/ml) were also tested for voriconazole by a broth microdilution (M38-A2) method (4). Isolates with MICs of ≥2 μg/ml were considered resistant (4).
TABLE 1.
Country of origin, source of isolation, susceptibility to itraconazole with MICs, and presence or absence of TR34/L98H mutations in the cyp51A gene of A. fumigatus isolates
| Serial no. | Isolate no. | Country of origin | Source of isolation | Susceptibility to ITR,d MIC (μg/ml) | TR34/L98H mutation |
|---|---|---|---|---|---|
| 1 | E-296 | Kuwait | Outdoor air | Susceptible, 0.5 | No/no |
| 2 | E-149 | Kuwait | Outdoor air | Susceptible, 0.38 | No/no |
| 3 | E-288 | Kuwait | Hospital indoor air | Susceptible, 0.75 | No/no |
| 4 | E-298 | Kuwait | Hospital indoor air | Susceptible, 0.38 | No/no |
| 5 | E-256 | Kuwait | Hospital floor swab | Susceptible, 0.75 | No/no |
| 6 | E-262 | Kuwait | Hospital indoor air | Susceptible, 0.38 | No/no |
| 7 | E-267 | Kuwait | Hospital indoor air | Susceptible, 0.38 | No/no |
| 8 | E-142 | Kuwait | Hospital floor swab | Susceptible, 0.38 | No/no |
| 9 | Kw2287/09 | Kuwait | Clinical, sputum | Susceptible, 0.75 | No/no |
| 10 | Kw3468/10 | Kuwait | Clinical, sputum | Susceptible, 0.047 | No/no |
| 11 | Kw2893/10 | Kuwait | Clinical, sputum | Susceptible, 0.75 | No/no |
| 12 | Kw3068/10 | Kuwait | Clinical, sputum | Susceptible, 0.75 | No/no |
| 13 | Kw1830/08 | Kuwait | Clinical, wound swab | Susceptible, 0.5 | No/no |
| 14 | Kw3916/10 | Kuwait | Clinical, sputum | Susceptible, 0.5 | No/no |
| 15 | E-307 | Kuwait | Hospital indoor air | Susceptible, 0.38 | No/no |
| 16 | Kw2351/11 | Kuwait | Clinical, bronchoalveolar lavage specimen | Susceptible, 0.38 | No/no |
| 17 | Kw3328/10 | Kuwait | Clinical, sputum | Susceptible, 0.75 | No/no |
| 18 | Kw2349/11 | Kuwait | Clinical, bronchoalveolar lavage specimen | Susceptible, 0.125 | No/no |
| 19 | R-1 | Kuwait | Outdoor, soil sample | Susceptible, 0.75 | No/no |
| 20 | R-2 | Kuwait | Outdoor, soil sample | Susceptible, 0.38 | No/no |
| 21 | R-3 | Kuwait | Outdoor, soil sample | Susceptible, 0.25 | No/no |
| 22 | R-4 | Kuwait | Outdoor, soil sample | Susceptible, 0.25 | No/no |
| 23 | R-5 | Kuwait | Outdoor, soil sample | Susceptible, 0.75 | No/no |
| 24 | R-6 | Kuwait | Outdoor, soil sample | Susceptible, 0.5 | No/no |
| 25 | R-7 | Kuwait | Outdoor, soil sample | Susceptible, 0.19 | No/no |
| 26 | R-8 | Kuwait | Outdoor, soil sample | Susceptible, 0.38 | No/no |
| 27 | E-1 | Kuwait | Outdoor air | Susceptible, 0.5 | No/no |
| 28 | E-143 | Kuwait | Outdoor air | Susceptible, 0.75 | No/no |
| 29 | E-290 | Kuwait | Outdoor air | Susceptible, 0.38 | No/no |
| 30 | E-320 | Kuwait | Hospital indoor air | Susceptible, 0.38 | No/no |
| 31 | E-335 | Kuwait | Outdoor air | Susceptible, 0.75 | No/no |
| 32 | E-286 | Kuwait | Hospital indoor air | Susceptible, 0.75 | No/no |
| 33 | E-106 | Kuwait | Hospital indoor air | Susceptible, 0.5 | No/no |
| 34 | Kw1431/10 | Kuwait | Clinical, sputum | Susceptible, 0.5 | No/no |
| 35 | Kw2285/09 | Kuwait | Clinical, endotracheal aspirate | Susceptible, 0.19 | No/no |
| 36 | Kw2941/11 | Kuwait | Clinical, sputum | Susceptible, 0.012 | No/no |
| 37 | Kw3862/10 | Kuwait | Clinical, sputum | Susceptible, 0.5 | No/no |
| 38 | Kw1724/09 | Kuwait | Clinical, ear swab | Susceptible, 0.25 | No/no |
| 39 | Kw1787/10 | Kuwait | Clinical, skin swab | Susceptible, 0.75 | No/no |
| 40 | Kw2881/11 | Kuwait | Clinical, bronchoalveolar lavage specimen | Susceptible, 0.38 | No/no |
| 41 | E-76 | Kuwait | Hospital indoor air | Resistant, >16 | Yes/yes |
| 42 | E-119 | Kuwait | Hospital floor swab | Resistant, >16 | Yes/yes |
| 43 | E-218 | Kuwait | Outdoor air | Resistant, >16 | Yes/yes |
| 44 | E-454 | Kuwait | Hospital floor swab | Resistant, >16 | Yes/yes |
| 45 | R-15 | Kuwait | Soil sample | Resistant, >16 | Yes/yes |
| 46 | R-18 | Kuwait | Soil sample | Resistant, >16 | Yes/yes |
| 47 | R-44 | Kuwait | Soil sample | Resistant, >16 | Yes/yes |
| 48 | 10-03-18-79 | India | Soil sample | Resistant, >16 | Yes/yes |
| 49 | 10-03-18-83 | India | Soil sample | Resistant, >16 | Yes/yes |
| 50 | 10-03-19-75 | India | Soil sample | Resistant, >16 | Yes/yes |
| 51 | 10-03-19-73 | India | Soil sample | Resistant, >16 | Yes/yes |
| 52 | 10-03-18-81 | India | Soil sample | Resistant, >16 | Yes/yes |
| 53 | 10-03-15-27 | France | Clinical, sputum | Resistant, >16 | No/noa |
| 54 | 10-03-18-82 | India | Soil sample | Resistant, >16 | Yes/yes |
| 55 | 10-03-15-38 | India | Clinical, sputum | Resistant, >16 | Yes/yes |
| 56 | 10-03-19-72 | India | Soil sample | Resistant, >16 | Yes/yes |
| 57 | 10-03-18-80 | India | Soil sample | Resistant, >16 | Yes/yes |
| 58 | 10-03-19-74 | India | Soil sample | Resistant, >16 | Yes/yes |
| 59 | 10-03-15-19 | France | Clinical, sputum | Resistant, >16 | No/nob |
| 60 | 10-03-19-70 | India | Soil sample | Resistant, >16 | Yes/yes |
| 61 | 10-03-19-71 | India | Soil sample | Resistant, >16 | Yes/yes |
| 62 | 10-04-15-05 | The Netherlands | Clinical, sputum | Resistant, >16 | Yes/yes |
| 63 | 10-01-02-62 | The Netherlands | Clinical, sputum | Resistant, >16 | Yes/yes |
| 64 | 10-01-02-27 | The Netherlands | Clinical, sputum | Resistant, >16 | Yes/yes |
| 65 | 10-04-15-16 | The Netherlands | Clinical, sputum | Resistant, >16 | Yes/yes |
| 66 | 10-01-13-76 | The Netherlands | Clinical, sputum | Resistant, >16 | No/noc |
| 67 | 10-01-12-86 | The Netherlands | Clinical, sputum | Resistant, >16 | Yes/yes |
| 68 | 10-01-13-15 | The Netherlands | Clinical, sputum | Resistant, >16 | Yes/yes |
| 69 | 10-04-15-12 | The Netherlands | Clinical, sputum | Resistant, >16 | Yes/yes |
| 70 | 10-04-15-03 | The Netherlands | Clinical, sputum | Resistant, >16 | Yes/yes |
| 71 | 10-01-04-26 | The Netherlands | Clinical, sputum | Resistant, >16 | Yes/yes |
| 72 | 10-01-04-22 | The Netherlands | Clinical, sputum | Resistant, >16 | Yes/yes |
| 73 | 10-03-16-61 | India | Soil sample | Resistant, >16 | Yes/yes |
| 74 | 10-01-13-23 | The Netherlands | Clinical, sputum | Resistant, >16 | Yes/yes |
| 75 | 10-03-16-63 | India | Clinical, sputum | Resistant, >16 | Yes/yes |
This isolate contained an M220R mutation in cyp51A.
This isolate contained a G54E mutation in cyp51A.
This isolate contained a G54W mutation in cyp51A.
ITR, itraconazole.
DNA from the isolates was prepared and the internal transcribed spacer (ITS) region of ribosomal DNA (rDNA) was amplified with AFUF2 and AFUR2 primers for the identification of A. fumigatus isolates as described previously (17). The presence or absence of TR34 in the promoter region was determined by PCR amplification by using AFCYPPF (5′-AATAATCGCAGCACCACTTC-3′) and AFCYPPR (5′-TGGTATGCTGGAACTACACCTT-3′) primers. PCR was carried out in a total volume of 50 μl containing 1× AmpliTaq PCR buffer I, 1 U AmpliTaq DNA polymerase, 4 pmol (each) of AFCYPPF and AFCYPPR primers, 2 μl of DNA, and 0.1 mM each deoxynucleoside triphosphate (dNTP). PCR cycling (total, 35 cycles) included denaturation at 95°C for 1 min, annealing at 60°C for 30 s, and extension at 72°C for 1 min. An initial denaturation step at 95°C for 5 min and a final extension step at 72°C for 10 min were also included, and the amplicons were detected by use of 2% agarose gels (16). A. fumigatus isolates containing TR34 in the cyp51A promoter region should yield an amplicon of 139 bp, while isolates containing the wild-type sequence (no tandem repeat) should yield an amplicon of 105 bp. For the detection of the wild-type sequence or the L98H mutation at cyp51A98 in A. fumigatus isolates, DNA was amplified by using AFCYP98F (5′-CAAGTTCTTCTTTGCGTGCAGA-3′) and AFCYP98R (5′-ATAAGTGGCACATGAGACTCT-3′) primers and the reaction and cycling conditions described above. A portion (5 μl) of PCR products was run on 2% agarose gels. Amplicons in the remaining sample (45 μl) were purified with a PCR product purification kit (Qiagen) according to instructions supplied with the kit. Purified DNA (5 μl) was digested with 5 units of AluI (New England Bio-Labs) in a final volume of 25 μl at 37°C for 5 h, and digested products were separated by 2% agarose gels to generate PCR-restriction fragment length polymorphism (PCR-RFLP) patterns. The 350-bp amplicon from A. fumigatus isolates containing the wild-type (L98) sequence (CTC, encoding Leu) in cyp51A98 should yield three DNA fragments of 189 bp, 90 bp, and 71 bp (with the 189-bp fragment serving as the diagnostic fragment), while isolates containing the L98H mutation (CAC, encoding His) in cyp51A98 should yield two DNA fragments of 279 bp and 71 bp (with the 279-bp fragment serving as the diagnostic fragment). PCR amplicons from all A. fumigatus isolates were also sequenced to confirm the results. Sequencing reactions were carried out and data were analyzed as described previously (16).
In agarose gels, PCR amplification of the promoter region yielded distinct 105-bp and 139-bp amplicons from reference A. fumigatus isolates CBS 113.26 (containing the wild-type sequence) and VPCI1042/09 (containing TR34 in the promoter region), respectively. Similarly, PCR-RFLP assay of reference A. fumigatus isolates CBS 113.26 (containing the wild-type sequence) and VPCI1042/09 (containing the L98H mutation at cyp51A98) yielded three (189-bp, 90-bp, and 71-bp) and two (279-bp and 71-bp) DNA fragments, respectively, as expected. Etest data showed that 40 A. fumigatus isolates were susceptible to itraconazole, while the remaining 35 isolates were resistant. All itraconazole-resistant isolates were also resistant to voriconazole according to the broth microdilution assay.
PCR amplification of the cyp51A98 region from all 40 itraconazole-susceptible and 35 itraconazole-resistant A. fumigatus isolates yielded an amplicon of ∼350 bp (data from 7 isolates are shown in Fig. 1a). When the amplicons were digested with AluI, 32 itraconazole-resistant isolates yielded two (279-bp and 71-bp) fragments (data from 8 isolates are shown in Fig. 1b, lanes 1 to 8), indicating the L98H mutation, while 3 isolates yielded a wild-type pattern in cyp51A98. The latter 3 isolates contained a mutation at either codon 54 (two isolates) or codon 220 (one isolate) (Table 1). The PCR-RFLP assay with all 40 itraconazole-susceptible isolates yielded three (189-bp, 90-bp, and 71-bp) fragments (data from 6 isolates are shown in Fig. 1b, lanes 9 to 14) indicating the wild-type sequence in cyp51A98. All 32 itraconazole-resistant isolates with the L98H mutation in cyp51A98 also yielded a 139-bp amplicon (data from 4 isolates are shown in Fig. 2, lanes 1, 2, 5, and 6) for the promoter region, indicating the presence of TR34, while the remaining 3 itraconazole-resistant isolates and all 40 itraconazole-susceptible isolates yielded 105-bp amplicons (data from 4 isolates are shown in Fig. 2, lanes 3, 4, 7, and 8), indicating the absence of the tandem-repeat sequence in the promoter region. The DNA sequencing data confirmed the results of PCR/PCR-RFLP assays for all the isolates analyzed in this study. None of the A. fumigatus isolates lacking TR34/L98H mutations yielded false-positive results by PCR/PCR-RFLP assays. It is important to emphasize here that the PCR-RFLP assay described in this study is designed for the detection of only the L98H mutation within the cyp51A gene. Since other cyp51A mutations are known to occur in triazole-resistant A. fumigatus isolates, the lack of detection of the L98H mutation by the PCR-RFLP assay does not rule out triazole resistance (4, 6, 18). Thus, there is a possibility of some triazole-resistant A. fumigatus isolates yielding false-negative results by exhibiting wild-type patterns for the promoter region and codon 98 of the cyp51A gene.
FIG 1.
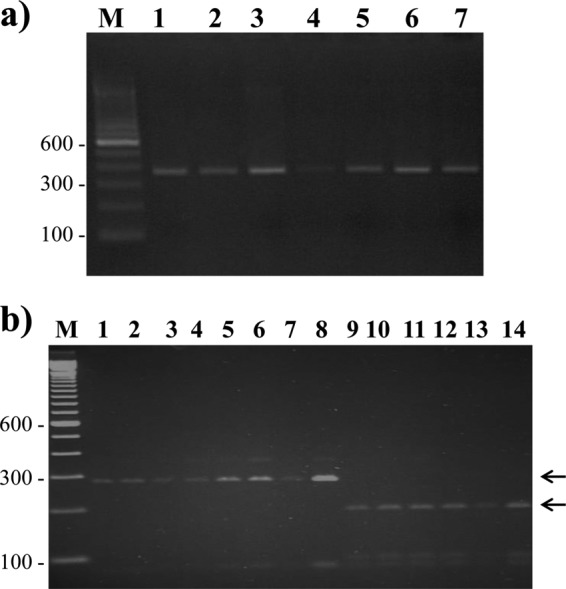
Agarose gels of PCR amplicons with AFCYP98F and AFCYP98R primers from 7 A. fumigatus isolates (lanes 1 to 7) (a) and RFLP patterns obtained from 8 triazole-resistant (lanes 1 to 8) and 6 triazole-susceptible (lanes 9 to 14) A. fumigatus isolates (b). The positions of migration of diagnostic fragments of 289 bp (for the L98H mutation) and 189 bp (for the wild-type codon at cyp51A98) are indicated by arrows. The minor band at 350 bp in some lanes represents the undigested full-length amplicon. In both panels, lane M is a 100-bp DNA ladder, and the positions of migration of 100-bp, 300-bp, and 600-bp fragments are marked.
FIG 2.
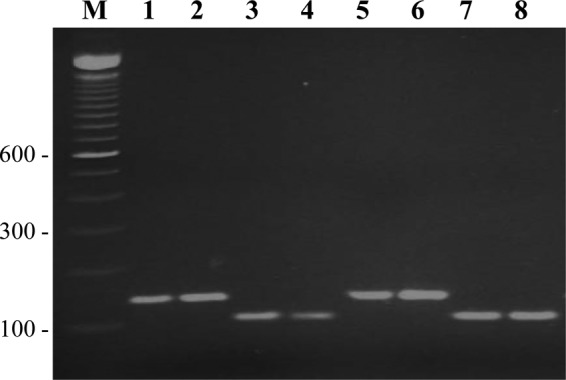
Agarose gel of PCR products obtained with AFCYPPF and AFCYPPR primers from 8 A. fumigatus isolates. The amplicon of ∼139 bp in lanes 1, 2, 5, and 6 indicates the presence of TR34, while the amplicon of ∼105 bp in lanes 3, 4, 7, and 8 indicates the absence of TR34 (wild-type sequence) in the promoter region of cyp51A. Lane M is a 100-bp DNA ladder, and the positions of migration of 100-bp, 300-bp, and 600-bp fragments are marked.
Previously, the L98H mutation has been detected by other rapid molecular techniques, such as two (L98-specific and L98H-specific) PCR assays (10), PCR amplification of the cyp51A98 region followed by DNA sequencing (6, 13, 18), real-time PCR with molecular beacons (12), and melting-curve analysis of specific probe primers with real-time PCR amplicons (11). TR34 has also been detected by a PCR or nested PCR assay followed by DNA sequencing (6, 13, 18), real-time PCR with molecular beacons (12), and melting-curve analysis of PCR amplicons (11). Most of these methods either are technically demanding or require expensive and sophisticated instruments/probes. A simple PCR-agarose gel assay similar to our protocol has been described previously (10); it generated amplicons of 188 bp and 222 bp from A. fumigatus isolates containing wild-type and TR34 sequences in the promoter region, respectively. However, the amplicons of 105 bp and 139 bp obtained in this study resolve better in 2% agarose gels than the amplicons of 222 bp and 188 bp, since the distance traveled by a DNA fragment in agarose gels is inversely proportional to the log of its molecular weight (19).
The PCR-based methods developed in this study for rapid identification of TR34/L98H mutations are simple to perform, use basic PCR and gel electrophoresis equipment that is readily available in most mycology laboratories, can be completed within 1 to 2 days, and cost ∼US$5 per sample (excluding the cost of culture and personnel time). The methods will help in determining the prevalence of TR34/L98H mutations in the cyp51A gene in clinical and environmental A. fumigatus isolates in resource-poor settings. The application of these methods will also help in identifying another mechanism (TR46/Y121F/T289A mutations) conferring resistance to triazoles in clinical and environmental A. fumigatus isolates described recently (20). This is because the L98H mutation without the TR34 mutation has not been described previously. Thus, the detection of A. fumigatus isolates containing a tandem repeat in the promoter region but lacking the L98H mutation would indicate the presence of Y121F/T289A in the cyp51A gene, which could be subsequently confirmed by other methods (20). Furthermore, the simple molecular assays developed in this study may also help in the rapid identification of TR34/L98H mutations in the cyp51A gene among clinical A. fumigatus isolates for proper management of patients with IA in developing countries. However, it should be emphasized here that the simple molecular assays described in this study are designed only for the detection of TR34/L98H mutations, and triazole-resistant A. fumigatus isolates harboring other mutations in the cyp51A gene will yield false-negative results.
ACKNOWLEDGMENTS
The study was supported by Kuwait University Research Sector grant no. MI 01/09.
We thank Ajmal Theyyathel and Leena Joseph for providing excellent technical assistance. We are grateful to Anuradha Chowdhary, Delhi, India, for providing azole-resistant A. fumigatus isolates.
J.F.M. has received grants from and been a consultant to Astellas, Basilea, and Merck and has received speaker's fees from Merck and Gilead. The other authors have no conflicts of interest to declare.
Footnotes
Published ahead of print 9 April 2014
REFERENCES
- 1.Walsh TJ, Anaissie EJ, Denning DW, Herbrecht R, Kontoyiannis DP, Marr KA, Morrison VA, Segal BH, Steinbach WJ, Stevens DA, van Burik JA, Wingard JR, Patterson TF, Infectious Diseases Society of America 2008. Treatment of aspergillosis: clinical practice guidelines of the Infectious Diseases Society of America. Clin. Infect. Dis. 46:327–360. 10.1086/525258 [DOI] [PubMed] [Google Scholar]
- 2.Howard SJ, Cesar D, Anderson MJ, Albarrag A, Fisher MC, Pasqualotto AC, Laverdiere M, Arendrup MC, Perlin DS, Denning DW. 2009. Frequency and evolution of azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg. Infect. Dis. 15:1068–1076. 10.3201/eid1507.090043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfaller M, Boyken L, Hollis R, Kroeger J, Messer S, Tendolkar S, Diekema D. 2011. Use of epidemiological cutoff values to examine 9-year trends in susceptibility of Aspergillus species to the triazoles. J. Clin. Microbiol. 49:586–590. 10.1128/JCM.02136-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgel PR, Baixench MT, Amsellem M, Audureau E, Chapron J, Kanaan R, Honoré I, Dupouy-Camet J, Dusser D, Klaassen CH, Meis JF, Hubert D, Paugam A. 2012. High prevalence of azole-resistant Aspergillus fumigatus in adults with cystic fibrosis exposed to itraconazole. Antimicrob. Agents Chemother. 56:869–874. 10.1128/AAC.05077-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camps SMT, van der Linden JWM, Li Y, Kuijper EJ, van Dissel JT, Verweij PE, Melchers WJ. 2012. Rapid induction of multiple resistance mechanisms in Aspergillus fumigatus during azole therapy: a case study and review of the literature. Antimicrob. Agents Chemother. 56:10–16. 10.1128/AAC.05088-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bader O, Weig M, Reichard U, Lugert R, Kuhns M, Christner M, Held J, Peter S, Schumacher U, Buchheidt D, Tintelnot K, Groß U, MykoLabNet-D Partners. 2013. cyp51A-based mechanisms of Aspergillus fumigatus azole drug resistance present in clinical samples from Germany. Antimicrob. Agents Chemother. 57:3513–3517. 10.1128/AAC.00167-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chowdhary A, Kathuria S, Xu J, Meis JF. 2013. Emergence of azole-resistant Aspergillus fumigatus strains due to agricultural azole use creates an increasing threat to human health. PLoS Pathog. 9:e1003633. 10.1371/journal.ppat.1003633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Badali H, Vaezi A, Haghani I, Yazdanparast SA, Hedayati MT, Mousavi B, Ansari S, Hagen F, Meis JF, Chowdhary A. 2013. Environmental study of azole-resistant Aspergillus fumigatus with TR34/L98H mutations in the cyp51A gene in Iran. Mycoses 56:659–663. 10.1111/myc.12089 [DOI] [PubMed] [Google Scholar]
- 9.Lockhart SR, Frade JP, Etienne KA, Pfaller MA, Diekema DJ, Balajee SA. 2011. Azole resistance in Aspergillus fumigatus isolates from the ARTEMIS global surveillance study is primarily due to the TR34/L98H mutation in the cyp51A gene. Antimicrob. Agents Chemother. 55:4465–4468. 10.1128/AAC.00185-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camps SM, Rijs AJ, Klaassen CH, Meis JF, O'Gorman CM, Dyer PS, Melchers WJ, Verweij PE. 2012. Molecular epidemiology of Aspergillus fumigatus isolates harboring the TR34/L98H azole resistance mechanism. J. Clin. Microbiol. 50:2674–2680. 10.1128/JCM.00335-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klaassen CH, de Valk HA, Curfs-Breuker IM, Meis JF. 2010. Novel mixed-format real-time PCR assay to detect mutations conferring resistance to triazoles in Aspergillus fumigatus and prevalence of multi-triazole resistance among clinical isolates in the Netherlands. J. Antimicrob. Chemother. 65:901–905. 10.1093/jac/dkq041 [DOI] [PubMed] [Google Scholar]
- 12.Denning DW, Park S, Lass-Florl C, Fraczek MG, Kirwan M, Gore R, Smith J, Bueid A, Moore CB, Bowyer P, Perlin DS. 2011. High-frequency triazole resistance found in nonculturable Aspergillus fumigatus from lungs of patients with chronic fungal disease. Clin. Infect. Dis. 52:1123–1129. 10.1093/cid/cir179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spiess B, Seifarth W, Merker N, Howard SJ, Reinwald M, Dietz A, Hofmann WK, Buchheidt D. 2012. Development of novel PCR assays to detect azole resistance-mediating mutations of the Aspergillus fumigatus cyp51A gene in primary clinical samples from neutropenic patients. Antimicrob. Agents Chemother. 56:3905–3910. 10.1128/AAC.05902-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chowdhary A, Kathuria S, Xu J, Sharma C, Sundar G, Singh PK, Gaur SN, Hagen F, Klaassen CH, Meis JF. 2012. Clonal expansion and emergence of environmental multiple-triazole-resistant Aspergillus fumigatus strains carrying the TR34/L98H mutations in the cyp51A gene in India. PLoS One 7:e52871. 10.1371/journal.pone.0052871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chowdhary A, Kathuria S, Randhawa HS, Gaur SN, Klaassen CH, Meis JF. 2012. Isolation of multiple-triazole-resistant Aspergillus fumigatus strains carrying the TR/L98H mutations in the cyp51A gene in India. J. Antimicrob. Chemother. 67:362–366. 10.1093/jac/dkr443 [DOI] [PubMed] [Google Scholar]
- 16.Al-Wathiqi F, Ahmad S, Khan ZU. 2013. Molecular characterization and antifungal susceptibility profile of Aspergillus flavus isolates recovered from clinical specimens in Kuwait. BMC Infect. Dis. 13:126. 10.1186/1471-2334-13-126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan ZU, Ahmad S, Theyyathel AM. 2008. Detection of Aspergillus fumigatus-specific DNA, (1-3)-β-d-glucan and galactomannan in serum and bronchoalveolar lavage specimens of experimentally infected rats. Mycoses 51:129–135. 10.1111/j.1439-0507.2007.01461.x [DOI] [PubMed] [Google Scholar]
- 18.Mellado E, Garcia-Effron G, Alcázar-Fuoli L, Melchers WJ, Verweij PE, Cuenca-Estrella M, Rodríguez-Tudela JL. 2007. A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob. Agents Chemother. 51:1897–1904. 10.1128/AAC.01092-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helling RB, Goodman HM, Boyer HW. 1974. Analysis of endonuclease R-EcoRI fragments of DNA from lambdoid bacteriophages and other viruses by agarose-gel electrophoresis. J. Virol. 14:1235–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Linden JW, Camps SM, Kampinga GA, Arends JP, Debets-Ossenkopp YJ, Haas PJ, Rijnders BJ, Kuijper EJ, van Tiel FH, Varga J, Karawajczyk A, Zoll J, Melchers WJ, Verweij PE. 2013. Aspergillosis due to voriconazole highly resistant Aspergillus fumigatus and recovery of genetically related resistant isolates from domestic homes. Clin. Infect. Dis. 57:513–520. 10.1093/cid/cit320 [DOI] [PubMed] [Google Scholar]


