Abstract
Molecular types of the Cryptococcus neoformans/Cryptococcus gattii species complex that infect dogs and cats differ regionally and with host species. Antifungal drug susceptibility can vary with molecular type, but the susceptibility of Cryptococcus isolates from dogs and cats is largely unknown. Cryptococcus isolates from 15 dogs and 27 cats were typed using URA5 restriction fragment length polymorphism analysis (RFLP), PCR fingerprinting, and multilocus sequence typing (MLST). Susceptibility was determined using a microdilution assay (Sensititre YeastOne; Trek Diagnostic Systems). MICs were compared among groups. The 42 isolates studied comprised molecular types VGI (7%), VGIIa (7%), VGIIb (5%), VGIIc (5%), VGIII (38%), VGIV (2%), VNI (33%), and VNII (2%), as determined by URA5 RFLP. The VGIV isolate was more closely related to VGIII according to MLST. All VGIII isolates were from cats. All sequence types identified from veterinary isolates clustered with isolates from humans. VGIII isolates showed considerable genetic diversity compared with other Cryptococcus molecular types and could be divided into two major subgroups. Compared with C. neoformans MICs, C. gattii MICs were lower for flucytosine, and VGIII MICs were lower for flucytosine and itraconazole. For all drugs except itraconazole, C. gattii isolates exhibited a wider range of MICs than C. neoformans. MICs varied with Cryptococcus species and molecular type in dogs and cats, and MICs of VGIII isolates were most variable and may reflect phylogenetic diversity in this group. Because sequence types of dogs and cats reflect those infecting humans, these observations may also have implications for treatment of human cryptococcosis.
INTRODUCTION
Cryptococcosis, caused by the yeasts Cryptococcus neoformans and Cryptococcus gattii, is a worldwide invasive mycosis causing severe disease in humans and animals. Cryptococcus is the most common systemic fungal pathogen of cats and can also cause severe disseminated disease in dogs (1). In humans, C. neoformans is opportunistic, usually infecting HIV/AIDS patients and other immunosuppressed individuals (2), whereas C. gattii is more likely to infect immunocompetent hosts (3). Cryptococcosis typically occurs in cats with no obvious underlying immunodeficiency, and infections are not associated with concurrent retroviral infection or other immunocompromised states (4, 5). Among dogs, purebred animals are most commonly affected, and an increased risk for breeds such as American cocker spaniels suggests an underlying genetic immunodeficiency (6). In general, cryptococcosis in humans is treated with a combination of amphotericin B and flucytosine (5FC), but high-dose (1,200 mg/day) fluconazole monotherapy is often used in resource-poor health care settings, such as among humans with HIV in sub-Saharan Africa (7). Similarly, cats and dogs with cryptococcosis are often treated with fluconazole monotherapy because of its relatively low cost, ease of administration, and favorable pharmacokinetic properties. Response to treatment can be slow or inadequate, and reported rates of successful disease control or cure range from 7 to 100% in cats and 27% in dogs (8–11).
Among C. gattii and C. neoformans strains, 8 molecular types have been identified by PCR fingerprinting and amplified fragment length polymorphism analysis (AFLP): VNI, VNII, VNIII, and VNIV for C. neoformans and VGI, VGII, VGIII, and VGIV for C. gattii (12). C. gattii strains that belong to molecular type VGII can be further categorized into a large number of subtypes. Of those VGIIa, VGIIb, and VGIIc have been implicated in disease in immunocompetent humans, cats, dogs, and other animal species in British Columbia and the Pacific Northwest of the United States (3, 13–15). An epidemiological study of dogs and cats with cryptococcosis in California showed that cats were most often infected with C. gattii molecular type VGIII, whereas dogs were more likely to be infected with C. neoformans, possibly reflecting host differences in susceptibility (6). Infections with strains of molecular type VGIII in dogs have not yet been described, but C. gattii molecular type VGIII has emerged as a cause of disease in immunocompromised humans in southern California (16), and molecular type VGIII was the predominant C. gattii molecular type identified in humans from California in another recent report. Two VGIII subclusters have been identified in these individuals based on the results of multilocus sequence typing, VGIIIa and VGIIIb (15, 16). In eastern Australia, molecular type VGI predominates among C. gattii isolates as a cause of disease in cats (17). VGIV is considered to be a rare molecular type but has been detected in humans and animals in sub-Saharan Africa (18).
The results of a number of studies have suggested that differences in in vitro drug susceptibility are correlated with different Cryptococcus species and molecular types (19–23). MICs of all azoles, particularly fluconazole, appear to be higher for molecular type VGII isolates than for other C. gattii molecular types, primarily VGI, and C. neoformans (19, 21, 23). There is also growing evidence that antifungal drug susceptibility may vary between geographical locations for the same molecular type (19, 21, 24).
To date, no studies have compared antifungal drug susceptibility among molecular types isolated solely from dogs or cats in North America, with a focus on isolates from California. If infecting species or molecular type correlates with resistance to certain antifungal drugs, the initial drug of choice may differ regionally and differ between cats and dogs. Identification of drug susceptibility patterns in isolates of Cryptococcus from dogs and cats may also have relevance to the human population, because molecular types of Cryptococcus that infect animals can reflect those infecting people that reside in the same geographic location (25). The objectives of this study were therefore to genetically characterize a group of Cryptococcus isolates from dogs and cats from North America using molecular methods and to determine the existence and strength of associations between antifungal drug MICs and Cryptococcus species and Cryptococcus molecular types. Susceptibilities were determined using the Sensititre YeastOne (SYO) method (YO-9; Trek Diagnostic Systems, Inc., Cleveland, OH), because it is widely used for yeast susceptibility testing in commercial diagnostic laboratories (26). We also sought to determine whether fluconazole area-under-the-curve (AUC)/MIC ratios for the isolates in this study would be likely to achieve or exceed the target ratio (≥389.3) to prevent progressive fungal growth in the central nervous system (CNS), which was previously determined using a mouse meningoencephalitis model (7).
MATERIALS AND METHODS
Fungal isolation.
Cryptococcal strains were isolated from swab specimens, biopsies, aspirates, or body fluids from dogs and cats with cryptococcosis. Isolates were obtained from animals with cryptococcosis that were evaluated by veterinarians at the University of California, Davis, veterinary medical teaching hospital (VMTH) or from specimens submitted to the investigators' laboratory by veterinarians or veterinary diagnostic laboratories between 2004 and 2011. Isolates not processed immediately were retrieved from storage at −80°C and grown for a minimum of 3 days on inhibitory mold agar (Hardy Diagnostics, Santa Maria, CA) and potato flake agar at 30°C until individual colonies were isolated. Yeast colonies were subcultured to Sabouraud dextrose agar. Identification of Cryptococcus spp. was based on morphology observed under light microscopy after Gram staining and use of a commercial yeast identification kit according to the manufacturer's instructions (API 20C AUX yeast identification kit; bioMérieux, Durham, NC). C. neoformans was differentiated from C. gattii with l-canavanine–glycine–bromthymol blue agar (27).
Medical record review.
Information obtained from medical records included each patient's geographic location, species, breed, sex, age at time of diagnosis, history of antifungal drug therapy, site of specimen collection, method of specimen collection, and the date that Cryptococcus spp. were isolated in culture. Geographic locations within California were divided into north coastal (cities west of Fairfield and north of Big Sur; climate zones 1, 2, 3, and 4), north central (cities east of Fairfield and north of Merced; climate zones 11 and 12), central (cities in the central valley that are south of Merced and north of Cahente; climate zone 13), and south coastal (cities west of Palm Springs and south of Big Sur; climate zones 8 to 10). Sites of organ or tissue involvement were categorized as nasal (within the nasal cavity or a mass protruding from the nasal cavity), cutaneous (dermal nodules, including those originating on the bridge of the nose or eyelid; other superficial dermal masses; and draining wounds of the lips), CNS (central nervous system) (involving brain, spinal cord, or meninges), eyes (including conjunctiva), lungs (including pleura or parenchyma), lymph nodes, urinary tract (kidneys and urine), or other tissues, as described previously (6). Disease was described as local or disseminated (>1 organ system involved), although the possibility of disseminated disease could not be ruled out in animals with local lesions that did not subsequently have a full necropsy performed.
Molecular genotyping.
High-molecular-weight genomic DNA was extracted and purified as previously described (28). Mating types were determined by PCR using the primers MfαU and MfαL (mating type α) and MFa2U and MFa2L (mating type a), as previously reported (29). The major molecular types of the C. neoformans/C. gattii species complex were initially identified by URA5 restriction fragment length polymorphism analysis (RFLP). The URA5 gene was first amplified via PCR with the primers URA5 (5′-ATGTCCTCCCAAGCCCTCGACTCCG-3′) and SJO1 (5′-TTAAGACCTCTGAACACCGTACTC-3′), as described previously, followed by a double digestion with the restriction enzymes Sau96I and HhaI (28). The molecular subtypes VGIIa and VGIIb were identified via triple digestion of the URA5 gene with the restriction enzymes HhaI, DdeI, and BsrGI, as described previously (30). Molecular subtypes VGIIa and VGIIb were also differentiated by means of PCR fingerprinting with the primer M13, as described previously (31). The patterns were assigned via visual comparison with patterns obtained for standard strains of the major molecular types of the C. neoformans/C. gattii species complex, including VNI (WM 148), VNII (WM626), VNIII (WM628), VNIV (WM629), VGI (WM179), VGII (WM178), VGIII (175), and VGIV (WM779), or for representative strains of the VGIIa (CDCR265) and VGIIb (CDCR272) molecular subtypes. MLST was performed on isolates using the ISHAM (International Society of Human and Animal Mycology) MLST consensus scheme for the C. neoformans/C. gattii species complex (12). Dendrograms showing the genetic relationships among C. gattii isolates and among C. neoformans isolates were constructed using a software package (MEGA 5.05; Center for Evolutionary Medicine and Informatics, Tempe, AZ) based on maximum likelihood analysis of the concatenated seven ISHAM consensus MLST loci. Additional C. gattii strains included in the MLST analysis are listed in Table S1 in the supplemental material (15, 28, 32–37).
Antifungal drug susceptibility testing.
The susceptibilities of the isolates in this study were determined by a commercially available colorimetric microdilution susceptibility test (Sensititre YeastOne YO-9; Trek Diagnostic Systems, Inc., Cleveland, OH) performed according to the manufacturer's instructions. Briefly, yeasts were subcultured on potato flake agar and incubated at 30°C for 24 h. Fungal colonies larger than 1 mm in diameter were collected using a sterile loop and added to 5 ml of sterile demineralized water and adjusted to a cell density of 0.5 McFarland standard (1 × 106 to 5 × 106 cells/ml). A total of 20 μl of yeast suspension was transferred into 11 ml of inoculum broth for a 1.5 × 103 to 8 × 103 viable CFU/ml. The purity of the diluted cell suspension was determined by plating the McFarland standard and the inoculum broth on defibrinated sheep blood agar. Colony counts were performed after 48 to 72 h of incubation to confirm purity and inoculum strength. The ranges of drug concentrations tested in 2-fold dilutions were as follows: amphotericin B (AMB), 0.008 to 16 μg/ml; fluconazole (FLC), 0.125 to 256 μg/ml; itraconazole (ITC), 0.008 to 16 μg/ml; flucytosine (5FC), 0.03 to 64 μg/ml; voriconazole (VRC), 0.008 to 16 μg/ml; posaconazole (POS), 0.008 to 8 μg/ml; and caspofungin, anidulafungin, and micafungin, 0.008 to 16 μg/ml.
A total of 100 μl of inoculum broth was placed in each well of the manufacturer's plate using an autoinoculator (Trek/Sensititre autoinoculator with nephelometer; Trek Diagnostic Systems, Inc., Cleveland, OH). Reference strains (Candida parapsilosis ATCC 22019 and Candida krusei ATCC 6258) were used for quality control purposes. Plates were incubated at 35°C in ambient air. Controls were read at 24 h, and Cryptococcus species MIC endpoints were read after 72 h of incubation or until the colonies grew to the 1 mm in diameter required to interpret the assay (up to 108 h of incubation) using a software program (Sensititre Trek/SWIN system with Sensitouch; Trek Diagnostic Systems, Inc., Cleveland, OH). Duplicate susceptibility tests were performed for isolates that required incubation times that exceeded 72 h in order to confirm assay results. Fluconazole AUC/MIC ratios were calculated for each isolate using previously reported pharmacokinetic data available for dogs and cats, which estimate a mean AUC of 375 mg/h/liter after an oral dose of 50 mg in cats (38) and 268 mg/h/liter after an oral dose of 10 mg/kg in dogs (39). The AUC/MIC ratios were compared with the target ratio to inhibit fungal growth that was identified in a mouse meningoencephalitis model (7).
Statistical methods.
The differences in MICs for each antifungal drug tested between two groups were compared using a Mann-Whitney test, with a null hypothesis that there were no differences between the groups. Group comparisons for MIC data included C. gattii versus C. neoformans, molecular type VGIII versus all C. gattii isolates that did not belong to the molecular type VGIII, molecular type VGIII versus VGII isolates, molecular type VGIII versus C. neoformans isolates, and isolates from animals with a history of antifungal drug therapy versus those that had not yet been treated with antifungal drugs. The F test was used to compare the distribution of MICs between each pair of groups. A chi-square analysis was used to compare the geographic distribution of isolates from cats to that of isolates from dogs. All analyses were performed with a statistical software package (MEGA 5.05; Center for Evolutionary Medicine and Informatics, Tempe, AZ) (GraphPad Prism, version 4.00; GraphPad, San Diego, CA). P values of <0.05 were considered significant.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the alleles in this study are KF667177 to KF667240 and KJ476370 to KJ476394 (see Tables S4 and S5 in the supplemental material).
RESULTS
Animals.
A total of 42 Cryptococcus isolates were obtained from 27 cats and 15 dogs. Twenty-two isolates were obtained from animals evaluated at the VMTH, and 20 isolates were obtained from dogs or cats evaluated by veterinarians outside the VMTH. Whether antifungal drug therapy had been administered before the isolate was obtained was known for 30 animals. Nine of the 30 animals had received antifungal drug therapy, and all had been treated for a week or longer without apparent response to treatment. Five animals had been treated with fluconazole, and 5 had been treated with itraconazole. Fourteen (52%) of the 27 cats were male, 12 (44%) were female, and all were neutered. Sex, age, and breed information was not available for one cat. The cats ranged in age from 1 to 12 years (median, 6 years). Twenty-two were of mixed breed, two were Siamese, one was a Maine coon, and one was a Persian. Disease sites identified included the nasal cavity, skin, CNS, lungs, myocardium, and mediastinum. The most common site of infection was the nasal cavity, in 11/27 (40%) of the cats. At least 8 of the 27 cats had disseminated disease.
Of the 15 dogs, 7 (47%) were male and 8 (53%) were female. One male and one female dog were intact, and the remaining dogs were neutered. The dogs ranged in age from 1.5 to 8 years (median, 3 years). Breeds represented were recorded for all but one dog, and included Labrador retrievers or their crosses (6), and one each of Shar-Pei, Shetland sheepdog, Rhodesian ridgeback, Doberman, basset hound, vizsla, German shepherd, and Yorkshire terrier. Sites of infection in the dogs included the nasal cavity, eye, CNS, kidneys, liver, lungs, gastrointestinal tract, and thyroid gland. The most common site of infection in dogs was the CNS, in 6/15 (40%) cases. At least 9 of the 15 dogs had disseminated disease.
Isolate epidemiology.
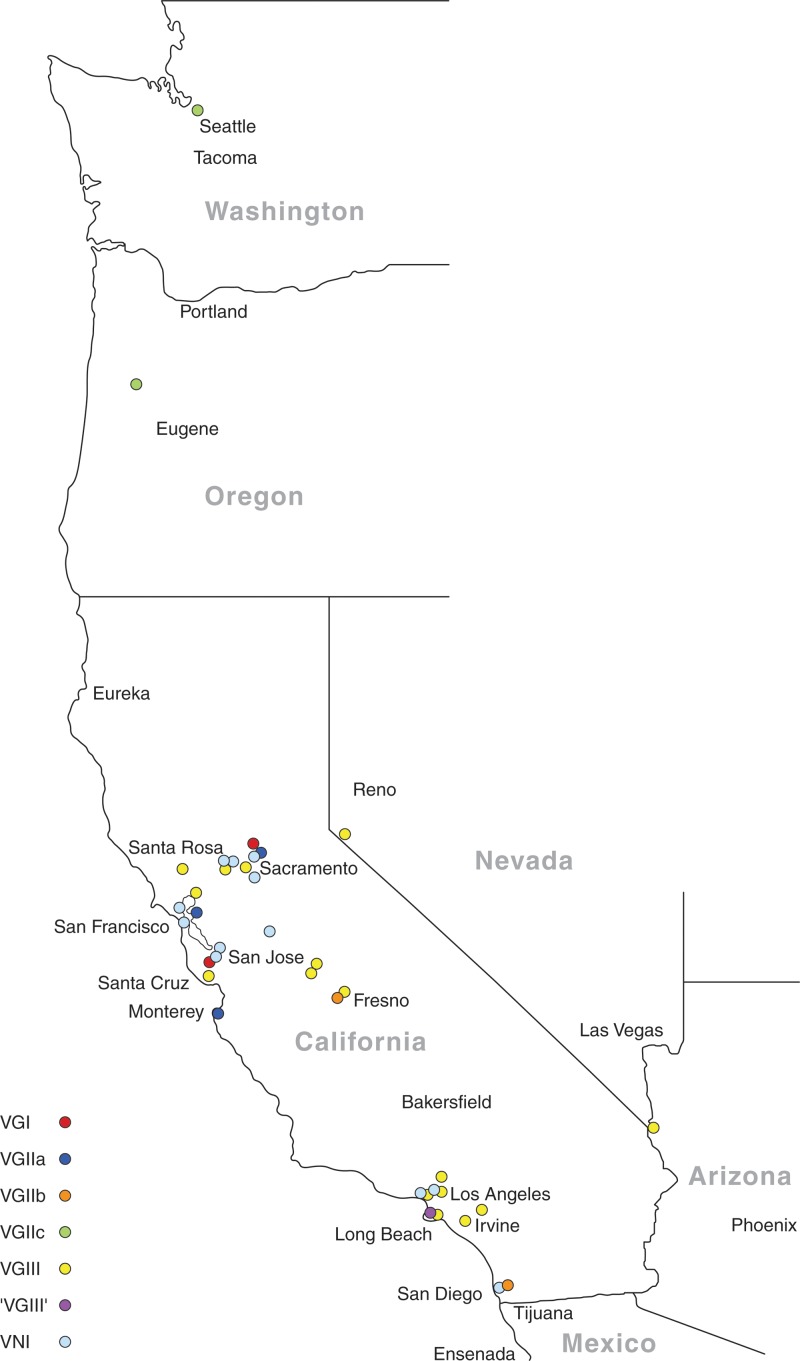
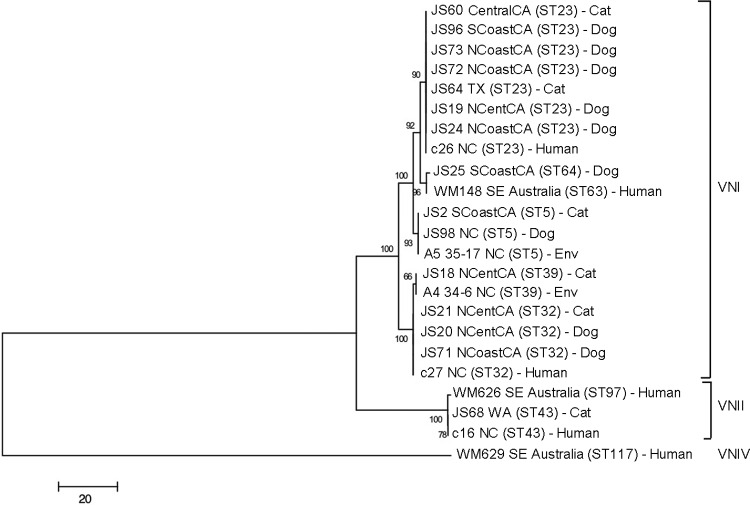
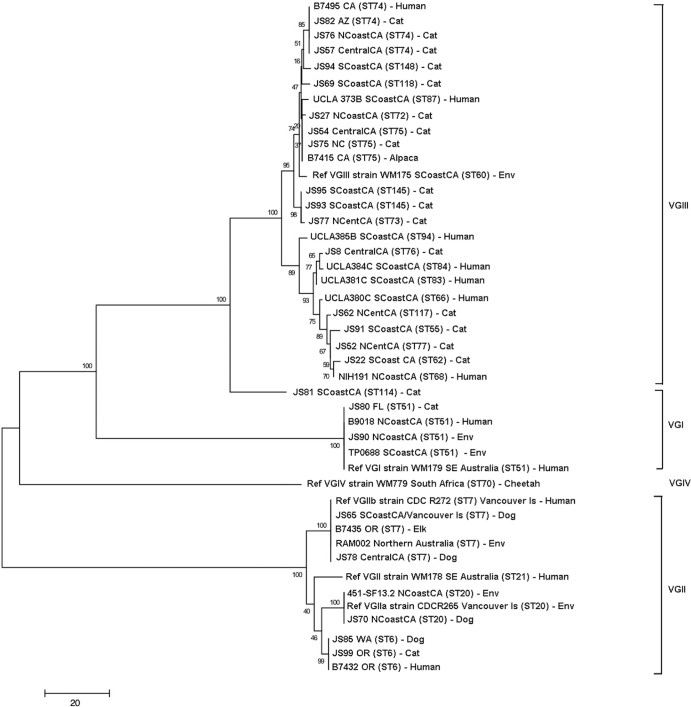
Twenty-seven isolates were identified as C. gattii, and 15 were C. neoformans. The molecular types isolated and their geographic origins are shown in Table 1, and the MLST results are shown in Tables S2 and S3 in the supplemental material. The most prevalent molecular types were VGIII (16/42 [38%]) and VNI (14/42 [33%]). All of the VGIII isolates originated from cats and were distributed throughout northern, central, and southern California (Fig. 1). The most common molecular type identified in dogs was VNI. There was no significant difference in the geographic location (by climate zone) for isolates obtained from dogs and those obtained from cats. All of the molecular types identified were found in California except VGIIc and VNII. With the exception of a VGII isolate from a dog that had recently traveled to southern California from Vancouver in British Columbia, Canada, VGII isolates were not identified south of Fresno (Fig. 1). Analysis of MLST results revealed that the C. gattii and C. neoformans isolates from dogs and cats clustered with isolates from human patients (Fig. 2 and 3). The most prevalent C. neoformans sequence type was ST23. VGIII isolates were genetically heterogeneous (Fig. 3; also, see Table S3 in the supplemental material), and both mating types α and a were identified. One of the canine isolates identified as VGIV by URA5 RFLP, strain JS81, was more closely related to VGIII according to MLST. Comparison of the URA5 gene sequence of this strain with that of the reference VGIII and VGIV strains revealed 3 SNPs (single-nucleotide polymorphisms) between strain JS81 and the reference VGIII strain and 30 SNPs between strain JS81 and the reference VGIV strain. The misclassification of this strain as a VGIV strain resulted from the existence of a SNP at the location of the Sau96I restriction site, generating an additional fragment, which made it identical to the VGIV restriction pattern.
TABLE 1.
Source and molecular types of Cryptococcus isolates in this study
| Host species | Molecular type | Isolate no. | Date of isolation | Region or statea | City |
|---|---|---|---|---|---|
| Feline | VNI | JS2 | Sept. 2009 | South coastal California | Los Angeles |
| JS18 | Aug. 2005 | North central California | Davis | ||
| JS21 | Oct. 2005 | North central California | Davis | ||
| JS60 | July 2010 | North central California | Modesto | ||
| JS64 | Aug. 2010 | Texas | Dallas | ||
| VNII | JS68 | Aug. 2010 | Washington | Vancouver | |
| VGI | JS53 | June 2010 | North central California | Rocklin | |
| JS80 | Nov. 2010 | Florida | Gainesville | ||
| VGIIa | JS74 | Feb. 2010 | North coastal California | El Cerrito | |
| VGIIc | JS99 | Apr. 2011 | Oregon | Corvallis | |
| VGIII | JS57 | July 2010 | Central California | Chowchilla | |
| JS8 | Dec. 2009 | Central California | Fresno | ||
| JS52 | Mar. 2010 | North central California | Davisb | ||
| JS54 | June 2010 | North central California | Sacramento | ||
| JS62 | July 2010 | North central California | Sacramento | ||
| JS77 | Dec. 2007 | North central California | St. Helena | ||
| JS27 | Mar. 2006 | North coastal California | American Canyon | ||
| JS76 | Jul. 2008 | North coastal California | Santa Cruz | ||
| JS22 | Apr. 2006 | South coastal California | Santa Clarita | ||
| JS69 | Aug. 2010 | South coastal California | Lomita | ||
| JS91 | Jan. 2011 | South coastal California | Riverside | ||
| JS93 | Feb. 2011 | South coastal California | Los Angeles | ||
| JS94 | Mar. 2011 | South coastal California | Irvinec | ||
| JS95 | Jan. 2011 | South coastal California | South Pasadena | ||
| JS75 | June 2008 | Nevada | Gardnerville | ||
| JS82 | Dec. 2010 | Arizona | Fort Mohave | ||
| “VGIII”d | JS81 | Nov. 2010 | South coastal California | Torrance | |
| Canine | VNI | JS19 | Aug. 2004 | North central California | Fair Oaks |
| JS20 | Oct. 2005 | North central California | Wilton | ||
| JS24 | Mar. 2006 | North coastal California | San Jose | ||
| JS71 | July 2007 | North coastal California | San Francisco | ||
| JS72 | June 2007 | North coastal California | Tiburon | ||
| JS73 | Sept. 2008 | North coastal California | San Jose | ||
| JS25 | Mar. 2006 | South coastal California | San Diego | ||
| JS96 | Apr. 2011 | South coastal California | Los Angeles | ||
| JS98 | Apr. 2011 | North Carolina | Charlotte | ||
| VGI | JS90 | Feb. 2011 | North coastal California | Campbell | |
| VGIIa | JS7 | Jan. 2010 | North central California | Sacramento | |
| JS70 | Sept. 2010 | North coastal California | Monterey | ||
| VGIIb | JS78 | Apr. 2008 | Central California | Fresno | |
| JS65 | Aug. 2010 | South coastal California | La Mesae | ||
| VGIIc | JS85 | Dec. 2010 | Washington | Seattle |
Location of the animal's residence at the time Cryptococcus was isolated.
Resided in southern California 3 years previously.
Resided in Arizona 7 months previously.
VGIV by URA5 RFLP but VGIII by MLST analysis.
Recently moved from Vancouver, Canada.
FIG 1.
Map showing the distribution of Cryptococcus molecular types identified in the current study from the western United States (n = 38). The 4 isolates from other states are not shown. ‘VGIII' indicates that the isolate was VGIV by URA5 RFLP but VGIII by MLST analysis.
FIG 2.
Dendrogram showing phylogenetic relationships between C. neoformans isolates from the current study and 8 reference strains. Bootstrap values (%) are shown at each branch point. A total of 4,004 bp were aligned. NCoastCA, north coastal California; NCentCA, north central California; SCoastCA, south coastal California; CentralCA, central California; Env, environmental; SE, southeastern.
FIG 3.
Dendrogram showing phylogenetic relationships between C. gattii isolates from the current study and 20 reference strains. Molecular type VGIII isolates are clustered at the top of the dendrogram and can be divided into two major subgroups. Bootstrap values (%) are shown at each branch point. A total of 4,196 bp were aligned. NCoastCA, north coastal California; NCentCA, north central California; SCoastCA, south coastal California; CentralCA, central California; Env, environmental; SE, southeastern; Ref, reference.
Antifungal susceptibility testing.
For 37 isolates, MICs could be determined at 72 h. Susceptibilities for four isolates (two VGIIa, one VGIIb, and one VGIII isolate) could not be clearly determined until 84 h of incubation, and susceptibilities for one VGIII isolate could not be clearly determined until 108 h of incubation.
The MIC ranges, MIC50, MIC90, and geometric mean and median MICs for C. gattii and C. neoformans of AMB, FLC, ITC, 5FC, VRC, and POS are shown in Table 2. Six isolates had FLC MICs of ≥16 μg/ml; three of these were VGIII isolates, and there was one isolate each of VGI, VGIIc, and VNI. Only two isolates had FLC MICs of 32 μg/ml, and both were VGIII isolates. Flucytosine MICs were lower among C. gattii than C. neoformans isolates (P < 0.001). The MICs of 5FC and ITC were lower among VGIII isolates than they were among C. neoformans isolates (P = 0.02 for both comparisons). Itraconazole MICs for molecular type VGIII isolates were lower than those for isolates of all other C. gattii molecular types (P = 0.01). Posaconazole MICs for molecular type VGIII isolates appeared to be lower than those for isolates of all other C. gattii molecular types and lower for VGIII isolates than for VGII isolates, but P values only approached the cutoff for significance for these observations (P = 0.052 and 0.056, respectively). There were no difference in MICs among isolates from dogs and cats and no difference in MICs of FLC or ITC between isolates from animals that had been treated with FLC or ITC and those that had not.
TABLE 2.
MICs for 27 C. gattii and 15 C. neoformans isolates from dogs and cats
| Drug and MIC measurement | MIC(s) (μg/ml) for isolates of molecular type (n) |
|||||
|---|---|---|---|---|---|---|
| VGI (3) | VGII (7) | VGIII (16) | “VGIII” (1)a | VNI (14) | VNII (1) | |
| Flucytosine | ||||||
| Range | 0.5–1 | 0.5–4 | 0.12–16b | 2 | 4–8 | 4 |
| Geometric mean | 0.79 | 2.00 | 1.83 | 5.12 | ||
| Median | 2 | 2 | 4 | |||
| MIC50, MIC90 | 2, 4 | 2, 8 | 4, 8 | |||
| Amphotericin B | ||||||
| Range | 0.25 | 0.25–0.5 | 0.25–0.5 | 0.5 | 0.25–0.5 | 0.5 |
| Geometric mean | 0.25 | 0.45 | 0.39 | 0.48 | ||
| Median | 0.5 | 0.5 | 0.5 | |||
| MIC50, MIC90 | 0.5, 0.5 | 0.25, 0.25 | 0.5, 0.5 | |||
| Fluconazole | ||||||
| Range | 2–16 | 4–16 | 0.5–32b | 2 | 2–16 | 2 |
| Geometric mean | 6.34 | 8.00 | 4.36 | 5.38 | ||
| Median | 8 | 4 | 6 | |||
| MIC50, MIC90 | 8, 8 | 4, 16 | 4, 8 | |||
| Itraconazole | ||||||
| Range | 0.03–0.25 | 0.03–0.25 | ≤0.015–0.25 | 0.03 | 0.03–0.25 | 0.12 |
| Geometric mean | 0.12 | 0.07 | 0.04 | 0.07 | ||
| Median | 0.06 | 0.03 | 0.06 | |||
| MIC50, MIC90 | 0.06, 0.06 | 0.03, 0.06 | 0.06, 0.12 | |||
| Posaconazole | ||||||
| Range | 0.06–0.25 | 0.06–0.25 | 0.015–0.25 | 0.03 | 0.03–0.12 | 0.06 |
| Geometric mean | 0.16 | 0.11 | 0.05 | 0.08 | ||
| Median | 0.12 | 0.06 | 0.06 | |||
| MIC50, MIC90 | 0.12, 0.12 | 0.06, 0.12 | 0.06, 0.12 | |||
| Voriconazole | ||||||
| Range | 0.03–0.12 | 0.015–0.25 | 0.015–0.12 | 0.015 | 0.015–0.06 | 0.03 |
| Geometric mean | 0.08 | 0.06 | 0.03 | 0.03 | ||
| Median | 0.06 | 0.03 | 0.03 | |||
| MIC50, MIC90 | 0.06, 0.06 | 0.03, 0.06 | 0.03, 0.03 | |||
VGIV by URA5 RFLP but VGIII by MLST analysis.
Variance significantly wider than for molecular types other than VGIII.
For all drugs except ITC, C. gattii exhibited a wider variation in MICs than C. neoformans isolates (P = 0.01, 0.04, 0.003, and 0.001 for AMB, 5FC, FLC, and POS, respectively, and P < 0.001 for VRC), which was also true for C. gattii molecular type VGIII compared with C. neoformans isolates (P = 0.02, 0.007, and 0.004 for AMB, 5FC, and POS, respectively, and P < 0.001 for both FLC and VRC). VGIII isolates exhibited a wider variation in MICs for FLC and 5FC than isolates of all other C. gattii molecular types (P < 0.001 and P = 0.01, respectively) and a wider variation in MICs for FLC, 5FC, and VRC than VGII isolates (P = 0.02, 0.02, and 0.01, respectively) (Table 2).
For cats, the median FLC AUC/MIC ratio for all C. neoformans isolates in this study was 70.3 (range, 23.4 to 187.5), and for dogs, it was 50.3 (range, 16.8 to 134). For cats, the median FLC AUC/MIC ratio for all C. gattii isolates in this study was 93.8 (range, 11.7 to 750), and for dogs, it was 50.3 (range, 8.4 to 536). A ratio that exceeded 389.3 was identified for only one isolate, which was from a cat.
DISCUSSION
In this study, we showed clearly that in California, the spectrum of major molecular types of the Cryptococcus neoformans/C. gattii species complex infecting cats differs from that infecting dogs. The most common molecular type to infect cats was C. gattii molecular type VGIII, whereas dogs were more commonly infected by molecular type VNI (C. neoformans var. grubii); no dogs were infected by molecular type VGIII. This is in sharp contrast with the Pacific Northwest region of the United States and British Columbia, where infection by C. gattii molecular type VGII has predominated regardless of host species (14, 34). Our study also confirmed the widespread distribution of C. gattii molecular type VGIII in cats from California. The reason for the predilection of this molecular type for cats and not dogs is unclear, but it may relate to differences in host immunity, virulence properties of this organism, and/or factors that put cats at increased risk of exposure to VGIII strains. Regional differences did not seem to be an explanation given that there was no difference in the geographic distribution of isolates from dogs and cats, although analysis of a larger number of isolates would be required to confirm this observation.
Based on our phylogenetic analysis, both C. neoformans and C. gattii strains isolated from dogs and cats in this study were closely related to those isolated from humans. The most prevalent C. neoformans sequence type, ST23, is a prevalent type found previously in the United States, Asia, Africa, and Europe in both environmental and clinical specimens (40–43). Other sequence types identified in our study have been also reported before from Asia (ST5 and ST32), Africa (ST5, ST23, ST39, and ST43), and the United States (ST32) (40–45). Molecular type VNI, VNII, VGI, and VGII isolates from dogs and cats in this study had sequence types that were identical or closely related to those of the reference VGI and VGII strains, respectively, supporting the more clonal relationship of these populations as described previously (15, 16).
Historically, VGIII has been one of the least prevalent molecular types found in humans and animals. In southeastern Australia, C. gattii isolates from dogs and cats are predominantly molecular type VGI, whereas molecular type VGII has been identified in dogs and cats from Western Australia (4, 10). The most prevalent molecular type found in human patients with HIV/AIDS is C. neoformans molecular type VNI (2). However, infections with C. gattii molecular type VGIII were recently identified in human patients with HIV/AIDS from southern California (16). A lower number of molecular type VGIII isolates have been isolated from humans in other states, including those in the Pacific Northwest, the south, Michigan, and Alaska (15), although the travel history of many of the infected individuals in these states was unknown. Analysis of VGIII isolates infecting humans in the United States has shown a genetically diverse population of isolates that includes both mating types α and a and that can be separated into VGIIIa and VGIIIb subclusters (15, 16). The strains infecting cats in this study also appeared to fall into two major subclusters, with both mating types α and a being represented. A small number of feline molecular type VGIII isolates in our study had matching sequence types (ST); these included 3 isolates from distant locations with ST74 (Arizona, coastal northern California, and central California) and 2 isolates with ST145 from southern California. One isolate from southern California that was identified as VGIV by URA5 RFLP (JS81) appeared to be more closely related to VGIII.
We chose to use the colorimetric SYO method as opposed to standard broth microdilution susceptibility testing using methods defined by the Clinical Laboratories Standards Institute (CLSI), because the SYO method is widely used by clinical laboratories for yeast susceptibility testing (26) and can be implemented in a commercial veterinary diagnostic laboratory setting because it has a convenient format and is affordable for pet owners. A comparison of the SYO method to the CLSI method for Cryptococcus found that essential agreements were 63.6% (FLC), 75% (ITC), 86.4% (VRC), 77.3% (POS), 88.6% (5FC), and 86.4% (AMB) (46). Given the relatively low number of isolates in this study, it was not possible to accurately determine epidemiological cutoff values (ECVs), but 95% of VNI isolates in our study had MICs of ≤0.12 μg/ml for POS and ITC, ≤0.06 μg/ml for VOR, and ≤8 μg/ml for FLC; this compared with statistical ECVs that included 95% of the wild-type isolates of 0.25 μg/ml for POS, ITC, and VOR and 8 μg/ml for FLC using broth microdilution (47). In our study, 95% of VGIII isolates had MICs of ≤0.12 μg/ml for ITC and 32 μg/ml for FLC; this compared with statistical ECVs of 0.5 μg/ml for ITC and 8 μg/ml for FLC (47). The 1- to 2-fold-dilution discrepancies in these numbers could reflect variations in methods used or true differences in susceptibility in the collection of isolates studied and/or reflect the relatively low numbers of isolates in our study. In the future, larger numbers of isolates should be used to determine ECVs using the SYO method, as has been done for Candida (26).
In general, for any of the tested antifungal drugs, C. gattii VGIII isolates had lower MICs than C. neoformans isolates or all other C. gattii isolates combined. Of perhaps greater significance, C. gattii isolates exhibited a significantly wider range of MICs than C. neoformans isolates. The same was true for the VGIII isolates compared with the isolates of all other C. gattii molecular types. Of the 6 isolates with FLC MICs of ≥16 μg/ml, 3 were VGIII isolates, and FLC MICs of 32 μg/ml were recorded only among VGIII isolates. In other studies, clinical and environmental isolates of C. gattii had higher MICs than C. neoformans isolates (48), and VGII isolates had higher MICs than the isolates of all other C. gattii molecular types (19). VGII isolates in this study appeared to have higher MICs for most antifungal drugs than the isolates of all other C. gattii molecular types, although the relatively low number of VGII isolates in this study limited our ability to document significance and to compare our results with those of other investigators.
Although combination therapy with AMB and FLC or 5FC has been recommended for animals with disseminated disease (1, 8), the use of AMB and 5FC has been limited by expense, and there is a high incidence of cutaneous adverse effects in dogs treated with 5FC (49). High FLC MICs identified in some strains of C. gattii might not correlate with treatment failure in vivo, and clinical breakpoints that define susceptibility versus resistance have not been clearly defined for Cryptococcus spp. in humans or animals. A history of FLC or ITC therapy (without clinical response) was not associated with higher MICs of these drugs in this study. Extrapolation of data from a mouse model of cryptococcal meningoencephalitis to HIV-infected humans suggested that high-dose FLC monotherapy might achieve or exceed the target AUC/MIC ratio (≥389.3) required to inhibit fungal growth across the expected distribution of MICs for C. neoformans in only two-thirds of treated patients (37). With the assumption that results obtained with the SYO method correlate with those using CLSI methods, we estimated that the FLC AUC/MIC ratio for cats and dogs with meningoencephalitis caused by the C. neoformans isolates in this study would also not be predicted to approach this target. If the same target is applied for C. gattii meningoencephalitis, progressive cryptococcal growth would be expected in all of the dogs and >90% of the cats in this study had they been treated with 10 mg/kg FLC daily. This target would also not have been attainable for 12 C. gattii isolates from dogs and cats for which susceptibilities were reported using CLSI methods (20). Thus, as suggested for humans, FLC monotherapy may not be an appropriate treatment, at least for animals with meningoencephalitis, and this might explain the poor responses to treatment in some patients in the face of relatively low MICs.
In summary, this study shows that (i) the molecular types that infect dogs and cats in California differ, but isolates from both dogs and cats genetically resemble those infecting humans; (ii) VGIII isolates that infect cats from California show a high genetic diversity and can be separated into two major subclusters (as documented in humans); (iii) significant differences in the distribution of MICs exist among cryptococcal molecular types that infect dogs and cats, with molecular type VGIII isolates exhibiting a wider range of MICs than other molecular types; and (iv) the use of fluconazole at currently employed dosing regimens may not be adequate to effectively inhibit growth of Cryptococcus in dogs and cats with meningoencephalitis. The variability in antifungal drug susceptibility identified among VGIII isolates may relate to the high level of genetic diversity that existed in this group. This is the first time that a possible correlation between genetic diversity and variable antifungal drug susceptibility within a Cryptococcus molecular type has been identified. The data reported in this study should also be useful for monitoring for the appearance of more resistant strains using the SYO method.
Supplementary Material
ACKNOWLEDGMENTS
We thank the following people for supplying strains listed in Table S1 in the supplemental material: Sharon C. A. Chen, June Kwon-Chung, Valerie Davis, David Ellis, Murray Fyfe, Texter Howard, Thomas Mitchell, and Tania J. Pfeiffer. We also thank LeAnn Lindsay for her technical support.
This work was supported by an NH&MRC project grant APP1031943 to Wieland Meyer and a grant from the University of California, Davis Center for Companion Animal Health (2010-34-R). Carolina Firacative was supported by a Ph.D. scholarship “Becas Francisco José de Caldas” from COLCIENCIAS Colombia.
George R. Thompson has consulted for Astellas, the manufacturer of isavuconazole.
Footnotes
Published ahead of print 2 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03392-13.
REFERENCES
- 1.Sykes JE, Malik R. 2014. Cryptococcosis, p 599–612 In Sykes JE. (ed), Canine and feline infectious diseases. Elsevier, St. Louis, MO [Google Scholar]
- 2.Fries BC, Cox GM. 2011. Cryptococcosis in AIDS, p 515–525 In Heitman J, Kozel TR, Kwon-Chung KJ, Perfect J, Casadevall A. (ed), Cryptococcus—from human pathogen to model yeast. ASM Press, Washington, DC [Google Scholar]
- 3.Bartlett KH, Kidd SE, Kronstad JW. 2008. The emergence of Cryptococcus gattii in British Columbia and the Pacific Northwest. Curr. Infect. Dis. Rep. 10:58–65. 10.1007/s11908-008-0011-1 [DOI] [PubMed] [Google Scholar]
- 4.O'Brien CR, Krockenberger MB, Wigney DI, Martin P, Malik R. 2004. Retrospective study of feline and canine cryptococcosis in Australia from 1981 to 2001: 195 cases. Med. Mycol. 42:449–460. 10.1080/13693780310001624547 [DOI] [PubMed] [Google Scholar]
- 5.Trivedi SR, Malik R, Meyer W, Sykes JE. 2011. Feline cryptococcosis: impact of current research on clinical management. J. Feline Med. Surg. 13:163–172. 10.1016/j.jfms.2011.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trivedi SR, Sykes JE, Cannon MS, Wisner ER, Meyer W, Sturges BK, Dickinson PJ, Johnson LR. 2011. Clinical features and epidemiology of cryptococcosis in cats and dogs in California: 93 cases (1988–2010). J. Am. Vet. Med. Assoc. 239:357–369. 10.2460/javma.239.3.357 [DOI] [PubMed] [Google Scholar]
- 7.Sudan A, Livermore J, Howard SJ, Al-Nakeeb Z, Sharp A, Goodwin J, Gregson L, Warn PA, Felton TW, Perfect JR, Harrison TS, Hope WW. 2013. Pharmacokinetics and pharmacodynamics of fluconazole for cryptococcal meningoencephalitis: implications for antifungal therapy and in vitro susceptibility breakpoints. Antimicrob. Agents Chemother. 57:2793–2800. 10.1128/AAC.00216-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Brien CR, Krockenberger MB, Martin P, Wigney DI, Malik R. 2006. Long-term outcome of therapy for 59 cats and 11 dogs with cryptococcosis. Aust. Vet. J. 84:384–392. 10.1111/j.1751-0813.2006.00040.x [DOI] [PubMed] [Google Scholar]
- 9.Sykes JE, Sturges BK, Cannon MS, Gericota B, Higgins RJ, Trivedi SR, Dickinson PJ, Vernau KM, Meyer W, Wisner ER. 2010. Clinical signs, imaging features, neuropathology, and outcome in cats and dogs with central nervous system cryptococcosis from California. J. Vet. Intern. Med. 24:1427–1438. 10.1111/j.1939-1676.2010.0633.x [DOI] [PubMed] [Google Scholar]
- 10.McGill S, Malik R, Saul N, Beetson S, Secombe C, Robertson I, Irwin P. 2009. Cryptococcosis in domestic animals in Western Australia: a retrospective study from 1995–2006. Med. Mycol. 47:625–639. 10.1080/13693780802512519 [DOI] [PubMed] [Google Scholar]
- 11.Lester SJ, Kowalewich NJ, Bartlett KH, Krockenberger MB, Fairfax TM, Malik R. 2004. Clinicopathologic features of an unusual outbreak of cryptococcosis in dogs, cats, ferrets, and a bird: 38 cases (January to July 2003). J. Am. Vet. Med. Assoc. 225:1716–1722. 10.2460/javma.2004.225.1716 [DOI] [PubMed] [Google Scholar]
- 12.Meyer W, Aanensen DM, Boekhout T, Cogliati M, Diaz MR, Esposto MC, Fisher M, Gilgado F, Hagen F, Kaocharoen S, Litvintseva AP, Mitchell TG, Simwami SP, Trilles L, Viviani MA, Kwon-Chung J. 2009. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med. Mycol. 47:561–570. 10.1080/13693780902953886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillece JD, Schupp JM, Balajee SA, Harris J, Pearson T, Yan Y, Keim P, DeBess E, Marsden-Haug N, Wohrle R, Engelthaler DM, Lockhart SR. 2011. Whole genome sequence analysis of Cryptococcus gattii from the Pacific Northwest reveals unexpected diversity. PLoS One 6:e28550. 10.1371/journal.pone.0028550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byrnes EJ, III, Li W, Lewit Y, Ma H, Voelz K, Ren P, Carter DA, Chaturvedi V, Bildfell RJ, May RC, Heitman J. 2010. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog. 6:e1000850. 10.1371/journal.ppat.1000850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lockhart SR, Iqbal N, Harris JR, Grossman NT, DeBess E, Wohrle R, Marsden-Haug N, Vugia DJ. 2013. Cryptococcus gattii in the United States: genotypic diversity of human and veterinary isolates. PLoS One 8:e74737. 10.1371/journal.pone.0074737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Byrnes EJ, III, Li W, Ren P, Lewit Y, Voelz K, Fraser JA, Dietrich FS, May RC, Chaturvedi S, Chaturvedi V, Heitman J. 2011. A diverse population of Cryptococcus gattii molecular type VGIII in southern Californian HIV/AIDS patients. PLoS Pathog. 7:e1002205. 10.1371/journal.ppat.1002205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell LT, Currie BJ, Krockenberger M, Malik R, Meyer W, Heitman J, Carter D. 2005. Clonality and recombination in genetically differentiated subgroups of Cryptococcus gattii. Eukaryot. Cell 4:1403–1409. 10.1128/EC.4.8.1403-1409.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Litvintseva AP, Thakur R, Reller LB, Mitchell TG. 2005. Prevalence of clinical isolates of Cryptococcus gattii serotype C among patients with AIDS in sub-Saharan Africa. J. Infect. Dis. 192:888–892. 10.1086/432486 [DOI] [PubMed] [Google Scholar]
- 19.Lockhart SR, Iqbal N, Bolden CB, DeBess EE, Marsden-Haug N, Worhle R, Thakur R, Harris JR. 2012. Epidemiologic cutoff values for triazole drugs in Cryptococcus gattii: correlation of molecular type and in vitro susceptibility. Diagn. Microbiol. Infect. Dis. 73:144–148. 10.1016/j.diagmicrobio.2012.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iqbal N, DeBess EE, Wohrle R, Sun B, Nett RJ, Ahlquist AM, Chiller T, Lockhart SR. 2010. Correlation of genotype and in vitro susceptibilities of Cryptococcus gattii strains from the Pacific Northwest of the United States. J. Clin. Microbiol. 48:539–544. 10.1128/JCM.01505-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chong HS, Dagg R, Malik R, Chen S, Carter D. 2010. In vitro susceptibility of the yeast pathogen cryptococcus to fluconazole and other azoles varies with molecular genotype. J. Clin. Microbiol. 48:4115–4120. 10.1128/JCM.01271-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trilles L, Meyer W, Wanke B, Guarro J, Lazéra M. 2012. Correlation of antifungal susceptibility and molecular type within the Cryptococcus neoformans/C. gattii species complex. Med. Mycol. 50:328–332. 10.3109/13693786.2011.602126 [DOI] [PubMed] [Google Scholar]
- 23.Hagen F, Illnait-Zaragozi MT, Bartlett KH, Swinne D, Geertsen E, Klaassen CH, Boekhout T, Meis JF. 2010. In vitro antifungal susceptibilities and amplified fragment length polymorphism genotyping of a worldwide collection of 350 clinical, veterinary, and environmental Cryptococcus gattii isolates. Antimicrob. Agents Chemother. 54:5139–5145. 10.1128/AAC.00746-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chowdhary A, Randhawa HS, Sundar G, Kathuria S, Prakash A, Khan Z, Sun S, Xu J. 2011. In vitro antifungal susceptibility profiles and genotypes of 308 clinical and environmental isolates of Cryptococcus neoformans var. grubii and Cryptococcus gattii serotype B from north-western India. J. Med. Microbiol. 60:961–967. 10.1099/jmm.0.029025-0 [DOI] [PubMed] [Google Scholar]
- 25.Duncan C, Schwantje H, Stephen C, Campbell J, Bartlett K. 2006. Cryptococcus gattii in wildlife of Vancouver Island, British Columbia, Canada. J. Wildl. Dis. 42:175–178. 10.7589/0090-3558-42.1.175 [DOI] [PubMed] [Google Scholar]
- 26.Cantón E, Pemán J, Iñiguez C, Hervás D, Lopez-Hontangas JL, Pina-Vaz C, Camarena JJ, Campos-Herrero I, García-García I, García-Tapia AM, Guna R, Merino P, Pérez del Molino L, Rubio C, Suárez A, FUNGEMYCA Study Group. 2013. Epidemiological cutoff values for fluconazole, itraconazole, posaconazole, and voriconazole for six Candida species as determined by the colorimetric Sensititre YeastOne method. J. Clin. Microbiol. 51:2691–2695. 10.1128/JCM.01230-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon-Chung KJ, Polacheck I, Bennett JE. 1982. Improved diagnostic medium for separation of Cryptococcus neoformans var. neoformans (serotypes A and D) and Cryptococcus neoformans var. gattii (serotypes B and C). J. Clin. Microbiol. 15:535–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer W, Castaneda A, Jackson S, Huynh M, Castañeda E. 2003. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg. Infect. Dis. 9:189–195. 10.3201/eid0902.020246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halliday CL, Bui T, Krockenberger M, Malik R, Ellis DH, Carter DA. 1999. Presence of alpha and a mating types in environmental and clinical collections of Cryptococcus neoformans var. gattii strains from Australia. J. Clin. Microbiol. 37:2920–2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacDougall L, Kidd SE, Galanis E, Mak S, Leslie MJ, Cieslak PR, Kronstad JW, Morshed MG, Bartlett KH. 2007. Spread of Cryptococcus gattii in British Columbia, Canada, and detection in the Pacific Northwest, U. S. A. Emerg. Infect. Dis. 13:42–50. 10.3201/eid1301.060827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer W, Mitchell TG. 1995. Polymerase chain reaction fingerprinting in fungi using single primers specific to minisatellites and simple repetitive DNA sequences: strain variation in Cryptococcus neoformans. Electrophoresis 16:1648–1656. 10.1002/elps.11501601273 [DOI] [PubMed] [Google Scholar]
- 32.Pfeiffer T, Ellis D. 1991. Environmental isolation of Cryptococcus neoformans gattii from California. J. Infect. Dis. 163:929–930. 10.1093/infdis/163.4.929 [DOI] [PubMed] [Google Scholar]
- 33.Carriconde F, Gilgado F, Arthur I, Ellis D, Malik R, van de Wiele N, Robert V, Currie BJ, Meyer W. 2011. Clonality and alpha-a recombination in the Australian Cryptococcus gattii VGII population—an emerging outbreak in Australia. PLoS One 6:e16936. 10.1371/journal.pone.0016936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, Macdougall L, Boekhout T, Kwon-Chung KJ, Meyer W. 2004. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc. Natl. Acad. Sci. U. S. A. 101:17258–17263. 10.1073/pnas.0402981101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer W, Mitchell TG, Freedman EZ, Vilgalys R. 1993. Hybridization probes for conventional DNA fingerprinting used as single primers in the polymerase chain reaction to distinguish strains of Cryptococcus neoformans. J. Clin. Microbiol. 31:2274–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwon-Chung KJ, Bennett JE, Theodore TS. 1978. Cryptococcus bacillisporus sp. nov.: serotype B-C of Cryptococcus neoformans. Int. J. Syst. Bacteriol. 28:616–620. 10.1099/00207713-28-4-616 [DOI] [Google Scholar]
- 37.Litvintseva AP, Thakur R, Vilgalys R, Mitchell TG. 2006. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics 172:2223–2238. 10.1534/genetics.105.046672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vaden SL, Heit MC, Hawkins EC, Manaugh C, Riviere JE. 1997. Fluconazole in cats: pharmacokinetics following intravenous and oral administration and penetration into cerebrospinal fluid, aqueous humour and pulmonary epithelial lining fluid. J. Vet. Pharmacol. Ther. 20:181–186. 10.1111/j.1365-2885.1997.tb00093.x [DOI] [PubMed] [Google Scholar]
- 39.Humphrey MJ, Jevons S, Tarbit MH. 1985. Pharmacokinetic evaluation of UK-49,858, a metabolically stable triazole antifungal drug, in animals and humans. Antimicrob. Agents Chemother. 28:648–653. 10.1128/AAC.28.5.648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cogliati M, Zamfirova RR, Tortorano AM, Viviani MA, Fimua Cryptococcosis Network 2013. Molecular epidemiology of Italian clinical Cryptococcus neoformans var. grubii isolates. Med. Mycol. 51:499–506. 10.3109/13693786.2012.751642 [DOI] [PubMed] [Google Scholar]
- 41.Khayhan K, Hagen F, Pan W, Simwami S, Fisher MC, Wahyuningsih R, Chakrabarti A, Chowdhary A, Ikeda R, Taj-Aldeen SJ, Khan Z, Ip M, Imran D, Sjam R, Sriburee P, Liao W, Chaicumpar K, Vuddhakul V, Meyer W, Trilles L, van Iersel LJ, Meis JF, Klaassen CH, Boekhout T. 2013. Geographically structured populations of Cryptococcus neoformans variety grubii in Asia correlate with HIV status and show a clonal population structure. PLoS One 8:e72222. 10.1371/journal.pone.0072222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi YH, Ngamskulrungroj P, Varma A, Sionov E, Hwang SM, Carriconde F, Meyer W, Litvintseva AP, Lee WG, Shin JH, Kim EC, Lee KW, Choi TY, Lee YS, Kwon-Chung KJ. 2010. Prevalence of the VNIc genotype of Cryptococcus neoformans in non-HIV-associated cryptococcosis in the Republic of Korea. FEMS Yeast Res. 10:769–778. 10.1111/j.1567-1364.2010.00648.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiesner DL, Moskalenko O, Corcoran JM, McDonald T, Rolfes MA, Meya DB, Kajumbula H, Kambugu A, Bohjanen PR, Knight JF, Boulware DR, Nielsen K. 2012. Cryptococcal genotype influences immunologic response and human clinical outcome after meningitis. mBio 3:e00196–12. 10.1128/mBio.00196-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mihara T, Izumikawa K, Kakeya H, Ngamskulrungroj P, Umeyama T, Takazono T, Tashiro M, Nakamura S, Imamura Y, Miyazaki T, Ohno H, Yamamoto Y, Yanagihara K, Miyzaki Y, Kohno S. 2013. Multilocus sequence typing of Cryptococcus neoformans in non-HIV associated cryptococcosis in Nagasaki, Japan. Med. Mycol. 51:252–260. 10.3109/13693786.2012.708883 [DOI] [PubMed] [Google Scholar]
- 45.Simwami SP, Khayhan K, Henk DA, Aanensen DM, Boekhout T, Hagen F, Brouwer AE, Harrison TS, Donnelly CA, Fisher MC. 2011. Low diversity Cryptococcus neoformans variety grubii multilocus sequence types from Thailand are consistent with an ancestral African origin. PLoS Pathog. 7:e1001343. 10.1371/journal.ppat.1001343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bertout S, Dunyach C, Drakulovski P, Reynes J, Mallié M. 2011. Comparison of the Sensititre YeastOne dilution method with the Clinical and Laboratory Standards Institute (CLSI) M27–A3 microbroth dilution reference method for determining MIC of eight antifungal agents on 102 yeast strains. Pathol. Biol. (Paris) 59:48–51. 10.1016/j.patbio.2010.07.020 [DOI] [PubMed] [Google Scholar]
- 47.Espinel-Ingroff A, Aller AI, Canton E, Castañón-Olivares LR, Chowdhary A, Cordoba S, Cuenca-Estrella M, Fothergill A, Fuller J, Govender N, Hagen F, Illnait-Zaragozi MT, Johnson E, Kidd S, Lass-Flörl C, Lockhart SR, Martins MA, Meis JF, Melhem MS, Ostrosky-Zeichner L, Pelaez T, Pfaller MA, Schell WA, St-Germain G, Trilles L, Turnidge J. 2012. Cryptococcus neoformans-Cryptococcus gattii species complex: an international study of wild-type susceptibility endpoint distributions and epidemiological cutoff values for fluconazole, itraconazole, posaconazole, and voriconazole. Antimicrob. Agents Chemother. 56:5898–5906. 10.1128/AAC.01115-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varma A, Kwon-Chung KJ. 2010. Heteroresistance of Cryptococcus gattii to fluconazole. Antimicrob. Agents Chemother. 54:2303–2311. 10.1128/AAC.00153-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malik R, Medeiros C, Wigney DI, Love DN. 1996. Suspected drug eruption in seven dogs during administration of flucytosine. Aust. Vet. J. 74:285–288. 10.1111/j.1751-0813.1996.tb13776.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.