Abstract
A beta-hemolytic Lancefield antigen A-, B-, C-, D-, F-, and G-positive Enterococcus durans strain was cultivated from the rectovaginal swab of a pregnant woman who underwent antenatal screening for Streptococcus agalactiae. The isolate raised concern as to what extent similar strains are misrecognized and lead to false diagnosis of group B streptococci.
TEXT
Streptococcus agalactiae (group B Streptococcus [GBS]) inhabits the human gut, from where it can then colonize the genitourinary tract (1). Up to 30% of pregnant women are asymptomatic GBS carriers, and neonates can therefore acquire the organism vertically at the time of birth unless penicillin or ampicillin is given intravenously as intrapartum antibiotic prophylaxis (IAP). Newborn colonization may establish in 50% of the cases and involves different mucosal and skin sites. One percent to 2% of neonates develop early GBS infection, including bacteremia and/or pneumonia and/or meningitis (1, 2). For these reasons, the CDC (Centers for Disease Control and Prevention) has suggested that women between the 35th and 37th weeks of gestation be screened for GBS carriage through rectovaginal cultures. As IAP has been shown to reduce the risk of vertical S. agalactiae transmission, all GBS-positive women receive prophylaxis as soon as labor begins or when rupture of the amniotic membranes occurs (1).
GBS screening is based on the recognition of Gram-positive catalase-negative cocci, mostly beta-hemolytic (on sheep blood agar), showing the Lancefield group B antigen. Nonhemolytic (gamma-hemolytic) variants have been described, however, and may be missed because they are not easily detected. Hence, the CDC recommends the use of chromogenic media that are designed to highlight GBS growth through species-specific coloration of colonies (that may vary depending on the manufacturer) (1).
A pregnant woman attending the Spirito Santo Hospital in Pescara, Italy, was screened for rectovaginal GBS colonization according to the CDC guidelines. On Trypticase soy agar (Liofilchem, Italy), beta-hemolytic (Fig. 1) catalase-negative colonies were grown. The isolate formed pairs and chains and showed group B antigen agglutination (detected with the Liofilchem Strepto B latex kit). It was therefore believed to be GBS. Nevertheless, it grew in purple colonies (Fig. 1) on Chromatic StrepB (Liofilchem), whereas this chromogenic medium is designed to support GBS growth in light blue-green colonies (based on the manufacturer's indication, the purple color we observed instead placed the isolate within the Enterococcus genus). Accordingly, the strain was found to produce a negative CAMP reaction (10) (mild enhancing of hemolysis was observed, however).
FIG 1.
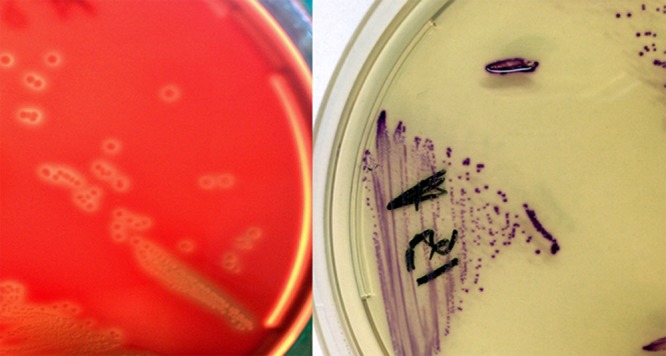
(Left) E. durans A51 (Ed135) beta-hemolytic colonies on Trypticase soy agar (Liofilchem); (Right) E. durans A51 (Ed135) growing on Chromatic StrepB (Liofilchem).
Identification as Enterococcus durans was provided by the Vitek 2 GP card (bioMérieux, France) and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) using Bruker Biotyper software 2.0 (Bruker Daltonics, Germany) and the Vitek MS v2.0 system (bioMérieux). Sequencing of a 16S rRNA gene 1,458-bp amplicon, analyzed using CLC DNA Workbench 5.5 and BLAST (see http://blast.ncbi.nlm.nih.gov/Blast.cgi), confirmed that the strain belonged to the E. durans species, with a BLAST 99.9% homology with E. durans strain CK1106 (GenBank accession no. AY902456.2). The organism was deposited in internal collections both at the Microbiology and Virology Laboratory, Spirito Santo Hospital of Pescara (Italy), and at the Istituto Zooprofilattico Sperimentale delle Regioni Lazio e Toscana, Rome (Italy), under the accession numbers A51 (Ed135) and 14070723 A51, respectively. Further investigation showed, surprisingly, that A51 (Ed135) also agglutinated Lancefield group A, C, D, F, and G antisera (with the Liofilchem Strep-Check kit). This result was confirmed by the Oxoid streptococcal grouping kit (Thermo Fisher, USA) and the bioMérieux Slidex Strepto Plus (Fig. 2).
FIG 2.
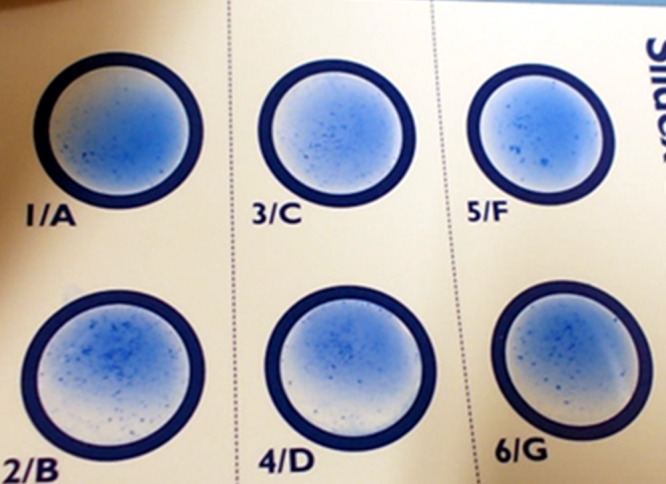
Lancefield group A, B, C, D, F, and G antisera agglutination by E. durans A51 (Ed135).
The woman was considered to be GBS negative. However, this experience was intriguing because, to our knowledge, E. durans had never been observed to agglutinate Lancefield group B antiserum. This organism (formerly Streptococcus durans) is an exceptional cause of human disease. It was anecdotally described as an agent of bacteremias (3), urinary tract infections (4), wound infections (5), and four endocarditis cases (6). Also, it has been found in dairy products and in human and animal milk microbiota (6–8). E. durans appears as elongated nonmotile cells that are mostly arranged in pairs and short chains; it forms circular, smooth, entire, and nonpigmented colonies when grown on a blood medium, showing the Lancefield group D and, occasionally, group G antigen (8, 9). The species may mimic Enterococcus faecium, from which it may be distinguished based on a few features, like the absence of growth at 50°C and a poor saccharolytic activity (compared to E. faecium). Finally, E. durans is mostly nonhemolytic and sometimes alpha-hemolytic; nevertheless, beta-hemolytic colonies are hardly ever described (8).
In general, S. agalactiae-like enterococci with beta-hemolysis (e.g., Enterococcus gallinarum, Enterococcus faecalis) may be easily recognized in pregnancy as not being GBS through Lancefield group D antigen detection (S. agalactiae belongs to group B). Instead, strain A51 (Ed135) unexpectedly agglutinated the B antiserum and was among the few beta-hemolytic E. durans isolates that have been reported in the literature. As no coccoid organisms other than GBS have ever been found to agglutinate group B, we initially labeled A51 (Ed135) as S. agalactiae. However, the Liofilchem chromogenic medium, along with the Vitek 2, MALDI-TOF MS, and 16S rRNA gene sequencing, allowed us to place it within the Enterococcus genus; accordingly, no CAMP reaction was observed.
Although falsely negative results have been known to be the major concern with antepartum GBS screening (1) (leading to an increased risk of neonatal infection), it appears from this experience that pregnant women may be erroneously labeled as GBS carriers as well.
The prevalence of group B antigen-agglutinating enterococci is unknown, as no previously published report exists, to our knowledge. Nevertheless, we wonder whether misidentification of beta-hemolytic enterococci as GBS has occurred and/or still occurs. Our observations seem to indicate that group B antigen detection, even in beta-hemolytic catalase-negative organisms, is not enough to diagnose GBS unless combined identification methodologies are used. In the future, therefore, evaluation of the prevalence of S. agalactiae-mimicking organisms should be required to prevent IAP abuse (due to false-positive reporting). The use of chromogenic media especially will strongly support this purpose. In particular, we suggest that all beta-hemolytic isolates be screened with Lancefield group A, B, C, D, F, and G antisera and that presumptive identification be reliably confirmed. This experience raised concern as to what extent similar Enterococcus strains may be misidentified in daily practice as group A/B/C/G streptococci, leading to the false diagnosis of GBS colonization and/or infection in pregnant women and newborns.
Nucleotide sequence accession number.
The strain A51 (Ed135) 16S rRNA sequence was deposited in GenBank under the accession number KJ631376.
ACKNOWLEDGMENTS
This project was partially supported by a grant from the Scientific Committee of the IRCCS Arcispedale Santa Maria Nuova, Reggio Emilia (to E.C.).
We have no conflicts of interests to declare.
Footnotes
Published ahead of print 26 March 2014
REFERENCES
- 1.Centers for Disease Control and Prevention. 2010. Prevention of perinatal group B streptococcal disease. Revised guidelines from CDC, 2010. MMWR Morb. Mortal. Wkly. Rep. 59:11–36 [Google Scholar]
- 2.Creti R, Berardi A, Baldassarri L, Imperi M, Pataracchia M, Alfarone G, Recchia S, Prevention Working Group Emilia-Romagna GBS, the Neonatal GBS Italian Network 2013. Characteristics of neonatal GBS disease during a multicentre study (2007-2010) and in the year 2012. Ann. Ist Super Sanita 49:370–375. 10.4415/ANN_13_04_09 [DOI] [PubMed] [Google Scholar]
- 3.Tan CK, Lai CC, Wang JY, Lin SH, Liao CH, Huang YT, Wang CY, Lin HI, Hsueh PR. 2010. Bacteremia caused by non-faecalis and non-faecium Enterococcus species at a medical center in Taiwan, 2000 to 2008. J. Infect. 61:34–43. 10.1016/j.jinf.2010.04.007 [DOI] [PubMed] [Google Scholar]
- 4.Miskeen PA, Deodhar L. 2002. Antimicrobial susceptibility pattern of Enterococcus species from urinary tract infections. J. Assoc. Physicians India. 50:378–381 [PubMed] [Google Scholar]
- 5.Ruoff KL, de la Maza L, Murtagh MJ, Spargo JD, Ferraro MJ. 1990. Species identities of enterococci isolated from clinical specimens. J. Clin. Microbiol. 28:435–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kenzaka T, Takamura N, Kumabe A, Takeda K. 2013. A case of subacute infective endocarditis and blood access infection caused by Enterococcus durans. BMC Infect. Dis. 13:594. 10.1186/1471-2334-13-594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeurink PV, van Bergenhenegouwen J, Jimenez E, Knippels LM, Fernandez L, Garssen J, Knol J, Rodriguez JM, Martin R. 2013. Human milk: a source of more life than we imagine. Benef. Microbes 4:17–30. 10.3920/BM2012.0040 [DOI] [PubMed] [Google Scholar]
- 8.Collins MD, Jones D, Farrow JAE, Kilpper-Balz R, Schleifer KH. 1984. Enterococcus avium nom. rev., comb. nov.; E. casseliflavus nom. rev., comb. nov.; E. durans nom. rev., comb. nov.; E. gallinarum comb. nov. and E. malodoratus sp. nova. Int. J. Syst. Bacteriol. 34:220–223. 10.1099/00207713-34-2-220 [DOI] [Google Scholar]
- 9.Harvey CL, McIllmurray MB. 1984. Streptococci with dual antigen specificity for Lancefield groups D and G. Eur. J. Clin. Microbiol. 3:526–530. 10.1007/BF02013612 [DOI] [PubMed] [Google Scholar]
- 10.Savini V, Kosecka M, Marrollo R, Carretto E, Miȩdzobrodzki J. 2013. CAMP test detected Staphylococcus delphini ATCC 49172 β-haemolysin production. Pol. J. Microbiol. 62:465–466 [PubMed] [Google Scholar]


