Abstract
A multipurpose high-throughput genotyping tool for the assessment of recent epidemiological data and evolutional pattern in Mycobacterium tuberculosis complex (MTBC) clinical isolates was developed in this study. To facilitate processing, 51 highly informative single nucleotide polymorphisms (SNPs) were selected for discriminating the clinically most relevant MTBC species and genotyping M. tuberculosis into its principle genetic groups (PGGs) and SNP cluster groups (SCGs). Because of the high flexibility of the DigiTag2 assay, the identical protocol and DNA array containing the identical set of probes were applied to the highly GC-rich mycobacterial genome. The specific primers with multiplex amplification and hybridization conditions based on the DigiTag2 principle were optimized and evaluated with 14 MTBC reference strains, 4 nontuberculous mycobacteria (NTM) isolates, and 322 characterized M. tuberculosis clinical isolates. The DNA chip that was developed revealed a 99.85% call rate, a 100% conversion rate, and 99.75% reproducibility. For the accuracy rate, 98.94% of positive calls were consistent with previous molecular characterizations. Our cost-effective technology was capable of simultaneously identifying the MTBC species and the genotypes of 96 M. tuberculosis clinical isolates within 6 h using only simple instruments, such as a thermal cycler, a hybridization oven, and a DNA chip scanner, and less technician skill was required than for other techniques. We demonstrate this approach's potential as a simple, flexible, and rapid tool for providing clearer information regarding circulating MTBC isolates.
INTRODUCTION
Tuberculosis (TB) is one of the most infectious and deadly global diseases, and it causes medical, social, and economic disasters worldwide. The effects of the genotype-to-genotype variations of the Mycobacterium tuberculosis complex (MTBC) causative agents on the pathogenesis, host immune response, and susceptible hosts have been elucidated (1, 2). A clear understanding of the bacterial variability associated with phenotypic properties is relevant in terms of the disease, its transmission potential, the immunological response, and its manifestation, and an awareness of the genetic diversity in circulating MTBC isolates is crucial (3).
Members of the MTBC comprise M. tuberculosis, M. bovis subsp. bovis, M. bovis subsp. caprae, M. bovis BCG, M. africanum (subtypes I and II), M. microti, M. pinnipedii, M. canettii, M. mungi (4), M. orygis (5), Dassie bacillus (6), and chimpanzee bacillus (7). Biochemical testing, phage typing, and serotyping methods for identifying the members of the MTBC have been described, as have several molecular techniques. However, the high costs and complicated interpretations limit the use of these techniques for routine applications. Current published data suggest that the MTBC and its sublineages attained genetic and biological diversity through a combination of discrete acquired single nucleotide polymorphisms (SNPs) fixed within the clonal population structure. The regions of the genome that are beneficial for identification of mycobacteria to the species level in this complex include gyrB, pncA, katG, pks15, and the 16S rRNA gene (8, 9). Molecular epidemiological studies have suggested that the trend for drug resistance acquisition in M. tuberculosis appears to depend on the infecting M. tuberculosis types, which are identified by DNA fingerprinting (10–12). Some related types appear to be strongly associated with specific geographic locations or sites of infection. Several studies described the prevalence and distribution of the Beijing (BJ) lineage and sublineage in many Asian countries, North America, and Russia (13). The East African-Indian (EAI) lineage strongly predominated in Kampala and Uganda (14).
High reproducibility, digital data properties; category congruence with the large-sequence polymorphism (LSP), spoligotyping, and the other SNP set; less chances of convergence evolution; and the ability to infer selective advantages of SNPs have been proven to be promising genetic markers for epidemiological studies (14). An extensive study of whole-genome comparisons recommended a minimal set of 45 synonymous SNPs (sSNPs) that categorized M. tuberculosis clinical isolates into 6 genetically related SNP cluster groups (SCGs) with 5 subgroups and 1 SCG clade for M. bovis and M. pinnipedii (15). The limited numbers of SNPs provided sufficient information for epidemiological and evolutionary studies (15, 16).
In recent years, several SNP detection methods were developed for characterizing the genetic diversity among MTBC clinical isolates. These include the hairpin primers assay (17), matrix-assisted laser desorption ionization−time of flight mass spectrometry (MALDI-TOF MS) analysis (18), the 5′ exonuclease-based assay (TaqMan), the SNaPshot primer extension method (19), the SNaPshot minisequencing-based assay (20), and multiplex ligation-dependent probe amplification (MLPA) (21), which have provided the ability for rapid responses with solutions. A multipurpose high-throughput genotyping platform with cost-effective technology is still needed for investigation of circulating MTBC isolates. The DigiTag2 assay using a universal DNA chip contains 24 samples per array, enabling up to 96 samples to be run in parallel against 96 targets. In this assay, Multiplex PCRs for selectively increasing the target of interest followed by ligation assays for reducing the complexity of the candidate SNPs are the main processes for successful genotyping, and this SNP typing assay is widely used for screening SNPs spanning the human chromosome (22, 23). We hypothesized that the human high-throughput genotyping platform could be applied as an MTBC molecular epidemiological tool using only general molecular laboratory equipment, such as a thermal cycler, a hybridization oven, and a chip scanner. We, therefore, describe here the development of a universal DNA chip based on a DigiTag2 assay for species identification and genotypic characterization of MTBC isolates with a robust and cost-effective operating platform.
MATERIALS AND METHODS
Bacterial strains and study population.
A total of 338 DNAs from mycobacterial strains belonging to 5 species of MTBC (n = 334) and 3 species of nontuberculous mycobacteria (NTM) (n = 4) were evaluated (Table 1). Thirteen previously well-characterized M. tuberculosis clinical isolates and 15 Mycobacterium reference strains (M. tuberculosis H37Rv [ATCC 27294], M. tuberculosis H37Ra [ATCC 25177], M. tuberculosis CDC1551, M. tuberculosis 14323, M. bovis ATCC 19210, M. bovis ATCC 35720, M. bovis BCG Danish [ATCC 35733], M. bovis BCG Pasteur [ATCC 35734], M. bovis BCG Tokyo [ATCC 35737], M. africanum ATCC 25420, M. microti ATCC 19422, M. marinum strain 987, M. avium ATCC 25291, M. avium IWGMT, and M. intracellulare ATCC 13950) were used for the optimization step (24). A total of 322 clinical strains of tuberculosis collected from 1996 to 2011 for evaluating the feasibility of the assay were retrieved from the Molecular Mycology and Mycobacteriology Laboratory (Drug-Resistant Tuberculosis Research Fund), Department of Microbiology, Faculty of Medicine Siriraj Hospital, Mahidol University, Thailand). The ethical and scientific committees of the Faculty of Medicine Siriraj Hospital, Mahidol University, approved the study (EC no. 759/2551 and EC no. 604/2555).
TABLE 1.
Genotyping results for 14 MTBC reference strains, 4 NTM isolates, and 322 well-characterized M. tuberculosis clinical isolates based on PGG, spoligotype, SCG, and ST
| Mycobacterium species | Strain no. | Lineage | SIT(s)a | PGG | SCG: ST(s) (no.) |
|---|---|---|---|---|---|
| M. africanum | ATCC 25420 | AFRI I | 859 | 1 | 7: NAb (1) |
| M. bovis BCG | ATCC 35733, 35734, 35737 | BOVIS_BCG | 482 | 1 | 7: 28 (3) |
| M. bovis | ATCC BAA-935, 19210 | BOVIS | 683, 670 | 1 | 7: NA (2) |
| M. microti | ATCC 19422 | MICROTI | NA | 1 | 7: NA (1) |
| M. tuberculosis | Mt14323 | Haarlem | 47 | 2 | 3b: NA (1) |
| M. tuberculosis | CDC1551 | X | 549 | 2 | 4: 39 (1) |
| M. tuberculosis | ATCC 27294, 25177 | T | 451 | 3 | 6a: 40 (2) |
| M. intracellulare | ATCC 13950 | NDc | ND | ND | ND (1) |
| M. marinum | Strain 927 | ND | ND | ND | ND (1) |
| M. avium | ATCC 25291, IWGMT | ND | ND | ND | ND (2) |
| M. africanum | Clinical isolate | NA | NA | NA | ND (1) |
| M. tuberculosis | Clinical isolate | Beijing | 1 | 1 | 2: 8 (5), 10 (108), 19 (29), 22 (27), 26 (3), 10 (1), STF (2), STK (3), NA (67) |
| M. tuberculosis | Clinical isolate | CAS | 89, 523, 1462, ND | 1 | 3a: NA (3) |
| M. tuberculosis | Clinical isolate | EAI | 19, 88, 139, 204, 474, 735, 951, ND | 1 | 1: NA (59) |
| M. tuberculosis | Clinical isolate | Haarlem | 49, 50, 742 | NA | NA: NA (4) |
| M. tuberculosis | Clinical isolate | T | 52, 53, 196, 393, 462, 943 | NA | NA: NA (10) |
| M. tuberculosis | Clinical isolate | Unclassified | ND | NA | NA: NA (1) |
SIT, Spoligotype International Type. The spoligotyping definition corresponds to the SpolDB4 database.
NA, not available.
ND, no data.
Mycobacterial characterization. (i) Identification of mycobacteria to the species level.
All the isolates were previously identified to the species level by a combination of phenotypic, biochemical, and molecular characterization tests. A one-tube multiplex PCR was used to screen species of slow-growing acid-fast bacilli (25). Briefly, the slow-growing dysgonic colonies that were positive for KS4 MTBC-specific and negative for mtp40 M. tuberculosis-specific DNA fragments and negative for the niacin accumulation test were defined as M. bovis or M. bovis BCG (26). Colonies of positive acid-fast bacilli with an absence of KS4 MTBC-specific and mtp40 M. tuberculosis-specific DNA fragments were defined as NTM isolates. PCR-restriction endonuclease analysis (PCR-REA) of the hsp65 gene and/or the rpoB gene was performed to differentiate the NTM species, and the 16S rRNA gene was sequenced if necessary.
(ii) Molecular genotyping.
All clinical isolates were typed by the spoligotyping method (27). The results of LSP typing, insertion of IS6110 at the NTF region (28), and the sequence type (ST) based on a minimal set of 10 SNPs (15) among the 253 M. tuberculosis isolates were obtained from the previous study (24).
(iii) SNP target selection.
To identify the members of the MTBC and define the genetic lineages based on the PGGs and STs among MTBC clinical isolates simultaneously, 51 well-characterized target SNPs were selected: SNP01, 1472307; SNP02, 6446; SNP03, 5752; SNP04, 5671; SNP05, 2154724; SNP06, 7585; SNP07, 37031; SNP08, 43945; SNP09, 92199; SNP10, 220050; SNP11, 311613; SNP12, 519806; SNP13, 797736; SNP14, 909166; SNP15, 918316; SNP16, 923065; SNP17, 949221; SNP18, 1068151; SNP19, 1163134; SNP20, 1191861; SNP21, 1294398; SNP22, 1477598; SNP23, 1548149; SNP24, 1692069; SNP25, 1692685; SNP26, 1884697; SNP27, 1892017; SNP28, 1952599; SNP29, 2158582; SNP30, 2223682; SNP31, 2376135; SNP32, 2462871; SNP33, 2532616; SNP34, 2627946; SNP35, 2825581; SNP36, 2891267; SNP37, 2990040; SNP38, 3207250; SNP39, 3438386; SNP40, 3440464; SNP41, 3440542; SNP42, 3450725; SNP43, 3455686; SNP44, 3544710; SNP45, 3783056; SNP46, 4024273; SNP47, 4119246; SNP48, 4137829; SNP49, 4254006; SNP50, 4255922; and SNP51, 4280708 as given by the M. tuberculosis H37Rv genome sequence (GenBank accession no. NC_000962) (8, 9, 15, 29). The first SNP located at the 16S rRNA gene was selected as the positive control for defining the MTBC. Three SNPs, SNP02 (gyrB 1450), SNP03 (gyrB 756), and SNP04 (gyrB 675), were used to differentiate the members of the MTBC, including TC1 (M. tuberculosis, M. africanum II, and M. canettii), TC2 (M. bovis, M. bovis BCG, and M. caprae), TC3 (M. pinnipedii and M. africanum I), and TC4 (M. microti) (8, 9). For characterizing three PGGs among the MTBC based on neutral mutations, the katG codon 463 (SNP05) and gyrA codon 95 (SNP06) were included (29). The last 45 SNP positions were selected from a minimal standard set of 45 SNPs based on whole-genome comparisons of M. tuberculosis H37Rv, CDC1551, and 210 plus M. bovis AF2122/97, which were capable of classifying M. tuberculosis, M. bovis, and M. pinnipedii into 7 distinct SCGs and 5 subgroups or 56 STs (15). This set was useful for subtyping the BJ genotype into the ancestral and modern subfamilies related to the presence or absence of an IS6110 insertion into the NTF region (24).
DigiTag2 assay framework.
The DigiTag2 assay is a multiplex SNP typing method, which is composed of 4 important steps: target preparation, encoding, labeling, and hybridization (22, 23). The SNP sequence type of each position was defined from fluorescence signal intensities on the DNA microarray. Each slide was composed of 24 separate areas for 24 tested samples. Each separated area had 100 immobilized probes, and only 52 positions were used in this study; the last position was originally designed as a hybridization control. Because of the high flexibility of the DigiTag2 assay, oligonucleotide primers and probes were newly designed by Visual OMP software to cover these 51 target SNPs for identifying species and typing the MTBC comprehensively. The DNA sequences of 51 designed primer pairs had relative lengths from 26 to 41 bp and synthesized DNA amplicons from 334 to 969 bp. For the high GC-rich genome of the Mycobacterium spp., the denaturation time and enhanced GC-rich polymerase enzyme, KAPA2G Robust HotStart DNA polymerase, are factors supportive of the reaction. Two-step multiplex PCRs were performed to increase 51 target SNPs and reduce nonspecific DNA amplification in the target preparation step. The 10-μl total volume PCR was composed of 1× KAPA2G Robust HotStart ready mix with dye (KAPA Biosystems, Woburn, MA) containing KAPA2G Robust HotStart DNA polymerase in an appropriate reaction buffer (including 2.25 mM Mg2+), an additional 2.25 mM Mg2+, 0.2 mM deoxynucleoside triphosphates (dNTPs), stabilizers, inert dyes with 25 nM each primer, and 20 to 40 ng of Mycobacterium spp. genomic DNA. The PCR denaturing time was extended to 15 min at 95°C, followed by 40 cycles of 95°C for 1.30 min and 68°C for 2 min using a TGradient thermal cycler (Biometra, Göttingen, Germany) (23). For the screening efficiency of each primer, a simplex PCR was performed by determining the presence of the PCR product with capillary electrophoresis (Agilent 2100 bioanalyzer; Agilent, Santa Clara, CA). Whereas the encoding, the labeling, and the hybridization mixture were prepared following the original assay, the reaction conditions were adjusted for the mycobacterial genome. The annealing temperature in the encoding reaction was increased as follows: 95°C for 5 min, 59°C for 15 min, and holding at 10°C, with an extended denaturation time for the labeling reaction; and initiation denaturation at 95°C for 15 min, 30 cycles of 95°C for 1 min, 55°C for 6 min, and 72°C for 30 s. Holding at 10°C using a TGradient thermal cycler revealed better genotyping results (23). The performance was estimated by call rate, conversion rate, accuracy, and reproducibility parameters. The call rate was defined as the number of genotype calls among the total number of samples examined. The conversion rate was defined as the proportion of successfully genotyped SNPs among the total number of SNPs examined. The accuracy rate was determined by a comparison with the spoligotyping result. Reproducibility was evaluated from duplication experiments which tested 24 DNA samples.
RESULTS
Determination of the feasibility of the modified DigiTag2 assay.
For development of a universal DNA chip for species identification of the MTBC and typing M. tuberculosis, the multiplex genotyping conditions for 51 target SNPs were optimized using well-characterized Mycobacterium spp. DNAs, which simultaneously covered the testing of 102 possible nucleotide polymorphisms. Four NTM and 20 MTBC isolates, including 13 M. tuberculosis (M. tuberculosis H37Rv, M. tuberculosis H37Ra, M. tuberculosis CDC1551, and M. tuberculosis strain 14323), 7 M. tuberculosis BJ genotype (ST8, n = 1; ST10, n = 3; ST19, n = 1; ST22, n = 1; and ST26, n = 1) (24, 30), and 2 M. tuberculosis T genotype (SCG3b, n = 1 and SCG5, n = 1), M. bovis BCG (n = 3), M. bovis (n = 2), M. africanum (n = 1), and M. microti (n = 1) were tested (Table 1). The results revealed successful genotyping for all of the 51 target SNPs spanning MTBC genomes, and the total running time of the assay was approximately 6 h (Fig. 1). A total of 322 previously characterized M. tuberculosis clinical isolates and 1 M. africanum clinical isolate were blindly used to evaluate the efficiency of the high-throughput assay. Perfect call rates, conversion rates, reproducibility when independently tested with 24 well-characterized DNA samples, and consistency with the previously defined MTBC level were shown at SNP01, indicating that this assay can be used to differentiate MTBC from NTM strains correctly. M. marinum strain 987, M. avium ATCC 25291, M. avium IWGMT, and M. intracellulare ATCC 13950 were distinctly genotyped to NTM. For screening of the species of MTBC members, SNP02 to SNP04, the genotype call rates among 334 MTBC isolates were 100%, conversion rates were 100% for these 3 SNP positions, and reproducibility was 100% for independently duplicating 24 well-characterized DNA samples. The results were completely concordant with the previous identification data. All of the M. tuberculosis isolates were obviously categorized to TC1, and the M. africanum clinical isolate was designed as M. africanum subtype II. Isolates of M. bovis and M. bovis BCG were grouped into TC2. M. africanum ATCC 25420, known as subtype I, and M. microti ATCC 19422 were defined to TC3 and TC4, respectively (9). The genotyping results for SNP05 and SNP06 defined as PGGs showed a 100% call rate, 100% conversion rate, and 100% reproducibility. The group of PGGs determined by spoligotyping data among the 334 MTBC isolates revealed perfect concordance with the genotyping data of a modified DigiTag2 assay. For accessing the genotype diversity among M. tuberculosis clinical isolates, SNP07 to SNP51 showed a 99.90% call rate, 100% conversion rate, and 99.75% reproducibility. Compared with the data for spoligotyping and 9 minimal SNPs typing, 98.94% accuracy was shown when 335 previously characterized MTBC isolates were analyzed (Table 2).
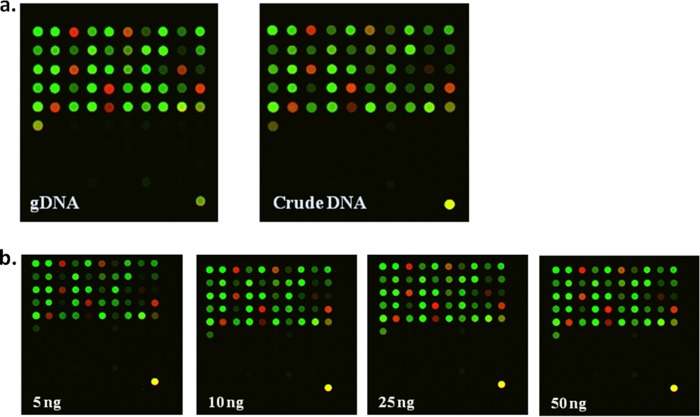
FIG 1.
(a) Fluorescence images of M. tuberculosis H37Rv for all 51 SNPs between high-quality DNA and crude DNA at 40 ng/reaction. (b) Fluorescence signals of high-quality DNA at 5 ng, 10 ng, 2 5 ng, and 50 ng/reaction, respectively. Each fluorescence signal, Alexa 647 (red) or Alexa 555 (yellow green), represents an SNP calling. gDNA, genomic DNA.
TABLE 2.
Feasibility of DNA chip based on modified DigiTag2 assay for species identification of MTBC and genotyping of M. tuberculosis
| Parameter | Results for SNP: |
||||
|---|---|---|---|---|---|
| 1 | 2–4 | 5–6 | 7–51 | All | |
| No. of calls | 338 | 1,002 | 668 | 15,007 | 17,009 |
| Call rate (%) | 100.00 | 100.00 | 100.00 | 99.89 | 99.90 |
| Conversion rate (%) | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| Accuracy (%) | 100.00 | 100.00 | 100.00 | 98.80 | 98.94 |
| Reproducibility (%) | 100.00 | 100.00 | 100.00 | 99.72 | 99.75 |
Comparison of genotyping results between crude DNA and high-quality DNA templates.
To explore the promising faster and easier processing qualities, a DNA template extracted by a boiling centrifugation DNA extraction procedure and enzymatic lysis extraction method were evaluated (31). Under the optimal conditions of the DigiTag2 assay, 40 ng of high-quality DNA and crude DNA samples could be used for genotyping M. tuberculosis with a 100% call rate, 100% accuracy, 100% conversion rate, and 100% reproducibility. The crude DNA sample revealed a slightly low signal intensity (fluorescence signal of <1,000 units), but genotype calling for each individual SNP was still acceptable for analysis (Fig. 1a). The comparisons among different concentrations of high-quality DNA showed that the appropriate concentration of M. tuberculosis H37Rv ranged between 25 ng and 50 ng per reaction. SNP19, using 5, 10, and 25 ng of DNA template, presented a signal intensity of <1,000 units, and a higher concentration of DNA template provided an acceptable signal (Fig. 1b).
Congruent genotyping application.
Previous genotyping data for M. tuberculosis isolates were used for evaluating the new technology and for studying the epidemiological aspects. The correlations of genotypes based on 45 SNPs were analyzed across the other acceptable genotypic markers for M. tuberculosis. Previously published data and the genotyping results of 183 M. tuberculosis isolates revealed results concordant with spoligotyping and PGG results and were related to the presence or absence of TbD1, RD105, and RD181 and the orientation of IS6110 at the NTF region (24, 32–35). Our 51 selected SNPs were able to differentiate ancestral MTBC strains (EAI [SCG1], M. bovis [SCG7], and M. africanum subtype I [SCG1]) related to the presence of TbD1 markers by using SNP08 or SNP41 (Table 3). The investigation of known spoligotype isolates, along with the previously published results, revealed that this SNP set was more informative for subgenotyping the BJ family (SCG2) (Table 4). Thirteen STs among SCG2 revealed concordant grouping with classification of the BJ genotype using RD105, RD181, and insertion of IS6110 at the NTF markers as expected. Previous genotyping results of all 13 STs revealed the absence of RD105 and a unique spoligotyping pattern of the BJ genotype. SCG2 could likely be directly differentiated from the other genotypes using SNP51. Among 103 well-characterized BJ isolates, ST19, K, and 26 were categorized into an ancestral BJ genotype, and ST8, 22, 10, and STF were grouped into the modern BJ genotype using the differences of SNP22. The BJ (W) genotype was classified into ST10, which was the modern BJ genotype. Our SNP set could not separate modern BJ from the BJ (W) genotype. While the presence or absence of the RD181 region is commonly used to divide the ancestral BJ genotype into early ancestral and late ancestral isolates, our results revealed perfect concordance of evolutionary events among SNP13 or SNP35 and the RD181 marker. Based on this position, ST26 was defined as an early ancestral BJ type, and STK and ST19 were defined as late ancestral BJ types (36). The results indicated that this genotyping platform can be used to classify MTBC strains into SNP-based genotypes and to provide information referring to the other acceptable markers.
TABLE 3.
SNP genotyping signature of SNP02, -03, -04, -08, and -41 and results for the absence or presence of TbD1 among MTBC isolates
| MTBC species and strain | SCG | ST(s) | Genotyping signature for: |
TbD1a | ||||
|---|---|---|---|---|---|---|---|---|
| SNP02: 6446b | SNP03: 5752b | SNP04: 5671b | SNP08: 43945b | SNP41: 3440542b | ||||
| M. tuberculosis H37Rv | 6a | 40 | G | G | C | A | A | − |
| M. tuberculosis CDC1551 | 4 | 39 | G | G | C | A | A | − |
| M. tuberculosis 210c | 2 | 8 | G | G | C | A | A | − |
| M. tuberculosis | 1 | 15 | G | G | C | G | G | + |
| M. tuberculosisc | 1 | 37 | G | G | C | G | G | + |
| MTBCc | 7 | 16, 28–33 | G | G | C | G | G | + |
| M. bovis BCGd | 7 | 28 | T | A | C | G | G | + |
| M. bovis AF2122/97c | 7 | 29 | T | A | C | G | G | + |
| M. bovise | 7 | 29 | T | A | C | G | G | + |
| M. africanum subtype II | 1 | 15 | G | G | C | G | G | + |
| M. africanum subtype I | 7 | 16var | T | G | C | G | G | + |
| M. microti 203 | 7 | 16var | T | G | T | G | G | + |
The presence (+) or absence (−) of TBD1 followed a previous study (35).
Coordinates as given by the M. tuberculosis H37Rv genome sequence (GenBank accession no. NC_000962).
Genotype data based on SNP markers were obtained from a previous publication (15).
M. bovis BCG Tokyo, M. bovis BCG Pasteur, and M. bovis BCG Danish.
M. bovis 19210 and M. bovis 35720.
TABLE 4.
Comparison of SNP genotyping signature of SNPs 13, 22, 35, and 51 and other markers for subtyping BJ genotyping
| SCG | ST (n) | TbD1a | PGG | Spoligotype | NTF insertionb | RD105c | RD181d | RD150e | RD142e | SNP13: 797736 | SNP22: 1477598 | SNP35: 2825581 | SNP51: 4280708 | BJ sublineagee |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 8 (4) | + | 1 | BJ | + | − | − | −/+ | 0 | T | T | G | A | Modern |
| 2 | 22 (96) | + | 1 | BJ | + | − | − | + | + | T | T | G | A | Modern |
| 2 | 10 (533) | + | 1 | BJ | + | − | − | −/+ | + | T | T | G | A | Modern |
| 2 | 10 (1) | + | 1 | BJ (W) | +/+ | − | − | + | + | T | T | G | A | Modern |
| 2 | 27 (0) | + | 1 | BJ | NAf | NA | NA | NA | NA | T | T | G | A | NA |
| 2 | F (10) | + | 1 | BJ | + | − | − | + | + | T | T | G | A | Modern |
| 2 | 19 (226) | + | 1 | BJ | − | − | − | + | + | T | C | G | A | Late ancestral |
| 2 | 25 (69) | + | 1 | BJ | − | − | − | + | + | T | C | G | A | Late ancestral |
| 2 | 17 (0) | + | 1 | BJ | NA | − | NA | NA | − | T | C | G | A | NA |
| 2 | K (114) | + | 1 | BJ | − | − | − | + | + | T | C | G | A | Late ancestral |
| 2 | 3 (192) | + | 1 | BJ | − | − | − | NA | NA | T | C | G | A | Late ancestral |
| 2 | 50 (0) | + | 1 | BJ | NA | − | − | − | − | T | C | G | A | NA |
| 2 | 26 (57) | + | 1 | BJ | − | − | + | + | + | C | C | T | A | Early ancestral |
| 2 | 11 (41) | + | 1 | BJ | − | − | + | + | + | C | C | T | A | Early ancestral |
The presence (+) or absence (−) of an M. tuberculosis-specific deletion (TbD1) followed reference 35.
Overall, 103 isolates in Thailand (24) and 74 isolates in Japan (30) were assigned to early ancestral BJ (−); modern BJ (+); or modern W-BJ (+/+).
A total of 103 isolates in Thailand (24) were assigned to a non-BJ genotype (+) or a BJ genotype (−).
A total of 103 isolates in Thailand (24), 74 isolates in Japan (30), 62 isolates in Korea (34), 187 isolates in China (37), and 268 isolates in Peru (32) were assigned to early ancestral (+) or late ancestral (−).
Designations of 103 isolates in Thailand (24) and 74 isolates in Japan (30) were referred to the presence (+) or absence (−) of RD150 or RD142, respectively.
NA, data not available.
DISCUSSION
To investigate genetic diversity among tuberculosis cases worldwide, an SNP set that is comprehensive for screening species of the MTBC to identify the lineage and subtypes of BJ genotypes and a usable high-throughput genotyping platform were combined in this study. Our results revealed successful optimization of a human high-throughput genotyping platform, DigiTag2, as a MTBC molecular epidemiological tool (23). Longer extension of the denaturing time and changing of the polymerase enzyme are the main factors for successfully amplifying the GC-rich genome of mycobacteria. A melting agent-tolerant polymerase enzyme, KAPA2G Robust DNA polymerase, which was genetically engineered for GC-rich PCR, was used to improve the success rate and reduce the cycling time. The turnaround time for the modified DigiTag2 version is approximately 6 h, which is 1½ h longer than the original version (22, 23). Only the thermal cycler, hybridization oven, and chip scanner are needed. The data showed the efficiency of the assay, with an easily extracted DNA template and high-quality DNA testing. To have an easily readable signal without false positives, a large amount of DNA template (40 ng/reaction) was required in our platform (Fig. 1). Further improvement in the sensitivity of the assay might provide the opportunity to characterize the MTBC in direct specimens. This multipurpose high-throughput genotyping platform has likely driven the MTBC molecular genotyping method to be faster and simpler than the IS6110 restriction fragment length polymorphism (RFLP), spoligotyping, LSP, or variable-number tandem repeat (VNTR) typing methods (18, 21, 27). Ninety-six strains can be identified and typed for 51 SNP markers within 6 working hours, and the cost is about US $3 per strain.
Compared with the acceptable molecular genotyping methods for the MTBC, our assay is capable of simultaneously screening species of the MTBC and genotyping M. tuberculosis isolates for epidemiological purposes. Our tool was able to distinguish nearly all members of the MTBC. The selected SNPs were capable of identifying the most common genotypes causing human tuberculosis (TB), such as EAI and Beijing, to the species level for epidemiological and treatment purposes. Specifically, M. bovis, which has the broadest host range of the MTBC, is naturally resistant to the antituberculous drug pyrazinamide (PZA), and is known to be the second most important causative agent of TB in humans, was differentiated from M. tuberculosis. Our technique differs markedly from that in an earlier study in its throughput and discrimination power. Bouakaze et al. applied the SNaPshot minisequencing-based assay for identifying members of the MTBC and determining the lineage of M. tuberculosis using 8 species-specific SNPs and 8 lineage-specific SNPs (18). Almost all of the human MTBC strains were differentiated; however, higher discrimination power and superior throughput were demonstrated in our study. Although the high-throughput genotyping assays based on the commercially available Sequenom MassArray MALDI-TOF platform and MagPix technology are reported to have rapid turnaround times and high flexibility, these techniques are more suited to specialized molecular laboratories because they require specially trained technicians and special laboratory equipment (18, 21), whereas our DNA chip can type M. tuberculosis BJ and EAI genotypes, which correspond to >80% of the human TB cases worldwide. The drawback of our DNA chip is that it has less discrimination for typing the EAI genotype than spoligotyping and for typing the Beijing genotype than the VNTR typing method. The concordant results among this SNP set and the presence or absence of TbD1, RD105, and RD181 and the insertion of IS6110 at the NTF region support the accuracy of this SNP set and benefit epidemiological data determination. Compared with the other minimal SNP set, the 59 genotype-specific SNPs reported by Homolka et al. revealed higher discrimination power in the Euro-American lineage; however, the discrimination power of the other genotypes was less than that of our selected markers (36). The addition of more significant SNPs for better subgenotyping of the SCG1 (EAI) genotype and the Euro-American lineage is needed. For further investigation, additional SNPs that could distinguish M. canettii, M. caprae, and M. pinnipedii from the other MTBC members should be tested for species identification (8). Further testing of an unbiased collection is one of the improvements to provide information for more efficient documentation in many nations worldwide. Moreover, the next-generation sequencing approach with reduced running costs for whole-genome sequencing might facilitate selection of specific candidate SNPs for other lineages of the MTBC. A modified DigiTag2 assay could be easily applied in diagnostic and research laboratories working with mycobacteria. We hope that this high-efficiency screening tool provides a clear representation for global phylogenetic studies and a better understanding of the underlying biological differences among clinical strains.
ACKNOWLEDGMENTS
We thank the Drug-Resistant Tuberculosis Research Fund Laboratory, the Siriraj Foundation, under the patronage of HRH Princess Galyani Vadhana KromLaung Narathiwas Rajnakarindth, and the Department of Microbiology and Faculty of Medicine Siriraj Hospital, Mahidol University for the isolates used for the study, the facilities, and the patient data. We sincerely thank the Department of Human Genetics of the Graduate School of Medicine at the University of Tokyo for research support in performing the experiments for 6 months and the Japan Anti-tuberculosis Association Research Institute of Tuberculosis (RIT/JATA) for providing the DNAs of M. africanum ATCC 25420, M. tuberculosis CDC1551, M. bovis ATCC 19210, M. bovis ATCC 35720, and M. microti ATCC 19422 and Prasit Palittapongarnpim for providing M. tuberculosis strain Mt14323.
This study was supported by the Thailand Research Fund through the Royal Golden Jubilee Ph.D. Program (Grant PHD/0315/2551) to P.S. (student) and A.C. (advisor) and by joint funding from the Japan Science and Technology Agency (JST) and National Science and Technology Development Agency (NSTDA), the Mahidol University Research Fund, a Siriraj Graduate Thesis Scholarship, and the Drug-Resistant Tuberculosis Research Fund of the Siriraj Foundation.
This work is dedicated to Karl Esser, Ph.D. adviser of Angkana Chaiprasert, on his 90th birthday, for his excellent scientific role model.
We declare no conflicts of interest.
Footnotes
Published ahead of print 26 March 2014
REFERENCES
- 1.Comas I, Coscolla M, Luo T, Borrell S, Holt KE, Kato-Maeda M, Parkhill J, Malla B, Berg S, Thwaites G, Yeboah-Manu D, Bothamley G, Mei J, Wei L, Bentley S, Harris SR, Niemann S, Diel R, Aseffa A, Gao Q, Young D, Gagneux S. 2013. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat. Genet. 45:1176–1182. 10.1038/ng.2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gutierrez MC, Brisse S, Brosch R, Fabre M, Omaïs B, Marmiesse M, Supply P, Vincent V. 2005. Ancient origin and gene mosaicism of the progenitor of mycobacterium tuberculosis. PLoS Pathog. 1:e5. 10.1371/journal.ppat.0010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gagneux S. 2012. Host-pathogen coevolution in human tuberculosis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367:850–859. 10.1098/rstb.2011.0316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexander KA, Laver PN, Michel AL, Williams M, van Helden PD, Warren RM, Gey van Pittius NC. 2010. Novel Mycobacterium tuberculosis complex pathogen, M. mungi. Emerg. Infect. Dis. 16:1296–1299. 10.3201/eid1608.100314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Ingen J, Rahim Z, Mulder A, Boeree MJ, Simeone R, Brosch R, van Soolingen D. 2012. Characterization of Mycobacterium orygis as M. tuberculosis complex subspecies. Emerg. Infect. Dis. 18:653–655. 10.3201/eid1804.110888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parsons S, Smith SG, Martins Q, Horsnell WG, Gous TA, Streicher EM, Warren RM, van Helden PD, Gey van Pittius NC. 2008. Pulmonary infection due to the dassie bacillus (Mycobacterium tuberculosis complex sp.) in a free-living dassie (rock hyrax—Procavia capensis) from South Africa. Tuberculosis (Edinb.) 88:80–83. 10.1016/j.tube.2007.08.012 [DOI] [PubMed] [Google Scholar]
- 7.Coscolla M, Lewin A, Metzger S, Maetz-Rennsing K, Calvignac-Spencer S, Nitsche A, Dabrowski PW, Radonic A, Niemann S, Parkhill J, Couacy-Hymann E, Feldman J, Comas I, Boesch C, Gagneux S, Leendertz FH. 2013. Novel Mycobacterium tuberculosis complex isolate from a wild chimpanzee. Emerg. Infect. Dis. 19:969–976. 10.3201/eid1906.121012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huard RC, Fabre M, de Haas P, Lazzarini LC, van Soolingen D, Cousins D, Ho JL. 2006. Novel genetic polymorphisms that further delineate the phylogeny of the Mycobacterium tuberculosis complex. J. Bacteriol. 188:4271–4287. 10.1128/JB.01783-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niemann S, Harmsen D, Rusch-Gerdes S, Richter E. 2000. Differentiation of clinical Mycobacterium tuberculosis complex isolates by gyrB DNA sequence polymorphism analysis. J. Clin. Microbiol. 38:3231–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drobniewski F, Balabanova Y, Nikolayevsky V, Ruddy M, Kuznetzov S, Zakharova S, Melentyev A, Fedorin I. 2005. Drug-resistant tuberculosis, clinical virulence, and the dominance of the Beijing strain family in Russia. JAMA 293:2726–2731. 10.1001/jama.293.22.2726 [DOI] [PubMed] [Google Scholar]
- 11.Yorsangsukkamol J, Chaiprasert A, Prammananan T, Palittapongarnpim P, Limsoontarakul S, Prayoonwiwat N. 2009. Molecular analysis of Mycobacterium tuberculosis from tuberculous meningitis patients in Thailand. Tuberculosis (Edinb.) 89:304–309. 10.1016/j.tube.2009.05.001 [DOI] [PubMed] [Google Scholar]
- 12.Yuen CM, Kurbatova EV, Click ES, Cavanaugh JS, Cegielski JP. 2013. Association between Mycobacterium tuberculosis complex phylogenetic lineage and acquired drug resistance. PLoS One 8:e83006. 10.1371/journal.pone.0083006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bifani PJ, Mathema B, Kurepina NE, Kreiswirth BN. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10:45–52. 10.1016/S0966-842X(01)02277-6 [DOI] [PubMed] [Google Scholar]
- 14.Gagneux S, Small PM. 2007. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect. Dis. 7:328–337. 10.1016/S1473-3099(07)70108-1 [DOI] [PubMed] [Google Scholar]
- 15.Filliol I, Motiwala AS, Cavatore M, Qi W, Hazbon MH, Bobadilla del Valle M, Fyfe J, Garcia-Garcia L, Rastogi N, Sola C, Zozio T, Guerrero MI, Leon CI, Crabtree J, Angiuoli S, Eisenach KD, Durmaz R, Joloba ML, Rendon A, Sifuentes-Osornio J, Ponce de Leon A, Cave MD, Fleischmann R, Whittam TS, Alland D. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 188:759–772. 10.1128/JB.188.2.759-772.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ford C, Yusim K, Ioerger T, Feng S, Chase M, Greene M, Korber B, Fortune S. 2012. Mycobacterium tuberculosis—heterogeneity revealed through whole genome sequencing. Tuberculosis (Edinb.) 92:194–201. 10.1016/j.tube.2011.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hazbón MH, Alland D. 2004. Hairpin primers for simplified single-nucleotide polymorphism analysis of Mycobacterium tuberculosis and other organisms. J. Clin. Microbiol. 42:1236–1242. 10.1128/JCM.42.3.1236-1242.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouakaze C, Keyser C, Gonzalez A, Sougakoff W, Veziris N, Dabernat H, Jaulhac B, Ludes B. 2011. Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based single nucleotide polymorphism genotyping assay using iPLEX gold technology for identification of Mycobacterium tuberculosis complex species and lineages. J. Clin. Microbiol. 49:3292–3299. 10.1128/JCM.00744-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutacker MM, Smoot JC, Migliaccio CA, Ricklefs SM, Hua S, Cousins DV, Graviss EA, Shashkina E, Kreiswirth BN, Musser JM. 2002. Genome-wide analysis of synonymous single nucleotide polymorphisms in Mycobacterium tuberculosis complex organisms: resolution of genetic relationships among closely related microbial strains. Genetics 162:1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bouakaze C, Keyser C, de Martino SJ, Sougakoff W, Veziris N, Dabernat H, Ludes B. 2010. Identification and genotyping of Mycobacterium tuberculosis complex species by use of a SNaPshot minisequencing-based assay. J. Clin. Microbiol. 48:1758–1766. 10.1128/JCM.02255-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bergval I, Sengstake S, Brankova N, Levterova V, Abadia E, Tadumaze N, Bablishvili N, Akhalaia M, Tuin K, Schuitema A, Panaiotov S, Bachiyska E, Kantardjiev T, de Zwaan R, Schurch A, van Soolingen D, van't Hoog A, Cobelens F, Aspindzelashvili R, Sola C, Klatser P, Anthony R. 2012. Combined species identification, genotyping, and drug resistance detection of Mycobacterium tuberculosis cultures by MLPA on a bead-based array. PLoS One 7:e43240. 10.1371/journal.pone.0043240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishida N, Tanabe T, Takasu M, Suyama A, Tokunaga K. 2007. Further development of multiplex single nucleotide polymorphism typing method, the DigiTag2 assay. Anal. Biochem. 364:78–85. 10.1016/j.ab.2007.02.005 [DOI] [PubMed] [Google Scholar]
- 23.Nishida N, Mawatari Y, Sageshima M, Tokunaga K. 2012. Highly parallel and short-acting amplification with locus-specific primers to detect single nucleotide polymorphisms by the DigiTag2 assay. PLoS One 7:e29967. 10.1371/journal.pone.0029967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faksri K, Drobniewski F, Nikolayevskyy V, Brown T, Prammananan T, Palittapongarnpim P, Prayoonwiwat N, Chaiprasert A. 2011. Genetic diversity of the Mycobacterium tuberculosis Beijing family based on IS6110, SNP, LSP and VNTR profiles from Thailand. Infect. Genet. Evol. 11:1142–1149. 10.1016/j.meegid.2011.04.007 [DOI] [PubMed] [Google Scholar]
- 25.Chaiprasert A, Prammananan T, Tingtoy N, Na-Ubol P, Srimuang S, Samerpitak K, Rangsipanuratn W. 2006. One-tube multiplex PCR method for rapid identification of Mycobacterium tuberculosis. Southeast Asian J. Trop. Med. Public Health 37:494–502 [PubMed] [Google Scholar]
- 26.Cole ST. 2002. Comparative and functional genomics of the Mycobacterium tuberculosis complex. Microbiology 148:2919–2928 [DOI] [PubMed] [Google Scholar]
- 27.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanekom M, van der Spuy GD, Streicher E, Ndabambi SL, McEvoy CR, Kidd M, Beyers N, Victor TC, van Helden PD, Warren RM. 2007. A recently evolved sublineage of the Mycobacterium tuberculosis Beijing strain family is associated with an increased ability to spread and cause disease. J. Clin. Microbiol. 45:1483–1490. 10.1128/JCM.02191-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sreevatsan S, Pan X, Stockbauer KE, Connell ND, Kreiswirth BN, Whittam TS, Musser JM. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. U. S. A. 94:9869–9874. 10.1073/pnas.94.18.9869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwamoto T, Fujiyama R, Yoshida S, Wada T, Shirai C, Kawakami Y. 2009. Population structure dynamics of Mycobacterium tuberculosis Beijing strains during past decades in Japan. J. Clin. Microbiol. 47:3340–3343. 10.1128/JCM.01061-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Soolingen D, Hermans PW, de Haas PE, Soll DR, van Embden JD. 1991. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J. Clin. Microbiol. 29:2578–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwamoto T, Grandjean L, Arikawa K, Nakanishi N, Caviedes L, Coronel J, Sheen P, Wada T, Taype CA, Shaw MA, Moore DA, Gilman RH. 2012. Genetic diversity and transmission characteristics of Beijing family strains of Mycobacterium tuberculosis in Peru. PLoS One 7:e49651. 10.1371/journal.pone.0049651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakanishi N, Wada T, Arikawa K, Millet J, Rastogi N, Iwamoto T. 2013. Evolutionary robust SNPs reveal the misclassification of Mycobacterium tuberculosis Beijing family strains into sublineages. Infect. Genet. Evol. 16:174–177. 10.1016/j.meegid.2013.02.007 [DOI] [PubMed] [Google Scholar]
- 34.Kang HY, Wada T, Iwamoto T, Maeda S, Murase Y, Kato S, Kim HJ, Park YK. 2010. Phylogeographical particularity of the Mycobacterium tuberculosis Beijing family in South Korea based on international comparison with surrounding countries. J. Med. Microbiol. 59:1191–1197. 10.1099/jmm.0.022103-0 [DOI] [PubMed] [Google Scholar]
- 35.Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, Eiglmeier K, Garnier T, Gutierrez C, Hewinson G, Kremer K, Parsons LM, Pym AS, Samper S, van Soolingen D, Cole ST. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. U. S. A. 99:3684–3689. 10.1073/pnas.052548299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Homolka S, Projahn M, Feuerriegel S, Ubben T, Diel R, Nubel U, Niemann S. 2012. High resolution discrimination of clinical Mycobacterium tuberculosis complex strains based on single nucleotide polymorphisms. PLoS One 7:e39855. 10.1371/journal.pone.0039855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo T, Yang C, Gagneux S, Gicquel B, Mei J, Gao Q. 2012. Combination of single nucleotide polymorphism and variable-number tandem repeats for genotyping a homogenous population of Mycobacterium tuberculosis Beijing strains in China. J. Clin. Microbiol. 50:633–639. 10.1128/JCM.05539-11 [DOI] [PMC free article] [PubMed] [Google Scholar]